Abstract
Background
Identification of biomarkers associated with the prognosis of different cancer subtypes is critical to achieve better therapeutic assistance. In colorectal cancer (CRC) the discovery of stable and consistent survival markers remains a challenge due to the high heterogeneity of this class of tumors. In this work, we identified a new set of gene markers for CRC associated to prognosis and risk using a large unified cohort of patients with transcriptomic profiles and survival information.
Results
We built an integrated dataset with 1273 human colorectal samples, which provides a homogeneous robust framework to analyse genome-wide expression and survival data. Using this dataset we identified two sets of genes that are candidate prognostic markers for CRC in stages III and IV, showing either up-regulation correlated with poor prognosis or up-regulation correlated with good prognosis. The top 10 up-regulated genes found as survival markers of poor prognosis (i.e. low survival) were: DCBLD2, PTPN14, LAMP5, TM4SF1, NPR3, LEMD1, LCA5, CSGALNACT2, SLC2A3 and GADD45B. The stability and robustness of the gene survival markers was assessed by cross-validation, and the best-ranked genes were also validated with two external independent cohorts: one of microarrays with 482 samples; another of RNA-seq with 269 samples. Up-regulation of the top genes was also proved in a comparison with normal colorectal tissue samples. Finally, the set of top 100 genes that showed overexpression correlated with low survival was used to build a CRC risk predictor applying a multivariate Cox proportional hazards regression analysis. This risk predictor yielded an optimal separation of the individual patients of the cohort according to their survival, with a p-value of 8.25e-14 and Hazard Ratio 2.14 (95% CI: 1.75–2.61).
Conclusions
The results presented in this work provide a solid rationale for the prognostic utility of a new set of genes in CRC, demonstrating their potential to predict colorectal tumor progression and evolution towards poor survival stages. Our study does not provide a fixed gene signature for prognosis and risk prediction, but instead proposes a robust set of genes ranked according to their predictive power that can be selected for additional tests with other CRC clinical cohorts.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5193-9) contains supplementary material, which is available to authorized users.
Keywords: Cancer, Colorectal cancer, Colon, Survival, Kaplan-Meier analysis, Gene marker, Bioinformatics, Transcriptomics, Gene Expression
Background
Colorectal cancer (CRC) is one of the most frequent tumors that causes great morbidity worldwide. It is the third most common cancer in men, the second most common cancer in women and the third leading cause of global cancer mortality (https://www.wcrf.org/). CRC is a heterogeneous disease since from one patient to another it differs in clinical presentation, molecular characteristics, and prognosis [1]. The heterogeneity of CRC increases the complexity of this tumoral pathology, making subtyping and stratification a difficult task for therapeutic decisions. In this way, personalized medicine for CRC is becoming increasingly needed, especially for targeted therapies where large variations between individual’s treatment responses exist [1, 2]. In this context, the need to find robust gene markers associated with specific subtypes of CRC led us to this study. Furthermore, the specific purpose of our work was to find consistent biomolecular targets that, together to facilitate samples stratification, could be related to the prognosis of the disease using survival data.
The genomic and transcriptomic profiling of human cancer samples has been demonstrated over the last decade as an excellent way to obtain a better molecular characterization of many tumor types and subtypes. While gene expression-based CRC classifications has been heavily approached [2], little consensus in CRC standalone gene bio-marking has been achieved. In fact, several studies have identified a broad variety of gene sets as gene expression profiles for classification and categorization of this malignant disorder [3, 4]. Moreover, several transcriptomic-based tests oriented towards prognosis have also been investigated. Some examples of these are: ColoLipidGene [5], ColoGuidePro [6] or ColoPrint [7]; that include gene signatures associated with CRC survival in some specific biological contexts. Despite these efforts, at present there is not a clear compendium of gene markers for CRC survival and it is quite difficult to find consistency in the literature.
In the clinic, patients are classified into four CRC stages based in the anatomo-pathologycal characteristics of their tumors. It is common to use the TNM Staging System (where T stands for tumor, N for lymph node, and M for metastasis). The disease “staging” also allows grouping the patients in 4 progressive cancer stages, indicated by roman numerals: I, II, III, and IV [8]. In this way, stages I and II correspond to cases which had not shown cancer cells beyond the tumor or blood. By contrast, stages III and IV correspond to individuals in where the cancer had diseminate to the lymph system or other organs in the body. This four stage categorization represents significantly distinctive patients groups for final outcome or disease relapse, but the stages do not predict the risk of each individual patient because they are not directly associated to survival [9].
Based on the described need and potential benefits to find survival marker genes correlated with high risk and poor prognosis in CRC; we investigated global gene expression profiles of colorectal tumors and its alteration throughout stages, to identify genes that could be levered as biomarkers of survival and prognosis for CRC in late stages (i.e., III and IV). To undertake this work we performed a deep analysis on a large cohort of human samples derived from a robust integration of several datasets that had transcriptomic and clinical survival data. The integration provided a homogeneous and well-standardized meta-dataset that includes 1273 human colorectal samples. The identification of candidate markers was performed using an initial contrast between the gene expression of the subset of patients with CRC allocated by their clinical features to stages I and II versus the patients with tumors corresponding to stages III and IV. Finally, after internal and external cross-validation, the genes selected as best survival markers were used to construct a risk predictor to allow stratification of the patients with respect to their relative risk.
Results
A large dataset of CRC samples including global expression and survival data
We first built a large cohort of CRC samples collected from individuals that had clinical record with survival data times, as well as genome-wide expression profiles of their colorectal primary tumors at diagnosis (i.e. before any drug treatment). Our aim was to achieve a meta-dataset with at least 1 thousand samples and to demonstrate a good integration of the global transcriptomic profiles of different samples sets avoiding the typical batch-effects that can alterate any unified analysis.
Table 1 presents the datasets of CRC samples that were collected to produce the integrated dataset analysed in this work. All the CRC samples included in this meta-dataset were tested for global gene expression profiling using the platform of high-density microarrays from Affymetrix: Human Genome U133 Plus 2.0. Using this platform, the probesets of the arrays were mapped to single genes (as indicated in Risueño et al.) [10] and, in this way, each microarray measured the expression signal of 20,079 human genes (using the mapping provided by the Chip Description File, CDF v.21 from: http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/).
Table 1.
Summary information about the series of colorectal cancer (CRC) samples that were collected to produce the integrated data set analyzed in this work
| GEO dataset | Sample Source | Sample Description | Total samples in dataset | PubMed PMID | Authors and Year | Samples discarded | Samples processed |
|---|---|---|---|---|---|---|---|
| GSE14333 | Royal Melbourne Hospital, Western Hospital and Peter MacCallum Cancer Center, AUSTRALIA. H Lee Moffitt Cancer Center, USA | primary colorectal cancers | 290 | 19996206 | Jorissen RN et al. (2009) | 64 | 226 |
| GSE17536 | Moffitt Cancer Center, USA | colorectal cancer patients | 177 | 19914252 | Smith JJ et al. (2010) | 0 | 177 |
| GSE31595 | Roskilde Hospital, DENMARK | patients with stage II and III colorectal cancer | 37 | – | Thorsteinsson M et al. (2011) | 0 | 37 |
| GSE33113 | Academic Medical Center in Amsterdam, NETHERLANDS | primary tumor resections from stage II colorectal patients | 90 | 22496204 | Kemper K et al. (2012) | 0 | 90 |
| GSE38832 | Vandervilt University Medical Center, USA | tumor samples collected from colorectal patients | 122 | 25320007 | Tripathi MK et al. (2014) | 0 | 122 |
| GSE39084 | Toulouse Hospital, FRANCE | sporadic early onset primary colorectal carcinomas | 70 | 25083765 | Kirzin S et al. (2014) | 1 | 69 |
| GSE39582 | Institut G. Roussy (Villejuif), Hosp. Saint Antoine (Paris), Hosp. G.Pompidou (Paris), Hosp. Hautepierre (Strasbourg), Hosp. Purpan (Toulouse), Institut P. Calmettes (Marseille), Centre Antoine Lacassagne (Nice), FRANCE | colorectal cancer samples | 566 | 23700391 | Marisa L et al. (2013) | 14 | 552 |
| Total number | 1352 | 1273 |
All the CRC samples were tested for global gene expression profiling using high-density microarrays Human Genome U133 Plus 2.0 from Affymetrix (that measure the signal of 20,141 human genes). The total collection included 1352 samples, but only 1273 were finally used. A group of 79 samples were discarded because they did not have survival data or they presented anomalous data distributions with respect to the other samples of the same series
As a whole, Table 1 includes 7 series that were obtained from the Gene Expression Omnibus repository (GEO, https://www.ncbi.nlm.nih.gov/geo/). These datasets included a total amount of 1352 CRC samples, but after collecting the clinical survival data and carrying out the integration and normalization protocols we finished with 1273 samples, since we filtered 79 samples that did not have survival data or did not show comparable data distributions after normalization. The phenotypic and clinical information about the final collection of 1273 samples, i.e., the available data about age, gender, survival time, location of the tumor, degree and TNM staging, presence of mutation in some cancer genes (TP53, KRAS, BRAF), etc.; is included in Additional file 1: Table S1. When information was not available for a given sample the table includes not assigned values (NA).
