Abstract
Background
Sesame is a major oilseed crop which is widely cultivated all around the world. Flowering, the timing of transition from vegetative to reproductive growth, is one of the most important events in the life cycle of sesame. Sesame is a typical short-day (SD) plant and its flowering is largely affected by photoperiod. However, the flowering mechanism in sesame at the molecular level is still not very clear. Previous studies showed that the CONSTANS (CO) gene is the crucial photoperiod response gene which plays a center role in duration of the plant vegetative growth.
Results
In this study, the CO-like (COL) genes were identified and characterized in the sesame genome. Two homologs of the CO gene in the SiCOLs, SiCOL1 and SiCOL2, were recognized and comprehensively analyzed. However, sequence analysis showed that SiCOL2 lacked one of the B-box motifs. In addition, the flowering time of the transgenic Arabidopsis lines with overexpressed SiCOL2 were longer than that of SiCOL1, indicating that SiCOL1 was more likely to be the potential functional homologue of CO in sesame. Expression analysis revealed that SiCOL1 had high expressed levels before flowering in leaves and exhibited diurnal rhythmic expression in both SD and long-day (LD) conditions. In total, 16 haplotypes of SiCOL1 were discovered in the sesame collections from Asia. However, the mutated haplotypes did not express under both SD and LD conditions and was regarded as a nonfunctional allele. Notably, the sesame landraces from high-latitude regions harboring nonfunctional alleles of SiCOL1 flowered much earlier than landraces from low-latitude regions under LD condition, and adapted to the northernmost regions of sesame cultivation. The result indicated that sesame landraces from high-latitude regions might have undergone artificial selection to adapt to the LD environment.
Conclusions
Our results suggested that SiCOL1 might contribute to regulation of flowering in sesame and natural variations in SiCOL1 were probably related to the expansion of sesame cultivation to high-latitude regions. The results could be used in sesame breeding and in broadening adaptation of sesame varieties to new regions.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1583-z) contains supplementary material, which is available to authorized users.
Keywords: Sesame, Photoperiod response, Flowering, Artificial selection, CONSTANS
Background
Flowering, the timing of transition from vegetative to reproductive development, is one of the critical developmental steps in plants. Previous research revealed that flowering is regulated by both genotype and environmental factors such as temperature, light spectrum, light intensity and day length (photoperiod). For example, winter wheat required several weeks at low temperature, also named vernalization to flower [1]. Flowering of rice is promoted in short photoperiod, and it is therefore regarded as a facultative SD plant [2], while flowering of Arabidopsis thaliana is mainly promoted by LD condition [3]. Recently, low red light to far-red light ratio was also reported to accelerate Arabidopsis flowering [4]. Flowering of Phalaenopsis is positively influenced by supplemental lighting during the inductive phase [5]. Among these environment factors that are related to plant flowering, photoperiod might be the critical signal that regulate the initiation of flowering in angiosperms since day length is a reliable indicator of the time of year for plants [6].
Flowering time is the key trait for geographical and seasonal adaptation in crops. For the worldwide cultivated crops, such as rice and soybean, the flowering time of varieties varied in a broad range and is related to yield. In a reasonable range, the late flowering, also means longer vegetative growth, contributes to the higher biomass and yield of the varieties [7, 8]. Usually, the varieties of SD plants from low latitude areas flower later than those from high latitude areas when the cultivars are planted under LD condition. To adapt the day-light conditions in different environments, the flowering time of crops would be selected during long-time breeding programs. Therefore, the flowering time and photoperiod sensitivity of crops are one of the primary improvement targets for crop breeding.
Sesame (Sesamum indicum L.), which belongs to the genus Sesamum in the family Pedaliaceae, is an important oilseed crop grown widely in various areas of the world. The harvest area of sesame has been doubled in the last four decades, and is still increasing gradually (http://www.fao.org/faostat/). Sesame was domesticated from wild relatives in the Indian subcontinent about 5000 years ago and it is therefore regarded as the most ancient oilseed crop [9, 10]. Nowadays, sesame is widely planted in more than 80 countries across the world, with a concentration in tropical and subtropical areas. Sesame flowering is mainly promoted by short photoperiod and is classified as a SD plant [11, 12]. The flowering time of sesame landraces ranges broadly from less than 30 d to more than 90 d [13]. However, few genetic researches of the sesame flowering have been reported and the genetic mechanism remains unknown.
The photoperiod gene, CO, encoding a B-box zinc-finger transcription factor, plays a central role in the photoperiod response and flowering regulation in Arabidopsis [3]. It acts between the circadian clock and florigen genes. The rhythmical expression of CO is regulated by GI, a circadian clock gene [14]. The peak expression of CO generally appears before dawn under SDs but at both afternoon and dawn under LDs [15]. Subsequently, CO activates expressions of the floral gene FLOWERING LOCUS T (FT), and promote flowering of Arabidopsis in LDs. It has been shown that CO protein binds to the FT promoter [16]. In LDs, the CO protein accumulates to higher levels due to the stability of the GI-FKF1 complex in the light which degrades CO repressor CDF1 [17]. Although CO was identified in the LD plant Arabidopsis, its homologous genes in SD plants were also found to be the key flowering regulators. Heading date 1 (Hd1), the homologous gene of CO in rice, contributes to photoperiod measurement and photoperiod-specific regulation of FT [18]. In contrast to CO, Hd1 appears to be a bifunctional regulator, promoting FT expression in SDs but repressing FT in LDs [19, 20]. Moreover, the homologs of CO have been investigated in a number of other species, such as wheat, maize, barley, cotton, rapeseed, soybean, potato, grapevine, apple and Pharbitis nil through various functional genomics analyses, showing its conserved function involved in regulating plant flowering [21–30].
Previous studies showed CO had plenty of allelic variations, mediating photoperiod-dependent flowering time in Arabidopsis [31]. Among the 51 flowering time loci in Arabidopsis, CO possessed the most significantly associated single nucleotide polymorphisms (SNPs) of flowering time [32]. Similarly, a highly degree of polymorphisms of Hd1 were the major determinant of the variation in flowering time diversity in rice [33]. Some variations of CO and Hd1 had been identified as the crucial mutations that strongly influenced flowering time in plants [34, 35]. COL genes were also identified from many plants [22, 23, 30, 36–40]. B-box motifs and CCT domain have been proved to be the conserved domains in COL genes [23, 29, 41, 42]. However, the functional variations of CO and COL genes in sesame have not been identified and investigated.
Genome sequencing and large-scale genome re-sequencing of sesame has been completed recently [13, 43–45], providing high-quality reference genome sequence and massive useful variations for the functional genomics research of sesame. In the present study, the sesame COL gene family was genome-wide identified and characterized from the sesame genome. Two homologous genes of CO in sesame, SiCOL1 and SiCOL2, were recognized. Functions of SiCOL1 were confirmed by the transgenic approach, expression pattern analysis, and haplotype analysis. Evolution analysis of SiCOL1 revealed that these genes had been selected to adapt to the photoperiod conditions in different areas. The results suggest that SiCOL1 is an important agronomic photoperiod response gene that significantly affected flowering time, contributing to the adaption of sesame to the high-latitude regions. Our results also shed light on the potential value of SiCOL1 in genetic improvement of sesame.
Results
Identification of COL genes in sesame
To identify the COL genes in sesame, the Hidden Markov Model (HMM) search was performed against the sesame protein database using the Zinc-finger B-box motif (PF00643) and CCT (CONSTANS, CONSTANS-like, TIMING OF CAB EXPRESSION 1) domain (PF06203). In total, 37 B-box Zinc-finger genes and 36 CCT domain-containing genes were identified in the sesame genome, respectively (Additional file 1: Table S1). The B-box Zinc-finger genes and the CCT domain-containing genes were then compared with each other and 13 genes of them were found to be the same. Therefore, the 13 genes which contained both Zinc-finger B-box motif and CCT domain were identified and named as sesame COL genes (Table 1). All of the Arabidopsis COL protein sequences were used as queries for the Basic Local Alignment Search Tool (BLAST) to identify sesame COL proteins. However, we had not identified any additional proteins containing both B-box motifs and CCT domain in the sesame genome. All B-box motif and CCT domain in the SiCOLs were validated by the CDD (http://www.ncbi.nlm.nih.gov/cdd/) and simple modular architecture research tool (SMART) analyses.
Table 1.
The COL gene family in sesame
| Gene | Linkage group | Position | Peptide length | Exon number | Group |
|---|---|---|---|---|---|
| SIN_1003782 | LG16 | 4,171,550–4,176,377 | 415 | 4 | III |
| SIN_1004896b | LG15 | 2,534,501–2,535,610 | 332 | 2 | I |
| SIN_1011796 | LG5 | 17,536,289–17,537,482 | 368 | 2 | I |
| SIN_1012499 | LG6 | 1,093,823–1,096,564 | 371 | 4 | III |
| SIN_1013665 | LG1 | 10,021,319–10,023,171 | 439 | 2 | II |
| SIN_1017370 | LG3 | 11,310,021–11,311,664 | 393 | 2 | II |
| SIN_1018340 | LG2 | 16,108,396–16,110,105 | 461 | 2 | II |
| SIN_1019767 | LG8 | 1,838,327–1,845,107 | 410 | 4 | III |
| SIN_1019889a | LG8 | 2,936,072-2,937,312 | 354 | 2 | I |
| SIN_1019954 | LG5 | 13,943,158–13,945,055 | 412 | 4 | III |
| SIN_1020629 | LG6 | 18,213,252–18,214,375 | 339 | 2 | I |
| SIN_1021657 | LG1 | 930,153–932,165 | 335 | 2 | I |
| SIN_1023877 | LG2 | 9,655,751–9,656,982 | 378 | 2 | I |
a SiCOL1
b SiCOL2
The SiCOL genes were not evenly distributed on the linkage groups (LGs) of the sesame genome: one gene on LG3, LG15 and LG16, and two genes on LG1, LG2, LG5, LG6 and LG8. The SiCOL proteins ranged from 332 (SIN_1004896) to 461 (SIN_1018340) amino acids (aa) in length, with an average length of approximately 385 aa. Moreover, no tandem duplicate genes were identified for these SiCOLs, although tandem duplication events had been observed in several other sesame gene families [46–48].
