Abstract
The overproduction of reactive oxygen species (ROS) in cells can lead to the development of diseases associated with aging. We have previously shown that C. elegans BRAP-2 (Brca1 associated binding protein 2) regulates phase II detoxification genes such as gst-4, by increasing SKN-1 activity. Previously, a transcription factor (TF) RNAi screen was conducted to identify potential activators that are required to induce gst-4 expression in brap-2(ok1492) mutants. The lipid metabolism regulator NHR-49/HNF4 was among 18 TFs identified. Here, we show that knockdown of nhr-49 suppresses the activation of gst-4 caused by brap-2 inactivation and that gain-of-function alleles of nhr-49 promote gst-4 expression. We also demonstrate that nhr-49 and its cofactor mdt-15 are required to express phase II detoxification enzymes upon exposure to chemicals that induce oxidative stress. Furthermore, we show that NHR-49 and MDT-15 enhance expression of skn-1a/c. These findings identify a novel role for NHR-49 in ROS detoxification by regulating expression of SKN-1C and phase II detoxification genes.
Keywords: C. elegans, NHR-49, oxidative stress, reactive oxygen species, SKN-1
In nature, cells may encounter both exogenous and endogenous stressors that can alter normal physiological processes. One such form of stress, oxidative stress, is caused by reactive oxygen species (ROS) that have an ability to threaten cell survival. The imbalance between ROS and protective detoxification enzymes can lead to extensive oxidative damage to macromolecules such as DNA, lipids, and proteins (Buonocore et al. 2010; Finkel 2011). To protect cells against oxidative stress, organisms have developed lines of defense to cope with changes in levels of ROS to maintain homeostasis. The regulation of detoxification genes frequently involves complex transcriptional regulatory networks and, as a result, increases the potential for cross talk between stress signaling pathways. The induction of detoxification genes could be the result of cooperation between multiple transcription factors (TFs) (Rahman 2007). Therefore, in order to understand the genetic regulatory network involved in maintaining cellular integrity, it is vital to identify the factors that regulate stress response genes and promote survival.
Like mammals, the nematode C. elegans has well-defined stress defense systems for protection from toxic compounds (Van Raamsdonk and Hekimi 2010). These signaling pathways, and their modes of regulation, share evolutionary conservation with their mammalian counterparts (Tissenbaum 2015). Thus, C. elegans offers a suitable model to dissect the gene regulatory network involved in the expression of stress response genes. In recent years, increased attention has been given to the conserved TFs DAF-16/FOXO and SKN-1/Nrf2 due to their associated roles in response to oxidative stress and lifespan extension in C. elegans (Kenyon et al. 1993; Murphy et al. 2003; An and Blackwell 2003; Blackwell et al. 2015). These factors regulate the transcription of essential detoxification genes such as sod-3 and gst-4 to promote resistance to oxidative stress (Oliveira et al. 2009; Wang et al. 2010; Shore and Ruvkun 2013). Although the signaling pathways and mechanisms that control the nuclear localization of both TFs have been revealed, the oxidative stressors that activate these pathways remain poorly understood.
Mammalian Brap2 (Brca1 associated binding protein 2 or Brap as listed in the HUGO database) was first identified as a Brca1 binding protein that occludes the Brca1 nuclear localization motif, preventing it from translocating to the nucleus, and subsequently been shown to act as a cytoplasmic retention protein for a number of proteins (Li et al. 1998; Asada et al. 2004; Chen et al. 2009; Davies et al. 2013). Brap2 is also a Ras-responsive E3 ubiquitin ligase that functions as a modulator of the Ras signaling pathway by facilitating activation of Erk upon cell stimulation (Ory and Morrison 2004; Matheny and White 2006, 2009). Work in C. elegans has shown that loss of functional brap-2 causes hypersensitivity to hydrogen peroxide or paraquat (Koon and Kubiseski 2010). Furthermore, we performed an RNAi screen and found that BRAP-2 regulates the TFs SKN-1 and ELT-3 for the induction of phase II detoxification genes (Hu et al. 2017). The study also revealed that NHR-49, a TF with a role in regulating expression of proteins involved in lipid synthesis, suppresses gst-4 expression in brap-2(ok1492).
Nuclear hormone receptors (NHRs) are TFs that are generally activated by lipophilic hormones (Antebi 2015). NHR-49 is a key regulator of the “fasting response” that leads to changes in fatty acid metabolism for both basal or starvation states (Van Gilst et al. 2005a; b; Ratnappan et al. 2014). Loss of nhr-49 causes an increase in body fat and stimulates an impaired nutritional response. Additionally, nhr-49(lf) mutants exhibit a shortened lifespan caused by an imbalance in lipid composition that leads to lipotoxicity (Pathare et al. 2012; Grants et al. 2015). NHR-49 functions together with the mediator MDT-15 and removal of mdt-15 fails to stimulate nhr-49-dependent fasting response genes (Taubert et al. 2006). MDT-15 also interacts with SKN-1 to facilitate oxidative metabolism and promote lifespan in an nhr-49 independent manner (Goh et al. 2014; Pang et al. 2014). NHR-49 is a homolog of hepatocyte nuclear factor 4 alpha (HNF4α), yet appears to function in a manner similar to that of the related proliferator-activated receptor alpha (PPARα) in regulating fatty acid uptake, lipoprotein transport, and mitochondrial and peroxisomal β-oxidation (Contreras et al. 2013; Ratnappan et al. 2014).