Evaluation of normalization procedures to integrate independent batches
We performed the integration and combined normalization of the CRC expression datasets using 5 different procedures. The procedures applied different normalization algorithms to provide a homogeneous signal matrix, avoiding bias due to batch effect on the global expression profile of the CRC samples. The procedures applied were: (i) Robust Multi-array Average (RMA) algorithm [11]; (ii) RMA plus Combatting Batch effects (ComBat) algorithm [12]; (iii) Frozen Robust Multi-array Average (fRMA) algorithm [13]; (iv) fRMA plus Combat; (v) fRMA plus scaling of the data using mean-centered expression values.
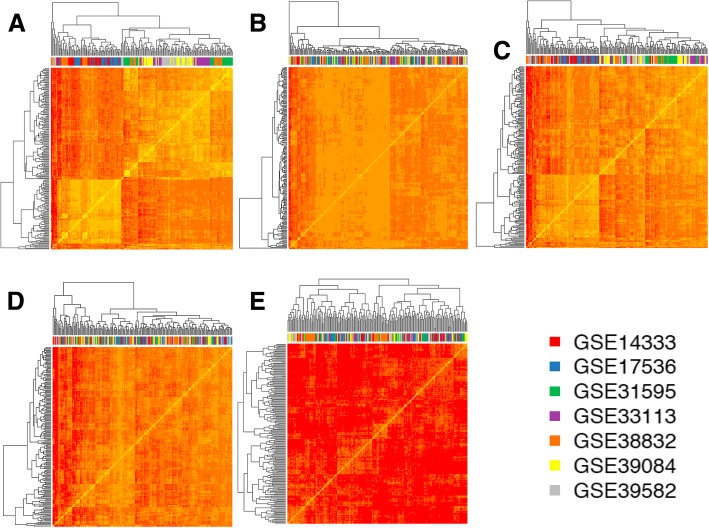
To evaluate and compare the results provided by each one of these 5 procedures we carried out several analyses. Figure 1 presents the heatmaps derived from an unsupervised clustering of the samples using in each case the expression data matrix derived from each one of the 5 procedures applied. Due to the fact that each series has a different number of samples (one with more than 500 and several other with less than 100), we did a random selection of an even number of samples for each dataset to be included in the cluster analysis: 30 samples from each one. In this way, each heatmap is composed of 210 samples (30 × 7): 30 samples from each one of the 7 datasets (identified by the ID number, GSE, from GEO). In Fig. 1 the samples of each batch are identified by a color that is indicated in the horizontal bar below the dendrograms. Each heatmap represents a different preprocessing and normalization method performed to merge the datasets in one meta-dataset. The results shown in these clustering analyses indicate that in the case of methods that gave the heatmaps A, C and E, several samples of the same color are grouped together showing that they have a common correlation profile within the global expression signature. By contrast, in the case of methods that gave the heatmaps B and D, there is a clearer shuffling of all the colors, which reflects a homogenous mix of the overall expression signal coming from different datasets.
Fig. 1.
Symmetric heatmaps representing the similarity between the overall gene expression signal of the samples compared with each other. Each heatmap is composed of 210 samples (30 × 7, 30 samples random selected from each batch, i.e. from each one of the 7 GSE datasets). The samples of each batch are identified by a color in the top bar below the top dendrograms (following the colors legend). Each heatmap represents a different preprocessing and normalization method performed to merge the datasets in one batch. The methods applied were: a RMA; b RMA plus ComBat; c fRMA; d fRMA plus ComBat; e fRMA plus scaling of the data using mean-centered expression values
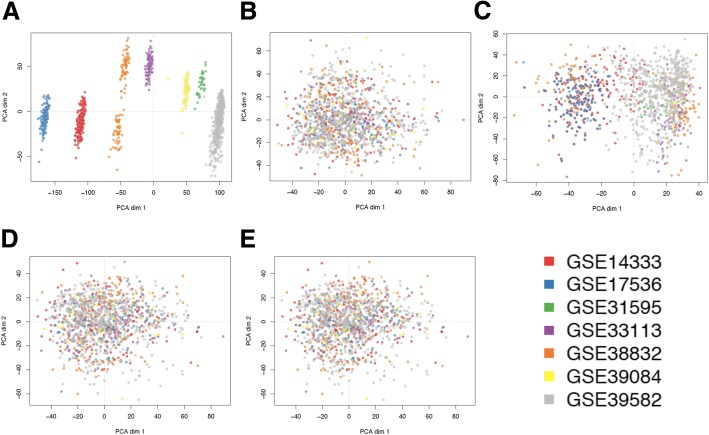
The clustering analysis presented in the symmetric heatmaps of Fig. 1 was done using, for each sample, a vector including the expression signals along all genes and calculating with these vectors the pair-wise Pearson correlations between samples and the pair-wise distance matrix derived from such correlations. This approach can reveal major effects associated to the global expression signal of the samples, but it is not very sensitive to detect minor changes in a small number of genes. For this reason we applied a second approach to compare the results provided by the 5 normalization procedures in order to select the one that produces the best unification of the 7 CRC datasets, preserving a good signal to noise ratio in the expression distributions. Algorithms of dimensionality reduction, such as PCA (Principal Component Analysis), allow exploring large datasets in an accurate way to identify factors that are relevant for the variance of studied variables (in our case the expression of the genes in the unified meta-dataset of 1273 samples). Figure 2 presents the plots derived from the PCA done over the 5 expression matrices (i.e. the signal of 20,079 genes in 1273 samples) obtained with 5 different normalization approaches. These results show very clearly that the RMA method (Fig. 2a) is not good to provide a proper normalization of different batches, since the samples keep a very strong signal associated to each batch. The fRMA method (Fig. 2c) neither is good, since some samples (specially the ones from the largest batch GSE39582) still keep a strong signal associated to their batch. By contrast, the analysis of the data provided by the other 3 procedures (RMA plus Combat, fRMA plus Combat and fRMA plus mean-centered scaling, Fig. 2b, d and e, respectively) showed an adequate mix of all the samples from different batches. Within these 3 procedures, the normalization is very similar keeping a good signal to noise ratio along the genes and a small signal reduction. We finally select option B, RMA plus Combat, because the heatmap in Fig. 1b showed the best mix between series and a better similarity between the samples (compared to options D or E).
Fig. 2.
Plots presenting the distribution of the 1273 samples from 7 datasets (GSEs) obtained by Principal Component Analysis (PCA) of the global gene expression profile of each sample; that converts the signal of each sample using an orthogonal transformation in linearly uncorrelated variables called principal components or dimensions. Each plot presents the values of the two main dimensions (dim 1 versus dim 2) and corresponds to the PCA results obtained using the expression data calculated with different preprocessing and normalization methods. The methods applied were: a RMA; b RMA plus ComBat; c fRMA; d fRMA plus ComBat; e fRMA plus scaling of the data using mean-centered expression values. The samples of each batch are identified by color dots following the colors legend
As a final testing to identify the best integration and normalization procedure of the 7 CRC expression datasets, we carried out a linear regression analyses on the global expression matrix considering as predictors 7 independent dummy variables or factors. These variables correspond to the series from which each sample comes from. In this way, if these factors have a significant influence in the expression signal distributions, the linear regression analysis will show a significant p-value and correlation. The results of this analysis are presented in Table 2, that reveals again that only the data matrices produced by the methods B and D (RMA plus Combat and fRMA plus Combat, respectively) do not show a significant effect attributed to belonging to one of the series. Finally, we choose B versus D as the final procedure applied because, despite being very similar, the application of RMA plus Combat provoked less dramatic changes with respect to the raw signal expression.
Table 2.