Phylogenetic analysis of the SiCOL genes
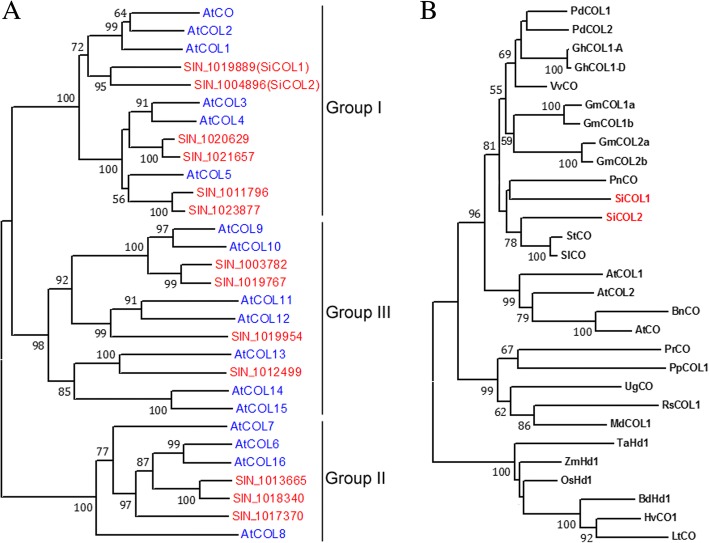
A phylogenetic tree was constructed using the neighbor-joining (NJ) method basing on multiple alignments of sesame and Arabidopsis COL genes (Fig. 1a). The 13 SiCOLs were classified into three groups (I, II, and III) and each group consisted of 6, 3, and 4 SiCOL proteins, respectively. Two SiCOL genes, SIN_1019889 and SIN_1004896 showed the closest relationship with the Arabidopsis CO gene. The Arabidopsis CO protein sequence was also used as query for the BLAST to identify the homologous genes. It showed that SIN_1019889 and SIN_1004896 were the only homologous genes of Arabidopsis CO gene in sesame. Thus, these two genes were referred as SiCOL1 (SIN_1019889) and SiCOL2 (SIN_1004896), respectively. We therefore concluded that these genes might be involved in the photoperiodic regulation of sesame flowering.
Fig. 1.
Phylogenetic analysis of the SiCOL genes. a A NJ phylogenetic tree of the COL proteins in sesame and Arabidopsis. The bootstrap values were inferred from 1000 replicates. b Phylogenetic relationship among COL proteins. The phylogram was generated from the multiple alignments of the deduced amino acid sequence from SiCOL1 and SiCOL2 and homologous proteins from other plant species. Bootstrap values from 1000 replicates were used to assess the robustness of the tree and the bootstrap values > 50% are showed
Phylogenetic analysis of SiCOL1, SiCOL2, CO and CO homologous proteins in the other 19 plant species was performed. CO homologous proteins from monocots and dicots were clustered into two groups. Both SiCOL1 and SiCOL2 proteins were divided into the dicots group. SiCOL1 protein (GeneBank ID: XP_011085568) displayed the highest similarity to PnCO protein (the CO protein in Pharbitis nil, 53% identity, AF300700) whereas it showed a 44% identity with CO protein from Arabidopsis (NP_197088). SiCOL2 protein (XP_011099077) displayed the highest identity to SlCO protein (60% identity, NP_001233839) and StCO protein (60% identity, ARU77840), which was higher than that of Arabidopsis CO protein (48% identity). However, SlCO was not involved in the control of flowering time of Solanum lycopersicum [49]. Previous research suggested that sesame was taxonomically close to Utricularia gibba, S. lycopersicum and S. tuberosum [43]. However, in this study, UgCO protein was not close to either SiCOL1 or SiCOL2 protein.
Conserved motifs and structure of the SiCOL genes
Using the SiCOL phylogenetic relationship data, we identified structural features of the sesame COLs, including conserved motifs and the locations of exons and introns (Additional file 1: Figure S1). The SiCOL genes of Group I and Group II had a simple gene structure -- one intron and two exons (Additional file 1: Figure S1b), while all genes in Group III had more exons and presented more complex gene structure than that of Group I and Group II. Multiple Em for Motif Elicitation (MEME) analysis confirmed the presence of the B-box motifs and CCT domains in SiCOL gene sequences. All genes in Group I and Group III had two B-box motifs except SiCOL2, which lacked one of the B-box motifs (Additional file 1: Figure S1c).
The protein sequences of SiCOL1 and SiCOL2 were further analyzed (Additional file 1: Figure S2). The result showed that they shared high similarity in amino acid sequence (61.7%), especially in the regions of B-box 2 motif (83.7%) and CCT domain (97.7%). SiCOL1 and SiCOL2 proteins had large differences in the B-box 1 motif region. Most amino acids of B-box 1 motif in SiCOL2 protein were lost. Even the remaining amino acids in B-box 1 motif of SiCOL2 protein were also quite different from that of SiCOL1. B-box motif plays an important role in the regulation of transcription and in mediating protein–protein interaction [50], and the missing of B-box 1 motif may cause loss of partial function of SiCOL2.
Overexpression of SiCOL1 and SiCOL2 in Arabidopsis
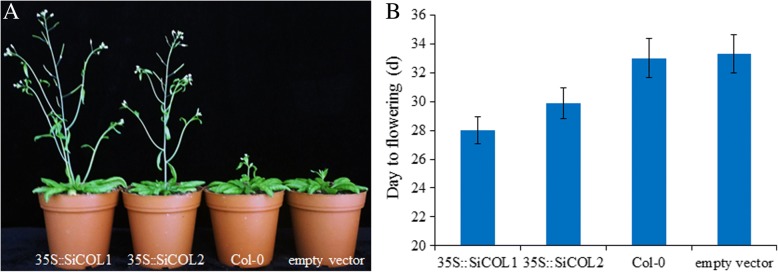
To explore the role of SiCOL1 and SiCOL2 in flowering, we constructed SiCOL1 and SiCOL2 overexpression vectors, and transferred into Arabidopsis Col-0 lines, respectively. Ten independent T0 transgenic lines were obtained for each gene. T1 generation transgenic lines planted in LD condition were about 3 days earlier flowering than the wild type. T2 generation plants were significantly earlier flowering (5 days of 35S::SiCOL1 on average, P < 0.001, and 3 days of 35S::SiCOL2 on average, P < 0.001) than the wild type (Fig. 2 and Additional file 1: Table S2). It is noteworthy that the T2 transgenic lines of 35S::SiCOL1 flowered earlier (2 days in average) than that of 35S::SiCOL2. This result might be caused by the loss of B-box 1 motif in SiCOL2 protein. Therefore, we concluded that SiCOL2 might lose partial function of flowering regulation and SiCOL1 was potential functional homologous gene of CO in sesame.
Fig. 2.
Days to flowering of transgenic Arabidopsis with overexpressed SiCOL1 and SiCOL2 under LD condition. a Flowering phenotype of T2 transgenic Arabidopsis lines with overexpressed SiCOL1 and SiCOL2. Photo was taken at 7 d after flowering of the SiCOL1 transgenic line. b Days to flowering of T2 transgenic Arabidopsis lines with overexpressed SiCOL1 and SiCOL2 under LD condition. The T2 transgenic Arabidopsis lines containing empty vector were used as control. For each test of 35S::SiCOL1, 35S::SiCOL2 and empty vector, days to flowering of ten lines (each contained 10 plants) were counted (Additional file 1: Table S2). The bar indicates standard deviation
To investigate the mechanism of action of SiCOL1 and SiCOL2 in Arabidopsis, we compared the expression patterns of flowering related genes FT in transgenic lines with wild type under LDs. Under LDs, FT is induced by CO and promotes flowering in Arabidopsis [51]. Comparing with the FT in wild type, FT in the transgenic lines expressed in an extremely high level (Additional file 1: Figure S3). The result suggested that SiCOL1 and SiCOL2 promoted Arabidopsis flowering by inducing the expression of FT. Moreover, expression of FT in T2 transgenic lines with 35S::SiCOL1 was much higher than that in the 35S::SiCOL2 transgenic lines, indicating SiCOL1 had higher induction efficiency of FT expression than SiCOL2.
Expression patterns of SiCOL1 and SiCOL2
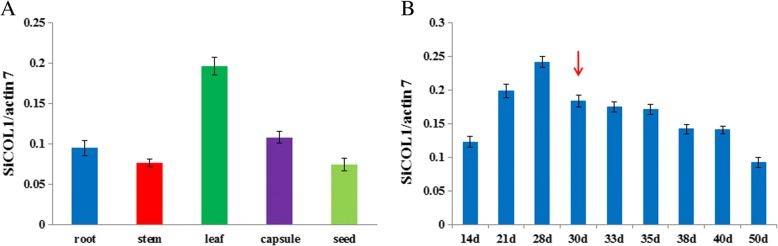
Five different tissues of sesame were collected from the widely cultivated sesame variety ‘Zhongzhi13’, including root, stem, leaf, capsule and seed. Quantitative real-time polymerase chain reaction (qRT–qPCR) was used to investigate the expression of SiCOL1 and SiCOL2 in these tissues. The result revealed that the expression of SiCOL1 and SiCOL2 in root, stem, capsule and seed were almost in the same level (Fig. 3a and Additional file 1: Figure S4a). However, both the expression levels of SiCOL1 and SiCOL2 in leaf were significantly higher than that in other tissues (P < 0.001).
