Abstract
Current lye processing for debittering California black table olives produces large amounts of caustic wastewater and destroys many of the beneficial phenolic compounds in the fruit. Herein, we propose using enzyme treatment in place of lye, potentially reducing the amount and causticity of wastewater produced. By specifically targeting the bitterness-causing compound, oleuropein, retention of other beneficial phenolics may be possible. A β-glucosidase from Streptomyces sp. was identified from a screen of 22 glycosyl hydrolases to completely degrade oleuropein in 24 h. Computational modeling was performed on this enzyme, and mutation C181A was found to improve the rate of catalysis by 3.2-fold. This mutant was tested in the context of the olive fruit and leaf extract. Degradation was observed in the olive leaf extract but not in the fruit matrix, suggesting that enzyme fruit penetration is a limiting factor. This work discovers and begins the refinement process for an enzyme that has the catalytic properties for debittering olives and provides direction for future engineering efforts required to make a product with commercial value.
Introduction
There is a need to replace the lye-curing method used by the California table olive industry to debitter olives. The current process involves multiple cycles of lye treatments and water rinses, which generates large quantities of caustic wastewater. A life cycle assessment performed for a three-step process estimated that 580–660 kg of wastewater is produced per 100 kg olives.1,2 The wastewater has a high pH (9.0–12.0), a high chemical oxygen demand (1500–3800 mgO2/L), and a high biological oxygen demand (390–1150 mgO2/L), which makes mitigation difficult and expensive for processors.3 Additionally, California-style processing removes many phenolic compounds from the olive fruit, which reduces potential health benefits of the finished product.4,5 Replacing lye-curing with a more efficient and targeted method could potentially relieve these problems.6
The secoiridoid oleuropein is primarily responsible for bitterness in olives.7 Selective degradation of oleuropein using an enzyme would accomplish the goal of debittering the olive while retaining the nutritional content of the fruit. Furthermore, an enzyme that is highly active against oleuropein could shorten processing time and reduce the amount and causticity of wastewater produced. Endogenous olive β-glucosidases and esterases contained in the fruit are known to degrade oleuropein during ripening, storage, or when the fruit is damaged.8−10 The purported reactions that take place are shown in Scheme 1 (could occur in either order), wherein a β-glucosidase cleaves the sugar moiety, resulting in oleuropein aglycone and glucose; oleuropein aglycone is hydrolyzed to nonbitter elenolic acid and hydroxytyrosol via esterase cleavage.11−13 We hypothesized that introducing one or both of these enzymes during olive processing (see Figure S1) in the place of lye could degrade oleuropein specifically, while retaining other phenolics in the fruit that would otherwise be destroyed.
Scheme 1. Oleuropein is Converted to the Nonbitter Glucose, Elenolic Acid, and Hydroxytyrosol via Endogenous β-Glucosidases and Esterases.

It has been reported that the ester hydrolysis can also occur spontaneously.18
Involving enzymes in olive debittering is not a novel idea and has been previously considered. Select Lactobacillus plantarum strains produce β-glucosidases with activity toward oleuropein, and these organisms have been shown to accelerate debittering of olives during fermentation.14,15 It was also found that adding a commercial β-glucosidase during fermentation with L. plantarum yielded slightly lower levels of oleuropein after 90 days compared with fermentation alone.16 However, debittering olives via fermentation can take several weeks to months and fermented olives have a different sensory profile than California-style olives. De Leonardis et al. found that treating olives with crude olive leaf protein extract (which exhibited β-glucosidase activity) resulted in faster oleuropein degradation than fermentation with L. plantarum, and an 87% reduction in oleuropein was achieved after 30 days.17 Unfortunately, this method exceeds the current processing time of California-style olives and oleuropein degradation was incomplete. In addition to looking for oleuropein degradation activity under olive storage conditions, in vitro characterization of enzymes has been explored as well. For example, a recombinant β-glucosidase from the organism Sulfolobus solfataricus, which has wide substrate specificity, was immobilized onto a chitosan matrix and used to convert commercially purchased oleuropein into hydroxytyrosol.18 Furthermore, degradation activity of a β-glucosidase extracted from almonds has also been reported.19,20 However, the utility of these methods for removing oleuropein in an actual olive fruit was not tested.
It is clear from previous research that naturally occurring β-glucosidases can degrade oleuropein and debitter olives. However, enzyme discovery and design efforts are yet to be conducted to discover and optimize an enzyme for this commercial activity. Here, we utilize a combination of genomic mining and computational protein design to identify leads and initiate efforts toward engineering an enzyme for this application. Specifically, we identify an enzyme capable of complete degradation of oleuropein to its nonbitter components and perform computationally guided engineering to improve the turnover rate. This enzyme is also tested in the context of the olive fruit and against olive leaf extract herein to define current limitations and provide direction for future engineering efforts.