Results of the linear regression analyses on the global expression matrix calculated for the 1273 samples from 7 datasets (GSEs) combined using 5 different preprocessing and normalization methods
| FACTORS considered | Estimated coefficients | std. error | t value | p.value | Factor effect |
|---|---|---|---|---|---|
| (A) RMA | |||||
| Intercept | 6.925 | 0.014 | 512.610 | <2e-16 | – |
| (GSE14333+) GSE17536 | 0.387 | 0.019 | 20.230 | <2e-16 | yes |
| GSE31595 | −1.212 | 0.019 | −63.440 | <2e-16 | yes |
| GSE33113 | −0.577 | 0.019 | −30.210 | <2e-16 | yes |
| GSE38832 | −0.355 | 0.019 | −18.570 | <2e-16 | yes |
| GSE39084 | −0.978 | 0.019 | −51.180 | <2e-16 | yes |
| GSE39582 | −1.375 | 0.019 | −71.970 | <2e-16 | yes |
| (B) RMA plus Combat | |||||
| Intercept | 6.219 | 0.013 | 473.582 | <2e-16 | – |
| (GSE14333+) GSE17536 | 0.000 | 0.019 | 0.001 | 0.999 | no |
| GSE31595 | 0.002 | 0.019 | 0.122 | 0.903 | no |
| GSE33113 | 0.001 | 0.019 | 0.051 | 0.959 | no |
| GSE38832 | −0.001 | 0.019 | −0.033 | 0.973 | no |
| GSE39084 | 0.002 | 0.019 | 0.092 | 0.927 | no |
| GSE39582 | 0.001 | 0.019 | 0.029 | 0.977 | no |
| (C) fRMA | |||||
| Intercept | 6.535 | 0.015 | 450.434 | <2e-16 | – |
| (GSE14333+) GSE17536 | −0.011 | 0.021 | −0.553 | 0.580 | no so much |
| GSE31595 | 0.089 | 0.021 | 4.329 | 0.000 | yes |
| GSE33113 | 0.071 | 0.021 | 3.455 | 0.001 | yes |
| GSE38832 | 0.054 | 0.021 | 2.641 | 0.008 | yes |
| GSE39084 | 0.096 | 0.021 | 4.695 | 0.000 | yes |
| GSE39582 | 0.089 | 0.021 | 4.336 | 0.000 | yes |
| (D) fRMA plus Combat | |||||
| Intercept | 6.590 | 0.014 | 457.338 | <2e-16 | – |
| (GSE14333+) GSE17536 | 0.000 | 0.020 | 0.001 | 1.000 | no |
| GSE31595 | 0.002 | 0.020 | 0.093 | 0.926 | no |
| GSE33113 | 0.001 | 0.020 | 0.072 | 0.942 | no |
| GSE38832 | 0.000 | 0.020 | 0.019 | 0.985 | no |
| GSE39084 | 0.002 | 0.020 | 0.089 | 0.929 | no |
| GSE39582 | 0.000 | 0.020 | 0.007 | 0.994 | no |
| (E) fRMA plus mean centered | |||||
| Intercept | 0.000 | 0.000 | −1.638 | 0.101 | – |
| (GSE14333+) GSE17536 | 0.000 | 0.000 | 1.264 | 0.206 | yes |
| GSE31595 | 0.000 | 0.000 | 0.288 | 0.773 | no so much |
| GSE33113 | 0.000 | 0.000 | 1.605 | 0.108 | yes |
| GSE38832 | 0.000 | 0.000 | 1.449 | 0.147 | yes |
| GSE39084 | 0.000 | 0.000 | −0.076 | 0.940 | no |
| GSE39582 | 0.000 | 0.000 | 1.395 | 0.163 | yes |
The methods applied were: (A) RMA; (B) RMA plus ComBat; (C) fRMA; (D) fRMA plus ComBat; (E) fRMA plus scaling of the data using mean-centered expression values. The linear regression is done to evaluate the “batch effect” (i.e. considering that the tested factors are the fact of “belonging” to a given dataset). Thus, when the p-value of the factors are significant (< 0.05), the “batch effect” remains on the overall expression signal. A marginal low significance was considered when p-values were < 0.20 in the case E
Identification of genes associated to advanced CRC that mark survival differences
Once we produced a large and well-integrated meta-dataset of CRC samples, having global expression profiles and clinical survival data for all cases, we proceed to the identification of the subset of genes that suffer significant changes with colorectal tumor progression. To do this, we explored the overall expression matrix to detect the genes that showed a significant expression change when comparing CRC tumors in early stages (stages I and II) versus CRC tumors in late or advanced stages (stages III and IV). This comparison was done applying LIMMA, differential expression algorithm, and retrieving all genes that gave a significant p-value (adjusted p < 0.05) in either direction (i.e., genes up-regulated with the progression of the disease, in late versus early CRC stages; or genes down-regulated with the progression of the disease). Such differential expression analysis gave a subset of 2707 human genes: 2524 corresponding to protein-coding genes and the rest to non-coding genes (in this work we focused only in the protein-coding genes).
Once we had the subset of genes that can be associated to advanced or progression of CRC, we perform a second analysis on these gene candidates to find out which ones can be correlated with the survival of the corresponding patient samples based on their expression signals. To do this, we carried out Kaplan-Meier (KM) analysis of the survival times of the set of 1273 colorectal cancer samples for each one of the 2524 genes found in the previous exploration. In this analysis, the genes were ranked considering the non-parametric log-rank test that evaluates the separation between the two KM curves for two prognostic groups: one with good survival and another with poor survival. To do this, our algorithm performs for each gene multiple splits of the sample cohort in two groups, and looks for the splitting that provides the best separation between groups (i.e. the best p-value). Then, a stringent cut-off value (adjusted p < 0.0003) was used to select the genes that are considered significant. This allowed the identification of 429 significant genes in which the overexpression correlated with low survival, plus 336 significant genes where the repression correlated with low survival. These analyses were done in a univariate mode, considering each gene as an independent factor.
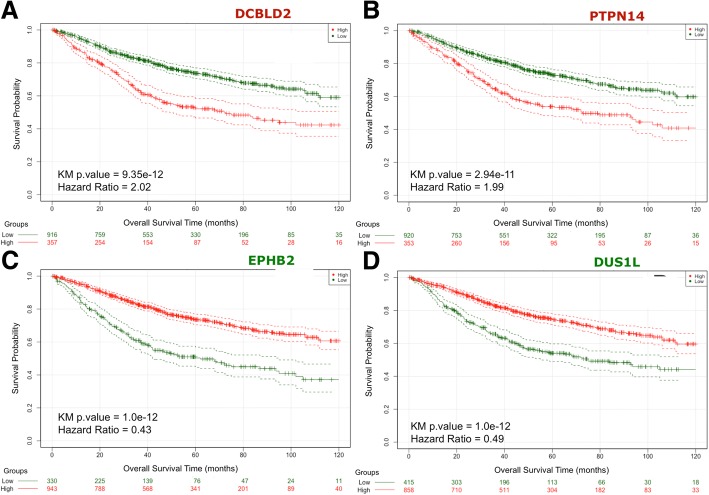
Figure 3 shows the Kaplan-Meier plots corresponding to the survival profiles of the two populations of individuals that were segregated according to the expression values of the gene tested. The 4 plots correspond to the top genes: DCBLD2 and PTPN14 with overexpression correlated to low survival; and EPHB2 and DUS1L with repression correlated to low survival. The separation of the two populations in both cases is very significant, with KM p-values < 1.0e-10 and Hazard Ratios (HR) around 2.0 for overexpression cases and around 0.45 for repression cases. These parameters were calculated using all the 1273 samples; however it was necessary to do an internal cross-validation of these results to assess how stable and reliable was the signal for each one of the selected genes.
Fig. 3.
Kaplan-Meier plots of the survival analysis of the set of 1273 samples from colorectal cancer (CRC) patients. The patients are separated in two groups (high in red and low in green) according to the expression profiles of 4 genes: a DCBLD2, b PTPN14, c EPHB2, d DUS1L. These genes provided the best split between patients of high and low risk based in their expression levels. In the case of genes DCBLD2 and PTPN14 (labelled in red) the over-expression is correlated with poor survival; and in the case of genes EPHB2 and DUS1L (labelled in green) the over-expression is correlated with good survival. In all cases the adjusted p.values of the analyses are very significant (as indicated inside each plot), indicating that the two populations represented by the two curves have a very clear difference in their overall survival
We carried out a cross-validation of the top-200 genes selected in any of the two conditions (i.e. selected as survival markers when they were up-regulated for the cases of poor survival or when they were up-regulated for the cases of better survival). This internal cross-validation was done using for each gene a resampling strategy that randomly selected 80% of the sample 100 times (i.e. doing 100 iterations). The results corresponding to the top 100 genes are included in Additional file 2: Table S2, for the case of up-regulation for poor survival, and the other top 100 genes in Additional file 3: Table S3, for the case up-regulation for better survival.
A short view of these data is shown in Table 3 that presents the 50 genes selected as best survival markers of CRC: the first part of the table corresponds to the top 25 genes, where up-regulation corresponds to shorter survival and higher risk (HR > 1); the second part of the table corresponds to the top 25 genes, where up-regulation corresponds to longer survival and lower risk (HR < 1). The genes were ranked by their KM p-values and the HR values calculated for the whole dataset (i.e. for all the 1273 samples, all-dt). As indicated, the stability and robustness of the gene survival markers was assessed via a resampling strategy with random selection of 80% of the dataset 100 times. For the final ranking of the genes included in these tables we also considered that they had to give a significant adjusted p-value in more than 80 out of 100 bootstrap iterations (i.e. N-sinf-in-100i > 80).
Table 3.