Fig. 3.
Relative expression of SiCOL1 in different tissues and development stages of sesame. a Relative expression of SiCOL1 in five tissues of sesame. b Relative expression of SiCOL1 in leaves of different development stages. The red arrow indicates that tiny flower buds begin to appear in the axil of sesame plants. Transcript abundance was quantified using qRT-PCR and expression levels were normalized using sesame actin7 as a reference gene. The bar indicates standard deviation
Expression of SiCOL1 and SiCOL2 in leaf at the different development stages (from 14 days to 50 days after seed sowing) of ‘Zhongzhi13’ was investigated. All samples were collected in the same time (8:00 am) during a day. Generally, the flower buds of the variety ‘Zhongzhi13’ appear in approximately 30 days and ‘Zhongzhi13’ flowers at about 40 days in the growing season at Wuhan, China. The SiCOL1 and SiCOL2 expression increased quickly from 14 to 28 days and reached the highest level in 28 days, which was the exactly time before the flower buds appeared in the axil of sesame (Fig. 3b and Additional file 1: Figure S4b). After the flower bud appeared, the expression of SiCOL1 moderately decreased (from 30 to 40 days). Although sesame is an indeterminate inflorescence species, the expression of SiCOL1 decreased noticeably after the plant flowered (50 days). However, the expression of SiCOL2 slightly increased after sesame flowering. The result suggested that the expression of SiCOL1 and SiCOL2 dynamic changed during the development of sesame floral organ.
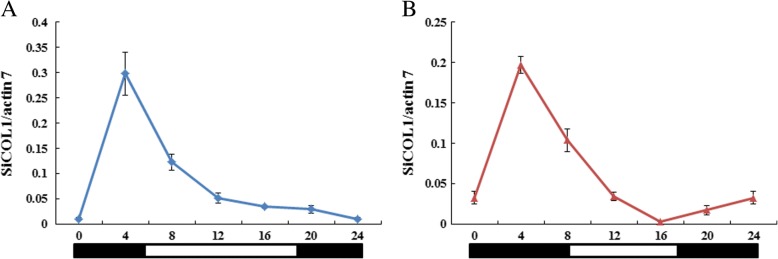
Individuals of ‘Zhongzhi13’ were grown in the LD (14 h light) and SD (9 h light) conditions, respectively. In about 3 days before the flower buds appeared, leaves from three individuals were collected during a 24 h period under LD and SD conditions, respectively. Expressions of SiCOL1 and SiCOL2 in the leaves under LD and SD conditions were detected. Although expression of SiCOL2 was higher than SiCOL1 in both LD and SD conditions, the expression patterns of these two genes were extremely similar. Both in LD and SD conditions, the expression of SiCOL1 and SiCOL2 increased during the darkness whereas decreased under light (Fig. 4 and Additional file 1: Figure S5). The peaks of transcript level of SiCOL1 and SiCOL2 in LD and SD conditions were both in the dawn. Under the SD condition, the lowest expression levels of SiCOL1 and SiCOL2 were both found at 1 h before dusk. Whereas, the valleys of the transcript levels for SiCOL1 and SiCOL2 under LD were different. Under LD condition, SiCOL1 and SiCOL2 had the lowest expression levels in 0 am and 8 pm, respectively. Therefore, as the homolog of CO in sesame, SiCOL1 and SiCOL2 exhibited significantly diurnal rhythmic expression and expressed in a high level before the flowering in leaves.
Fig. 4.
Relative diurnal expression of SiCOL1 under LD and SD conditions. a Relative expression of SiCOL1 under LD condition. b Relative expression of SiCOL1 under SD condition. White boxes below the graphs indicate light periods and dark boxes indicate darkness. The expression data was normalized by sesame actin7. The bar indicates standard deviation
Haplotype variation of SiCOL1 and SiCOL2
In order to analyze the haplotype variations of SiCOL1 and SiCOL2, SNPs of SiCOL1 and SiCOL2 in 132 landrace genomes were obtained from the SesameHapMap database (http://www.ncgr.ac.cn/SesameHapMap/). These landraces were collected from South Asia, Southeast Asia, East Asia and Central Asia. These regions are the main producing regions of sesame with rich germplasm resources. Among these regions, South Asia is also the geographic origin area of sesame [9, 52]. All samples were planted in the summer of Wuhan, China from 2015 to 2017 and their flowering dates were recorded. Previous study revealed that sesame accessions could be divided into south group and north group by the latitude 32°N [13]. In the present study, samples were also divided into south and north groups according to their geographic origin (Additional file 1: Table S3).
In total, 25, 23 and 2 SNPs were found in the promoter, coding region and intron of SiCOL1, respectively (Fig. 5). Among the 23 SNPs in the coding region, 13 SNPs were the synonymous mutations while the other 10 SNPs were the nonsynonymous mutations, which led to amino acid substitutions and might cause functional polymorphism of the SiCOL1 protein. Only one SNP and three SNPs were detected in the CCT domain and Zinc-finger domain, respectively.
Fig. 5.
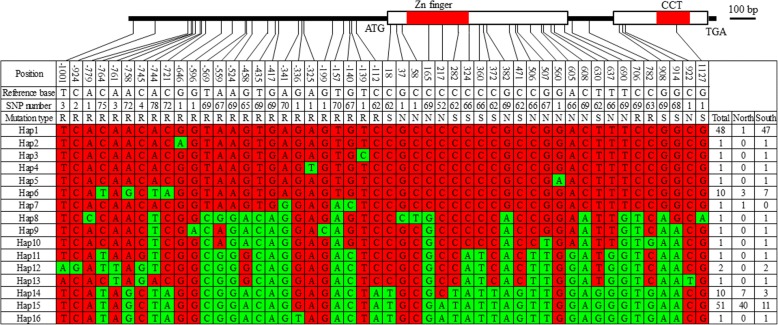
Haplotypes of SiCOL1 among landraces from Asia. Reference base is the base in reference genome ‘Zhongzhi13’. SNP number is the mutation number among the 132 landraces. R, S and N in mutation type indicate replacement, synonymous SNP and nonsynonymous SNP, respectively. Numbers in the right column are numbers of cultivars represented in every haplotypes. Total, South and North indicate total landraces, landraces from south group and landraces from north group, respectively. Variations that different from the reference bases are shown in green
Based on the identified SNPs, 16 haplotypes of SiCOL1 were detected in the tested sesame accessions. All bases in Hap1 (Haplotype 1) were the same as the reference genome ‘Zhongzhi13’ [43]. The bases in Hap1 that were different from other haplotypes ranged from 1 to 35. Six of the haplotypes (Hap2 to Hap7) were similar to Hap1 while the other nine haplotypes (Hap8 to Hap16) were quite different from Hap1. There was only one SNP in Hap 2, Hap3, Hap4 and Hap5. But in Hap 14, Hap 15 and Hap16, the different bases reached 33, 34 and 35, respectively.
The variety ‘Baizhima’ (S054 in Additional file 1: Table S3), which had the SiCOL1 of Hap15 was selected and the expression of SiCOL1 and SiCOL2 was investigated. SiCOL2 showed diurnal rhythmic expression in ‘Baizhima’ under both LD and SD conditions (Additional file 1: Figure S5). However, the expression of SiCOL1 was not detected in ‘Baizhima’ under both LD and SD conditions, suggesting that mutated SiCOL1 did not express and might lose the function of photoperiod response in sesame flowering.
Totally, 15 SNPs were identified in SiCOL2, including seven SNPs in promoter, six SNPs in coding regions and two SNPs in intron (Additional file 1: Figure S6). Four SNPs in the coding regions were the nonsynonymous mutations. However, these SNPs were identified in a few samples, indicating that SiCOL2 was more conserved than SiCOL1. Using the 15 SNPs, SiCOL2 was clustered into 12 haplotypes. The haplotypes contained more than 7 accessions (5.30% of the total samples) were regarded as major haplotypes. Therefore, Hap1, Hap3 and Hap8 were identified to be the three major haplotypes. Among these haplotypes, Hap1 was the biggest haplotype, containing 65.2% of the total samples.
To valid the truth of the SNPs in SiCOL1 and SiCOL2, ten accessions were selected and sequenced. All SNPs identified in SiCOL1 and SiCOL2 of the ten samples were the same as them in SesameHapMap. The result suggested that all SNPs of these genes were true and could be used in the haplotype analysis. However, a 6 bp deletion (from 421 bp to 426 bp) in the coding region, which resulted in an Aspartic acid and a Glutamic acid deletion in protein, was detected in Hap15 of SiCOL1 (Additional file 1: Figure S7). Previous study showed that a 36 bp deletion in the coding region of Hd1 was the crucial mutation that led function divergence of Hd1 in rice [2]. This deletion might have potential influence of gene function in the Hap15 of SiCOL1.
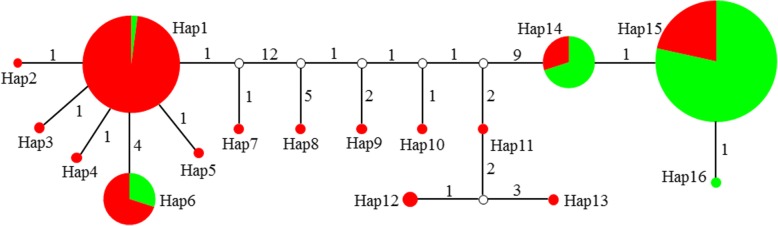
As shown in Fig. 6, a network of all haplotypes was constructed. The haplotype number of landraces from south group (15) was much more than that of north group (5), suggesting that SiCOL1 had highly polymorphisms in the landraces of south group. There were four haplotypes contained landraces from both south and north group: Hap1, Hap6, Hap14 and Hap15. These four haplotypes were also the largest haplotypes in number, containing 90.2% (119 of 132 landraces) of the samples. The landraces belonging to south group were concentrated in Hap 1 and Hap6 (54 of 80 landraces), while most of the landraces from north group were in Hap14 and Hap15 (47 of 52 landraces).