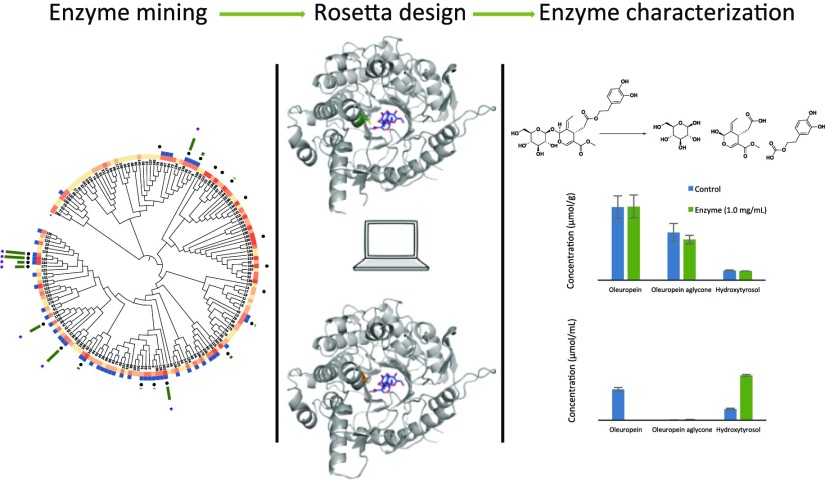
Results and Discussion
Inspired by the natural degradation pathway, we chose to select a panel of enzymes from the β-glucosidase enzyme class (EC 3.2.1.21) for initial oleuropein degradation screening. The chemistry performed by this EC consists of cleavage of a terminal, nonreducing, β-d-glucosyl residue with the release of β-d-glucose from a wide variety β-d-glucosides21 (i.e. oleuropein). Twenty-two β-glucosidases were chosen from a pool of 170 family 1 glycosyl hydrolases (GH1s; all sequences contained in the Supporting Information) gifted by the Joint Genome Institute (with 2 genes constructed in-house; GH1s are characterized based on their canonical eight-fold α/β barrel motif).22 This panel was chosen based on β-glucosidase activity on cellobiose and soluble expression levels in Escherichia coli according to experimental data provided by JGI.22Figure 1 depicts the phylogenetic tree that was built based on sequence similarity containing all 170 GH1s in the pool. β-Glucosidases that were tested are indicated on the tree and have an average pairwise sequence identity of 32%. Activity was analyzed by monitoring hydroxytyrosol formation by high-performance liquid chromatography (HPLC; μM hydroxytyrosol formation in 24 h) and is shown on the tree (green bar graph). Eight enzymes (starred on the tree and have a pairwise sequence identity of 41%) were found to degrade oleuropein to hydroxytyrosol in significant amounts (>200 μM hydroxytyrosol formation) after a 24 h, room temperature incubation. The oleuropein aglycone, formed in situ following glycosyl cleavage, spontaneously degrades to hydroxytyrosol without the need of further treatment with an esterase. This instability of oleuropein aglycone and rapid conversion to hydroxytyrosol has been observed in other work.18
Figure 1.
Phylogenetic tree based on sequence similarity of 170 GH1s. Expression level and cellobiose activity are indicated (salmon heat map and blue binary, respectively). Absence of color in the expression field indicates in-house gene construction and expression (data contained in the Supporting Information). The 22 GH1s tested (black dot) are shown along with activity (green bar graph in μM hydroxytyrosol produced at 24 h). The eight β-glucosidases showing significant activity are starred (forming >200 μM hydroxytyrosol after a 24 h incubation at room temperature).
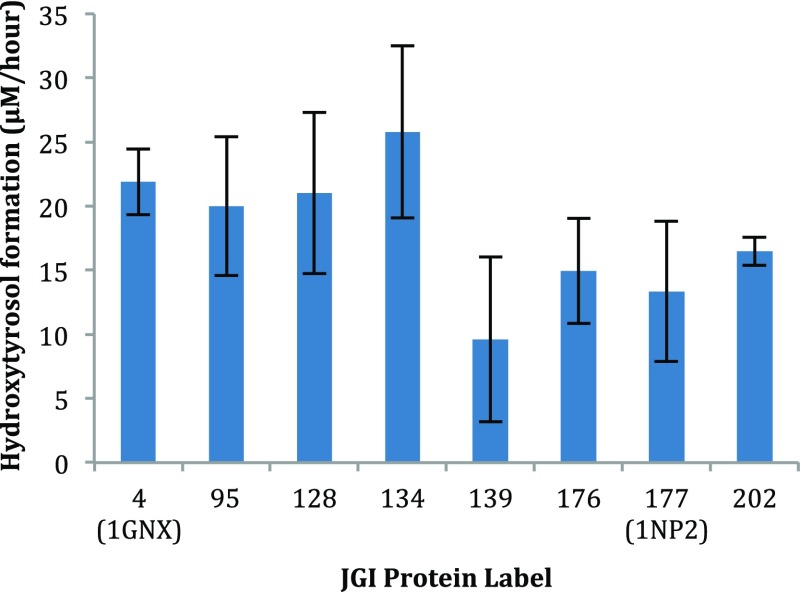
Following the initial screening, a time-point study of these eight candidates was conducted in order to identify the enzymes that produced hydroxytyrosol from oleuropein at a relatively faster rate. Figure 2 shows the rate of hydroxytyrosol formation for each selected candidate (indicated as the protein number and PDB code if available). It is also important to note that hydroxytyrosol, once cleaved from oleuropein, degrades over time, most likely due to oxidation and further dimerization of the catechol moiety via an o-quinone intermediate.23 On the basis of this time-point study and availability of structural data to simplify the modeling process, β-glucosidase from Streptomyces sp.24 (JGI 4) was chosen as the starting point for engineering (PDB accession code: 1GNX, uniprotKB AC: Q59976), which degraded 84% of oleuropein in 24 h.
Figure 2.