Here we report that NHR-49 is essential for the SKN-1 dependent expression of phase II detoxification genes. The elucidation of SKN-1 co-activators, such as NHR-49 and MDT-15, for the control of this transcriptional regulation provides further insight in the complex gene regulatory network that controls stress gene expression.
Materials and Methods
C. elegans Strains
All C. elegans strains were maintained as described by Brenner (Brenner 1974). Double mutant strains were generated according to standard protocols. Unless stated otherwise, worm strains were provided by the Caenorhabditis Genetics Center (CGC, University of Minnesota) and the National Bioresource Project (Tokyo, Japan). Strains used in this study were as follows: Bristol strain N2, dvIs19 (CL2166), brap-2(ok1492) (YF15), nhr-49(ok2165) (YF127), brap-2(ok1492);nhr-49(ok2165) (YF126), mdt-15(tm2182) (XA7702), brap-2(ok1492);mdt-15(tm2182) (YF131), nhr-49(nr2041) (STE68), nhr-49(et7) (QC120), nhr-49(et8) (QC121), nhr-49(et13) (QC126) and wdr-23(tm1817) (YF208).
RNAi Treatment
RNAi was performed as described previously (MacNeil et al. 2015; Hu et al. 2017). Bacteria expressing dsRNA was grown on nematode growth medium (NGM) containing 0.4 mM IPTG, 100 µg/mL ampicillin, and 12.5 µg/mL tetracycline. Synchronized worms were grown on RNAi plates. Animals were collected at the L4 stage and analyzed for GFP expression by confocal microscope or used for RNA isolation.
Fluorescence microscopy
Live L4 gst-4p::gfp expressing worms were anesthetized using 2 mM Levamisole (Sigma L9756) and mounted on 2% agarose pad. Images of fluorescent worms were taken using a Zeiss LSM 700 confocal laser-scanning microscope with Zen 2010 Software.
Paraquat/Arsenite/Acrylamide Treatment
For each strain, synchronized worms were grown on NGM plates and were collected in M9 buffer at the L4 stage. Sodium arsenite (Sigma #35000) or paraquat (Sigma #856177) was diluted in M9 buffer to a final concentration of 5 mM and 100 mM, respectively. Collected worms were treated with each drug at room temperature for 2 hr followed by RNA isolation and qPCR. Each experiment was completed in triplicate. For acrylamide treatment, three independent lines of N2 or nhr-49(ok2165) synchronized L1-stage animals were grown on acrylamide (500 mg/L) or control plates (without acrylamide) for 48 hr at 20° as described (Hasegawa et al. 2008). L4 animals were then collected using M9 buffer and stored at -80° until RNA isolation.
Quantitative PCR
Quantitative PCR was used to measure mRNA levels as described previously (Hu et al. 2017). qPCR data were derived from 3 independent replicates and were analyzed using the comparative method (ΔΔCt). Results were graphed and the relative expression of each strain was compared to N2. The endogenous control used for normalization was act-1. Primer sequences were previously described (Hu et al. 2017).
Statistics
Statistical significance was determined using unpaired student’s t-test when two means were compared and corrected for multiple comparisons using the Holm-Sidak method. P values of <0.05 were taken to indicate statistical significance. Error bars represent +/− standard error of the mean.
Data availability
Strains are available upon request or through the Caenorhabditis Genetics Center (CGC). All the data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
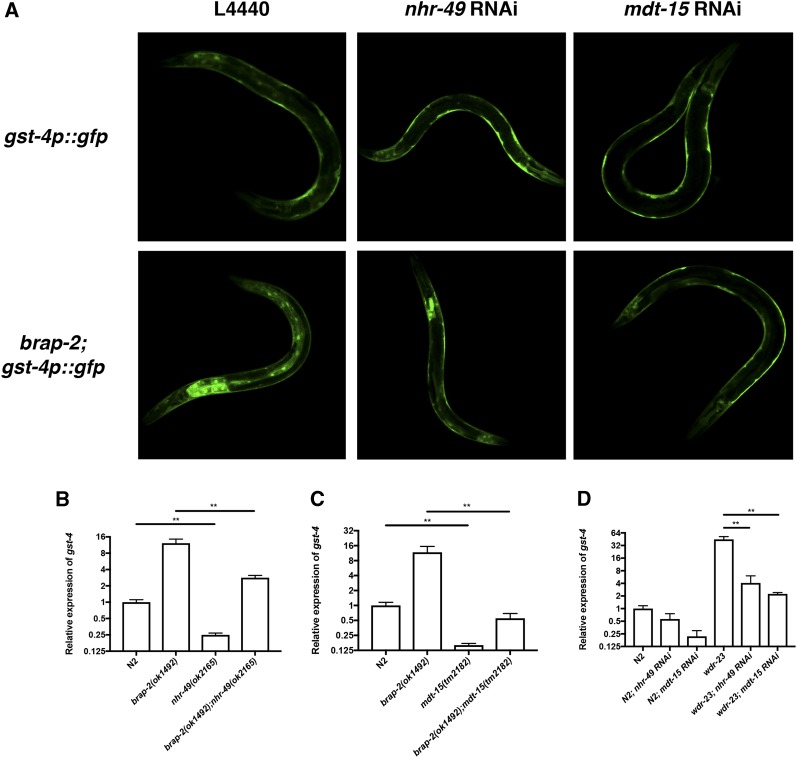
The TF NHR-49 and its mediator subunit MDT-15 are required for phase II detoxification gene expression in brap-2(ok1492) mutants. Previously, we showed that C. elegans BRAP-2 is required to regulate the TF SKN-1 to induce phase II detoxification gene expression. To further the study of this regulatory network, a TF specific RNAi screen was conducted to identify regulators of gst-4 expression in brap-2(ok1492) mutants (Hu et al. 2017). Our screen identified 18 TFs that decreased GFP expression in brap-2(ok1492) mutants, including nhr-49. To validate this result, L4 brap-2(ok1492) mutants carrying the gst-4p::gfp transgene were fed nhr-49 RNAi and GFP expression was examined. The nhr-49 RNAi treated brap-2(ok1492);gst-4p::gfp animals displayed lower GFP expression compared to the RNAi vector control (Figure 1A). We also examined gst-4 expression by qPCR in brap-2(ok1492);nhr-49(ok2165) double mutant and found an ∼75% reduction of gst-4 mRNA (Figure 1B). Taken together, this indicates that nhr-49 is required for gst-4 expression.
