Abstract
The Eurasian perch (Perca fluviatilis) is the most common fish of the Percidae family and is widely distributed across Eurasia. Perch is a popular target for professional and recreational fisheries, and a promising freshwater aquaculture species in Europe. However, despite its high ecological, economical and societal importance, the available genomic resources for P. fluviatilis are rather limited. In this work, we report de novo assembly and annotation of the whole genome sequence of perch. The linked-read based technology with 10X Genomics Chromium chemistry and Supernova assembler produced a draft perch genome ∼1.0 Gbp assembly (scaffold N50 = 6.3 Mb; the longest individual scaffold of 29.3 Mb; BUSCO completeness of 88.0%), which included 281.6 Mb of putative repeated sequences. The perch genome assembly presented here, generated from small amount of starting material (0.75 ng) and a single linked-read library, is highly continuous and considerably more complete than the currently available draft of P. fluviatilis genome. A total of 23,397 protein-coding genes were predicted, 23,171 (99%) of which were annotated functionally from either sequence homology or protein signature searches. Linked-read technology enables fast, accurate and cost-effective de novo assembly of large non-model eukaryote genomes. The highly continuous assembly of the Eurasian perch genome presented in this study will be an invaluable resource for a range of genetic, ecological, physiological, ecotoxicological, functional and comparative genomic studies in perch and other fish species of the Percidae family.
Keywords: Perca fluviatilis, whole genome sequencing, de novo assembly, 10X Genomics Chromium linked-read, fish
The Eurasian perch, Perca fluviatilis (NCBI Taxon ID: 8168, Fishbase ID: 358) is a common medium-size predatory fish of the Percidae family that is widely distributed across northern Eurasia. It can live in an extremely broad range of habitats, from estuarine lagoons and lakes of all types to rivers and medium-sized streams. Perch is an important commercially exploited fresh- and brackish water fish species, and also a very popular target among recreational fisherman. As the current demand of perch exceeds fisheries production, it has been introduced as a new aquaculture species in many European countries (Policar et al. 2015).
Although a number of phylogeographic, population genetic and genomic studies have been conducted in perch using mtDNA, microsatellites and RAD-seq (e.g., Nesbø et al. 1999; Gerlach et al. 2001; Bergek et al. 2010; Bergek and Björklund 2009; Olsson et al. 2011; Pukk et al. 2013; Pukk et al. 2015; Pukk et al. 2016), current genomic resources of P. fluviatilis are rather limited. Recently, several RNAseq studies have been performed on perch to generate de novo transcriptome assemblies (Pasquier et al. 2016; Chen et al. 2017) and a de novo genome draft assembly of perch was published among those of 66 teleost fishes (Malmstrøm et al. 2017). However, this genome draft assembly is very fragmented and incomplete (scaffold N50 = 5,973 bp) which severely limits its usefulness for subsequent genomic work.
Modern technologies of next-generation sequencing enable the generation of billions of short reads with high accuracy for a relatively low price (Levy and Myers 2016). However, de novo assembly of a genome using short reads is challenging; obtaining long continuous scaffolds is difficult, as short reads perform poorly for resolving repetitive structures or GC-biased regions (Sohn and Nam 2018). These challenges arising from the complexity of genome structure can be overcome by using single-molecule long reads, but the error rate and the sequencing cost for these long-read technologies remain high (Sohn and Nam 2018). A library preparation technology developed by 10X Genomics incorporates unique molecular barcodes into individual high molecular weight DNA molecules, after which libraries undergo standard Illumina short-read sequencing (Zheng et al. 2016). A phased assembly strategy algorithm implemented in Supernova software then uses these barcodes to tag short-reads that come from the same long DNA fragment (linked-reads), enabling the construction of highly continuous scaffolds (Weisenfeld et al. 2017). 10X Genomics linked-read sequencing has been successfully applied to generate de novo genome assemblies of several organisms including plants (Hulse-Kemp et al. 2018; Liu et al. 2018), amphibians (Hammond et al. 2017), mollusks (Li et al. 2018) and mammals (Jones et al. 2017; Mohr et al. 2017).
Here, we report a high-quality, highly continuous, and nearly complete assembly of the Eurasian perch genome generated using 10X Genomics linked-read sequencing, which will serve as a backbone for future genetic, genomic, ecological and evolutionary studies of perch and other fish species of the Percidae family.
Materials and Methods
Samples, library preparation and sequencing
A single female perch from the small humic lake Loosalu, Estonia (58.932°N 25.080°E; lake surface area 35.2 ha) was caught by gill-net on 19.06.2017 and killed by an overdose of tricaine methanesulfonate (MS-222) before sampling. A blood (350 µl) sample was collected, mixed with 15 µl of K2EDTA, and kept on ice during transportation to the laboratory. Buffered blood was kept at +4° and DNA isolation was carried out on the third day after sample collection. Transcriptome characterization was performed using a different female perch, caught one year earlier (16.09.2016) from the same lake and killed as described above. A whole left eyeball was dissected from this perch and immediately stored in liquid nitrogen. The permits for sample collection were issued by the Estonian Ministry of the Environment (no. 54/2016; 37/2017).