Activity of eight best candidates (protein number) displayed as the rate of hydroxytyrosol formation (μM/h; enzyme concentrations were normalized to 0.1 mg/mL; error bars represent standard deviation with n = 3; PDB accession codes are indicated if structural information is available).
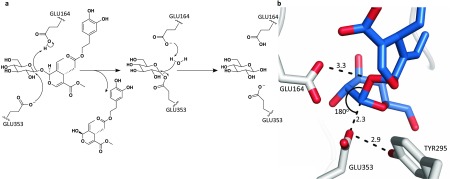
Following this lead candidate, computational modeling was performed using the Rosetta molecular modeling suite to guide our engineering efforts to enhance activity of this enzyme. The catalytic triad of 1GNX was identified by aligning the crystal structure with another β-glucosidase, PDB 2JIE (BglB), which is in complex with a covalently bound inhibitor (2-deoxy-2-fluoro-α-d-glucopyranose) and has catalytic residues elucidated.25 The oleuropein ligand was constrained in the active site according to known β-glucosidase chemistry. The purported catalytic mechanism performed on oleuropein is shown in Figure 3a and is initiated as GLU353 nucleophilically attacks the anomeric carbon of the β-d-glucosyl residue, displacing the oleuropein aglycone in an SN2 fashion. GLU164 works as an acid/base conduit, protonating the leaving group (oleuropein aglycone) and subsequently deprotonating a water molecule, creating a hydroxide nucleophile that displaces GLU353 and releases β-d-glucose with retention of stereochemistry. TYR295 orients the nucleophilic GLU353 to preorganize the active site for attack.25 Conformers of oleuropein, containing a transition state-like geometry at the anomeric carbon of the initial SN2 attack, were built and minimized using Spartan.26 This conformational library was then used in a Rosetta docking and design protocol with functional constraints for the enzyme defined.27 The electrophilic carbon was constrained as the center of a trigonal bipyramidal geometry, wherein the attacking and leaving group oxygen atoms would be placed at the two apexes, giving an angle between these three atoms of 180° (Onucleophile–Celectrophile–Oleaving: 180°).28 The constraints were further defined to include distances, angles, and dihedrals between the catalytic residues and the oleuropein substrate and also between catalytic residues in an intramolecular fashion (detailed constraint and modeling files are included in the Supporting Information). Figure 3b illustrates an example of a low-energy docking result of oleuropein in the active site with the functional constraints implemented.
Figure 3.
(a) Mechanistic depiction of the hydrolysis of oleuropein to oleuropein aglycone and β-d-glucose showing the catalytic roles of GLU353 and GLU164. (b) Example of a docking result with functional constraints implemented. The distances are shown in Å. Figure generated using PyMol.
The results from the design protocol were filtered based on the total energy and constraint scores as calculated by Rosetta. The top 20 designs with the lowest Rosetta energy and constraint scores were visually inspected in order to revert mutations that did not appear to be chemically sound (i.e. those that did not seem to improve hydrophobic, electrostatic, or hydrogen bonding interactions or remove steric clashing). Eleven mutants (each with 1–2 mutations) were chosen, constructed, and tested for oleuropein degradation activity using the HPLC assay described in the Methods section. Figure 4a shows the activity of these mutants given as μM hydroxytyrosol formation after 2 h. Through this, one mutant showed enhanced activity, degrading roughly 2.5-fold more oleuropein than the native enzyme [wild type (WT)]. This new enzyme has one cysteine to alanine mutation at amino acid position 181 and is depicted in Figure 4c in comparison to the WT (4b). This mutation from cysteine to alanine results in decreased system energy of 1 Rosetta energy unit, predicted to be primarily from relieving steric hindrance between the protein and ligand. A Michaelis–Menten kinetic analysis was performed on both the WT and the C181A mutant, and the resulting constants are shown in Figure 4d.
Figure 4.
(a) Mutant activity given as μM hydroxytyrosol formation after 2 h with 5 mM oleuropein substrate (experiment performed once; n = 1). Enzyme concentrations were normalized to 0.1 mg/mL. (b,c) Interaction of the oleuropein ligand with WT vs C181A, respectively, showing relief of steric clashing (Rosetta residue scores indicated with a lower score representing lower energy). (d) Enzyme kinetic constants, including kcat, Km, and kcat/Km, comparing the WT and the C181A mutant.
Interestingly, while kcat was improved by 3.2-fold with the mutant, the pseudobinding constant, Km, increased by 4.6-fold, which results in an overall decrease in catalytic efficiency. The Km value is quite comparable to a literature value of 7.9 mM for this enzyme on cellobiose.24 Although kcat was not reported, an activity of 0.6 μmol min–1 mg–1 is listed,24 which is less than the calculated activity, 2.5 μmol min–1 mg–1, of the engineered enzyme on oleuropein under similar conditions. However, there is room for improvement on catalytic efficiency through further iterations of computational engineering, given that the average efficiency of enzymes are often on the order of 103 to 105 M–1 s–1.29
Following these specific and kinetic activity analyses, mutant C181A was chosen to be our best candidate for assessing oleuropein degradation activity in fresh olives and olive leaf extract in order to investigate how our enzyme performs within a fruit and vegetative matrix. The enzyme was first tested at a concentration of 0.1 mg/mL in whole, pitted, and crushed olives. The olive fruit had an initial oleuropein content of 2.02 ± 0.13 μmol/g (wet olive weight). After 24 h of incubation, the concentration of oleuropein was not significantly different between control and enzyme-treated samples in any matrix (Figure 5a–c). Whole olives contained the highest amount of phenolics, followed by pitted olives and finally crushed olives, which retained almost nothing. Diffusion of phenolics from the fruit into solution likely occurred at a higher rate as the surface area of the olive increased. Phenolics may also have been degraded by endogenous enzymes such as β-glucosidase, esterase, and polyphenol oxidase, which activate when the drupe flesh is damaged.30
Figure 5.