Fig. 6.
Haplotype network of SiCOL1. Haplotypes are showed by colored solid circles. Circle size is proportional to the quantity of samples within a given haplotype. Hollow circles indicate the assumed haplotypes. Lines between haplotypes represent mutational steps between alleles. The numbers next to the lines indicate the nucleotide difference existed between the linked haplotypes. The red color and green color indicate landraces from south group and north group, respectively
The landraces from India presented in Hap1, Hap5, Hap6, Hap8, Hap9, Hap11, Hap12 and Hap13, indicating a high genetic diversity of SiCOL1 in India sesame landraces. If we take all landraces from South Asia (India, Bangladesh, Pakistan and Nepal) into account, more haplotypes could be found, including Hap4, Hap7, Hap10, Hap 15 and Hap16. Therefore, landraces from South Asia could be found in 13 haplotypes totally. For Southeast Asia, East Asia and Central Asia, the haplotypes of landraces from these regions were Hap7, Hap5 and Hap2, respectively. The haplotypes of landraces from South Asia were much more than haplotypes including landraces from other regions, suggesting that South Asia was the genetic diversity center of SiCOL1. This observation is consistent with previous suggestion that crop cultivars from the geographic origin areas tend to have higher genetic diversity [53, 54].
A network of all SiCOL2 haplotypes was also constructed (Additional file 1: Figure S8). Landraces from south group and north group were detected in twelve and five haplotypes, respectively. In the network of SiCOL1, two major haplotypes, Hap14 and Hap15 were dominated by the landraces from north group. However, landraces from south group were more than that from north group in all major haplotypes of SiCOL2 (Hap1, Hap3 and Hap8).
SiCOL1 haplotypes were related to sesame flowering
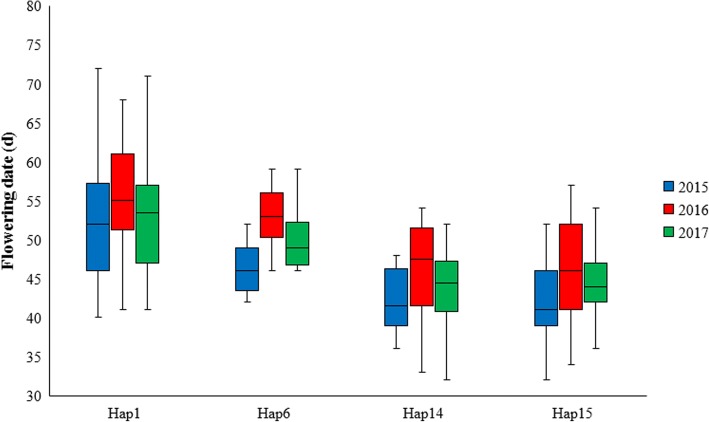
The flowering date of the 132 landraces from 2015 to 2017 in Wuhan, China (114°33′ E, 30°34′ N) was recorded and analyzed to further examine the relationship between SiCOL1 haplotypes and sesame flowering (Additional file 1: Table S3). The day light in the summer of Wuhan is a standard LD, sustaining from 13 h to 14.5 h. Under LDs, sesame landraces from north group flowering obviously earlier than that from south group. The box-plot showed the flowering date of landraces in Hap1, Hap 6, Hap14 and Hap15 from 2015 to 2017 (Fig. 7). As we described previously, Hap1 and Hap6 mainly contained sesame accessions from south group, while Hap14 and Hap15 included most sesame accessions from north group. Days to flowering time of the samples in Hap1 and Hap6 were significant more than that in Hap14 and Hap15 (Mann-Whitney test, P < 10− 9). Taking flowering time in 2016 for example, the average flowering date of accessions in Hap1, Hap6, Hap14 and Hap15 was 58.5, 53, 46.2 and 46.3 d, respectively. The Pearson correlation coefficient was used to test the correlation between SiCOL1 haplotypes and flowering date. Significant correlations were identified in all 3 years: 2015 (R2 = 0.32, R = 0.56, P = 3.10 × 10− 11), 2016 (R2 = 0.28, R = 0.53, P = 5.38 × 10− 10) and 2017 (R2 = 0.30, R = 0.55, P = 7.80 × 10− 11). The results suggested that SiCOL1 variations were strongly related to the flowering time of sesame.
Fig. 7.
Box-plot of the flowering date of sesame landraces in major haplotypes. The major haplotypes, Hap1, Hap6 Hap14 and Hap15 contained 48, 10, 10 and 51 sesame accessions, respectively. All sesame landraces were planted from May to October at Wuhan, China in every year. The detailed information of the flowering date of the sesame landraces was provided in Additional file 1: Table S3
Geographic distribution of SiCOL1 haplotype
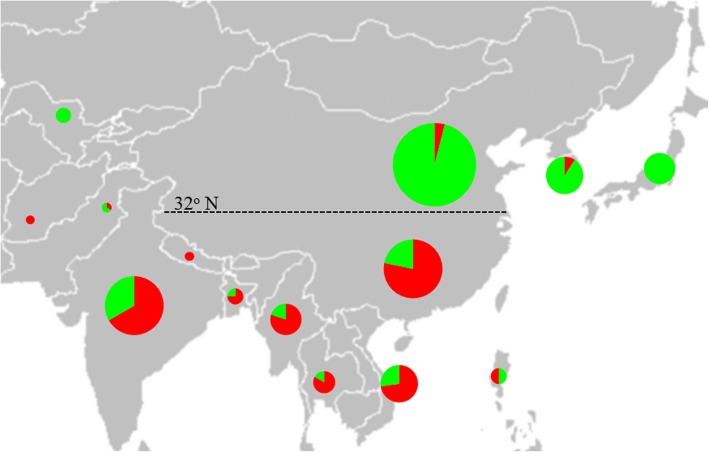
Comparing to Hap1 of SiCOL1, Hap15 had one 6 bp deletion in the coding region (Additional file 1: Figure S7) and many SNPs in the promoter as well as coding regions (Fig. 5). In addition, Hap15 did not express under both LD and SD conditions. Therefore, Hap15 of SiCOL1 was regarded as nonfunctional allele. Based on the similarity of haplotypes, we divided the 16 haplotypes of SiCOL1 into two groups, south haplotypes with functional alleles and north haplotypes with nonfunctional alleles. The south haplotypes included Hap1 to Hap7 while the north haplotypes contained Hap 8 to Hap 16. To investigate the relationship between the geographic origin and haplotypes of the sesame landraces, a map of Asia was downloaded from Wikimedia Commons (http://commons.wikimedia.org/wiki/Main_Page) and the distribution information of SiCOL1 haplotypes was showed in the map (Fig. 8). The map clearly showed that south haplotypes mainly existed in the south of 32°N while north haplotypes were concentrated in the north of 32°N. For the 13 countries, the proportion of the north haplotypes ranged from 0 (Nepal and Afghanistan) to 100% (Japan and Uzbekistan).
Fig. 8.
SiCOL1 protein type distribution among countries in Asia. Red solid circles indicate SiCOL1 protein types from Hap1 to Hap7, while the green solid circles represent SiCOL1 protein types from Hap8 to Hap16. The size of the circles is proportional to the quantity of sesame landraces. The latitude 32°N is indicated by dotted line. The original map was downloaded and adapted from “https://commons.wikimedia.org/wiki/File:BlankMap-Asia.png“(Bytebear at the English language Wikipedia). This original map is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license, which allows us to share and adapt for free with proper attribution
Since alleles contained in north haplotypes all were nonfunctional and very few landraces in the north haplotypes were from the geographic origin center of sesame, north haplotypes were regarded as the domesticated haplotypes of SiCOL1. The frequency of domesticated alleles is an indicator of artificial selection, so the proportion of the north haplotypes was used to examine the domestication and spread of sesame. North haplotypes were in the minority of Southern Asia, Southeast Asia and South China, but they were the dominant haplotypes in Northern China, Northeast Asia and Central Asia. Therefore, the result suggested that SiCOL1 had been strongly selected and might be the important domesticated gene that contributed to the spread of sesame from low-latitude regions to high-latitude regions.
Expression patterns of SiFT in two varieties with different SiCOL1 haplotypes
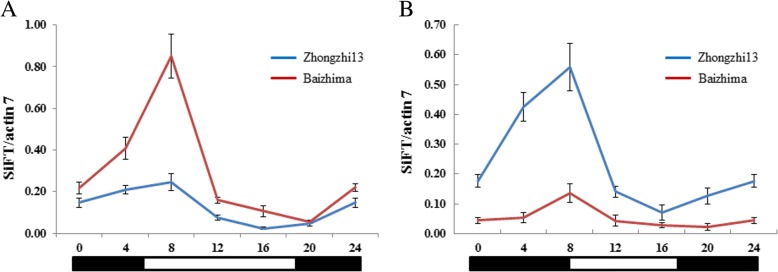
The homolog of FT in sesame, SiFT (SIN_1009320), was identified by BLAST [55]. Expression of SiFT was detected in ‘Zhongzhi13’ (with SiCOL1 of Hap1) and ‘Baizhima’ (with SiCOL1 of Hap15) under LD and SD conditions. The diurnal rhythmic expression pattern of SiFT was quite similar to that of SiCOL1 under both LD and SD conditions (Fig. 9), indicating that the expression of SiFT might be induced by SiCOL1. Although the expression pattern of SiFT in ‘Zhongzhi13’ and ‘Baizhima’ was similar, the expression level of SiFT in these two varieties was quite different under both LD and SD conditions. These significantly variant expression levels of SiFT in ‘Zhongzhi13’ and ‘Baizhima’ might result from the non-expression of SiCOL1 in ‘Baizhima’.