High molecular weight genomic DNA (gDNA) was isolated from blood using the MagAttract HMW DNA Kit (Qiagen, Halden, Germany) according to manufacturer instructions with a few modifications. As fish red blood cells contain a nucleus, we used only 1 µl of buffered blood (instead of recommended 200 µl). In addition, to avoid fragmentation of high molecular weight DNA, we applied very gentle vortexing and mixing during the DNA isolation procedure. Total DNA was eluted in 80 µl of double distilled water. The quantity of gDNA was measured by Qubit Fluorometric Quantitation (Life Technologies) and the average length of the gDNA fragments was determined using the Agilent 2200 TapeStation system using Genomic DNA ScreenTape (cat. 5067-5365) and Genomic DNA Reagents (cat. 5067-5366). The average fragment size of gDNA was > 60 Kb. Genomic DNA was adjusted to a concentration of 0.6 ng/µl and 0.75 ng of template gDNA was loaded on a Chromium Genome Chip. Whole genome sequencing libraries were prepared using Chromium Genome Library & Gel Bead Kit v.2 (10X Genomics, cat. 120258), Chromium Genome Chip Kit v.2 (10X Genomics, cat. 120257), Chromium i7 Multiplex Kit (10X Genomics, cat. 120262) and Chromium controller according to manufacturer’s instructions. Briefly, gDNA was combined with Master Mix, a library of Genome Gel Beads, and partitioning oil to create Gel Bead-in-Emulsions (GEMs) on a Chromium Genome Chip. The GEMs were isothermally amplified with primers containing an Illumina Read 1 sequencing primer, a unique 16-bp 10x bar-code and a 6-bp random primer sequence, and bar-coded DNA fragments were recovered for Illumina library construction. The amount and fragment size of post-GEM DNA was quantified prior to library construction using a Bioanalyzer 2100 with an Agilent High sensitivity DNA kit (Agilent, cat. 5067-4626). Quantitative polymerase chain reaction (qPCR) using KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems, cat. KK4873) was performed to assess library yield. The library size range and distribution was determined using the Fragment Analyzer Automated CE System (AATI) with a High Sensitivity NGS Fragment Analysis Kit (Advanced Analytical, cat. DNF-474-1000). The library was sequenced on two lanes of an Illumina HiSeq 2500 sequencer in rapid run mode, using paired-end sequencing to generate 580.55 M linked-reads with a mean read length of 139.5 bp after trimming. The weighted mean molecule size was estimated as 63.18 Kb and mean read coverage was ∼68x. The WGS library preparation and sequencing was performed in the Finnish Functional Genomics Centre (Turku, Finland).
For transcriptome characterization, the frozen eyeball was mechanically crushed in liquid nitrogen using a mortar and pestle to produce a fine powder. Total RNA was extracted from the whole homogenized eyeball (30 mg of tissue), using a NucleoSpin RNA extraction kit (MACHEREY-NAGEL, Duren, Germany). The quality of the total RNA sample was evaluated using Bioanalyzer 2100 (Agilent) electrophoresis and sample concentration was measured with a Nanodrop ND‐2000 (Thermo Scientific). The library was prepared from 300 ng of total RNA according to the Illumina TruSeq Stranded mRNA Sample Preparation Guide (part no. 15031047) to generate a 300-bp insert size library. The library was sequenced using an Illumina HiSeq 3000 (2 × 75 bp configuration, half a lane) in the Finnish Functional Genomics Centre (Turku, Finland).
Evaluation of the genome metrics based on raw reads
K-mer counting of quality and barcode trimmed Illumina reads was performed using Jellyfish v.2.2.6 (Marçais and Kingsford 2011), producing k-mer frequency distributions of 17-, 21- and 25-mers (jellyfish histo -h 3000000). These histograms were processed using GenomeScope (Vurture et al. 2017; high frequency k-mer cutoff = 10,000), and findGSE (Sun et al. 2018) to estimate genome size, heterozygosity and repeat content.
