Abstract
In light of the recent success of artificial intelligence (AI) in computer vision applications, many researchers and physicians expect that AI would be able to assist in many tasks in digital pathology. Although opportunities are both manifest and tangible, there are clearly many challenges that need to be overcome in order to exploit the AI potentials in computational pathology. In this paper, we strive to provide a realistic account of all challenges and opportunities of adopting AI algorithms in digital pathology from both engineering and pathology perspectives.
Keywords: Artificial intelligence, deep learning, digital pathology
INTRODUCTION
We are witnessing a transformation in pathology as a result of the widespread adoption of whole slide imaging (WSI) in lieu of traditional light microscopes.[1,2] Depicting microscopic pathology characteristics digitally presents new horizons in pathology. Access to digital slides facilitates remote primary diagnostic work, teleconsultation, workload efficiency and balancing, collaborations, central clinical trial review, image analysis, virtual education, and innovative research. Leveraging WSI technology, the computer vision and artificial intelligence (AI) communities have offered additional computational pathology possibilities including deep learning algorithms and image recognition. Artificial neural networks (ANNs) have witnessed tremendous progress mainly as a result of deep learning. Diverse deep architectures have been trained with large image datasets (e.g., The Cancer Genome Atlas or TCGA and ImageNet) to yield novel biomedical informatics discoveries[3] and perform impressive object recognition tasks. Whether AI will eventually replace, or how it can best assist pathologists, has emerged as a provocative topic.[4,5,6,7] In this article, we discuss key challenges and opportunities related to exploiting digital pathology in the up-and-coming AI era.
CHALLENGES
In spite of the enthusiasm and amassed impressive results shared to date,[8,9,10,11] there are clear obstacles that limit easy employment of AI methods in digital pathology. We discuss several compelling challenges that need to be tackled.
Challenge #1: Lack of labeled data
Most AI algorithms require a large set of good quality training images. These training images must ideally be “labeled” (i.e., annotated). This generally means that a pathologist needs to manually delineate the region of interest (i.e., anomalies or malignancy) in all images. Annotation is ideally best performed by experts. Besides the time constraint involved, manual annotations often also pose a financial bottleneck to app development. Crowdsourcing may be cheaper and quicker but has the potential to introduce noise. For pathologists, detailed annotation of large numbers of images may not only be boring but also can be particularly challenging when working with low resolution or blurry images, slow networks, and ambiguity of features. Active learning applied to annotation may alleviate this taxing task. At present, there are a small number of publicly available datasets that contain labeled images that can be employed for this purpose. For instance, with the Medical Image Computing and Computer Assisted Intervention Society 2014 brain tumor digital pathology challenge, digital histopathology image data of brain tumors were provided. For this competition, when classifying brain tumors, the target was to distinguish images of glioblastoma multiforme (GBM) from low-grade glioma (LGG). The training set had 22 LGG images and 23 GBM images, and the testing (validation) set had 40 images.[12] Another example is the Camelyon dataset that contains many digital slides (i.e., slides with pixel-level annotations and unlabeled slides as a test set) for automated detection of breast carcinoma metastases in hematoxylin and eosin (H and E)-stained whole slide images of lymph node sections.[13] Fortunately, datasets that emphasize the multiclass nature of tissue recognition are slowly emerging.[14] In addition, there are several methods that computational scientists can leverage to maximize limited training data such as data augmentation (i.e., artificially transforming original training images[15]).
Challenge #2: Pervasive variability
There are several basic types of tissue (e.g., epithelium, connective tissue, nervous tissue, and muscle). However, the actual number of patterns derived from these tissues from a computational perspective is nearly infinite if the histopathology images are to be “understood” by computer algorithms. Several tissue types build an organ that is also reflected in new textural variation of the basic tissue types. This extreme polymorphism makes recognizing tissues by image algorithms exceptionally challenging.[16,17] Thus, the inherent architecture of deep AI requires many training cases for each variation. This, however, may not be readily available, especially as labeled data.
Challenge #3: Non-boolean nature of diagnostic tasks
Many published research papers deal with classification problems in digital pathology that deal largely with binary variables, having just two possible values such as “yes” or “no” (e.g., benign or malignant). This is a drastic simplification of the complex nature of diagnosis in pathology.[18] A pathology diagnosis employs several processes including cognition, understanding clinical context, perception, and empirical experience. Sometimes, pathologists use cautious language or descriptive terminology for difficult and rare cases. Such language has ramifications for potential monitoring and treatment.[19] Hence, binary language may only be desirable in easy, obvious cases. This is rarely the case in clinical practice.