(a–c) Concentrations of oleuropein and degradation products in (a) whole olives (b) pitted olives and (c) crushed olives following 24 h of incubation with 0.1 mg/mL enzyme (candidate C181A). Concentrations are expressed in μmol per gram of olive, wet weight. (d,e) Concentrations of oleuropein and degradation products in (d) pitted olives and (e) olive leaf extract following 24 h of incubation with 1.0 mg/mL enzyme (candidate C181A). For pitted olives, concentrations are expressed in μmol per gram of olive, wet weight. For olive leaf extract, concentrations are expressed in μmol/mL of extract. Error bars represent standard deviation with n = 3.
From this initial analysis, we hypothesized that either the enzyme was unable to diffuse into the olive flesh to degrade oleuropein or other olive-related matrix components may be inhibitory. Therefore, a second round of experiments was conducted to test these two hypotheses. The first was increasing concentration of the enzyme by 10-fold, the second was to compare pitted olives (maximizing flesh exposure while not destroying the product) versus olive leaf extract which are high in oleuropein.31 In the pitted olives, there was again no significant difference in oleuropein between the control and enzyme-treated samples (Figure 5d). However, in the olive leaf extract, the enzyme yielded complete degradation of oleuropein and a significant increase in hydroxytyrosol (Figure 5e). The molar amounts of oleuropein, oleuropein aglycone, and hydroxytyrosol combined are equal in both samples. These results further support that diffusion into the olive flesh is the limiting factor for enzyme activity. It is hypothesized that oleuropein is located within the vacuoles of olive plant cells32 and the β-glucosidase is likely not able to gain contact with the oleuropein. In order to achieve debittering in the short time frame necessary for California-style olives, further enzyme optimization may be necessary to improve cell penetration. Addition of cell-penetrating peptides to macromolecules, such as proteins, has been shown to facilitate translocation across cell membranes in plants. Specifically, cellular internalization of fluorescently labeled pVEC and transportan has been reported in plant cells.33 Arginine-rich intracellular delivery peptide has been shown to translocate fused fluorescent protein into plant tissue of both tomatoes and onions.34 Pretreating olives with heat or even a single lye rinse can disrupt the integrity of cell walls and potentially improve enzyme diffusion as well.35,36
In its current form, the enzyme has potential applications in olive leaf tea, which is consumed in Mediterranean countries for its antioxidant, antihypertensive, and antimicrobial health benefits.37 The tea is very bitter, and degradation of oleuropein may increase palatability and widen the range of consumers. Additionally, previous research showed that the olive leaf extract concentrated in hydroxytyrosol had higher antiglycative and antioxidant activity than the olive leaf extract concentrated in oleuropein.38 Therefore, increasing the hydroxytyrosol content of olive leaf tea could potentially increase its health benefits. Briante et al.39 explored using recombinant β-glucosidase immobilized in a chitosan support to produce hydroxytyrosol from olive leaf extract on an industrial scale. It is possible that our engineered β-glucosidase could be similarly optimized for commercial applications.
In conclusion, through genomic mining of β-glucosidase sequence space, a unique enzyme from Streptomyces sp. was discovered as being efficient at degrading the bitterness-causing compound in olives, oleuropein. In addition, initial computational protein design efforts to optimize activity have resulted in a mutant with a 3.2-fold increase in the turnover rate. This engineered enzyme was further tested in the context of the fruit and leaf extract matrix. While degradation of oleuropein in the leaf extract was successful, penetration of the enzyme into the fruit proved difficult and further protein engineering is needed to overcome this issue.
Methods
Materials
Oleuropein and hydroxytyrosol were purchased from Sigma-Aldrich (St. Louis, MO, USA). Purity was stated as ≥98%. Manzanilla olives and olive leaves were hand-picked from the UC Davis Orchard in November, 2016.
Phylogenetic Tree Generation
The 170 sequences (listed in the Supporting Information Tables S1–S3) were aligned with MUSCLE (default settings).40 The phylogenetic tree was built using the Geneious tree builder with default settings. The resulting graphic was built and exported from iTOL.41,42
Enzyme Modeling
Conformers of the oleuropein ligand were built in Spartan and minimized using default settings.26 This ligand was built as a transition state-like structure of the initial SN2 attack. Rosetta enzyme docking and design protocols were run with default settings.27 An example of Rosetta enzyme design is provided in the Rosetta molecular modeling suite demos. The oleuropein ligand was constrained in the active site to the catalytic residues according to previously reported chemistry.25 All files used in modeling are contained in the Supporting Information (S1 code).