de novo genome assembly
The linked-read data were assembled using Supernova v.2.0.1 assembler (Weisenfeld et al. 2017) with default settings. The assembler software was run for 25 days on a 28 core and 240 Gb RAM CSC – IT Center for Science cPouta virtual private server, based on Intel Xeon CPU E5-2680 v.4 2.4 GHz processors. The initial draft genome assembly was presented in pseudohaplotype format, and contained 1,024.4 Mb of scaffold sequence (37,560 scaffolds ≥ 1 Kb), of which 111.5 Mb represented unknown bases. GenomeTools sequniq v.1.5.10 (Gremme et al. 2013) was applied to remove duplicated scaffolds (1,374 scaffolds) and only scaffolds with more than 10% of unique sequence were retained (36,169 scaffolds). The redundancy of the genome assembly was further reduced in two steps. First, all scaffolds < 2 Mb were clustered using CD-HIT v.4.7 package (Fu et al. 2012; Li and Godzik 2006). When two or more scaffolds showed ≥ 99% similarity, all but the longest scaffold were removed to generate a non-redundant set of scaffolds < 2 Mb. This resulted in removal of 4,396 potentially redundant scaffolds from the assembly. Second, to further reduce potential redundancy, the assembly including non-redundant set of < 2 Mb scaffolds was self-aligned using LAST v.926 (Kiełbasa et al. 2011; identity ≥ 99%, coverage of query sequence ≥ 95%), resulting in exclusion of 668 additional scaffolds. In total, 66,188,489 bp were removed from the initial assembly due to potential duplication or redundancy. It should be noted that the size of the majority of potentially redundant scaffolds did not exceed 10 Kb, varying from 1 Kb to 621.4 Kb (Figure S1).
The final perch genome assembly included 31,105 scaffolds. The assembly was screened for vectors and contaminants using a Kraken v.1.0 (Wood and Salzberg 2014) customized database, which included standard Kraken viral, bacterial, archaeal, plasmid and human databases, additional genomes of Trypanosoma brucei (GCF_000210295.1, Jackson et al. 2010) and seven fish species (Cyprinus carpio GCF_000951615.1, Li J.-T., Chinese Academy of Fishery Science; Danio rerio GCF_000002035.6, Howe et al. 2013; Esox lucius GCF_000721915.3, Rondeau et al. 2014; Lates calcarifer GCF_001640805.1, Vij et al. 2016; Nothobranchius furzeri GCF_001465895.1, Senf et al., Leibniz Institute for Age Research – Fritz Lipmann; Oncorhynchus mykiss GCF_002163495.1, Lien et al., Norwegian University of Life Sciences; and Takifugu rubripes GCF_000180615.1, Kai et al. 2011). In total, 71 and 472 scaffolds were detected as potentially contaminated by unicellular organisms or human DNA, respectively. NCBI’s blastn v.2.6.0 (Boratyn et al. 2013) was further applied to align those scaffolds to viral, bacterial, trypanosoma or to human refseq gene sequences. As the majority of the significant hits did not cover more than 1% of query sequence, all of the scaffolds were considered as non-contaminated and were retained for further analyses.
QUAST v.4.5 (Gurevich et al. 2013) was utilized to generate metrics for genome assembly and to compare it with the previously published P. fluviatilis assembly by Malmstrøm et al. (2017). Genome assembly completeness was estimated with BUSCO v.3.0 (Simão et al. 2015) using a ray-finned fishes (Actinopterygii obd9) database consisting of 4,584 orthologs from 20 fish species.
Transcriptome assembly
To assist in the subsequent genome annotation, we performed RNA sequencing and de novo transcriptome assembly, which was used to complement a perch transcriptome assembly published earlier (Pasquier et al. 2016). A total of 526 M reads were generated. Short (< 50 bp) and low quality reads (average quality ≤ 25) were trimmed using Trimmomatic v.0.35 (Bolger et al. 2014; SLIDINGWINDOW:5:25 MINLEN:50). rCorrector (Song and Florea 2015) was applied to correct random sequencing errors and remove erroneous k-mers from Illumina paired-end reads. Further, to reduce bias in downstream analyses due to over ribo-depletion (Lahens et al. 2014) the corrected trimmed reads were mapped to an rRNA database (SILVA Release 128; Pruesse et al. 2007). Finally, 419 M filtered reads were assembled de novo using Trinity v.2.3.2 (Haas et al. 2013) with default parameters. As our de novo transcriptome assembly was based only on eye tissue and its estimated BUSCO completeness was 79.1%, we combined it with the multi-tissue transcriptome assembly of perch published earlier (Pasquier et al. 2016) following the protocol described in Cerveau and Jackson (2016). The redundancy of the combined transcriptome assembly was further reduced using CD-HIT v.4.7 (Fu et al. 2012; Li and Godzik 2006). When two or more transcripts showed 90% or higher similarity all but the longest transcript were removed to generate non-redundant set of transcripts.
Repeat-content analysis
To identify repeats in the genome assembly, a de novo repeat library was first built based on the large scaffolds (≥ 10 Kb) using RepeatModeler v.1.0.11 (Smit and Hubley 2008-2015) with default parameters. The screening for repeats and low complexity sequences in the assembly was performed in RepeatMasker v.4.0.7 (Smit et al. 2013-2015) using de novo repeat library in combination with Dfam consensus 20171107 (Hubley et al. 2016) and RepBase 20170127 (Bao et al. 2015) repeat libraries.