Genes selected as top-50 best survival markers of colorectal cancer (CRC)
| Number | GENE ENSEMBL_ID | GENE Symbol | KM.p.value (all-dt) | HR (all-dt) | N-signf-in-100i (KM.p.value) | HR (mean-in-100i) | GENE HGNC_ID | GENE DESCRIPTION |
|---|---|---|---|---|---|---|---|---|
| 1 | ENSG00000057019 | DCBLD2 | 0.0000000000 | 2.02 | 99 | 2.106 | 24627 | discoidin; CUB and LCCL domain containing 2 [HGNC:24627] |
| 2 | ENSG00000152104 | PTPN14 | 0.0000000000 | 1.99 | 99 | 2.082 | 9647 | protein tyrosine phosphatase; non-receptor type 14 |
| 3 | ENSG00000125869 | LAMP5 | 0.0000000000 | 1.99 | 93 | 2.046 | 16097 | lysosomal associated membrane prot.member 5 [HGNC:16097] |
| 4 | ENSG00000169908 | TM4SF1 | 0.0000000001 | 1.96 | 93 | 2.031 | 11853 | transmembrane 4 L six family member 1 [HGNC:11853] |
| 5 | ENSG00000113389 | NPR3 | 0.0000000002 | 1.95 | 97 | 2.136 | 7945 | natriuretic peptide receptor 3 [HGNC:7945] |
| 6 | ENSG00000186007 | LEMD1 | 0.0000000003 | 1.95 | 85 | 1.937 | 18,725 | LEM domain containing 1 [HGNC:18725] |
| 7 | ENSG00000135338 | LCA5 | 0.0000000003 | 1.89 | 97 | 2.021 | 31,923 | LCA5; lebercilin [HGNC:31923] |
| 8 | ENSG00000169826 | CSGALNACT2 | 0.0000000008 | 1.91 | 92 | 1.974 | 24,292 | chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| 9 | ENSG00000059804 | SLC2A3 | 0.0000000014 | 1.93 | 89 | 1.993 | 11,007 | solute carrier family 2 member 3 [HGNC:11007] |
| 10 | ENSG00000099860 | GADD45B | 0.0000000018 | 1.92 | 97 | 2.074 | 4096 | growth arrest and DNA damage inducible beta [HGNC:4096] |
| 11 | ENSG00000136155 | SCEL | 0.0000000018 | 1.88 | 87 | 1.928 | 10,573 | sciellin [HGNC:10573] |
| 12 | ENSG00000100625 | SIX4 | 0.0000000019 | 1.89 | 91 | 1.951 | 10,890 | SIX homeobox 4 [HGNC:10890] |
| 13 | ENSG00000131016 | AKAP12 | 0.0000000028 | 1.85 | 95 | 2.092 | 370 | A-kinase anchoring protein 12 [HGNC:370] |
| 14 | ENSG00000158270 | COLEC12 | 0.0000000028 | 1.84 | 92 | 1.941 | 16,016 | collectin subfamily member 12 [HGNC:16016] |
| 15 | ENSG00000154553 | PDLIM3 | 0.0000000047 | 1.84 | 91 | 1.985 | 20,767 | PDZ and LIM domain 3 [HGNC:20767] |
| 16 | ENSG00000082781 | ITGB5 | 0.0000000049 | 1.82 | 88 | 1.911 | 6160 | integrin subunit beta 5 [HGNC:6160] |
| 17 | ENSG00000144366 | GULP1 | 0.0000000050 | 1.81 | 88 | 1.911 | 18,649 | engulfment adaptor PTB domain containing 1 [HGNC:18649] |
| 18 | ENSG00000171951 | SCG2 | 0.0000000051 | 1.81 | 93 | 2.034 | 10,575 | secretogranin II [HGNC:10575] |
| 19 | ENSG00000185567 | AHNAK2 | 0.0000000066 | 1.80 | 87 | 1.896 | 20,125 | AHNAK nucleoprotein 2 [HGNC:20125] |
| 20 | ENSG00000138061 | CYP1B1 | 0.0000000075 | 1.84 | 85 | 1.884 | 2597 | cytochrome P450 family 1 subfamily B member 1 [HGNC:2597] |
| 21 | ENSG00000184304 | PRKD1 | 0.0000000451 | 1.74 | 87 | 1.872 | 9407 | protein kinase D1 [HGNC:9407] |
| 22 | ENSG00000152583 | SPARCL1 | 0.0000000471 | 1.74 | 85 | 1.863 | 11,220 | SPARC like 1 [HGNC:11220] |
| 23 | ENSG00000147883 | CDKN2B | 0.0000000717 | 1.73 | 84 | 1.847 | 1788 | cyclin dependent kinase inhibitor 2B [HGNC:1788] |
| 24 | ENSG00000213190 | MLLT11 | 0.0000001989 | 1.70 | 84 | 1.813 | 16,997 | myeloid/lymphoid or mixed-lineage leukemia; translocated to 11 |
| 25 | ENSG00000135218 | CD36 | 0.0000002751 | 1.69 | 85 | 1.891 | 1663 | CD36 molecule [HGNC:1663] |
| 1 | ENSG00000133216 | EPHB2 | 0.0000000000 | 0.43 | 100 | 0.426 | 3393 | EPH receptor B2 [HGNC:3393] |
| 2 | ENSG00000169718 | DUS1L | 0.0000000000 | 0.49 | 98 | 0.481 | 30,086 | dihydrouridine synthase 1 like [HGNC:30086] |
| 3 | ENSG00000163545 | NUAK2 | 0.0000000001 | 0.51 | 96 | 0.495 | 29,558 | NUAK family kinase 2 [HGNC:29558] |
| 4 | ENSG00000158169 | FANCC | 0.0000000002 | 0.51 | 95 | 0.498 | 3584 | Fanconi anemia complementation group C [HGNC:3584] |
| 5 | ENSG00000277972 | CISD3 | 0.0000000002 | 0.51 | 87 | 0.511 | 27,578 | CDGSH iron sulfur domain 3 [HGNC:27578] |
| 6 | ENSG00000099800 | TIMM13 | 0.0000000003 | 0.53 | 95 | 0.511 | 11,816 | translocase of inner mitochondrial membrane 13 [HGNC:11816] |
| 7 | ENSG00000116771 | AGMAT | 0.0000000005 | 0.52 | 95 | 0.515 | 18,407 | agmatinase [HGNC:18407] |
| 8 | ENSG00000118513 | MYB | 0.0000000006 | 0.52 | 93 | 0.508 | 7545 | MYB proto-oncogene. Transcription factor [HGNC:7545] |
| 9 | ENSG00000016391 | CHDH | 0.0000000006 | 0.53 | 90 | 0.520 | 24,288 | choline dehydrogenase [HGNC:24288] |
| 10 | ENSG00000137460 | FHDC1 | 0.0000000008 | 0.52 | 96 | 0.505 | 29,363 | FH2 domain containing 1 [HGNC:29363] |
| 11 | ENSG00000132846 | ZBED3 | 0.0000000009 | 0.52 | 88 | 0.522 | 20,711 | zinc finger BED-type containing 3 [HGNC:20711] |
| 12 | ENSG00000162408 | NOL9 | 0.0000000015 | 0.54 | 92 | 0.527 | 26,265 | nucleolar protein 9 [HGNC:26265] |
| 13 | ENSG00000109534 | GAR1 | 0.0000000017 | 0.50 | 99 | 0.479 | 14,264 | GAR1 ribonucleoprotein [HGNC:14264] |
| 14 | ENSG00000133477 | FAM83F | 0.0000000019 | 0.54 | 93 | 0.518 | 25,148 | family with sequence similarity 83 member F [HGNC:25148] |
| 15 | ENSG00000100348 | TXN2 | 0.0000000036 | 0.53 | 88 | 0.527 | 17,772 | thioredoxin 2 [HGNC:17772] |
| 16 | ENSG00000108479 | GALK1 | 0.0000000036 | 0.55 | 88 | 0.525 | 4118 | galactokinase 1 [HGNC:4118] |
| 17 | ENSG00000110917 | MLEC | 0.0000000045 | 0.55 | 96 | 0.476 | 28,973 | malectin [HGNC:28973] |
| 18 | ENSG00000114738 | MAPKAPK3 | 0.0000000048 | 0.55 | 92 | 0.520 | 6888 | mitogen-activated protein kinase-activated 3 [HGNC:6888] |
| 19 | ENSG00000137752 | CASP1 | 0.0000000180 | 0.56 | 87 | 0.523 | 1499 | caspase 1 [HGNC:1499] |
| 20 | ENSG00000131844 | MCCC2 | 0.0000000183 | 0.57 | 93 | 0.516 | 6937 | methylcrotonoyl-CoA carboxylase 2 [HGNC:6937] |
| 21 | ENSG00000178409 | BEND3 | 0.0000000193 | 0.55 | 88 | 0.529 | 23,040 | BEN domain containing 3 [HGNC:23040] |
| 22 | ENSG00000114737 | CISH | 0.0000000216 | 0.55 | 87 | 0.508 | 1984 | cytokine inducible SH2 containing protein [HGNC:1984] |
| 23 | ENSG00000011376 | LARS2 | 0.0000000239 | 0.55 | 91 | 0.528 | 17,095 | leucyl-tRNA synthetase 2; mitochondrial [HGNC:17095] |
| 24 | ENSG00000164045 | CDC25A | 0.0000000481 | 0.57 | 90 | 0.539 | 1725 | cell division cycle 25A [HGNC:1725] |
| 25 | ENSG00000154655 | L3MBTL4 | 0.0000000606 | 0.54 | 90 | 0.506 | 26,677 | l(3)mbt-like 4 (Drosophila) [HGNC:26677] |
The first part of the table corresponds to the top-25 genes where up-regulation corresponds to shorter survival and higher risk (i.e., HR > 1); the second part of the table corresponds to the top-25 genes where UP-regulation corresponds to longer survival and lower risk (HR < 1). The genes were ranked by their KM adjusted p values and the Hazard Ratio values calculated for the whole dataset, i.e. for all the 1273 samples (all-dt). The stability and robustness of the gene survival markers was assessed by cross-validation, applying to each gene a resampling strategy with random selection of 80% of the samples 100 times (i.e. doing 100 iterations). For the ranking we also considered that the genes had to give a significant adjusted p-value in more than 80 iterations (N-sinf-in-100i > 80)
External validation of prognostic markers with a CRC cohort studied using RNA-seq
The analyses done so far provided a ranked collection of genes found as robust markers of survival in CRC. The consistency of the results obtained with the internal cross-validation gives strong support to the top genes found (presented in Table 3), but we had to consider the value of using other external independent CRC cohorts to corroborate these findings. As far as we could investigate we did not find other large CRC datasets (i.e., sets with more than one thousand samples) that included global gene expression data plus survival as part of the clinical characterization of samples. Despite this limitation, we look for independent datasets and found in The Cancer Genome Atlas (TCGA, http://tcga-data.nci.nih.gov/docs/publications/coadread_2012/) a well-characterized cohort of 276 colorectal carcinomas that had been studied with several genome-scale technologies (including RNA-seq gene expression profiling) and that had survival data for 269 samples [14]. We used these data to validate the top genes found as best survival markers in our previous analysis. The results indicated a good performance in more than two thirds of the genes tested. In Additional file 4: Table S4 we present the KM p-values and HR of the genes that were validated from the top 10 previously found: 7 genes of the top 10 for the case of up-regulation associated with poor survival (PTPN14, LAMP5, TM4SF1, LCA5, CSGALNACT2, SLC2A3 and GADD45B) and 6 genes of the top 10 previously found for the case of up-regulation associated with good survival (EPHB2, DUS1L, NUAK2, FANCC, MYB and CHDH).