Mutagenesis, Protein Expression, and Purification
The plasmids encoding the 20 β-glucosidase enzymes tested were gifted to us from the Joint Genome Institute. Two plasmids were constructed in-house. For these, GeneArt custom gene fragments were ordered from Life Technologies and codon optimized for E. coli. Isothermal Gibson assembly43 was used to clone the strings into the pET-29b(+) vector (New England Biolabs, Inc) at the NdeI and XhoI restriction cloning sites. Successful assembly was confirmed by DNA sequencing (Eurofins MWG Operon). The mutant plasmids were constructed using site-directed mutagenesis according to the Kunkel protocol.44 Oligonucleotides that were used in the Kunkel protocol were purchased from Eurofins MWG Operon (Louisville, KY). The mutations were confirmed by DNA sequencing (Eurofins MWG Operon). The WT and mutant plasmids were transformed into the BLR(DE3) strain of E. coli (New England Biolabs, Ipswich, MA). 2 mL of terrific broth containing 50 μg/mL carbenicillin (Fisher Scientific, Hampton, NH) was inoculated with each transformant and was grown for 12 h at 37 °C while shaking at 300 rpm. This 2 mL culture was then expanded to 500 mL (terrific broth with 50 μg/mL carbenicillin) and grown at 37 °C while shaking at 300 rpm until the optical density (OD) reached 0.7–0.9 (OD determined by measuring absorbance at 600 nm). Isopropyl β-d-1-thiogalactopyranoside (IPTG Fisher reagent) to induce protein expression was then added at a final concentration of 1 mM. The cultures continued to grow for an additional 30 h at 18 °C while shaking at 300 rpm. At this point, the cultures were centrifuged at 4700 rpm and the media decanted. The cell pellet was resuspended in 30 mL 1× phosphate buffered saline (1× PBS) with 30 mM imidazole at pH 7.4. To lyse the cells, the suspension was sonicated on ice (with a Fisher Scientific model 705 sonic dismembrator) for a total of 2 min:30 s at an amplitude of 30 and a 30 s break, which is then repeated. The lysate was then clarified by centrifugation at 4700 rpm for 1 h. The supernatant was passed over 1 mL of HisPur Ni-NTA (nickel ion affinity) resin 50% slurry (Fisher Scientific) in a gravity column. The resin was then washed with 10 mL of 1× PBS with 30 mM imidazole six times. The protein was eluted with 10 mL 1× PBS with 200 mM imidazole into Corning Spin-X UF concentrators with a 10 kD molecular weight cutoff. The concentrators were centrifuged at 4700 rpm until the volume reached about 1 mL. Protein concentration was detected by measuring the absorbance at 280 nm with a BioTek spectrophotometer. Protein purity was assessed with a Coomassie stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (NuPAGE bis-tris, Fisher Scientific) and assayed against oleuropein within three days of purification (gel images contained in the Supporting Information Figures S2 and S4).
Assay Conditions and HPLC Method
Once the β-glucosidase WT and mutant enzymes were produced and verified via gel electrophoresis, the assay against oleuropein was performed. The enzymes were normalized to a concentration of 0.1 mg/mL by dilution with 1× PBS up to 95 μL. 100 mM oleuropein (5 μL) in dimethyl sulfoxide was added to initiate the reaction (5 mM final concentration of oleuropein). The reactions were incubated at room temperature for 5–24 h and quenched by the addition of 300 μL acetonitrile. The resulting solutions were centrifuged at 15 000g for 30 min, and the supernatant was pipetted away from the pelleted protein and added to a HPLC vial. An Agilent 1100 series HPLC with variable wavelength UV detection was used to detect hydroxytyrosol, oleuropein, and intermediate products of the reaction at 280 nm. A Phenomenex C18 column (specifications: 2.6 μm C18 100 Å, LC column 50 × 4.6 mm) was used to separate these products with a gradient mobile phase of water (with 0.1% formic acid)/acetonitrile (with 0.1% acetic acid) over a 15 min method (specifically: 0–4 min at 100% water phase; 4–10 min gradient increase of acetonitrile phase to 95%; 10–12 min held at 95% acetonitrile phase; 12–15 min gradient decrease of acetonitrile phase back to 0%).
Kinetic Analysis of the WT and C181A Mutant
To obtain steady-state kinetic data, the WT and C181A mutant were assayed against varying concentrations of oleuropein (7, 5, 3, 2, 1, 0.75, 0.5, and 0.25 mM) at varying time points (1, 2, and 3 h). The concentration of enzyme in all cases was normalized to 0.1 mg/mL (0.9 μM). Reaction buffer, quenching procedure, and HPLC method were conducted as outlined above. From this, kobs (μM hydroxytyrosol min–1 [μM enzyme]−1) per concentration substrate was elucidated and fit to a Michaelis–Menten kinetics curve using R computing software. Km and kcat were also calculated in R using the Michaelis–Menten equation (all raw data, R scripts, and curve fits are contained in the Supporting Information S2 code, Figures S3, S5, and S6).
Assessment of Oleuropein Degradation in Olive Matrices
Enzyme candidate C181A was tested in the olive fruit and olive leaf extract to assess oleuropein degradation in the matrix. In the first experiment, ten whole olives, ten hand-pitted olives, or ten olives crushed in a food processor were soaked in 60 mL of 1× PBS. In the treatment samples, enzyme candidate C181A was added to a final concentration of 0.1 mg/mL. Control samples received an equivalent volume of elution buffer (1× PBS with 200 mM imidazole) without enzyme. All conditions were prepared in triplicate. After 24 h of incubation at room temperature, the fruit was drained. Oleuropein and its degradation products were extracted from the olive fruit using a previously described method45 and quantified using LC–mass spectrometry (MS)/MS.