Gene prediction and annotation
Gene models were constructed with MAKER v.2.31.8 (Holt and Yandell 2011), which incorporates ab initio gene prediction, homology-based prediction and RNA-seq assisted prediction. Prior to ab initio gene prediction, repeat regions of the perch genome were masked based on repeat annotation results. A total of three MAKER runs were performed. First, protein sequences from 11 other fish species from the Ensembl 91 database and the combined set of perch transcripts were aligned to the genome in an initial MAKER run as evidence to retrain Augustus v.3.2.2 (Stanke et al. 2006) and SNAP v.2006-07-28 (Korf 2004) ab initio gene prediction tools. The second and third runs of MAKER included gene models trained from the first (and then second) runs with ab initio gene prediction tools. Augustus was retrained within the BUSCO v.3.0 pipeline using genomic regions containing mRNA annotations from initial (and then second) MAKER run (including additional 1,000 bp on each side). BUSCO runs were performed using the –long option to optimize the HMM settings of the raw zebrafish HMM (–sp zebrafish; first run) or trained perch HMM (second run) and to generate the final trained perch HMM. Retraining of SNAP was performed using gene models from the initial (and then second) MAKER run with an annotation edit distance (AED) ≤ 0.25 and a length of amino acids ≥ 50. AED ranges from 0 to 1 and quantifies the congruency between a gene annotation and its supporting evidence (EST, protein and mRNA-seq alignments). Lower AED values imply higher congruency between the intron-exon coordinates of an annotation and its aligned evidence, whereas AED = 1 indicates no evidence for support of predicted genes. Only sequences with AED < 0.5 and CDS ≥ 90 bp were retained in the final set of predicted genes.
NCBI’s blastp v.2.6.0 (Boratyn et al. 2013; -evalue 1e-10, -soft_masking true, -lcase_masking, and a hit fraction filter to include only hits of > 70% target length, -qcov_hsp_perc 70) was used to functionally annotate the genes against vertebrate sequences in the NCBI non-redundant database. Further, non-annotated sequences were searched against all sequences in the NCBI non-redundant database. In addition, protein motifs, domains and signatures present in the predicted protein sequences were annotated using Interproscan v.5.24 (Jones et al. 2014) by searching against publicly available databases, including Pfam (Finn et al. 2014), PRINTS (Attwood et al. 2012), PROSITE (Sigrist et al. 2013), SMART (Letunic et al. 2012), SUPERFAMILY (de Lima Morais et al. 2011), and TIGRFAMs (Haft et al. 2013).
Data availability
Short Illumina linked-reads are available in the NCBI Sequence Read Archive (SRA; SRR7091761), and the Whole Genome Assembly has been deposited at DDBJ/EMBL/GenBank under the accession QFAT00000000, both under BioProject PRJNA450919. Transcriptome reads are available in the NCBI SRA (SRR7091762), and the eye Transcriptome Shotgun Assembly has been deposited at DDBJ/EMBL/GenBank under accession number GGNF00000000, as part of BioProject PRJNA450919. The combined transcriptome assembly from multiple tissues and Figure S1 have been uploaded as supplementary file to Figshare: https://doi.org/10.25387/g3.7156304.
Results and Discussion
Genome characteristics
Genome size estimates from GenomeScope ranged from 851.7 to 928.2 Mb, whereas estimates based on findGSE (fitted and original counts with corrected k-mer coverage) were higher and ranged from 1,050.9 Mb to 1,172.8 Mb (Table 1). Genome size estimates from both methods were in reasonable agreement with those determined earlier using cell flow cytometry (880.2 – 1,193.2 Mb; Vialli 1957; Vinogradov 1998). The analysis using GenomeScope indicated low heterozygosity (0.24–0.28%) in comparison with other species (e.g., Kajitani et al. 2014; Vurture et al. 2017). Low heterozygosity of the sequenced individual is also consistent with population genetic data from 16 perch populations screened using microsatellite markers, where lake Loosalu perch population showed the lowest level of genetic diversity (A. Vasemägi, unpublished results). Similar to other freshwater Perciformes (Yuan et al. 2018), the estimated proportion of repeats in perch was relatively high, ranging from 33.1% (k = 25, GenomeScope) to 55.0% (k = 17, findGSE).