External validation of prognostic markers using multivariate survival analysis
Up to now the search to find gene survival markers associated to the prognosis of CRC have been done using univariate analysis that look for the value and influence of each singular gene. The results presented provided multiple parameters to allow a proper statistical assessment and ranking of each gene survival markers proposed (Table 3). To provide extra support to these results we did another external validation using a second independent cohort of CRC samples from the platform SurvExpress [15]. The CRC dataset selected was called “Colon-Metabase-Uniformized” and it included 482 samples with overall survival data and genome-wide expression determined with Affymetrix microarrays. We performed several multivariate survival analyses (OS, overall survival) on this dataset using combinations of the top genes proposed in Table 3. As an example of these analyses we present the KM plot (Additional file 5: Figure S1) corresponding to the multivariate survival study done using the top 5 genes found up-regulated for poor survival (DCBLD2, PTPN14, LAMP5, TM4SF1 and NPR3). It can be seen that the combination of these genes provides a very good separation of two CRC populations: one group of high-risk, associated to the overexpression (or up-regulation) of the genes; and another group of low-risk, associated to the lower expression (or down-regulation) of these genes (Additional file 5: Figure S1). This analysis was repeated with several other combinations of the top up-regulated genes associated with poor survival (present in Table 3), resulting in similar results. For example, combining DCBLD2, LAMP5, TM4SF1, NPR3 and GADD45B the separation of the high and low-risk groups improved a bit: KM p-value = 2.21e-07 and HR = 2.23 (95% confidence interval, CI: 1.65–3.02). Another combination that provided very good separation was using genes DCBLD2, LAMP5, TM4SF1, NPR3 and AKAP12: KM p-value = 2.51e-10 and HR = 2.74 (95% CI: 2.00–3.74).
Gene expression profiles of CRC tumor samples versus normal colorectal samples
All the integrated datasets, so far presented in this study corresponded to CRC samples, because we want to provide genes that are disease markers present in the transformed tumor cells of the intestinal epithelium, and genes that mark the progression and aggravation of this type of cancer. In addition, we can only have survival information about patients since in healthy individuals survival time cannot be related to disease and there are not disease-associated events. Despite this obvious consideration, it is interesting to explore what would be the level of expression of the genes, that we identified as survival markers, when they are analysed in normal colorectal tissue. Exploring back on the experimental series used to create our meta-dataset of 1273 CRC samples, we found in series GSE33113 and GSE39582 a collection of 25 samples that corresponded to normal colorectal tissue. We took these samples and included then with our CRC dataset using the same normalization protocol. After this integration, we could explore the expression level of the top up-regulated genes (identified as markers of poor survival), comparing the expression distribution on a set of cancer samples versus a set of normal tissue samples. In both cases the number of samples compared were 25, since this is the number of normal samples that we had. We did this comparison 20 times, random selecting each time a different subset of 25 cancer samples. The results were always very similar and the boxplots of the expression distributions for the top 10 genes are presented in Additional file 6: Figure S2. These results indicate that the gene markers, identified in our survival studies, are most of the times also up-regulated in CRC tumors with respect to normal colorectal tissue.
Risk predictor score based in the multivariate analysis of candidate survival markers
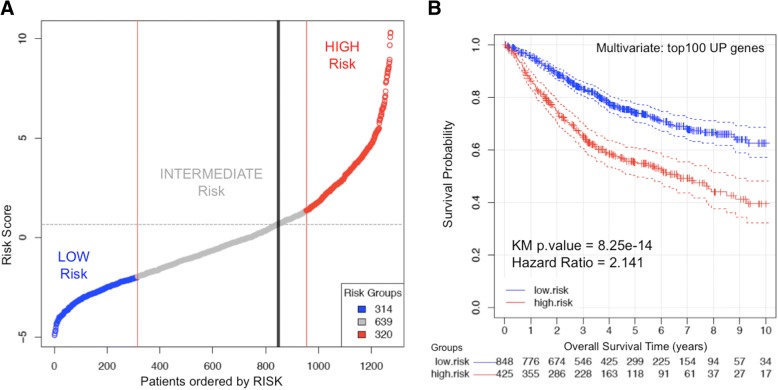
Finally, to obtain a more accurate evaluation of the prognostic value of all the genes selected as best candidates (reported in Additional files 2 and 3, Table S2 and Table S3), we performed another analysis of the candidate markers using a regularized multivariate Cox proportional-hazards regression with L1 norm penalty [16], with the scope of building a multigenic “risk predictor”. This analysis was done on the cohort of 1273 samples of CRC patients, using for the multivariate analysis the top 100 genes that showed up-regulation correlated with poor prognosis (i.e. overexpressed in low survival cases). The results are presented in Fig. 4 that shows a graph ordering the patients according to their risk score, from low-risk (blue) to high-risk (red), including also an intermediate region (grey) (Fig. 4a). A recursive algorithm using 10-fold cross-validation was applied to find the value of risk score. The threshold (marked with a vertical black line) is obtained by maximizing the separability between the survival curves for the resulting groups. Therefore, it allows the best splitting of the cohort in two groups. A Kaplan-Meier plot showing the separation of these two groups is also presented (Fig. 4b); dividing the population into a high risk group including 425 individuals and a low risk group including 848 individuals. As shown, the division is significant (p-value = 8.25e-14) and allows an optimal separation of individuals according to their survival. The analysis of the beta factors assigned by the regression to each of the top 100 genes, i.e. to each variable within the multivariate vector (data included in Additional file 7: Table S5), allows the identification of the genes that were the most influential factors in this risk analysis and therefore it facilitated the selection of the best “gene survival markers”. As indicated in previous sections, the top 100 genes included in the construction of this multigenic risk predictor score were selected from the list of best markers found during the survival test with single genes.
Fig. 4.
Risk prediction done for the cohort of 1273 patients of CRC based in the multivariate analysis using the top 100 genes that showed up-regulation correlated with poor prognosis (i.e. overexpressed in low survival cases). a Plot presenting the patients according to their risk score, from Low (blue) to High (red) risk. A recursive algorithm using 10-fold cross-validation finds the value of risk score (marked with a vertical black line) that allows the best splitting of the cohort in two groups. b Kaplan-Meier plot showing the separation of these two groups: a high-risk group including 425 individuals (in red) and a low-risk group including 848 individuals (in blue). The analysis has been done using a multivariate Cox proportional-hazards regression. As shown, the division is very significant (p-value = 8.25e-14) and allows an optimal separation of individuals according to their survival. The analysis of the beta factors assigned by the regression to each of the top 100 genes (i.e. to each variable within the multivariate vector) allows the identification of the genes that are the most influential factors in this risk analysis and therefore it helps in the selection of the best “gene survival markers”
Discussion
CRC is a complex disease composed of biologically and clinically diverse subtypes, which can originate in different ways provoking multiple clinical scenarios [1, 2]. This complexity causes the molecular characterization of CRC to remain deficient, with a lack of clear gene markers associated to specific CRC subtypes and to the prognosis of the disease [17–19]. In fact, current molecular phenotyping of colorectal tumors is usually linked to the traditional determination of somatic mutations in well-known oncogenes such as KRAS and BRAF [20].
The recent advance of genomic and transcriptomic technologies applied to the study of clinical samples did open the way to obtain genome-wide expression profiles of multiple patient cohorts and correlate the expression of certain genes with different disease subtypes, disease stages and progression [21, 22]. This approach had been widely applied in cancer research in the last decade and is very powerful when the identification of marker genes is associated with survival time. The correlation between gene expression and survival is an excellent tool to investigate prognosis of the disease and to build risk predictors that will be applicable to individual patients.