Figure 1.
nhr-49 and mdt-15 are essential for enhanced gst-4 expression. (A) gst-4p::gfp and brap-2(ok1492);gst-4p::gfp worms were treated with nhr-49 or mdt-15 RNAi followed by examination of GFP expression using confocal microscopy. Representative GFP images show brap-2(ok1492);gst-4p::gfp worms grown in either nhr-49 or mdt-15 RNAi causes a reduction of gst-4p::gfp expression. Twenty worms were examined and one representative worm shown. (B, C) Following RNA extraction, gst-4 mRNA levels were measured by qPCR. gst-4 expression is reduced in both (B) brap-2(ok1492);nhr-49(ok2165) and (C) brap-2(ok1492);mdt-15(tm2181). (D) wdr-23(tm1817) worms grown in either nhr-49 or mdt-15 RNAi showed a significant decrease in gst-4 mRNA levels as measured by qPCR. P < 0.05*, P < 0.01**, P < 0.001***.
NHR-49 requires the mediator MDT-15 to modulate target gene expression and lipid composition (Taubert et al. 2006). MDT-15 contributes to detoxification gene induction and, together with its interacting partner SKN-1, is required for the oxidative stress response (Taubert et al. 2008; Goh et al. 2014; Grants et al. 2015). To determine if MDT-15 is also needed to up-regulate gst-4 expression in brap-2(ok1492), we knocked down mdt-15 in brap-2(ok1492);gst-4p::gfp animals and monitored effects on GFP expression. Additionally, a brap-2(ok1492);mdt-15(tm2182) double mutant was generated and gst-4 mRNA levels were measured using qPCR. As expected, loss of mdt-15 resulted in a reduction in gst-4 levels (Figure 1A and 1C), indicating that MDT-15 also plays a role in gst-4 regulation in the brap-2(ok1492) mutant, consistent with previous studies showing that MDT-15 is required for oxidative stress response. The C. elegans protein WDR-23 functions to prevent the accumulation of SKN-1 in the nucleus by targeting it for degradation (Choe et al. 2009). Since gst-4 expression is increased in wdr-23 mutants, and mdt-15 is required for this increase (Goh et al. 2014; Wu et al. 2016), we wanted to determine if nhr-49 was also required in this context. We knocked down nhr-49 using RNAi in the wdr-23(tm1817) mutant and again found a significant reduction in gst-4 mRNA levels (Figure 1D).
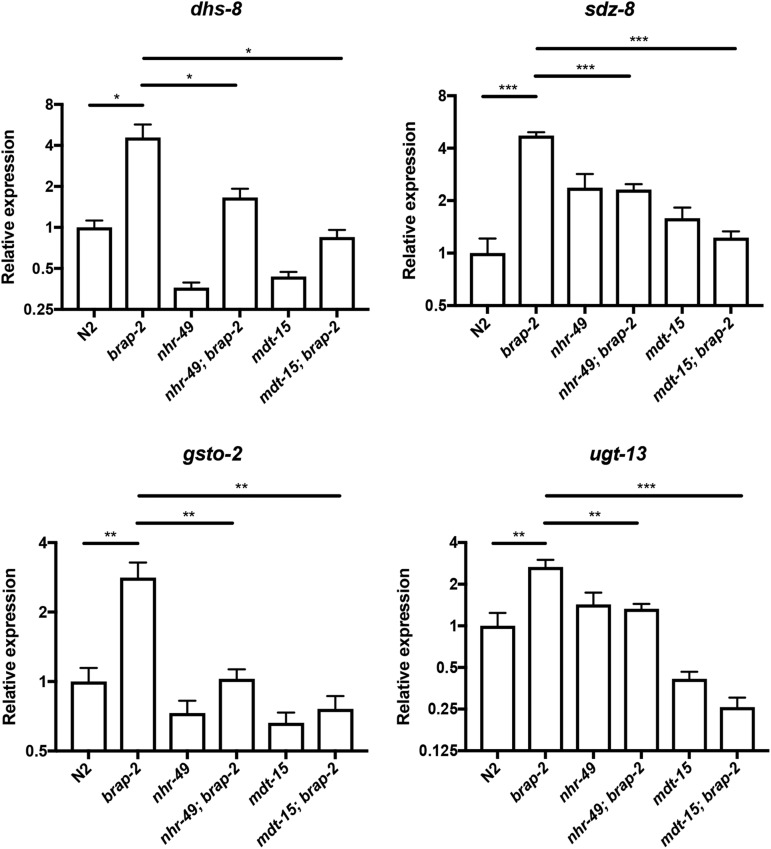
To confirm the requirement of nhr-49 and mdt-15 in phase II detoxification in brap-2(ok1492), we examined four additional phase II genes (dhs-8, sdz-8, gsto-2 and ugt-13). The expression of all four genes was significantly decreased in brap-2(ok1492) when either nhr-49 or mdt-15 was absent (Figure 2). Therefore, NHR-49 and MDT-15 are required for activation of phase II detoxification genes in brap-2(ok1492) animals.