Table 1. Genome size, heterozygosity and repeat content as estimated by GenomeScope and findGSE software.
| Genome characteristics | k-mer size | ||
|---|---|---|---|
| k = 17 | k = 21 | k = 25 | |
| GenomeScope | |||
| Genome haploid length (Mb) | 851.7 | 894.8 | 928.2 |
| Genome repeat length (Mb) | 426.9 | 306.9 | 307.4 |
| Genome unique length (Mb) | 424.8 | 587.9 | 620.8 |
| Heterozygosity, % | 0.28 | 0.26 | 0.24 |
| Estimated repetitive ratio,% | 50.1 | 34.3 | 33.1 |
| Read error rate, % | 0.14 | 0.18 | 0.19 |
| findGSE | |||
| Genome haploid length (Mb) | 1,050.9 | 1,163.8 | 1,172.8 |
| Genome repeat length (Mb) | 578.1 | 529.8 | 503.2 |
| Estimated repetitive ratio, % | 55.0 | 45.5 | 42.9 |
Genome assembly
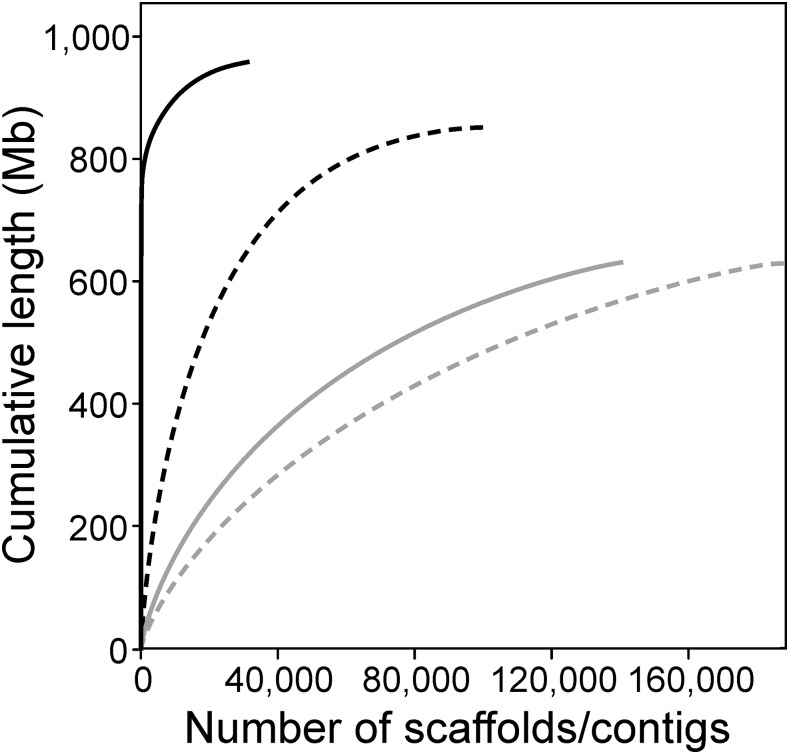
The total length of the assembly was 958.2 Mb, which included 106.6 Mb of unknown bases. The high number of unknown bases is typical for the Supernova assembler (e.g., Mohr et al. 2017; Hulse-Kemp et al. 2018), as it estimates gap sizes rather than introducing an arbitrary value of Ns during scaffolding (Weisenfeld et al. 2017). In the presented perch genome assembly, repeat regions were estimated to account for 32.72% (281.6 Mb). The contig N50 and scaffold N50 sizes were 18.2 Kb and 6.3 Mb, respectively (Table 2). More than 80% of the assembly was covered by the 516 longest scaffolds (≥ 50 Kb; 1.7% of all scaffolds). Compared to the draft perch genome assembly published by Malmstrøm et al. (2017) the contig and scaffold continuity (N50) metrics were improved by four and 1048 times, respectively (Figure 1, Table 2). The overall assembly size increased from 630.6 Mb to 958.2 Mb in comparison with the genome assembly by Malmstrøm et al. (2017) and was close to the estimates based on k-mer frequency distributions or cell flow cytometry.
Table 2. Eurasian perch genome assembly statistics.
| 10X Genome assembly* | Genome assembly by Malmstrøm et al. (2017)* | |
|---|---|---|
| Contig statistics | ||
| Number of contigs | 100,796 | 181,537 |
| Total contig size (bp) | 851,640,084 | 626,588,998 |
| Contig N50 size (bp) | 18,196 | 4,140 |
| Largest contig (bp) | 241,857 | 46,493 |
| Scaffold statistics | ||
| Number of scaffolds | 31,105 | 139,898 |
| Total scaffold size (bp) | 958,225,764 | 630,583,430 |
| Scaffold N50 size (bp) | 6,260,519 | 5,973 |
| Largest scaffold (bp) | 29,260,448 | 73,288 |
| GC/N (%) | 40.9/11.1 | 40.6/0.6 |
| BUSCO genome completeness | ||
| Complete | 4,033 (88.0%) | 2,144 (46.8%) |
| Complete and single copy | 3,933 (85.8%) | 2,105 (45.9%) |
| Complete and duplicated | 100 (2.2%) | 39 (0.9%) |
| Fragmented | 323 (7.0%) | 1246 (27.2%) |
| Missing | 228 (5.0%) | 1194 (26.0%) |
| Annotation | ||
| Number of protein-coding genes | 23,397 | |
| Number of functionally-annotated proteins | 23,171 | |
| Mean protein length (interquartile range, aa) | 506 (224-614) | |
| Longest protein (aa) | 8,907 (nesprin-1) | |
| Average number (length, interquartile range of length) of exon per gene | 9 (228, 89-189 bp) | |
| Average number (length, interquartile range of length) of intron per gene | 8 (1,224, 150-1,340 bp) |
Minimum scaffold length is 1 Kb.