Fig. 9.
Relative diurnal expression of SiFT under LD and SD conditions. a Relative expression of SiFT under LD condition. b Relative expression of SiFT under SD condition. White boxes below the graphs indicate light periods and dark boxes indicate darkness. The expression data was normalized by sesame actin7. The bar indicates standard deviation
The peak of SiFT expression appeared later than that of SiCOL1. This phenomenon was in line with the homologue genes, Hd3a and Hd1, in the SD plant rice. Although Hd1 had the expression peak in dark, Hd3a had the highest expression level after dawn under both LD and SD conditions [56].
Discussion
SiCOL1 might be involved in the photoperiod response and contributing to flowering
Photoperiod pathway is one of the crucial regulation factors of high plant flowering [57]. CO, one of the first identified plant photoperiod gene, plays an important role in the photoperiod response and flowering regulation of Arabidopsis [3]. The CO homolog of sesame has not been identified and the flowering mechanism of sesame largely remains unknown. In this study, molecular function, gene expression and sequence variations of the CO homolog in sesame, SiCOL1, were comprehensively analyzed.
Phylogenetic analysis showed SiCOL1 was one of the most similar genes of CO in sesame. Overexpression of SiCOL1 in the transgenic Arabidopsis lines significantly promoted flowering of Arabidopsis under LD condition. Both under LD and SD conditions, SiCOL1 showed diurnal rhythmic expression and had the peak expression at the dawn. Compared with the SiCOL1 expression in root, stem, capsule and seed, SiCOL1 had a higher expression level in leaf. The expression pattern of SiCOL1 was extremely similar to that of AtCO1 and AtCOL2 [58]. Although AtCOL1 and AtCOL2 doesn’t have major role in the control of flowering in Arabidopsis, homologous genes with similar dawn expression peaks have been shown to control FT expression and flowering time in other species including soybean and strawberry [59, 60]. The diurnal rhythmic expression pattern of SiFT was similar to that of SiCOL1, indicating that SiCOL1 might induce the expression of SiFT. In ‘Zhongzhi13’, SiFT had a higher expression level under SD condition than that under LD condition. It was consistent with the early flowering of ‘Zhongzhi13’ under SD condition. By analyzing the days to flowering of 132 landraces, we found that the north haplotypes of SiCOL1 harboring nonfunctional alleles flowered much earlier than that of other landraces under LD condition. In the variety ‘Baizhima’, which had Hap15 of SiCOL1, SiFT had a high expression level under LD condition. Therefore, the early flowering of sesame landraces, which had the nonfunctional haplotypes of SiCOL1, might result from the highly expression of SiFT under LD conditions. Since SiCOL1 did not express in these landraces, we concluded that SiCOL1 might repressed the expression of SiFT under LD condition. Hd1 functions in the promotion of rice flowering under SD condition and in inhibition under LD condition [2]. We speculated that SiCOL1 might have similar function of Hd1 in the photoperiod response and contributing to flowering. Because the transgenic approach of sesame had not been invented, it is hard to perform the transgenic experiments to validate the function of SiCOL1 in sesame. This will be addressed in future studies.
Besides SiCOL1, there were 12 other SiCOL genes that were identified from sesame genome. Function of these genes had not been reported yet. In the last decades, COL gene family has been studied in several plants [22, 23, 30, 36–39, 41]. In Arabidopsis, 17 COL genes were identified totally. Due to the differences of B-box motifs and introns, the AtCOLs were divided into three groups [41], Group I contained CO and COL1 to COL5 with two B-box motifs and one intron. Group II includes COL6 to COL8 and COL16 with one B-box motif and one intron. Group III incorporates COL9 to COL15 with two B-box motifs and three introns [23]. Similar result had been recognized in sesame COL genes. However, SiCOL2 which belonged to Group I lacked B-box 1 motif, indicating the possible divergence of SiCOLs and AtCOLs. Previous studies showed that Arabidopsis COL genes not only regulated flowering time, but also participated in plant architecture, development, and stresses tolerance [61–64]. The SiCOLs might be also involved in diverse molecular and genetic processes of sesame.
SiCOL1 rather than SiCOL2 is more likely to be the functional homologous gene of CO in sesame
Phylogenetic analysis of SiCOL1, SiCOL2, CO and CO homologues of 19 plant species showed SiCOL1 was close to PnCO, whereas SiCOL2 was close to SlCO and StCO. Previous study showed that PnCO could promote flowering of P. nil [25], a typical SD plant. However, there is no evidence shows that SlCO can regulate flowering of Solanum lycopersicum [49], a day-neutral plant. Comparison of SiCOL1 and SiCOL2 protein sequences and motifs revealed differences in the Zinc-finger domain that could be the underlying reason for differences in function. When we overexpressed SiCOL1 and SiCOL2 into Arabidopsis, the flowering time promoted by SiCOL2 was less than SiCOL1. In addition, the expression pattern of SiCOL2 was different from that of SiFT in the variety ‘Baizhima’ (Additional file 1: Figure S5). These results suggested that SiCOL1 rather than SiCOL2 was more likely to be the functional CO homologous gene in sesame.
Much fewer SNPs were detected in the coding region (6 SNPs) and domains (2 SNPs) of SiCOL2 than that of SiCOL1 (Additional file 1: Figure S6). Two SNPs were detected in the B-box motif and CCT domain of SiCOL2. But only one was nonsynonymous SNP and few of the landraces contained this mutation in SiCOL2. Totally, there were 12 haplotypes of SiCOL2 and most sesame accessions were concentrated in Hap1, Hap3, Hap5 and Hap8 (Additional file 1: Figure S8). The SiCOL2 haplotype network showed that the major haplotypes had an extremely close relationship and landraces from south and north group were mixed in the haplotypes. The results indicated that variations of SiCOL2 might not affect the flowering of sesame and SiCOL2 had not been significantly selected.
Genome research uncovered an independent whole genome duplication (WGD) event in sesame genome at approximately 71 ± 19 million years ago [43]. The paralogs, SiCOL1 and SiCOL2, may be the duplicated genes. Functional divergence of these paralogs might result from the loss of B-box 1 motif in SiCOL2. Redundant genes resulting from WGD are thought to be lost or acquire new functions [65]. SiCOL2 might lose its gene function after WGD.
Artificial selection of SiCOL1 might have contributed to sesame spread to a wide range of latitudes
Gene sequence analysis of SiCOL1 revealed that two nonsynonymous mutations which caused amino acid residues replacement were in the Zinc-finger domain. In addition, one 6 bp deletion in the coding region was detected in the haplotypes harboring this mutation. The amino acid residues replacement in Zinc-finger domain, the 6 bp deletion, and multiple SNPs in the coding regions as well as promoter might result in the loss of function of SiCOL1. Landraces which contained these mutations mainly distributed in high-latitude regions and flowered early in LD conditions. In contrast, most landraces from low-latitude regions, especially South Asia, which was the domestication center of sesame, did not have these mutations and flowered late in LD conditions. Photoperiod genes in the wild relatives of crops, such as rice, maize and soybean are generally functional and photoperiod genes tend to be selected during the crop spread [66–68]. Therefore, the functional SiCOL1 in the samples from South Asia was more likely to be the ancestral haplotypes. Further haplotype analysis of SiCOL1 in the Asia sesame collections revealed that the landraces from north group containing nonfunctional SiCOL1 alleles distributed across Northern China, Northeast Asia and Central Asia. Northeast Asia is in the northern-limit regions of sesame, with more than 15 h mean day length during the short growing season. Almost all sesame landraces in Northeast Asia had a few haplotypes with nonfunctional mutations and flowered early under LD condition. The haplotypes of landraces from Northeast Asia was significantly less than that from South Asia. Additionally, the landraces from Northeast Asia harbored nonfunctional haplotypes of SiCOL1. The results suggested SiCOL1 in the landraces from Northeast Asia might undergo positive selection or strong domestication and SiCOL1 played a significant role in sesame adaptation to high-latitude regions by reducing photoperiod sensitivity. Domestication and selection on SiCOL1 might be one of the critical events that contribute to adapt sesame to different cultivation areas and cropping seasons, resulting in sesame from a local crop in India to the global oilseed crop.
Several studies of the rice photoperiod genes have reported that selection of the flowering genes mainly contributed to the expansion of rice from tropical and subtropical areas to temperate areas, resulting in rice changing from a regional plant to a worldwide plant [33, 69–71]. Domestication of the photoperiod genes, such as Hd1, Ehd1, Hd3a, Ghd7, Ghd8, and DTH2, caused function loss and decreased the photoperiod sensitivity, leading to early-heading phenotypes. The artificial selection and domestication of rice flowering genes successfully extended the northern-limit regions of rice cultivation. In this study, similar phenomenon was observed in the sesame photoperiod gene SiCOL1. Artificial selection and domestication of SiCOL1 might contribute the early flowering of nonfunctional haplotypes and involved in the spread of sesame from low-latitude area (South Asia) to high-latitude areas (Northeast Asia and Central Asia). Since the function of different haplotypes had not been completely demonstrated by the sesame transformation approach, and there was weak population structure in the sesame landraces [13], this conclusion still need more evidences to support.