Analysis was performed on a PerkinElmer Series 200 autosampler and quaternary pump (PerkinElmer, Waltham, MA, USA) coupled to a SCIEX API 2000 triple-quadropole MS system (SCIEX, Framingham, MA, USA) using negative electrospray ionization. Separation of compounds was achieved using an Agilent C18 Eclipse Plus column (5 μm, 4.6 × 250 mm). The mobile phase was 60% water with formic acid (0.05%, v/v) and 40% methanol/acetonitrile (50:50 v/v) with formic acid (0.05%, v/v). The flow rate was 1.0 mL/min through the column but was split before analysis, directing 400 μL/min to the detector and 600 μL/min to waste. Total run time was 20 min. Focusing potential, ion spray voltage, entrance potential, and ion spray temperature were −400, −4500, −10 V, and 400 °C, respectively. Curtain gas, ion source gas 1, ion source gas 2, and collision gas were 50, 60, 40, and 6 psi, respectively. Compounds were identified using multiple reaction monitoring with the following conditions: oleuropein, transition of m/z 539 to m/z 377, declustering potential (DP) of −32 V, collision energy (CE) of −19 V, collision exit potential (CXP) of −11 V; oleuropein aglycone, transition m/z 377 to m/z 307, DP of −20 V, CE of −20 V, CXP of −15 V; hydroxytyrosol, transition m/z 153 to m/z 123, DP of −17 V, CE of −16 V, CXP of −7.5 V. Scan time was 200 ms. Oleuropein and hydroxytyrosol were quantified using six point calibration curves made from reference standards. Oleuropein aglycone was quantified using the calibration curve for hydroxytyrosol based on structural similarities.
The pitted olive trial was repeated in a subsequent experiment with the enzyme concentration at 1.0 mg/mL. This increased enzyme concentration was also tested on olive leaf extract. To prepare the olive leaf extract, 10 g of olive leaves were crushed in a food processor and extracted with 100 mL of 70 °C water for 10 min. The extract was cooled to room temperature and enzyme was added to a final concentration of 1.0 mg/mL. Following 24 h of incubation at room temperature, oleuropein and products were analyzed as previously described.
An independent t-test was performed using OriginPro 2016 b9.226 statistical software (OriginLab Corporation, Northampton, MA, USA) to determine if there was a difference in phenolics between the control and enzyme samples. A p-value of 0.05 was considered significant and equal variance between populations was assumed.
Acknowledgments
We are grateful for the funding and support from UC Davis and the CDFA.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b02169.
The authors declare no competing financial interest.
Supplementary Material
References
- Cappelletti G.; Nicoletti G.; Russo C.. Wastewater from table olive industries. Waste Water-Evaluation and Management; InTech, 2011; Vol. 2.1, pp 351–376. [Google Scholar]
- Cappelletti G. M.; Russo C.; Nicoletti G. M. Life Cycle Assessment (LCA) used to compare two different methods of ripe table olive processing. Grasas Aceites 2010, 61, 136–142. 10.3989/gya.087509. [DOI] [Google Scholar]
- Fernández A. G.; Adams M. R.; Fernández-Díez M.. Table Olives: Production and Processing, 1st ed.; Springer Science & Business Media, 1997; pp 1–496. [Google Scholar]
- Campestre C.; Marsilio V.; Lanza B.; Iezzi C.; Bianchi G.. Phenolic compounds and organic acids change in black oxidized table olives. IV International Symposium on Olive Growing, 2000; Vol. 586, pp 575–578.
- Marsilio V.; Campestre C.; Lanza B. Phenolic compounds change during California-style ripe olive processing. Food Chem. 2001, 74, 55–60. 10.1016/s0308-8146(00)00338-1. [DOI] [Google Scholar]
- Frankel E. A critical literature review on the processing of table olives. Lipid Technol. 2011, 23, 223–226. 10.1002/lite.201100145. [DOI] [Google Scholar]
- Panizzi L.; Scarpati J. Structure of the bitter glycoside oleuropein. Gazz. Chim. Ital. 1960, 90, 1449–1485. [Google Scholar]
- Hbaieb R. H.; Kotti F.; García-Rodríguez R.; Gargouri M.; Sanz C.; Pérez A. G. Monitoring endogenous enzymes during olive fruit ripening and storage: Correlation with virgin olive oil phenolic profiles. Food Chem. 2015, 174, 240–247. 10.1016/j.foodchem.2014.11.033. [DOI] [PubMed] [Google Scholar]
- Brenes M.; Garcia P.; Duran M. C.; Garrido A. Concentration of phenolic compounds change in storage brines of ripe olives. J. Food Sci. 1993, 58, 347–350. 10.1111/j.1365-2621.1993.tb04272.x. [DOI] [Google Scholar]