Figure 1.
Cumulative length of the assembly represented by scaffolds (solid line) and contigs (dashed line). De novo perch genome assembly obtained using linked-reads (this study, black lines) and recently published genome assembly using Illumina short reads (Malmstrøm et al. 2017, gray lines).
Our perch genome assembly covered 88.0% complete and 7.0% partial ray-finned fishes BUSCOs, showing a substantial increase in completeness compared to the genome assembly by Malmstrøm et al. (2017) (46.8% complete and 27.2% partial BUSCOs).
Transcriptome assembly
The final concatenated perch transcriptome assembly based on multiple tissues consisted of 36,431 transcripts covering 96.2% complete and 1.3% partial ray-finned fish benchmarking universal single-copy orthologs (BUSCOs). The total transcriptome size was 108.7 Mb and the N50 transcript size was 3.9 Kb (Table 3).
Table 3. Eurasian perch transcriptome assembly statistics.
| Combined transcriptome assembly (multiple tissues) | |
|---|---|
| Transcript statistics | |
| Number of transcripts | 36,431 |
| Total transcript size (bp) | 108,727,847 |
| Transcript N50 size (bp) | 3,962 |
| Largest transcript (bp) | 78,856 |
| BUSCO transcriptome completeness | |
| Complete | 4,411 (96.2%) |
| Complete and single copy | 3,644 (79.5%) |
| Complete and duplicated | 767 (16.7%) |
| Fragmented | 58 (1.3%) |
| Missing | 115 (2.5%) |
Genome annotation
The final annotation of the P. fluviatilis genome from the MAKER annotation pipeline included 23,397 protein-coding genes (Table 2). NCBI’s blastp resulted in putative function annotation of 21,997 proteins (94.0%) based on homology. Further, Interproscan detected motifs, domains and signatures for 22,426 proteins (95.8%). As a result, 23,171 genes were annotated by at least one of the two methods (blastp 94.0%, InterProScan 95.8%), accounting for about 99.0% of the genes of P. fluviatilis (Table 2).
Conclusions
10X Genomics linked-read technology combined with low error rate short-read sequencing enabled accurate and more continuous de novo assembly of the genome of Eurasian perch. A large set of annotated genes with known homology revealed in our study (21,997) will ease further gene ontology and functional genomic analyses. In addition, improved scaffold length will facilitate detection of SNPs and structural variants, such as large insertions/deletions and copy number variations, potentially responsible for adaptation of P. fluviatilis to various environments. While a relatively large proportion of repeat regions in the perch genome still remain unresolved, generated short reads will be useful for future analyses of repetitive DNA elements. Taken together, the highly continuous assembly of the Eurasian perch genome presented in this study will serve as an invaluable resource for a range of genetic, ecological, physiological, ecotoxicological, functional and comparative genomic studies in perch and other fish species of the Percidae family.
Acknowledgments
This study was funded by the Academy of Finland (Academy fellow grant 266321 to A.V.), the Ella and Georg Ehrnrooth foundation (to F.A. and M.O.), the Estonian Ministry of Education and Research (institutional research funding project IUT8-2 to R.G.) and the Institute of Technology, University of Tartu (to V.K.). The authors wish to acknowledge CSC – IT Center for Science, Finland, for generous computational resources, and Finnish Functional Genomics Centre for their technical support. RNA extraction was carried out by Katja Salminen from the Center of Evolutionary Applications, University of Turku. Brian Desany from the 10X Genomics support team helped to solve software issues of the earlier versions of Supernova assembler. We thank Victoria Pritchard (University of Turku) for a manuscript language check.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7156304.