To date, more than 700 quantitative trait loci (QTLs) and 30 photoperiod genes had been identified in rice [72]. Natural variations that related to rice flowering were found in 14 genes. In the present study, we found that some extremely early-heading landraces contained nonfunctional alleles of SiCOL1. For example, landraces containing the SiCOL1 of a nonfunctional haplotype (Hap14) flowers at 43.6 d in average under LD condition. But the accession ‘Baizhima’ (S049 in Additional file 1: Table S3) from Northeast China (125°8′ E, 45°51′ N), which also harboring SiCOL1 of Hap14, flowers quite earlier (30.7 ± 1.3 d) than other accessions. These finding suggested that sesame domestication in the northernmost regions might have been achieved by artificial selection of SiCOL1, as well as domestication in other photoperiod genes. Furthermore experiments needed to be carried out to recognize these photoperiod genes in sesame.
SiCOL1 identified in this research could be used in the sesame improvement and molecular breeding. Because of the simple and efficient of sesame artificial hybridization, any favorable allele of SiCOL1 in landraces can easily be transferred to commercial varieties for adapting to different light conditions. The gene editing technology, CRISPR/Cas9, had been successfully used in the editing of tomato flowering gene SELF PRUNING 5G, causing 2 weeks earlier flowering [73]. Using the CRISPR/Cas9 method on editing of photoperiod genes, such as SiCOL1, geographical range of sesame could be extended. Sesame might be grown in latitudes more northerly than currently possible, which could also bring more plantings per growing season and thus higher yield of sesame.
Conclusions
Flowering and photoperiod sensitivity are fundamental traits that determine sesame, an important oilseed crop, adaptation to a wide range of geographic environments. Whereas the flowering mechanism of sesame is still not clear. In the present study, we identified sesame COL gene family and focused on functional analysis of the CO homologous gene, SiCOL1. Phylogenetic analysis and sequence comparison revealed that SiCOL1 might be the homolog of the CO gene in sesame. Overexpression of the SiCOL1 in transgenic Arabidopsis significantly promoted flowering of Arabidopsis under LD conditions. Expression analysis revealed that SiCOL1 had highly expressed levels in leaf before flowering and exhibited a diurnal rhythmic expression under both SD and LD conditions. Moreover, SiCOL1 might induce the expression of SiFT under both SD and LD conditions. In the Asia sesame collections, different haplotype alleles of SiCOL1 were found. However, the mutated haplotype (Hap15) of SiCOL1 did not express under both SD and LD conditions. The similar haplotypes of Hap15 were regarded as nonfunctional alleles of SiCOL1. Notably, the sesame varieties from high-latitude regions harboring nonfunctional alleles of SiCOL1 flowered extremely early, and were adapted to the northernmost regions of sesame cultivation. The results suggested that SiCOL1 was the potential functional homolog of CO and haplotype variations of SiCOL1 enables sesame to adapt to different day-lengths characteristic of different latitudes. Moreover, the domestication and artificial selection of SiCOL1 might have contributed to the spread of sesame from low-latitude regions to high-latitude regions. Our results could be useful in both understanding the flowering mechanism and the molecular breeding of sesame.
Methods
Identification of the COL gene family in sesame
All sesame protein sequences were obtained from the sesame genome database (http://ocri-genomics.org/Sinbase/) [74]. The Arabidopsis thaliana AtCOL gene sequences were downloaded from TAIR (https://www.arabidopsis.org/). The HMM profile for the Zinc-finger B-box domain (PF00643) and CCT domain (PF06203) were downloaded from the PFAM protein families database (http://pfam.xfam.org) [75] and used to identify COL genes from the sesame genome with HMMER 3.0 [76]. BLAST analysis with all the Arabidopsis COLs was used to check the predicted COLs from the sesame database [55]. The CDD (http://www.ncbi.nlm.nih.gov/cdd/) [77] and the simple modular architecture research tool (SMART) [78] were used to validate all the potential sesame COL genes identified by HMM and BLAST if they contained the B-box motifs and CCT domains.
Phylogenetic and sequence analyses of the COL gene family in sesame
Clustal X 2.0 [79] was used to align the aa sequences of the sesame and Arabidopsis COL proteins. A unrooted NJ phylogenetic tree [80] of these genes was constructed by MEGA 6.0 [81]. The nodes of the NJ tree were evaluated by bootstrap analysis for 1000 replicates. Branches with less than 50% bootstrap values were collapsed.
Twenty eight protein sequences of CO and Hd1 homologs in plant species were download from NCBI, including AtCO (A. thaliana, X94937), AtCOL1 (AED92215), AtCOL2 (AEE73800), BnCO (Brassica napus, AY290868), BdHd1 (Brachypodium distachyon, XP_003563958), GhCOL1-A (Gossypium hirsutum, ASA69414), GhCOL1-D (ASA69421), GmCOL1a (Glycine max, NP_001235828/ Glyma.08G255200.1, the gene ID in G. max genome Wm82.a2.v1), GmCOL1b (NP_001235843/Glyma.18G278100.1), GmCOL2a (XP_003541197/Glyma.13G050300.1), GmCOL2b (NP_001278944/Glyma.19G039000.1), HvCO1 (Hordeum vulgare, AF490468), LtCO (Lolium temulentum, AY553297), MdCOL1 (Malus domestica, AAC99309), OsHd1 (Oryza sativa, AB041838), PdCOL1 (Populus deltoids, AAS00054), PdCOL2 (AAS00055), PnCO (P. nil, AF300700), PpCOL1 (Physcomitrella patens, BAD89084), PrCO (Pinus radiate, AF001136), RsCOL1 (Raphanus sativus, AF052690), SiCOL1 (S. indicum, XP_011085568), SiCOL2 (S. indicum, XP_011099077), SlCO (S. lycopersicum, NP_001233839), StCO (S. tuberosum, ARU77840), TaHd1 (Triticum aestivum, AB094490), VvCO (Vitis vinifera, CBI16899), ZmHd1 (Zea mays, ABW82153). Another CO homolog, UgCO (Scf02496.g25887.t1) was identified by BLAST with AtCO from the genome of U. gibba [82], which had taxonomically close relationship with sesame. Phylogenetic tree of CO and 21 CO homologs, including SiCOL1 and SiCOL2, was constructed by NJ method, with the 1000 replications bootstrap analysis.
The conserved motifs in the full-length COL proteins were identified using the MEME program (http://alternate.meme-suite.org/tools/meme) [83]. The parameters employed in the analysis were as follows: maximum number of motifs = 3; optimum width of motifs = 15–60. The exon/intron structures of the SiCOL genes were determined by comparing their predicted coding sequence (CDS) with genomic sequences using the gene structure display server web-based bioinformatics tool (http://gsds.cbi.pku.edu.cn/) [84].
Plant samples and treatments
The photoperiod-sensitive sesame variety ‘Zhongzhi13’ was selected and used for the gene expression analysis. It is widely cultivated in China and has been used in the genome sequencing of the sesame [43]. The materials were planted in summer of 2015 at Wuhan, China (114°33′ E, 30°34′ N). At the flowering stage, the roots, stems, leaves, capsules and developing seeds were collected from three plants of the variety ‘Zhongzhi13’ from 8:00 to 9:00 am during the day. After collection, these organs were immediately frozen in liquid nitrogen and stored at − 80 °C prior to further analysis. Leaves at 10 development stage of the variety ‘Zhognzhi13’ were collected, including 7, 14, 21, 28, 30, 32, 36, 38, 40 and 50 d. These leaves were collected at 8:00 am every time using the described method previously.
For the LD and SD treatments, sesame plants of ‘Zhongzhi13’ and ‘Baizhima’ (S054 in Additional file 1: Table S3) were firstly planted in pots at natural light condition for 1 week. Then the LD and SD treatment plants were planted under LD (14 h light from 5:00 to 17:00, 10 h darkness) and SD (9 h light from 8:00 to 17:00, 15 h darkness) conditions, respectively. Leaf samples from at least three sesame plants were collected every 4 h during a 24 h period (at 0:00, 4:00, 8:00, 12:00 16:00 and 20:00 every day) at the last week before flowering. The leaves were frozen in liquid nitrogen and total RNA was isolated immediately.
Totally, 132 sesame landraces from 13 counties in South Asia, Southeast Asia, East Asia and Central Asia were selected from sesame core-collections and planted in summer of Wuhan, China from 2015 to 2017 (Additional file 1: Table S3). The flowering date of each landrace was recorded. All the sesame samples were provided by the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan, China.
Overexpression of SiCOL1 and SiCOL2 in transgenic Arabidopsis
The binary vector pBI121 was digested by restriction enzymes Sma I and Sac I. We combined the amplified cDNA of SiCOL1 and SiCOL2 with the linear vector pBI121 using one step cloning kit (ClonExpress, Vazyme), and then transformed it into Agrobacterium tumefaciens. Arabidopsis was then transformed by the floral dip method [85]. Plasmid isolation was performed using the Plasmid DNA mini kit (Omega). The nucleotide sequencing was determined by Tsingke Company (Wuhan, China). The analysis of nucleotide sequence was done by the BioEdit [86] and DNAstar Lasergene (http://www.dnastar.com/t-dnastar-lasergene.aspx).
Kanamycin-resistant transgenic Arabidopsis T0 plants were regenerated, allowed to self-fertilize and T1 seeds were sown on medium containing kanamycin. Ten independently transformed kanamycin-resistant lines were self-fertilized and T2 seed collected from each individual. Then ten individuals of T2 generation were grown in LD condition (22 °C, 14 h photoperiod). Flowering time was measured as the number of days from sowing to the appearance of flower buds in the center of the plant rosette. In about 1 week before flowering, leaves of T2 lines were collected from the wild type Arabidopsis and individuals with overexpressed SiCOL1 and SiCOL2.