Figure 2.
The mRNA levels of four phase II detoxification genes (log2 scale) were compared between wild type and brap-2(ok1492) mutants in the presence and absence of nhr-49(ok2165) or mdt-15(tm2182) alleles. A significant decrease in mRNA levels was found in all four phase II genes tested (dhs-8, sdz-8, gsto-2, and ugt-13) upon nhr-49 or mdt-15 mutation in brap-2(ok1492). P < 0.05*, P < 0.01**, P < 0.001***.
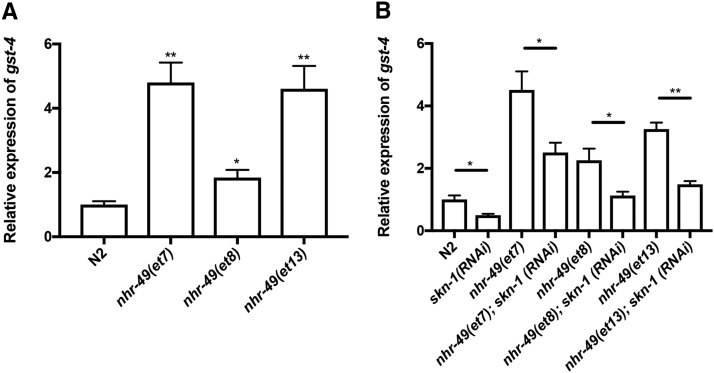
Since nhr-49 is required for the expression of the phase II detoxification gene gst-4, we hypothesized that an increase in gst-4 transcript levels would be seen with gain-of-function (gof) alleles of nhr-49 (Lee et al. 2016). Indeed, we observed a 1.8 to 4.8-fold increase in gst-4 expression in nhr-49(gof) mutants (Figure 3A). We next asked if this increase in gst-4 expression required SKN-1. skn-1 was knocked down in nhr-49(gof) strains and gst-4 mRNA levels were measured using qPCR. The depletion of skn-1 caused a decrease in gst-4 expression when compared to the untreated RNAi control (Figure 3B). Taken together, these observations are consistent with NHR-49 and SKN-1 being required to promote the induction of phase II detoxification genes.
Figure 3.
skn-1 is required to regulate gst-4 expression in gain-of-function nhr-49 worms. RNA was extracted from synchronized L4 worms followed by quantification of gst-4 transcript levels using qPCR. (A) Three gain-of-function nhr-49 strains were used to examine gst-4 mRNA expression. Results display an increase in gst-4 mRNA by at least twofold. (B) The nhr-49 gain-of-function strains were treated with control (L4440) or skn-1 RNAi and gst-4 mRNA levels were quantified. A reduction of gst-4 was seen with skn-1 knockdown compared to strains fed on the L4440 control; P < 0.01**, P < 0.05* vs. N2 in (A); P < 0.001***, P < 0.01** vs. L4440 control in (B).