Communicating editor: D. Macqueen
Literature Cited
- Attwood T. K., Coletta A., Muirhead G., Pavlopoulou A., Philippou P. B., et al. , 2012. The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database (Oxford) 2012: bas019 10.1093/database/bas019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kojima K. K., Kohany O., 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergek S., Björklund M., 2009. Genetic and morphological divergence reveals local subdivision of perch (Perca fluviatilis L.). Biol. J. Linn. Soc. Lond. 96: 746–758. 10.1111/j.1095-8312.2008.01149.x [DOI] [Google Scholar]
- Bergek S., Sundblad G., Björklund M., 2010. Population differentiation in perch Perca fluviatilis: environmental effects on gene flow? J. Fish Biol. 76: 1159–1172. 10.1111/j.1095-8649.2010.02565.x [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., et al. , 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 (Web Server issue):W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveau N., Jackson D. J., 2016. Combining independent de novo assemblies optimizes the coding transcriptome for nonconventional model eukaryotic organisms. BMC Bioinformatics 17: 525 10.1186/s12859-016-1406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wang J., Qian L., Gaughan S., Xiang W., et al. , 2017. Domestication drive the changes of immune and digestive system of Eurasian perch (Perca fluviatilis). PLoS One 12: e0172903 10.1371/journal.pone.0172903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima Morais D. A., Fang H., Rackham O. J., Wilson D., Pethica R., et al. , 2011. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 39: D427–D434. 10.1093/nar/gkq1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W., 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach G., Schardt U., Eckmann R., Meyer A., 2001. Kin-structured subpopulations in Eurasian perch (Perca fluviatilis L.). Heredity (Edinb) 86: 213–221. 10.1046/j.1365-2540.2001.00825.x [DOI] [PubMed] [Google Scholar]
- Gremme G., Steinbiss S., Kurtz S., 2013. GenomeTools: A Comprehensive Software Library for Efficient Processing of Structured Genome Annotations. IEEE/ACM Trans. Comput. Biol. Bioinformatics 10: 645–656. 10.1109/TCBB.2013.68 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G., 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H., Selengut J. D., Richter R. A., Harkins D., Basu M. K., et al. , 2013. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 41: D387–D395. 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. A., Warren R. L., Vandervalk B. P., Kucuk E., Khan H., et al. , 2017. The North American bullfrog draft genome provides insight into hormonal regulation of long noncoding RNA. Nat. Commun. 8: 1433 10.1038/s41467-017-01316-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., et al. , 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. 10.1038/nature12111 (Erratum: Nature 505: 248. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R., Finn R. D., Clements J., Eddy S. R., Jones T. A., et al. , 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res. 44: D81–D89. 10.1093/nar/gkv1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse-Kemp A. M., Maheshwari S., Stoffel K., Hill T. A., Jaffe D., et al. , 2018. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5: 4 10.1038/s41438-017-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Sanders M., Berry A., McQuillan J., Aslett M. A., et al. , 2010. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl. Trop. Dis. 4: e658 10.1371/journal.pntd.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H. Y., Fraser M., Li W., et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. J. M., Taylor G. A., Chan S., Warren R. L., Hammond S. A., et al. , 2017. The Genome of the Beluga Whale (Delphinapterus leucas). Genes (Basel) 8: 378 10.3390/genes8120378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai W., Kikuchi K., Tohari S., Chew A. K., Tay A., et al. , 2011. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol. Evol. 3: 424–442. 10.1093/gbe/evr041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., et al. , 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. 10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S. M., Wan R., Sato K., Horton P., Frith M. C., 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21: 487–493. 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahens N. F., Kavakli I. H., Zhang R., Hayer K., Black M. B., et al. , 2014. IVT-seq reveals extreme bias in RNA sequencing. Genome Biol. 15: R86 10.1186/gb-2014-15-6-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P., 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. E., Myers R. M., 2016. Advancements in Next-Generation Sequencing. Annu. Rev. Genomics Hum. Genet. 17: 95–115. 10.1146/annurev-genom-083115-022413 [DOI] [PubMed] [Google Scholar]
- Li W., Godzik A., 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Li C., Liu X., Liu B., Ma B., Liu F., et al. , 2018. Draft genome of the Peruvian scallop Argopecten purpuratus. Gigascience 7: giy031 10.1093/gigascience/giy031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Chang S., Hartman G. L., Domier L. L., 2018. Assembly and annotation of a draft genome sequence for Glycine latifolia, a perennial wild relative of soybean. Plant J. 95: 71–85. 10.1111/tpj.13931 [DOI] [PubMed] [Google Scholar]
- Malmstrøm M., Matschiner M., Tørresen O. K., Jakobsen K. S., Jentoft S., 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci. Data 4: 160132 10.1038/sdata.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D. W., Naguib A., Weisenfeld N., Kumar V., Shah P., et al. , 2017. Improved de novo genome assembly: Linked-read sequencing combined with optical mapping produce a high quality mammalian genome at relatively low cost. bioRxiv: 128348 10.1101/128348 [DOI] [Google Scholar]