Expression analysis
Total mRNA was extracted using the RNA extraction kit EASYspin Plus Plant RNA Kit (Aidlab Biotechnologies, Beijing, China) according to the manufacturer’s instructions. The RNA was reverse-transcribed into cDNA using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, USA). The quantitative real-time PCR (qRT-PCR) experiments were performed with gene-specific primers in the reaction system of SYBR Green Supermix (Bio-rad, USA) on the CFX384 Real-Time System (Bio-Rad) according to the manufacturer’s instructions. The qRT–PCR assay was performed in triplicate with independent individuals and the actin (At3g18780) and sesame actin7 gene (SIN_1006268) were used as internal controls for Arabidopsis and sesame genes, respectively. The expression data of SiCOL1, SiCOL2, SiFT (SIN_1009320) and FT were quantified by the 2-ΔΔCT method [87]. qRT-PCR primers used for sesame and transgenic Arabidopsis were listed in Additional file 1: Table S4. All primers were synthesized by Tsingke Company (Wuhan, China).
Haplotype and network analyses
SNPs of SiCOL1 in 132 landrace genomes were selected and downloaded from the SesameHapMap database (http://www.ncgr.ac.cn/SesameHapMap/) [13]. The SiCOL1 sequence regions included coding region, promoter and intron. These 132 landraces were selected from 13 Asian countries, containing Afghanistan, Bangladesh, Burma, China, India, Japan, Nepal, Pakistan, Philippines, South Korea, Thailand, Uzbekistan, and Vietnam. Haplotypes of SiCOL1 and SiCOL2 in these landraces were generated by DNASP version 6.0 [88].
Ten accessions of the landraces were selected and their SiCOL1 and SiCOL2 genes were sequenced. The information of the accessions was available at Additional file 1: Table S3. There were two haplotypes of SiCOL1 in these accessions -- Hap1 (S012, S016, S060, S062 and S075) and Hap15 (S050, S053, S054, S057 and S115 was Hap15). There were five haplotypes of SiCOL2 in these accessions. The haplotype of SiCOL2 in S050, S053, S057, S060 and S062 was Hap1. The haplotypes of SiCOL2 in S012, S016, S054, S075 and S112 included Hap2, Hap3, Hap4 and Hap8. SNPs and Indels of these genes in the accessions were identified by aligning with ClustalX 2.0 [79]. Primers used in the PCR were provided in Additional file 1: Table S4.
The haplotype networks of SiCOL1 and SiCOL2 were constructed by mutational steps with NETWORK 4.6 [89]. The networks represented the genetic distance of DNA sequences or alleles and were mainly consist of circles of different sizes and colors as well as lines that linked the circles. The circle size was proportional to the number of samples within a given haplotype, and the lines between the haplotypes represented mutational steps between the alleles.
The distribution of SiCOL1 haplotypes was showed in a map of Asia. The original map was downloaded and adapted from “https://commons.wikimedia.org/wiki/File:BlankMap-Asia.png“(Bytebear at the English language Wikipedia). This original map is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license, which allows us to share and adapt for free with proper attribution. SiCOL1 haplotypes was indicated by different colors. The size of the circles was proportional to the number of sesame landraces.
Additional file
Figure S1. Phylogenetic relationships and structures of SiCOL proteins. Figure S2. Comparison of SiCOL1, SiCOL2 and CO protein sequences. Figure S3. Relative expression of FT in leaves of T2 transgenic Arabidopsis lines with overexpressed SiCOL1 and SiCOL2. Figure S4. Relative expression of SiCOL2 in different tissues and development stages of sesame. Figure S5. Relative diurnal expression of SiCOL2 under LD and SD conditions. Figure S6. Nucleotide changes in the coding region of SiCOL2 among cultivated sesame. Figure S7. Sequences of SiCOL1 in ten sesame landraces. Figure S8. Haplotype network of SiCOL2. Table S1. Information of B-box gene family and CCT-containing gene family in sesame genome. Table S2. Days to flowering of Arabidopsis samples. Table S3. Information of the sesame landraces from Asia used in the present study. Table S4. Primers used in the qRT-PCR. (PDF 634 kb)
Acknowledgements
We are grateful to Prof. Xuehui Huang from Shanghai Normal University for the help of language editing. We thank Ms. Lixia Huang, Ms. Yiting Wu, Ms. Yuan Li and Ms. Yuan Gao from Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences for their help in the experiments.
Funding
This work was funded by the National Natural Science Foundation of China (31671282), Technology Innovation Project of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI), Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700), and Hubei Chenguang Talented Youth Development Foundation to XW.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files; Sesame genome sequence in this article is offered in the sesame genome database (http://ocri-genomics.org/Sinbase/); The protein sequences of CO and CO homologs can be found from NCBI (https://www.ncbi.nlm.nih.gov/); The SNPs of SiCOL1 and SiCOL2 are available at SesameHapMap (http://www.ncgr.ac.cn/SesameHapMap/); The Arabidopsis thaliana gene sequences in this article were downloaded from TAIR (https://www.arabidopsis.org/). All plant materials were selected from sesame germplasm provided by the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan, China.
Abbreviations
- Aa
Amino acid
- BLAST
Basic local alignment search tool
- CCT
CONSTANS, CONSTANS-like, TIMING OF CAB EXPRESSION 1
- CDD
Conserved domain database
- COL
CONSTANS-like
- FT
FLOWERING LOCUS T
- GSDS
Gene structure display server
- HMM
Hidden Markov model
- LD
Long day
- LGs
Linkage groups
- MEME
Multiple Em for Motif Elicitation
- NJ
Neighbor-joining
- qRT-PCR
quantitative real-time polymerase chain reaction
- QTLs
Quantitative trait loci
- SD
Short day
- SMART
Simple modular architecture research tool
- SNP
Single nucleotide polymorphism
- WGD
Whole genome duplication
Authors’ contributions
XW and ZX conceived and designed the experiments. RZ, PL and DL performed the experiments. XW, RZ and PL analyzed the data. XW and RZ wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rong Zhou, Email: rongzzzzzz@126.com.
Pan Liu, Email: liupan91040220@163.com.
Donghua Li, Email: ldh360681@163.com.
Xiurong Zhang, Email: zhangxr@oilcrops.cn.
Xin Wei, Email: xwei@shnu.edu.cn.
References
- 1.Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 2003;100(10):6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell. 1995;80(6):847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 4.Halliday KJ, Koornneef M, Whitelam GC. Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiol. 1994;104(4):1311–1315. doi: 10.1104/pp.104.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paradiso R, De Pascale S. Effects of plant size, temperature, and light intensity on flowering of Phalaenopsis hybrids in Mediterranean greenhouses. Sci World J. 2014;2014:420807. doi: 10.1155/2014/420807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson SD. Plant responses to photoperiod. New Phytol. 2009;181(3):517–531. doi: 10.1111/j.1469-8137.2008.02681.x. [DOI] [PubMed] [Google Scholar]
- 7.Schiessl S, Iniguez-Luy F, Qian W, Snowdon RJ. Diverse regulatory factors associate with flowering time and yield responses in winter-type Brassica napus. BMC Genomics. 2015;16:737. doi: 10.1186/s12864-015-1950-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Cahill J, Van Hautegem T, Feys K, Whipple C, Novak O, Delbare S, Versteele C, Demuynck K, De Block J, et al. Altered expression of maize PLASTOCHRON1 enhances biomass and seed yield by extending cell division duration. Nat Commun. 2017;8:14752. doi: 10.1038/ncomms14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedigian D. Evolution of sesame revisited: domestication, diversity and prospects. Genet Resour Crop Ev. 2003;50(7):779–787. doi: 10.1023/A:1025029903549. [DOI] [Google Scholar]
- 10.Fuller DQ. Further evidence on the prehistory of sesame. Asian Agri History. 2003;7(2):127–137. [Google Scholar]
- 11.Kumazaki T, Yamada Y, Karaya S, Tokumitsu T, Hirano T, Yasumoto S, Katsuta M, Michiyama H. Effects of day length and air temperature on stem growth and flowering in sesame. Plant Prod Sci. 2008;11(2):178–183. doi: 10.1626/pps.11.178. [DOI] [Google Scholar]
- 12.Sinha SK, Tomar DPS, Deshmukh PS. Photoperiodic response and yield potential of sesamum genotypes. Indian J Genet Plant Breed. 1973;33:293–296. [Google Scholar]
- 13.Wei X, Liu K, Zhang Y, Feng Q, Wang L, Zhao Y, Li D, Zhao Q, Zhu X, Zhu X, et al. Genetic discovery for oil production and quality in sesame. Nat Commun. 2015;6:8609. doi: 10.1038/ncomms9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285(5433):1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 15.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 16.Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187(1):57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 17.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brambilla V, Fornara F. Molecular control of flowering in response to day length in rice. J Integr Plant Biol. 2013;55(5):410–418. doi: 10.1111/jipb.12033. [DOI] [PubMed] [Google Scholar]
- 19.Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002;16(15):2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43(10):1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 21.Almada R, Cabrera N, Casaretto JA, Ruiz-Lara S, Villanueva EG. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 2009;28(8):1193–1203. doi: 10.1007/s00299-009-0720-4. [DOI] [PubMed] [Google Scholar]
- 22.Cai D, Liu H, Sang N, Huang X. Identification and characterization of CONSTANS-like (COL) gene family in upland cotton (Gossypium hirsutum L.) PLoS One. 2017;12(6):e0179038. doi: 10.1371/journal.pone.0179038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths S, Dunford RP, Coupland G, Laurie DA. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003;131(4):1855–1867. doi: 10.1104/pp.102.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong DH, Sung SK, An G. Molecular cloning and characterization of constans-like cDNA clones of the Fuji apple. J Plant Biol. 1999;42(1):23–31. doi: 10.1007/BF03031143. [DOI] [Google Scholar]