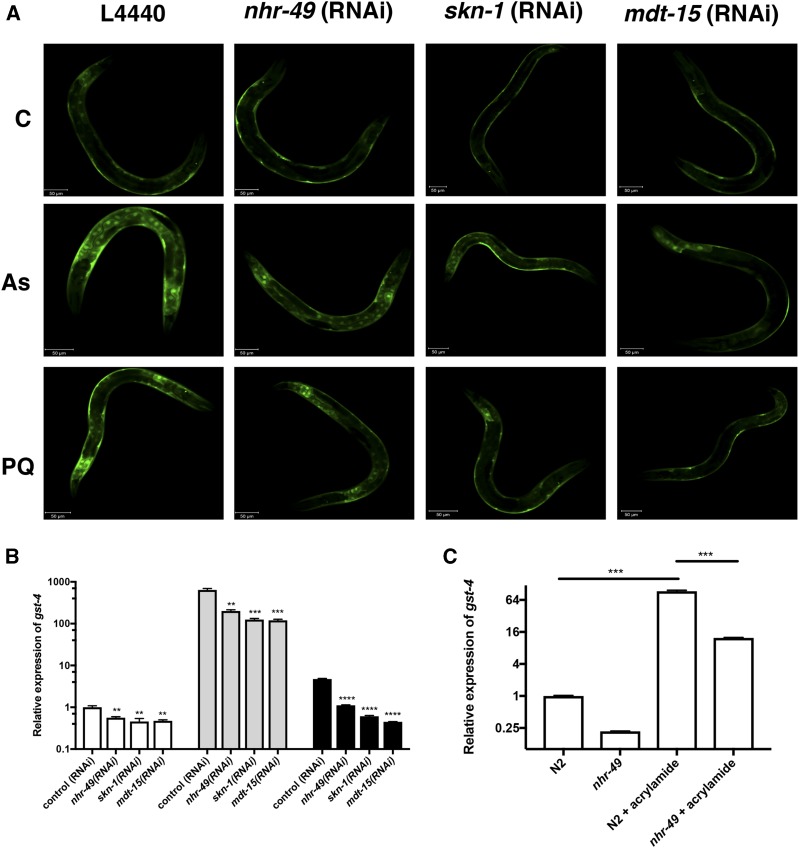
In C. elegans, oxidative stress can be induced using sodium arsenite or paraquat, both of which significantly increase the expression of phase II detoxification enzymes in wild type worms (Oliveira et al. 2009). Previously, it was shown that skn-1 and mdt-15 are required to upregulate phase II detoxification genes upon arsenite induction (Goh et al. 2014). Therefore, we asked if NHR-49 activates gst-4 in response to oxidative stress. We grew synchronized wild type worms containing gst-4p::gfp, knocked down nhr-49, mdt-15 or skn-1, and exposed the animals to 5 mM sodium arsenite or 100 mM paraquat for two hours after which GFP expression was examined using confocal microscopy. In wild type animals, gst-4p::gfp expression was increased upon exposure to arsenite or paraquat and this increase was reduced upon nhr-49, mdt-15 or skn-1 RNAi (Figure 4A). qPCR was performed to quantify levels of gst-4 and a reduction in mRNA was observed in nhr-49, skn-1, and mdt-15 RNAi treated animals (Figure 4B). We also examined gst-4 expression following acrylamide exposure over 48 hr and found a significant decrease in gst-4 mRNA levels in the nhr-49(ok2165) strain relative to wild type (Figure 4C). These results demonstrate an important role for nhr-49, mdt-15, and skn-1 in the regulation of ROS detoxification genes upon exposure to oxidative stress.
Figure 4.
skn-1, mdt-15, and nhr-49 are essential to induce the expression of the arsenite, paraquat, and acrylamide responsive gene gst-4 in L4 worms. (A) Synchronized gst-4p::gfp worms were grown in the control (L4440), skn-1 RNAi, nhr-49 RNAi or mdt-15 RNAi followed by exposure to M9 buffer (C), 5 mM sodium arsenite (As) or 100 mM paraquat (PQ) for 2 hr at L4 stage. Worms were recovered on NGM plates for 1 hr and the GFP expression was examined using confocal microscopy. Results show a reduction in GFP levels in RNAi treated worms. Twenty worms were examined and figures depict one worm. (B) Synchronized L4 stage worms were collected after RNAi exposure and drug treatment, followed by RNA extraction. gst-4 mRNA transcript levels were quantified using qPCR. Values are relative to the control (RNAi) and normalized to the endogenous control act-1. The knock down of either nhr-49, skn-1 or mdt-15 exhibit a decrease in gst-4 mRNA expression after mock treatment (white bars), arsenite (gray bars) or paraquat (black bars) treatment in comparison to the N2 drug-treated control. (C) Synchronized wild type and nhr-49(ok2165) animals were grown for 48 hr at 20°C on seeded plates with or without acrylamide and harvested for RNA extraction. gst-4 mRNA transcript levels were quantified using qPCR. P < 0.001*** P < 0.01** vs. untreated and treated controls in (B), P < 0.001*** vs. wild type in (C).