- Segovia-Bravo K. A.; Jarén-Galán M.; García-García P.; Garrido-Fernández A. Browning reactions in olives: mechanism and polyphenols involved. Food Chem. 2009, 114, 1380–1385. 10.1016/j.foodchem.2008.11.017. [DOI] [Google Scholar]
- Ramírez E.; Brenes M.; García P.; Medina E.; Romero C. Oleuropein hydrolysis in natural green olives: Importance of the endogenous enzymes. Food Chem. 2016, 206, 204–209. 10.1016/j.foodchem.2016.03.061. [DOI] [PubMed] [Google Scholar]
- García A.; Romero C.; Medina E.; García P.; De Castro A.; Brenes M. Debittering of olives by polyphenol oxidation. J. Agric. Food Chem. 2008, 56, 11862–11867. 10.1021/jf802967y. [DOI] [PubMed] [Google Scholar]
- Charoenprasert S.; Mitchell A. Factors influencing phenolic compounds in table olives (Olea europaea). J. Agric. Food Chem. 2012, 60, 7081–7095. 10.1021/jf3017699. [DOI] [PubMed] [Google Scholar]
- Zago M.; Lanza B.; Rossetti L.; Muzzalupo I.; Carminati D.; Giraffa G. Selection of Lactobacillus plantarum strains to use as starters in fermented table olives: Oleuropeinase activity and phage sensitivity. Food Microbiol. 2013, 34, 81–87. 10.1016/j.fm.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Kaltsa A.; Papaliaga D.; Papaioannou E.; Kotzekidou P. Characteristics of oleuropeinolytic strains of Lactobacillus plantarum group and influence on phenolic compounds in table olives elaborated under reduced salt conditions. Food Microbiol. 2015, 48, 58–62. 10.1016/j.fm.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Tuna S.; Akpinar-Bayizit A. The Use of β-Glucosidase Enzyme in Black Table Olives Fermentation. Not. Bot. Horti Agrobot. Cluj-Napoca 2009, 37, 182. 10.15835/nbha3723145. [DOI] [Google Scholar]
- De Leonardis A.; Testa B.; Macciola V.; Lombardi S. J.; Iorizzo M. Exploring enzyme and microbial technology for the preparation of green table olives. Eur. Food Res. Technol. 2016, 242, 363–370. 10.1007/s00217-015-2546-3. [DOI] [Google Scholar]
- Briante R.; La Cara F.; Febbraio F.; Barone R.; Piccialli G.; Carolla R.; Mainolfi P.; De Napoli L.; Patumi M.; Fontanazza G. Hydrolysis of oleuropein by recombinant β-glycosidase from hyperthermophilic archaeon Sulfolobus solfataricus immobilised on chitosan matrix. J. Biotechnol. 2000, 77, 275–286. 10.1016/s0168-1656(99)00219-9. [DOI] [PubMed] [Google Scholar]
- Mazzei R.; Giorno L.; Piacentini E.; Mazzuca S.; Drioli E. Kinetic study of a biocatalytic membrane reactor containing immobilized β-glucosidase for the hydrolysis of oleuropein. J. Membr. Sci. 2009, 339, 215–223. 10.1016/j.memsci.2009.04.053. [DOI] [Google Scholar]
- Capasso R.; Evidente A.; Visca C.; Gianfreda L.; Maremonti M.; Greco G. Production of glucose and bioactive aglycone by chemical and enzymatic hydrolysis of purified oleuropein fromOlea Europea. Appl. Biochem. Biotechnol. 1997, 61, 365–377. 10.1007/bf02787808. [DOI] [Google Scholar]
- Singhania R. R.; Patel A. K.; Sukumaran R. K.; Larroche C.; Pandey A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013, 127, 500–507. 10.1016/j.biortech.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Heins R. A.; Cheng X.; Nath S.; Deng K.; Bowen B. P.; Chivian D. C.; Datta S.; Friedland G. D.; D’Haeseleer P.; Wu D.; Tran-Gyamfi M.; Scullin C. S.; Singh S.; Shi W.; Hamilton M. G.; Bendall M. L.; Sczyrba A.; Thompson J.; Feldman T.; Guenther J. M.; Gladden J. M.; Cheng J.-F.; Adams P. D.; Rubin E. M.; Simmons B. A.; Sale K. L.; Northen T. R.; Deutsch S. Phylogenomically Guided Identification of Industrially Relevant GH1 β-Glucosidases through DNA Synthesis and Nanostructure-Initiator Mass Spectrometry. ACS Chem. Biol. 2014, 9, 2082–2091. 10.1021/cb500244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra-Gómez A.; Luzón-Toro B.; Capel-Cuevas S.; Morales J. C. Stability of hydroxytyrosol in aqueous solutions at different concentration, temperature and with different ionic content: a study using UPLC-MS. Food Nutr. Sci. 2011, 02, 1114–1120. 10.4236/fns.2011.210149. [DOI] [Google Scholar]
- Perez-Pons J. A.; Cayetano A.; Rebordosa X.; Lloberas J.; Guasch A.; Querol E. A β-glucosidase gene (bgl3) from Streptomyces sp. strain QM-B814. FEBS J. 1994, 223, 557–565. 10.1111/j.1432-1033.1994.tb19025.x. [DOI] [PubMed] [Google Scholar]
- Isorna P.; Polaina J.; Latorre-García L.; Cañada F. J.; González B.; Sanz-Aparicio J. Crystal Structures of Paenibacillus polymyxa β-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases. J. Mol. Biol. 2007, 371, 1204–1218. 10.1016/j.jmb.2007.05.082. [DOI] [PubMed] [Google Scholar]