The identification of molecular biomarkers with prognostic value in CRC has been a challenging task [23–26]. Molecular prognosis of colorectal tumor samples by transcriptional profiling started about 15 years ago (see review [24]), and in more recent years several specific gene signatures associated with CRC survival have been published [5–7, 27–31]. Despite these efforts, at present there is not a clear compendium of gene markers for CRC survival and it is quite difficult to find consistency in the literature [24]. A clear limitation comes from the fact that, in most of previous studies, the number of tumor samples used to select the genes that enter into the construction of the prognostic predictors is small (i.e., the size of the patient cohorts rarely it is greater than a few hundred individuals). For example, ColoPrint is a 18-gene signature for prognosis prediction of stage II and III CRC, that was identified using as training set tumor samples from 188 patients [7, 27]; a 113-gene expression signature for predicting prognosis in patients with CRC was built using 145 samples as dicovery set [28]; a 7-gene signature to predict overall survival of CRC patients was based in an initial training set of 67 samples [29]; a recurrence-associated CRC signature of 13 genes was developed using a screening set of 145 samples [30]; a 15-gene signature for prediction of CRC recurrence and prognosis was elaborated using for the gene selection a set of 55 patients [31]. In conclusion, we can say that as far as it is reflected in the current literature, the size of the initial training sets used to identify candidate gene markers for CRC survival is small and the overlap between the published gene signatures is very reduced and inconsistent. To address these critical problems, we constructed a large, well-standardized, integrated data set of 1273 tumor samples with survival information, which was used to identify genes that had a clear change in expression in the middle and late stages of CRC and were consistent markers of the disease-outcome and patient-risk.
With respect to the specific genes proposed as CRC survival markers, we want to underline that our study does not pretend to provide a fixed gene signature for prognosis and risk prediction, like the reported signatures of 7-genes, 15-genes or 113-genes [28, 29, 31]; but instead we propose a robust set of genes ranked according to their predictive power of CRC survival. In this way, an ordered list of 200 genes including the best survival markers is presented: 100 genes for which up-regulation marks “poor survival” and 100 genes for which up-regulation marks “good survival”. We think that this approach is more useful, since it allows an open selection of different number of genes for further purposes or investigations (for example, for additional tests with other CRC clinical cohorts). In fact, we used the 100 most significant genes, up-regulated with the progression of CRC, to build the risk predictor (presented in Fig. 4); and we used the top 5 or top 10 genes of this list for the external validations with different independent datasets.
Another relevant comment is that, as reminded above, we constructed the risk predictor using the genes that showed up-regulation correlated with poor prognosis. This was done because in the selection of biomarkers it is better to use the ones that provide a positive signal (i.e. “gain-of-function” factors) than the ones that provide a negative signal. Therefore, all the gene survival markers that we proposed were detectable as overexpressed in the CRC patients with high risk. The fact that they give a positive signal will also make easier their detection by standard biomolecular protocols (PCR, ELISA, immunohistochemistry, etc).
Finally, we are investigating the biological meaning of the genes found as best predictive and prognostic markers. We are focusing our efforts in the top 10 for which up-regulation marked poor survival: DCBLD2, PTPN14, LAMP5, TM4SF1, NPR3, LEMD1, LCA5, CSGALNACT2, SLC2A3, GADD45B. The analysis of the literature reveals some relevant observations. For example, the transmembrane protein DCBLD2 (ESDN), member of a family of neuropilin-like proteins, is a novel regulator of mitotic and metabolic effects of insulin, and it modulates signal transduction through regulation of the insulin receptor interaction with its adaptor proteins [32]. The importance of insulin regulation in the function of our digestive system is clear, and this adds extra value to the proposal of DCBLD2 as a CRC survival marker. Other genes within the top rank have been recently involved in cancer progression, like the case of SLC2A3 (GLUT3) a glucose transporter that mediates glucose utilization and glycogenolysis, which is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation [33]. Recent publications have also proposed the role of some other genes found as prognostic markers, like the case of LAMP5 that has been included in a multigenic assay to predict recurrence for gastric cancer patients after surgery [34]. As a final example, GADD45B (growth arrest and DNA-damage-inducible 45 beta) is a gene that responds to environmental stresses, associated with cell growth control, apoptosis and DNA damage repair response. GADD45B overexpression has been recently correlated with shorter overall survival in colorectal carcinoma [35]. Moreover, a recent integrative analysis of multiple colon cancer gene-expression-based subtype classifiers reported that one of the three highest scoring genes included in several classifiers was GADD45B [36].
Despite all these positive findings that correspond to the biological value and the support of the genes identified as most significant markers of CRC survival, there are some possible limitations of the results, beginning with the general observation about the frequent heterogeneity of the colorectal tumors [1, 17]. In fact, it is clear from the anatomical pathology that CRC can affect quite different regions of the digestive tract: ascending colon, transverse colon, descending colon, sigmoid colon and rectum. The causal genes that drive tumors in these different regions may not be the same, and most CRC studies do not enter into a detailed separation of these regions [19]. The variability due to the different staging of the tumors is another factor that can bring limitations to any CRC study; but in this case we clearly indicated that our work searched for genes that were candidate prognostic markers for CRC in stages III and IV. A final reason for the limitations of the results may be an over-adjustment to the tested data sets. To avoid this kind of limitations, we built a large well-normalized data set with more than a thousand samples, performed a cross-validation analysis on that set, and also explored the validity of the gene markers in two other independent sets.
Conclusions
In conclusion, we consider that the results presented in this work provide strong support and a solid rationale for the prognostic value of a new set of genes in CRC and for their potential to predict colorectal tumor progression and evolution towards stages III and IV. The final proposed set of gene survival markers includes an open list of one hundred up-regulated genes, with a robust statistical estimation of the value of each one. In this way the set of genes is clearly ranked, being the top in the list the ones that provide best prognostic strength and the ones that can be introduced to build smaller predictors. In fact, our results showed that a selection of the top 5 genes applied to independent external cohorts provided very good separation of CRC samples in two distinct groups of high and low risk.
Methods
Genome-wide expression data sets
In this study, we have analysed and integrated seven data sets of CRC samples (Table 1). All data sets are available at GEO repository, corresponding to 7 series with the following accession numbers: GSE14333, GSE17536, GSE31595, GSE33113, GSE38832, GSE39084 and GSE39582. All these series included the raw expression signal and correspond to data obtained with the microarrays expression platform: Affymetrix GeneChip U133 Plus 2.0 for Homo sapiens. The phenotypic information corresponding to all these series was analysed in order to select only the samples that included information regarding: the cancer stage and the Overall Survival (OS). The samples that did not have any survival information were discarded from the study. In all cases only primary tumors samples were considered for our analysis; in this way individuals who had received preoperative chemotherapy and/or radiotherapy were also discarded.
For the external validation we used two independent datasets. A cohort of 276 colorectal carcinomas that had been studied using RNA-seq gene expression profiling, and that had survival data for 269 samples [14] (which can be found in http://tcga-data.nci.nih.gov/docs/publications/coadread_2012/). A second cohort of CRC samples from the platform SurvExpress [15]. This second dataset selected, called “Colon-Metabase-Uniformized”, included 482 CRC samples with overall survival data and genome-wide expression determined with Affymetrix microarrays (see the website http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp).
Expression data sets exploration and integrative normalization
Previously, to make the best use of the information obtained from the microarrays, we have considered the importance to ascertain the quality of the data. To assess the validity of generated microarray information we have performed a wide variety of quality assessment methods, both in raw and pre-processed information. In this way, several explanatory data analysis were applied for the detection of problematic arrays. We used the R function image to create chip images of the raw intensities to discover spatial artefacts in the samples. We have also look at the distribution of probes intensities across all arrays, using the boxplot method available for the Affybatch class. We also applied to the samples the Normalized Unscaled Standard Error (NUSE) algorithm. This quality assessment tool requires a previous PLM fitting procedure applied on the raw expression data. We have used the function fitPLM provided in the AffyPLM package to create the PLMset class object used as the input in the elaboration of the NUSE analysis. After applying the referred quality assessment methods, we discarded 79 of the initial samples collected and proceed with the remaining 1273 (Table 1).
To create a table with all the phenotypic characteristics of the patients selected which involved all samples GSM accession numbers and related clinic variables in a consistent and homogenize way, we used getGEO and pData functions from GEOquery package (this table is provided as Additional file 1: Table S1). We made use of regular expressions and common text manipulation R functions to solve the issue of formatting heterogenic data. Finally, we created a binary variable to label the patients and select them in a proper way during the hypothesis contrasts and statistical modeling.
Batch effect removal
Batch effect is one of the main problems when several datasets are combined to be studied together, because different batches usually add large unwanted variability to the data. To avoid this effect we tested a combination of different pre-processing and normalization algorithms: Robust Multi-array Average (RMA) algorithm [11]; Combatting Batch effects (ComBat) algorithm [12]; Frozen Robust Multi-array Average (fRMA) algorithm [13]. For the fRMA algorithm application, we constructed the frozen parameter vector using a training dataset in where we distributed randomly selected samples proportionally to each labelled group to obtain a balanced sample from the 7 batches of microarrays.