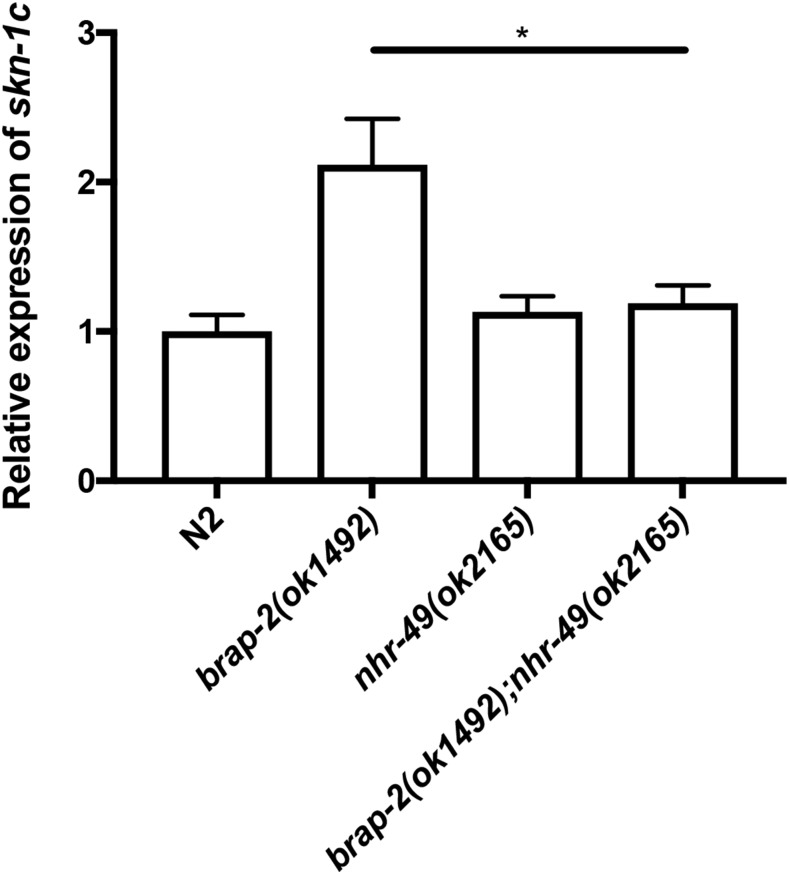
NHR-49 is required to activate skn-1 transcription. Previously, we found that brap-2(ok1492) mutants have higher levels of skn-1 mRNA than wild type animals (Hu et al. 2017). We asked if NHR-49 is required for the increased skn-1 expression observed in brap-2 mutants. We measured skn-1 mRNA levels in brap-2;nhr-49 deletion strains and observed a twofold increase of skn-1, and the loss of nhr-49 in brap-2(ok1492) decreases the amount of skn-1 mRNA, restoring it to wild type levels (Figure 5). This indicates that nhr-49 plays a role in inducing skn-1 expression in the brap-2(ok1492) strain under oxidative stress conditions yet is not required for its basal expression.
Figure 5.
Functional NHR-49 and MDT-15 are required to promote skn-1 transcriptional activity. (A) Relative skn-1 mRNA expression was quantified in nhr-49(ok2165) and brap-2(ok1492);nhr-49(ok2165) mutant strains using qPCR. The worms displayed a reduction of skn-1c in the double mutant to wild type levels. P < 0.001***.
Discussion
In this study we further investigated the role of BRAP-2 and SKN-1 in the C. elegans oxidative stress response. We show that NHR-49 (the C. elegans PPARα/HNF4 homolog) and the mediator MDT-15 (MED15 homolog) are essential to regulate the SKN-1 dependent stress response in brap-2(ok1492). We have found that NHR-49, MDT-15, and SKN-1 co-regulate the induction of gst-4 and skn-1. By investigating mRNA expression of designated target genes in null mutants, we were able to provide evidence that NHR-49 not only participates in fat metabolism but is also a key player in the oxidative stress response. Interestingly, neither null mutations of nhr-49 nor mdt-15 result in the complete loss of gst-4 (or phase II detoxification gene) expression in the brap-2(ok1492) strain, indicating that additional regulators exist.
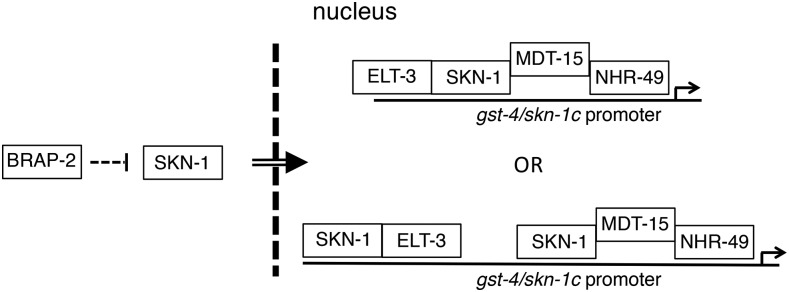
Although we do not show a direct interaction of these regulators with the gst-4 or skn-1 promoters, it has been reported that MDT-15 can interact with both SKN-1 and NHR-49 independently (Goh et al. 2014). This suggests that it is possible that MDT-15 acts as a bridge between SKN-1 and NHR-49 (Figure 6). Previously, we showed that SKN-1 and the GATA factor ELT-3 heterodimerize and promote expression of phase II detoxification genes (Hu et al. 2017). It will be of interest to determine whether MDT-15 interacting with SKN-1 is independent of ELT-3, and if a transcriptional complex consisting of ELT-3, SKN-1, MDT-15, and NHR-49 forms to induce gst-4 in brap-2 mutant animals.
Figure 6.
Proposed model of NHR-49 in the regulation of phase II detoxification gene gst-4. The induction of oxidative stress in brap-2(ok1492) mutant worms activates PMK, which then phosphorylates SKN-1 for nuclear translocation. SKN-1 then binds to NHR-49/MDT-15 in the nucleus to induce gst-4 expression. This complex can also create a feed-forward loop through skn-1c promoter binding to up regulate its own transcription, thereby enhancing both the SKN-1C response to oxidative stress and its target genes.