- Shao Y.; Molnar L. F.; Jung Y.; Kussmann J.; Ochsenfeld C.; Brown S. T.; Gilbert A. T. B.; Slipchenko L. V.; Levchenko S. V.; O’Neill D. P.; DiStasio R. A. Jr.; Lochan R. C.; Wang T.; Beran G. J. O.; Besley N. A.; Herbert J. M.; Lin C. Y.; Van Voorhis T.; Chien S. H.; Sodt A.; Steele R. P.; Rassolov V. A.; Maslen P. E.; Korambath P. P.; Adamson R. D.; Austin B.; Baker J.; Byrd E. F. C.; Dachsel H.; Doerksen R. J.; Dreuw A.; Dunietz B. D.; Dutoi A. D.; Furlani T. R.; Gwaltney S. R.; Heyden A.; Hirata S.; Hsu C.-P.; Kedziora G.; Khalliulin R. Z.; Klunzinger P.; Lee A. M.; Lee M. S.; Liang W.; Lotan I.; Nair N.; Peters B.; Proynov E. I.; Pieniazek P. A.; Rhee Y. M.; Ritchie J.; Rosta E.; Sherrill C. D.; Simmonett A. C.; Subotnik J. E.; Woodcock H. L. III; Zhang W.; Bell A. T.; Chakraborty A. K.; Chipman D. M.; Keil F. J.; Warshel A.; Hehre W. J.; Schaefer H. F. III; Kong J.; Krylov A. I.; Gilla P. M. W.; Head-Gordon M. Advances in Methods and Algorithms in a Modern Quantum Chemistry Program Package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. 10.1039/B517914A. [DOI] [PubMed] [Google Scholar]
- Richter F.; Leaver-Fay A.; Khare S. D.; Bjelic S.; Baker D. De novo enzyme design using Rosetta3. PloS One 2011, 6, e19230 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin D. A.; Caster R. W.; Wang X.; Betzenderfer S. A.; Chen C. X.; Duong V. M.; Ryklansky C. V.; Alpekin A.; Beaumont N.; Kapoor H. Kinetic characterization of 100 glycoside hydrolase mutants enables the discovery of structural features correlated with kinetic constants. PloS One 2016, 11, e0147596 10.1371/journal.pone.0147596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A.; Noor E.; Savir Y.; Liebermeister W.; Davidi D.; Tawfik D. S.; Milo R. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 2011, 50, 4402–4410. 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- Ramírez E.; Medina E.; Brenes M.; Romero C. Endogenous enzymes involved in the transformation of oleuropein in Spanish table olive varieties. J. Agric. Food Chem. 2014, 62, 9569–9575. 10.1021/jf5027982. [DOI] [PubMed] [Google Scholar]
- Ranalli A.; Contento S.; Lucera L.; Di Febo M.; Marchegiani D.; Di Fonzo V. Factors Affecting the Contents of Iridoid Oleuropein in Olive Leaves (Olea europaeaL.). J. Agric. Food Chem. 2006, 54, 434–440. 10.1021/jf051647b. [DOI] [PubMed] [Google Scholar]
- Mazzuca S.; Spadafora A.; Innocenti A. M. Cell and tissue localization of β-glucosidase during the ripening of olive fruit (Olea europaea) by in situ activity assay. Plant Sci. 2006, 171, 726–733. 10.1016/j.plantsci.2006.07.006. [DOI] [Google Scholar]
- Chugh A.; Eudes F. Cellular uptake of cell-penetrating peptidespVEC and transportan in plants. J. Pept. Sci. 2008, 14, 477–481. 10.1002/psc.937. [DOI] [PubMed] [Google Scholar]
- Chang M.; Chou J.-C.; Lee H.-J. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant Cell Physiol. 2005, 46, 482–488. 10.1093/pcp/pci046. [DOI] [PubMed] [Google Scholar]
- Marsilio V.; Lanza B.; Campestre C.; De Angelis M. Oven-dried table olives: textural properties as related to pectic composition. J. Sci. Food Agric. 2000, 80, 1271–1276. . [DOI] [Google Scholar]
- Jimenez A.; Guillen R.; Sanchez C.; Fernandez-Bolanos J.; Heredia A. Changes in Texture and Cell Wall Polysaccharides of Olive Fruit during “Spanish Green Olive” Processing. J. Agric. Food Chem. 1995, 43, 2240–2246. 10.1021/jf00056a051. [DOI] [Google Scholar]
- El S. N.; Karakaya S. Olive tree (Olea europaea) leaves: potential beneficial effects on human health. Nutr. Rev. 2009, 67, 632–638. 10.1111/j.1753-4887.2009.00248.x. [DOI] [PubMed] [Google Scholar]
- Navarro M.; Morales F. J.; Ramos S. Olive leaf extract concentrated in hydroxytyrosol attenuates protein carbonylation and the formation of advanced glycation end products in a hepatic cell line (HepG2). Food Funct. 2017, 8, 944–953. 10.1039/c6fo01738j. [DOI] [PubMed] [Google Scholar]
- Briante R.; Patumi M.; Febbraio F.; Nucci R. Production of highly purified hydroxytyrosol from Olea europaea leaf extract biotransformed by hyperthermophilic β-glycosidase. J. Biotechnol. 2004, 111, 67–77. 10.1016/j.jbiotec.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A. III; Smith H. O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. 1985, 82, 488–492. 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L. M.; Holstege D. M.; Wang S. C. High-throughput extraction method for phenolic compounds in olive fruit (Olea europaea). J. Food Compos. Anal. 2018, 66, 136–144. 10.1016/j.jfca.2017.12.013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.