Another important issue addressed was the fact that the Affymetrix probe-sets included in the expression microarrays many times do not correspond to singular genes and some probes inserted in the defined probe-sets are ambiguous or inaccurate [10]. Affymetrix GeneChip is a popular and usefull platform for gene expression profiling, but the use of its probes and probe-sets mapping has multiple inconveniences. In fact, the probe-sets for the Affymetrix Human Genome U133 Plus 2.0 Array are based on UniGene database (Build 133, April 20, 2001) and considering how rapidly human genome has evolved many probes on the array are not correctly assigned. To avoid this problem, we used the updated probe alignment and gene mapping that is provided by the Chip Definition File (CDF): hgu133plus2hsensgcdf (downloaded from http://brainarray.mbni.med.umich.edu/).
Batch effect removal evaluation
We performed unsupervised hierarchical clustering to observe unlikely clustering based on batches in those expression value matrixes where batch effects remained after pre-processing. We used a 30-random sampling per batch, identifying each batch by a different color (Fig. 1). The batch effect was also investigated using principal components analysis (PCA) (Fig. 2). A linear regression of average gene expression on array batch per pre-processing method was the final approach fulfilled to assure removal (Table 2).
Differential expression analysis
For the identification of gene whose altered expression achieved statistical significance we used the R algorithm Linear Models for Microarrays (LIMMA package). We applied LIMMA to the expression data matrix fixing an adjusted p-value threshold of FDR ≤ 0.01 to select significant genes. The comparison was done separating the samples according to their clinical and pathological stage (comparing CRC stages I and II versus III and IV). In this way we found a set of 2707 candidates genes, corresponding to 2524 protein-coding genes that were tested in the survival analysis (the rest were non-coding genes). In this work we focus only on the genes that encode proteins because we wanted to find CRC survival markers that later can be tested at protein level using, for example, immunohistochemistry (IHC) analysis.
Survival analysis
Our intention in this research was to identify genes whose relative expression level affect survival and prognosis in CRC, once we had made a preselection in its behavior through stage evolution of 2524 protein-coding genes.
The first step for the survival analysis was to define for each gene two separated distributions of high and low expression along the sample dataset investigated. This separation based in expression level determined the explanatory variable. We used the Surdiff function in the Survival package to address the issue. By sorting all the samples in ascending order, we performed Surdiff hypothesis testing, splitting the group of samples for each gene and every sample between quantile 25% and 75% to obtain its Chi-square associated p-value. Then we selected minimum p-value to perform final group assignation of high and low expression. Once we had the two groups clearly defined, we used the Coxph model to obtain each associated p-value and hazard ratio (HR) from every candidate gene. In this way, the survival analysis along the two groups also allowed estimating hazard ratios (HR) or, what is the same, tried to measure how the expression, in terms of high and low relative expression for each candidate gene, altered the hazard function. Finally, for computing the time to event, the response variable in the models was the Overall Survival (OS) time. All the data sets that we integrated in our analyses had OS information. In some cases for some individuals, Disease Specific Survival (DSS) times or Relapse Free Survival (RFS) times were also provided with the original data, but we did not considered these time-events since we wanted to focus on OS to achieve a homogeneous analysis.
Additional files
Table S1. Phenotypic and clinical information about the collection of 1273 colorectal cancer samples that has been integrated in this work. The table includes the IDs of the samples in GEO and all the available data about age, gender, survival time, location of the tumor, degree and TNM staging, presence of mutation in some cancer genes (TP53, KRAS, BRAF), etc. When information was not available for a given sample the table includes NA (not available values). (XLSX 272 kb)
Table S2. Top-100 best survival marker genes for colorectal cancer (CRC) that are up-regulated when survival is poor and the risk is higher (i.e., HR > 1). This table is an expension of the data in Table 3. The genes were ranked by their KM adjusted p-values and the HR values calculated for the whole dataset (i.e. for all the 1273 samples = all-dt). The stability of each survival marker gene was assessed by cross-validation (100 iterations). The table also includes the number of times that a survival marker was significant in the iterations (N-sinf-in-100i). (XLSX 73 kb)
Table S3. Top-100 best survival marker genes for colorectal cancer (CRC) that are down-regulated when survival is poor and the risk is higher (i.e., HR < 1). This table is an expension of the data in Table 3. The genes were ranked by their KM adjusted p-values and the HR values calculated for the whole dataset (i.e. 1273 samples = all-dt). The stability of each survival marker gene was assessed by cross-validation (100 iterations). The table also includes the number of times that a survival marker was significant in the iterations (N-sinf-in-100i). (XLSX 70 kb)
Table S4. Validation of the survival data done in an independent set of samples taken from The Cancer Genome Atlas (TCGA), that included 269 colorectal carcinomas with survival information and RNA-seq global expression profiling. The table includes the KM p-values and HR of the genes that were validated from the top-10 survival marker genes previously found presented in Table 3. Of the top-10 for the case of up-regulation associated with poor survival, 7 were validated (PTPN14, LAMP5, TM4SF1, LCA5, CSGALNACT2, SLC2A3 and GADD45B). Of the top-10 found for down-regulation associated with poor survival, 6 genes were validated (EPHB2, DUS1L, NUAK2, FANCC, MYB and CHDH). (XLSX 51 kb)
Figure S1. Survival multivariate analysis of an independent set of 482 samples of CRC patients carried out considering the expression profiles of 5 genes: DCBLD2, PTPN14, LAMP5, TM4SF1 and NPR3. (A) Kaplan-Meier plot presenting the patients divided in two groups according their risk score: High risk (red) and Low risk (green). (B) Box plots showing the distributions of global expression corresponding to these 5 genes. For each gene, the dataset of 482 samples was divided in the two groups of patients indentified as High risk (red) and Low risk (green). (PDF 356 kb)
Figure S2. Comparison of the distributions of the expression signal corresponding to ten genes in 25 samples from normal colorectal epithelium (green boxplots) versus 25 samples from CRC (red boxplots). The genes selected for this analysis were the top-10 best survival marker genes found up-regulated for poor prognosis (i.e. markers up-regulated when there is low CRC survival): DCBLD2, PTPN14, LAMP5, TM4SF1, NPR3, LEMD1, LCA5, CSGALNACT2, SLC2A3 and GADD45B. The tumor samples were not selected by stage (i.e. they were selected from any CRC stage: I, II, III or IV) and this comparison was done 20 times with different subsets of 25 CRC samples to check the stability of the signal. The plots of all the other comparisons were very similar to the plot here presented. (PDF 46 kb)
Table S5. Beta factors assigned by regression analysis to each of the top-100 survival marker genes. These genes are taken as variables within the multivariate Kaplan-Meier survival analysis included in Fig. 4b. The factors allowed the identification of the genes that were the most influential variables in this risk analysis (i.e. the higher the better) and therefore facilitate an additional evaluation of each survival marker gene. (XLSX 62 kb)
Acknowledgements
We acknowledge the funding provided to JDLR research group by the Spanish Government with grants of the ISCiii co-funded by FEDER (references PI15/00328 and AC14/00024). We also acknowledge a PhD research grant to SBF (from the Program “Ayudas a la Contratación de Personal Investigador”) provided by the “Junta de Castilla y Leon” (JCyL) with the support of the “Fondo Social Europeo” (FSE). The funding boards had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Funding
The publication costs for this article were funded by the research grants AC14/00024 and PI15/00328, from the Instituto de Salud Carlos III (ISCiii) co-funded by the “Fondo Europeo de Desarrollo Regional” (FEDER).
Availability of data and materials
All the data presented in this study is provided free and open to be used, included in the Supplementary Files that are quoted and described along the manuscript.
About this supplement
This article has been published as part of BMC Genomics Volume 19 Supplement 8, 2018: Selected articles from the IV Colombian Congress on Bioinformatics and Computational Biology & VIII International Conference on Bioinformatics SoIBio 2017. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-19-supplement-8.
Abbreviations
- CDF
Chip definition file
- CRC
Colorectal cancer
- DSS
Disease specific survival
- GEO
Gene expression omnibus database
- GSE
GEO Series (set of sample files that together form a single experiment)
- HR
Hazard ratio
- IHC
Immunohistochemistry
- KM
Kaplan-Meier hazard ratio
- LIMMA
Linear models for microarray data analysis
- OS
Overall survival
- PCA
Principal component analysis
- RFS
Relapse free survival
- RMA
Robust multi-array average algorithm
- TCGA
The cancer genome atlas
Authors’ contributions
JMR carried out the data collection, the databases construction and together with SBF the R progamming developments, the computational analyses and the datasets comparisons. They also contributed to write the manuscript. JDLR designed the study with the support of ARM and MMM. JDLR devised and designed the study, identified the experimental data sets used in the tests and validations, supervised the R programming, wrote the manuscript and managed the authors’ collaboration. ARM and MMM also contributed to the design of the work and help in the preparation of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval and consent to participate is “not applicable”, because this work does not include samples from new patients or donors. All the information and data of human samples used in this work come from data sets already public in open repositories and corresponded to Anonymized Patient Level Data (APLD). Moreover, the Ethical Committees of our Research Centers (CiC-IBMCC and IMDEA-Food) supervised the adequate use of the data corresponding to human samples.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Linnekamp JF, Wang X, Medema JP, Vermeulen L. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Cancer Res. 2015;75:245–249. doi: 10.1158/0008-5472.CAN-14-2240. [DOI] [PubMed] [Google Scholar]
- 2.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, Zhang W, Liu ZQ, Zhou HH. Associating transcriptional modules with colon cancer survival through weighted gene co-expression network analysis. BMC Genomics. 2017;18:361. doi: 10.1186/s12864-017-3761-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargas T, Moreno-Rubio J, Herranz J, Cejas P, Molina S, González-Vallinas M, et al. ColoLipidGene: signature of lipid metabolism-related genes to predict prognosis in stage-II colon cancer patients. Oncotarget. 2015;6:7348–7363. doi: 10.18632/oncotarget.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sveen A, Ågesen TH, Nesbakken A, Meling GI, TO R, Liestøl K, et al. ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res. 2012;18:6001–6010. doi: 10.1158/1078-0432.CCR-11-3302. [DOI] [PubMed] [Google Scholar]
- 7.Kopetz S, Tabernero J, Rosenberg R, Jiang ZQ, Moreno V, Bachleitner-Hofmann T, et al. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist. 2015;20:127–133. doi: 10.1634/theoncologist.2014-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The American Cancer Society medical and editorial content team. Colorectal Cancer Stages. https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/staged.html. Accessed 06 Oct 2017.