The focus of NHR-49 research in C. elegans has been to explore its role in regulating fatty acid metabolism. In addition, a study suggested NHR-49 also helps to promote lifespan in animals lacking a germline by controlling lipid metabolic pathways (Ratnappan et al. 2014). nhr-49 mutants are hypersensitive to various stress inducing molecules including arsenite and tert-butyl hydroperoxide (Goh et al. 2014; Pang et al. 2014). NHR-49 promotes fatty acid β-oxidation, increases acetyl CoA levels and enhances activity of the electron transport chain, effects that are expected to increase ROS levels. Here we show that NHR-49 has a complementary role in oxidative stress to combat the expected increase in ROS levels that occurs during fatty acid catabolism, preventing oxidation of cellular components. The human homologs of NHR-49, HNF-4α and PPARα, are well known regulators of energy metabolism, fatty acid uptake, lipoprotein transport, and mitochondrial and peroxisomal β-oxidation (Contreras et al. 2013). Knockdown of HNF4α in Caco-2 cells demonstrated increased lipid peroxidation and decreased antioxidant enzyme expression (Marcil et al. 2010), indicating that the role of NHR-49 in the oxidative stress response we describe is conserved between C. elegans and humans.
The genetic regulatory network uses positive and negative mechanisms to alter transcription, while at the same time coping with changes in the intracellular environment. We found that nhr-49 was required for the increased expression of skn-1c observed in brap-2 mutant animals, suggesting that nhr-49 regulates influences SKN-1 levels and activity. It is possible that the SKN-1/MDT-15/NHR-49 complex is able to create a feed-forward mechanism that ensures SKN-1 is continuously produced in response to oxidative stress for the further downstream amplification of gst-4 and phase II response genes (Figure 6). Our data provides insight into NHR-49 and its role in the oxidative stress response, where it influences stress response activation through co-regulation with SKN-1, an activation that requires a functional MDT-15. Although we were not able to detect direct binding between SKN-1 and NHR-49, we have shown that NHR-49 is required to coordinate with SKN-1 to induce the expression of stress genes.
In conclusion, we have established a new role for NHR-49 in the oxidative stress response in C. elegans. This result is in accord with recent studies published during the review/revision process (Qi et al. 2017; Goh et al. 2018) that nhr-49 is required for the skn-1 dependent oxidative stress response. Our work provides a framework for the continued study of stress genes and the ways in which they are regulated to maintain cell integrity and prevent damage caused by ROS.
Acknowledgments
A number of strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Stefan Taubert for strains QC120, QC121 and QC126. A.J.M.W and L.T.M were supported by the National Institutes of Health grants DK068429 and GM082971. D.R.D was a recipient of the Ontario Graduate Scholarship. Q.H., D.R.D and T.J.K were supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Communicating editor: M. Zetka
Literature Cited
- An J. H., Blackwell T. K., 2003. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17: 1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A., 2015. Nuclear receptor signal transduction in C. elegans. WormBook Online Rev. C Elegans Biol. 1–49. 10.1895/wormbook.1.64.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada M., Ohmi K., Delia D., Enosawa S., Suzuki S., et al. , 2004. Brap2 functions as a cytoplasmic retention protein for p21 during monocyte differentiation. Mol. Cell. Biol. 24: 8236–8243. 10.1128/MCB.24.18.8236-8243.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., Isik M., 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88: 290–301. 10.1016/j.freeradbiomed.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore G., Perrone S., Tataranno M. L., 2010. Oxygen toxicity: chemistry and biology of reactive oxygen species. Semin. Fetal Neonatal Med. 15: 186–190. 10.1016/j.siny.2010.04.003 [DOI] [PubMed] [Google Scholar]
- Chen J., Hu H. Y., Zhang S., He M., Hu R. M., 2009. Brap2 facilitates HsCdc14A Lys-63 linked ubiquitin modification. Biotechnol. Lett. 31: 615–621. 10.1007/s10529-009-9914-7 [DOI] [PubMed] [Google Scholar]
- Choe K. P., Przybysz A. J., Strange K., 2009. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 29: 2704–2715. 10.1128/MCB.01811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A. V., Torres N., Tovar A. R., 2013. PPAR-α as a key nutritional and environmental sensor for metabolic adaptation. Adv. Nutr. Bethesda Md 4: 439–452. 10.3945/an.113.003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. G., Wagstaff K. M., McLaughlin E. A., Loveland K. L., Jans D. A., 2013. The BRCA1-binding protein BRAP2 can act as a cytoplasmic retention factor for nuclear and nuclear envelope-localizing testicular proteins. Biochim. Biophys. Acta BBAMol. Cell Res. 1833: 3436–3444. 10.1016/j.bbamcr.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Finkel T., 2011. Signal transduction by reactive oxygen species. J. Cell Biol. 194: 7–15. 10.1083/jcb.201102095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y. S., Martelli K. L., Parhar K. S., Kwong A. W. L., Wong M. A., et al. , 2014. The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell 13: 70–79. 10.1111/acel.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y. S., Winter J. J., Bhanshali F., Doering K. R. S., Lai R., et al. , 2018. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell 17: e12743 10.1111/acel.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grants J. M., Goh G. Y. S., Taubert S., 2015. The Mediator complex of Caenorhabditis elegans: insights into the developmental and physiological roles of a conserved transcriptional coregulator. Nucleic Acids Res. 43: 2442–2453. 10.1093/nar/gkv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Isomura K., Tsutsumiuchi K., Taniguchi H., et al. , 2008. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol. Sci. Off. J. Soc. Toxicol. 101: 215–225. 10.1093/toxsci/kfm276 [DOI] [PubMed] [Google Scholar]