- 25.Liu J, Yu J, McIntosh L, Kende H, Zeevaart JA. Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 2001;125(4):1821–1830. doi: 10.1104/pp.125.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Garcia JF, Virgos-Soler A, Prat S. Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc Natl Acad Sci U S A. 2002;99(23):15211–15216. doi: 10.1073/pnas.222390599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller TA, Muslin EH, Dorweiler JE. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta. 2008;227(6):1377–1388. doi: 10.1007/s00425-008-0709-1. [DOI] [PubMed] [Google Scholar]
- 28.Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 2003;36(1):82–93. doi: 10.1046/j.1365-313X.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- 29.Robert LS, Robson F, Sharpe A, Lydiate D, Coupland G. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol Biol. 1998;37(5):763–772. doi: 10.1023/A:1006064514311. [DOI] [PubMed] [Google Scholar]
- 30.Wu FQ, Price BW, Haider W, Seufferheld G, Nelson R, Hanzawa Y. Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One. 2014;9(1):e85754. doi: 10.1371/journal.pone.0085754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, Gerard S, Chou L, Bhambhra N, Parks JD, et al. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun. 2014;5:3651. doi: 10.1038/ncomms4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrenreich IM, Hanzawa Y, Chou L, Roe JL, Kover PX, Purugganan MD. Candidate gene association mapping of Arabidopsis flowering time. Genetics. 2009;183(1):325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc Natl Acad Sci U S A. 2009;106(11):4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujino K, Wu J, Sekiguchi H, Ito T, Izawa T, Matsumoto T. Multiple introgression events surrounding the Hd1 flowering-time gene in cultivated rice, Oryza sativa L. Mol Gen Genomics. 2010;284(2):137–146. doi: 10.1007/s00438-010-0555-2. [DOI] [PubMed] [Google Scholar]
- 35.Wei X, Qiao W, Yuan N, Chen Y, Wang R, Cao L, Zhang W, Yang Q, Zeng H. Domestication and association analysis of Hd1 in Chinese mini-core collections of rice. Genet Resourc Crop Evol. 2014;61(1):121–142. doi: 10.1007/s10722-013-0019-8. [DOI] [Google Scholar]
- 36.Chia TYP, Muller A, Jung C, Mutasa-Gottgens ES. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J Exp Bot. 2008;59(10):2735–2748. doi: 10.1093/jxb/ern129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou ML, Shih MC, Chan MT, Liao SY, Hsu CT, Haung YT, Chen JJ, Liao DC, Wu FH, Lin CS. Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid Erycina pusilla. Planta. 2013;237(6):1425–1441. doi: 10.1007/s00425-013-1850-z. [DOI] [PubMed] [Google Scholar]
- 38.Fu J, Yang L, Dai S. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium. Mol Gen Genomics. 2015;290(3):1039–1054. doi: 10.1007/s00438-014-0977-3. [DOI] [PubMed] [Google Scholar]
- 39.Song N, Xu Z, Wang J, Qin Q, Jiang H, Si W, Li X. Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene. 2018;673:1–11. doi: 10.1016/j.gene.2018.06.032. [DOI] [PubMed] [Google Scholar]
- 40.Zobell O, Coupland G, Reiss B. The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 2005;7(3):266–275. doi: 10.1055/s-2005-865621. [DOI] [PubMed] [Google Scholar]
- 41.Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28(6):619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 42.Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. The Arabidopsis B-box zinc finger family. Plant Cell. 2009;21(11):3416–3420. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Yu S, Tong C, Zhao Y, Liu Y, Song C, Zhang Y, Zhang X, Wang Y, Hua W, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15(2):R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei X, Zhu X, Yu J, Wang L, Zhang Y, Li D, Zhou R, Zhang X. Identification of sesame genomic variations from genome comparison of landrace and variety. Front Plant Sci. 2016;7:1169. doi: 10.3389/fpls.2016.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang HY, Miao HM, Wang L, Qu LB, Liu HY, Wang Q, Yue MW. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 2013;14(1):401. doi: 10.1186/gb-2013-14-1-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Liu P, Yu J, Wang L, Dossa K, Zhang Y, Zhou R, Wei X, Zhang X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017;17(1):152. doi: 10.1186/s12870-017-1099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang YY, Zhang YJ, Zhou R, Dossa K, Yu JY, Li DH, Liu AL, Mmadi MA, Zhang XR, You J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS One. 2018;13(7):e0200850. doi: 10.1371/journal.pone.0200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Wang L, Guo H, Liao B, King G, Zhang X. Genome evolutionary dynamics followed by diversifying selection explains the complexity of the Sesamum indicum genome. BMC Genomics. 2017;18(1):257. doi: 10.1186/s12864-017-3599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46(3):462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 50.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2014;19(7):460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 52.Bedigian D. Characterization of sesame (Sesamum indicum L.) germplasm: a critique. Genet Resour Crop Ev. 2010;57(5):641–647. doi: 10.1007/s10722-010-9552-x. [DOI] [Google Scholar]
- 53.Huang XH, Kurata N, Wei XH, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490(7421):497. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A. 2006;103(45):16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422(6933):719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- 57.Komeda Y. Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:521–535. doi: 10.1146/annurev.arplant.55.031903.141644. [DOI] [PubMed] [Google Scholar]
- 58.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26(1):15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- 59.Cao D, Li Y, Lu SJ, Wang JL, Nan HY, Li XM, Shi DN, Fang C, Zhai H, Yuan XH, et al. GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 2015;56(12):2409–2422. doi: 10.1093/pcp/pcv152. [DOI] [PubMed] [Google Scholar]
- 60.Kurokura T, Samad S, Koskela E, Mouhu K, Hytonen T. Fragaria vesca CONSTANS controls photoperiodic flowering and vegetative development. J Exp Bot. 2017;68(17):4839–4850. doi: 10.1093/jxb/erx301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Datta S, Hettiarachchi GHCM, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18(1):70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min JH, Chung JS, Lee KH, Kim CS. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J Integr Plant Biol. 2015;57(3):313–324. doi: 10.1111/jipb.12246. [DOI] [PubMed] [Google Scholar]
- 63.Ordonez-Herrera N, Trimborn L, Menje M, Henschel M, Robers L, Kaufholdt D, Hansch R, Adrian J, Ponnu J, Hoecker U. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018;176(2):1327–1340. doi: 10.1104/pp.17.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci U S A. 2017;114(1):172–177. doi: 10.1073/pnas.1616459114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci U S A. 2015;112(48):14918–14923. doi: 10.1073/pnas.1507669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF, et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci U S A. 2012;109(28):E1913–E1921. doi: 10.1073/pnas.1203189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu H, Li Q, Xing Y. Genes contributing to domestication of rice seed traits and its global expansion. Genes. 2018;9(10). [DOI] [PMC free article] [PubMed]
- 68.Zhang SR, Wang H, Wang Z, Ren Y, Niu L, Liu J, Liu B. Photoperiodism dynamics during the domestication and improvement of soybean. Sci China Life Sci. 2017;60(12):1416–1427. doi: 10.1007/s11427-016-9154-x. [DOI] [PubMed] [Google Scholar]
- 69.Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153(4):1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, Wu C, Wang HJ, Wang Q, Zhou K, et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci U S A. 2013;110(8):2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 72.Hori K, Matsubara K, Yano M. Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor Appl Genet. 2016;129(12):2241–2252. doi: 10.1007/s00122-016-2773-4. [DOI] [PubMed] [Google Scholar]
- 73.Soyk S, Muller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jimenez-Gomez JM, Lippman ZB. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet. 2017;49(1):162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- 74.Wang L, Yu J, Li D, Zhang X. Sinbase: an integrated database to study genomics, genetics and comparative genomics in Sesamum indicum. Plant Cell Physiol. 2015;56(1):e2. doi: 10.1093/pcp/pcu175. [DOI] [PubMed] [Google Scholar]
- 75.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 80.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 81.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ibarra-Laclette E, Lyons E, Hernandez-Guzman G, Perez-Torres CA, Carretero-Paulet L, Chang TH, Lan T, Welch AJ, Juarez MJ, Simpson J, et al. Architecture and evolution of a minute plant genome. Nature. 2013;498(7452):94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 86.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 87.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 88.Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 89.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic relationships and structures of SiCOL proteins. Figure S2. Comparison of SiCOL1, SiCOL2 and CO protein sequences. Figure S3. Relative expression of FT in leaves of T2 transgenic Arabidopsis lines with overexpressed SiCOL1 and SiCOL2. Figure S4. Relative expression of SiCOL2 in different tissues and development stages of sesame. Figure S5. Relative diurnal expression of SiCOL2 under LD and SD conditions. Figure S6. Nucleotide changes in the coding region of SiCOL2 among cultivated sesame. Figure S7. Sequences of SiCOL1 in ten sesame landraces. Figure S8. Haplotype network of SiCOL2. Table S1. Information of B-box gene family and CCT-containing gene family in sesame genome. Table S2. Days to flowering of Arabidopsis samples. Table S3. Information of the sesame landraces from Asia used in the present study. Table S4. Primers used in the qRT-PCR. (PDF 634 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files; Sesame genome sequence in this article is offered in the sesame genome database (http://ocri-genomics.org/Sinbase/); The protein sequences of CO and CO homologs can be found from NCBI (https://www.ncbi.nlm.nih.gov/); The SNPs of SiCOL1 and SiCOL2 are available at SesameHapMap (http://www.ncgr.ac.cn/SesameHapMap/); The Arabidopsis thaliana gene sequences in this article were downloaded from TAIR (https://www.arabidopsis.org/). All plant materials were selected from sesame germplasm provided by the Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan, China.