- 9.Tauriello DVF, Batlle E. Targeting the microenvironment in advanced colorectal Cancer. Trends Cancer. 2016;2:495–504. doi: 10.1016/j.trecan.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Risueño A, Fontanillo C, Dinger ME, De Las Rivas J. GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs. BMC Bioinformatics. 2010;11:221. doi: 10.1186/1471-2105-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe-level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Stein CK, Qu P, Epstein J, Buros A, Rosenthal A, Crowley J, et al. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinformatics. 2015;16:63. doi: 10.1186/s12859-015-0478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui J, Li H. Penalized cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 17.Sameer AS. Colorectal cancer: molecular mutations and polymorphisms. Front Oncol. 2013;3:114. doi: 10.3389/fonc.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fessler E, Medema JP. Colorectal Cancer subtypes: developmental origin and microenvironmental regulation. Trends Cancer. 2016;2(9):505–518. doi: 10.1016/j.trecan.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Bijlsma MF, Sadanandam A, Tan P, Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2017;14(6):333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 20.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep. 2015;3(4):269–276. doi: 10.1093/gastro/gov046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aibar S, Fontanillo C, Droste C, Roson-Burgo B, Campos-Laborie FJ, Hernandez-Rivas JM, et al. Analyse multiple disease subtypes and build associated gene networks using genome-wide expression profiles. BMC Genomics. 2015;16(Suppl 5):S3. doi: 10.1186/1471-2164-16-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aibar S, Abaigar M, Campos-Laborie FJ, Sánchez-Santos JM, Hernandez-Rivas JM, De Las Rivas J. Identification of expression patterns in the progression of disease stages by integration of transcriptomic data. BMC Bioinformatics. 2016;17(Suppl 15):432. doi: 10.1186/s12859-016-1290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno V, Sanz-Pamplona R. Altered pathways and colorectal cancer prognosis. BMC Med. 2015;13:76. doi: 10.1186/s12916-015-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz-Pamplona R, Berenguer A, Cordero D, Riccadonna S, Solé X, Crous-Bou M, et al. Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One. 2012;7(11):e48877. doi: 10.1371/journal.pone.0048877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George B, Kopetz S. Predictive and prognostic markers in colorectal cancer. Curr Oncol Rep. 2011;13(3):206–215. doi: 10.1007/s11912-011-0162-3. [DOI] [PubMed] [Google Scholar]
- 26.Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. 2017;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 27.Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29(1):17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen MN, Choi TG, Nguyen DT, Kim JH, Jo YH, Shahid M, et al. CRC-113 gene expression signature for predicting prognosis in patients with colorectal cancer. Oncotarget. 2015;6(31):31674–31692. doi: 10.18632/oncotarget.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Sun X, Ge W, Qian Y, Bai R, Zheng S. A seven-gene signature predicts overall survival of patients with colorectal cancer. Oncotarget. 2016;8(56):95054–95065. doi: 10.18632/oncotarget.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian X, Zhu X, Yan T, Yu C, Shen C, Hu Y, et al. Recurrence-associated gene signature optimizes recurrence-free survival prediction of colorectal cancer. Mol Oncol. 2017;11(11):1544–1560. doi: 10.1002/1878-0261.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G, Zhang M, Zhu H, Xu J. A 15-gene signature for prediction of colon cancer recurrence and prognosis based on SVM. Gene. 2017;604:33–40. doi: 10.1016/j.gene.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Jung JJ, Nie L, Razavian M, Zhang J, Samuel V, et al. The neuropilin-like protein ESDN regulates insulin signaling and sensitivity. Am J Physiol Heart Circ Physiol. 2016;310:H1184–H1193. doi: 10.1152/ajpheart.00782.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masin M, Vazquez J, Rossi S, Groeneveld S, Samson N, Schwalie PC, et al. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer Metab. 2014;2:11. doi: 10.1186/2049-3002-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One. 2014;9:e90133. doi: 10.1371/journal.pone.0090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Xiao X, Li D, Chi Y, Wei P, Wang Y, Ni S, Tan C, Zhou X, Du X. Abnormal expression of GADD45B in human colorectal carcinoma. J Transl Med. 2012;10:215. doi: 10.1186/1479-5876-10-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sztupinszki Z, Győrffy B. Colon cancer subtypes: concordance, effect on survival and selection of the most representative preclinical models. Sci Rep. 2016;6:37169. doi: 10.1038/srep37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Phenotypic and clinical information about the collection of 1273 colorectal cancer samples that has been integrated in this work. The table includes the IDs of the samples in GEO and all the available data about age, gender, survival time, location of the tumor, degree and TNM staging, presence of mutation in some cancer genes (TP53, KRAS, BRAF), etc. When information was not available for a given sample the table includes NA (not available values). (XLSX 272 kb)
Table S2. Top-100 best survival marker genes for colorectal cancer (CRC) that are up-regulated when survival is poor and the risk is higher (i.e., HR > 1). This table is an expension of the data in Table 3. The genes were ranked by their KM adjusted p-values and the HR values calculated for the whole dataset (i.e. for all the 1273 samples = all-dt). The stability of each survival marker gene was assessed by cross-validation (100 iterations). The table also includes the number of times that a survival marker was significant in the iterations (N-sinf-in-100i). (XLSX 73 kb)
Table S3. Top-100 best survival marker genes for colorectal cancer (CRC) that are down-regulated when survival is poor and the risk is higher (i.e., HR < 1). This table is an expension of the data in Table 3. The genes were ranked by their KM adjusted p-values and the HR values calculated for the whole dataset (i.e. 1273 samples = all-dt). The stability of each survival marker gene was assessed by cross-validation (100 iterations). The table also includes the number of times that a survival marker was significant in the iterations (N-sinf-in-100i). (XLSX 70 kb)
Table S4. Validation of the survival data done in an independent set of samples taken from The Cancer Genome Atlas (TCGA), that included 269 colorectal carcinomas with survival information and RNA-seq global expression profiling. The table includes the KM p-values and HR of the genes that were validated from the top-10 survival marker genes previously found presented in Table 3. Of the top-10 for the case of up-regulation associated with poor survival, 7 were validated (PTPN14, LAMP5, TM4SF1, LCA5, CSGALNACT2, SLC2A3 and GADD45B). Of the top-10 found for down-regulation associated with poor survival, 6 genes were validated (EPHB2, DUS1L, NUAK2, FANCC, MYB and CHDH). (XLSX 51 kb)
Figure S1. Survival multivariate analysis of an independent set of 482 samples of CRC patients carried out considering the expression profiles of 5 genes: DCBLD2, PTPN14, LAMP5, TM4SF1 and NPR3. (A) Kaplan-Meier plot presenting the patients divided in two groups according their risk score: High risk (red) and Low risk (green). (B) Box plots showing the distributions of global expression corresponding to these 5 genes. For each gene, the dataset of 482 samples was divided in the two groups of patients indentified as High risk (red) and Low risk (green). (PDF 356 kb)
Figure S2. Comparison of the distributions of the expression signal corresponding to ten genes in 25 samples from normal colorectal epithelium (green boxplots) versus 25 samples from CRC (red boxplots). The genes selected for this analysis were the top-10 best survival marker genes found up-regulated for poor prognosis (i.e. markers up-regulated when there is low CRC survival): DCBLD2, PTPN14, LAMP5, TM4SF1, NPR3, LEMD1, LCA5, CSGALNACT2, SLC2A3 and GADD45B. The tumor samples were not selected by stage (i.e. they were selected from any CRC stage: I, II, III or IV) and this comparison was done 20 times with different subsets of 25 CRC samples to check the stability of the signal. The plots of all the other comparisons were very similar to the plot here presented. (PDF 46 kb)
Table S5. Beta factors assigned by regression analysis to each of the top-100 survival marker genes. These genes are taken as variables within the multivariate Kaplan-Meier survival analysis included in Fig. 4b. The factors allowed the identification of the genes that were the most influential variables in this risk analysis (i.e. the higher the better) and therefore facilitate an additional evaluation of each survival marker gene. (XLSX 62 kb)
Data Availability Statement
All the data presented in this study is provided free and open to be used, included in the Supplementary Files that are quoted and described along the manuscript.