- Nesbø C. L., Fossheim T., Vøllestad L. A., Jakobsen K. S., 1999. Genetic divergence and phylogeographic relationships among european perch (Perca fluviatilis) populations reflect glacial refugia and postglacial colonization. Mol. Ecol. 8: 1387–1404. 10.1046/j.1365-294x.1999.00699.x [DOI] [PubMed] [Google Scholar]
- Olsson J., Mo K., Florin A. B., Aho T., Ryman N., 2011. Genetic population structure of perch Perca fluviatilis along the Swedish coast of the Baltic Sea. J. Fish Biol. 79: 122–137. 10.1111/j.1095-8649.2011.02998.x [DOI] [PubMed] [Google Scholar]
- Pasquier J., Cabau C., Nguyen T., Jouanno E., Severac D., et al. , 2016. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17: 368 10.1186/s12864-016-2709-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policar T., Samarin A. M., Mélard C., 2015. Culture Methods of Eurasian Perch During Ongrowing, pp. 417–435 in Biology and Culture of Percid Fishes: Principles and Practices, edited by Kestemont P., Dabrowski K., Summerfelt R. C. Springer Netherlands, Dordrecht: 10.1007/978-94-017-7227-3_16 [DOI] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., et al. , 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukk L., Ahmad F., Hasan S., Kisand V., Gross R., et al. , 2015. Less is more: extreme genome complexity reduction with ddRAD using Ion Torrent semiconductor technology. Mol. Ecol. Resour. 15: 1145–1152. 10.1111/1755-0998.12392 [DOI] [PubMed] [Google Scholar]
- Pukk L., Gross R., Vetemaa M., Vasemägi A., 2016. Genetic discrimination of brackish and freshwater populations of Eurasian perch (Perca fluviatilis L.) in the Baltic Sea drainage: implications for fish forensics. Fish. Res. 183: 155–164. 10.1016/j.fishres.2016.05.027 [DOI] [Google Scholar]
- Pukk L., Kuparinen A., Järv L., Gross R., Vasemägi A., 2013. Genetic and life-history changes associated with fisheries-induced population collapse. Evol. Appl. 6: 749–760. 10.1111/eva.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau E. B., Minkley D. R., Leong J. S., Messmer A. M., Jantzen J. R., et al. , 2014. The genome and linkage map of the northern pike (Esox lucius): Conserved synteny revealed between the salmonid sister group and the Neoteleostei. PLoS One 9: e102089 10.1371/journal.pone.0102089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J., de Castro E., Cerutti L., Cuche B. A., Hulo N., et al. , 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41: D344–D347. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., 2008–2015. RepeatModeler Open-1.0. Available at: http://www.repeatmasker.org. Accessed: June 6, 2018.
- Smit A. F. A., Hubley R., Green P., 2013–2015. RepeatMasker Open-4.0. Available at: http://www.repeatmasker.org. Accessed: June 6, 2018
- Sohn J.-i., Nam J.-W., 2018. The present and future of de novo whole-genome assembly. Brief. Bioinform. 19: 23–40. 10.1093/bib/bbw096 [DOI] [PubMed] [Google Scholar]
- Song L., Florea L., 2015. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. Gigascience 4: 48 10.1186/s13742-015-0089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Schöffmann O., Morgenstern B., Waack S., 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7: 62 10.1186/1471-2105-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Ding J., Piednoel M., Schneeberger K., 2018. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics 34: 550–557. 10.1093/bioinformatics/btx637 [DOI] [PubMed] [Google Scholar]
- Weisenfeld N. I., Kumar V., Shah P., Church D. M., Jaffe D. B., 2017. Direct determination of diploid genome sequences. Genome Res. 27: 757–767. 10.1101/gr.214874.116 (Erratum: ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialli M., 1957. Volume et contenu en ADN par noyau. Exp. Cell Res. Suppl. 4: 284–293. [PubMed] [Google Scholar]
- Vij S., Kuhl H., Kuznetsova I. S., Komissarov A., Yurchenko A. A., et al. , 2016. Chromosomal-level assembly of the Asian Seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet. 12: e1005954 10.1371/journal.pgen.1005954 (Erratum: PLOS Genetics 12: e1006500. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov A. E., 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry 31: 100–109. [DOI] [PubMed] [Google Scholar]
- Wood D. E., Salzberg S. L., 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15: R46 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurture G. W., Sedlazeck F. J., Nattestad M., Underwood C. J., Fang H., et al. , 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33: 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Liu S., Zhou T., Tian C., Bao L., et al. , 2018. Comparative genome analysis of 52 fish species suggests differential associations of repetitive elements with their living aquatic environments. BMC Genomics 19: 141 10.1186/s12864-018-4516-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. X. Y., Lau B. T., Schnall-Levin M., Jarosz M., Bell J. M., et al. , 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34: 303–311. 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Short Illumina linked-reads are available in the NCBI Sequence Read Archive (SRA; SRR7091761), and the Whole Genome Assembly has been deposited at DDBJ/EMBL/GenBank under the accession QFAT00000000, both under BioProject PRJNA450919. Transcriptome reads are available in the NCBI SRA (SRR7091762), and the eye Transcriptome Shotgun Assembly has been deposited at DDBJ/EMBL/GenBank under accession number GGNF00000000, as part of BioProject PRJNA450919. The combined transcriptome assembly from multiple tissues and Figure S1 have been uploaded as supplementary file to Figshare: https://doi.org/10.25387/g3.7156304.