- Hu Q., D’Amora D. R., MacNeil L. T., Walhout A. J. M., Kubiseski T. J., 2017. The Oxidative Stress Response in Caenorhabditis elegans Requires the GATA Transcription Factor ELT-3 and SKN-1/Nrf2. Genetics 206: 1909–1922. 10.1534/genetics.116.198788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Koon J. C., Kubiseski T. J., 2010. Developmental arrest of Caenorhabditis elegans BRAP-2 mutant exposed to oxidative stress is dependent on BRC-1. J. Biol. Chem. 285: 13437–13443. 10.1074/jbc.M110.107011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Goh G. Y. S., Wong M. A., Klassen T. L., Taubert S., 2016. Gain-of-Function Alleles in Caenorhabditis elegans Nuclear Hormone Receptor nhr-49 Are Functionally Distinct. PLoS One 11: e0162708 10.1371/journal.pone.0162708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ku C. Y., Farmer A. A., Cong Y. S., Chen C. F., et al. , 1998. Identification of a novel cytoplasmic protein that specifically binds to nuclear localization signal motifs. J. Biol. Chem. 273: 6183–6189. 10.1074/jbc.273.11.6183 [DOI] [PubMed] [Google Scholar]
- MacNeil L. T., Pons C., Arda H. E., Giese G. E., Myers C. L., et al. , 2015. Transcription Factor Activity Mapping of a Tissue-Specific in vivo Gene Regulatory Network. Cell Syst. 1: 152–162. 10.1016/j.cels.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcil V., Seidman E., Sinnett D., Boudreau F., Gendron F.-P., et al. , 2010. Modification in Oxidative Stress, Inflammation, and Lipoprotein Assembly in Response to Hepatocyte Nuclear Factor 4α Knockdown in Intestinal Epithelial Cells. J. Biol. Chem. 285: 40448–40460. 10.1074/jbc.M110.155358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny S. A., White M. A., 2006. Ras-sensitive IMP modulation of the Raf/MEK/ERK cascade through KSR1. Methods Enzymol. 407: 237–247. 10.1016/S0076-6879(05)07020-5 [DOI] [PubMed] [Google Scholar]
- Matheny S. A., White M. A., 2009. Signaling threshold regulation by the Ras effector IMP. J. Biol. Chem. 284: 11007–11011. 10.1074/jbc.R800082200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., et al. , 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Oliveira R. P., Porter Abate J., Dilks K., Landis J., Ashraf J., et al. , 2009. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8: 524–541. 10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S., Morrison D. K., 2004. Signal transduction: implications for Ras-dependent ERK signaling. Curr. Biol. 14: R277–R278. 10.1016/j.cub.2004.03.023 [DOI] [PubMed] [Google Scholar]
- Pang S., Lynn D. A., Lo J. Y., Paek J., Curran S. P., 2014. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5: 5048 10.1038/ncomms6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare P. P., Lin A., Bornfeldt K. E., Taubert S., Van Gilst M. R., 2012. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 8: e1002645 10.1371/journal.pgen.1002645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Gutierrez G. E., Gao X., Dixon H., McDonough J. A., et al. , 2017. The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell 16: 1125–1135. 10.1111/acel.12651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman K., 2007. Studies on free radicals, antioxidants, and co-factors. Clin. Interv. Aging 2: 219–236. [PMC free article] [PubMed] [Google Scholar]
- Ratnappan R., Amrit F. R. G., Chen S.-W., Gill H., Holden K., et al. , 2014. Germline signals deploy NHR-49 to modulate fatty-acid β-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet. 10: e1004829 10.1371/journal.pgen.1004829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. E., Ruvkun G., 2013. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 23: 409–420. 10.1016/j.tcb.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Van Gilst M. R., Hansen M., Yamamoto K. R., 2006. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20: 1137–1149. 10.1101/gad.1395406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Hansen M., Van Gilst M. R., Cooper S. B., Yamamoto K. R., 2008. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 4: e1000021 10.1371/journal.pgen.1000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H. A., 2015. Using C. elegans for aging research. Invertebr. Reprod. Dev. 59: 59–63. 10.1080/07924259.2014.940470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Jolly A., Yamamoto K. R., 2005a. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53 10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R., 2005b. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA 102: 13496–13501. 10.1073/pnas.0506234102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk J. M., Hekimi S., 2010. Reactive Oxygen Species and Aging in Caenorhabditis elegans: Causal or Casual Relationship? Antioxid. Redox Signal. 13: 1911–1953. 10.1089/ars.2010.3215 [DOI] [PubMed] [Google Scholar]
- Wang J., Robida-Stubbs S., Tullet J. M. A., Rual J.-F., Vidal M., et al. , 2010. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 6: e1001048 10.1371/journal.pgen.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-W., Deonarine A., Przybysz A., Strange K., Choe K. P., 2016. The Skp1 Homologs SKR-1/2 Are Required for the Caenorhabditis elegans SKN-1 Antioxidant/Detoxification Response Independently of p38 MAPK. PLoS Genet. 12: e1006361 10.1371/journal.pgen.1006361 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains are available upon request or through the Caenorhabditis Genetics Center (CGC). All the data necessary for confirming the conclusions presented in the article are represented fully within the article.