Challenge #4: Dimensionality obstacle
WSI deals with gigapixel digital images of extremely large dimensions. Image sizes larger than 50,000 by 50,000 pixels are quite common. Deep ANNs, however, operate on much smaller image dimensions (i.e., not larger than 350 by 350 pixels). “Patching” [i.e., dividing an image into many small tiles, Figure 1] is a potential solution for not just AI algorithms but also for general computer vision methods. However, even for patches, one generally needs to downsample them in order to be able to feed them into a deep network. A region smaller than 1.5 μm2 may not be suitable for many diagnostic purposes and this is, most of the time, at least 1000 by 1000 pixels. Downsampling these patches may result in loss of crucial information. On the other hand, deep nets with larger input sizes would need much deeper topology and much larger number of neurons making them even more difficult and perhaps impossible to train. Of note, patch-based ANNs have been shown to outperform image-based ANNs.[20]
Figure 1.
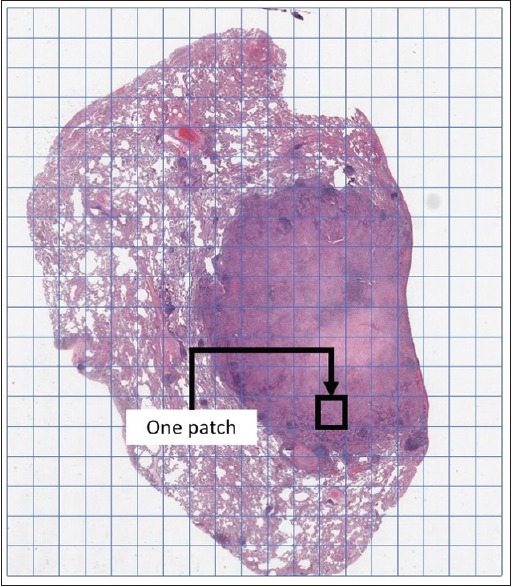
Patching is generally used to represent large scans. For instance, every patch could be a 1000 pixel ×1000 pixel image at ×20
Challenge #5: Turing test dilemma
Alan Turing, one of the most renowned pioneers of AI, suggested measuring the intelligence of machines using a human evaluator.[21] The level of machine intelligence, according to Turing, is inversely proportional to the time that human evaluators would need to figure out that the answers to one's questions are coming from a machine and not from a human operator if the source of the answers is concealed to the evaluator. The Turing test declares that human evolution is the ultimate validation of AI; a machine is as intelligent as a human only if it can successfully and infinitely impostor a human.[22] In digital pathology, we may not know the Turing test explicitly, but everybody adheres to its core statement, namely that the pathologist is the ultimate evaluator if AI solutions are deployed into clinical workflow. Thus, full automation is probably neither possible, it seems, nor wise as the Turing test postulates.
Challenge #6: Uni-task orientation of weak artificial intelligence
Weak AI is a form of AI focused on a specific task. What we speak of today is mostly “weak AI,”[23] representing a collection of specialized algorithms that can perform a given task with high accuracy, provided we can feed them with a large set of training data. In contrast, with strong AI, also called artificial general intelligence (AGI),[24,25] we expect algorithms with human-level intelligence, multitasking, and even consciousness as well as ethical cognition. Of course, the latter is still within scope in the distant future. Deep ANNs belong to the class of weak AI algorithms, as they are designed to perform only one task. That means we would need to separately train multiple AI solutions for tasks such as segmentation, classification, and search. Even for a given task like classification (the major domain for AI algorithms), one would need to design, develop, and train solutions for many anatomical sites. Needless to say, this would require tremendous resources.
Challenge #7: Affordability of required computational expenses
Deep AI solutions are heavily dependent on using Graphical Processing Units (GPUs), highly specialized electronic circuits for fast processing of pixel-based data (i.e., digital images and graphics). Training and using deep solutions on ordinary computers with Central Processing Units is prohibitively sluggish and hence impractical.[26] It is obvious that having access to GPU clusters is a must to deploy deep networks in practice.[27,28,29] Pathology laboratories, however, are already under immense financial pressure to adopt WSI technology, and acquiring and storing gigapixel histopathological scans is a formidable challenge to the adoption of digital pathology. Asking for GPUs, as a prerequisite for training or using deep AI solutions, is consequently going to be financially limiting in the foreseeable future.
Challenge #8: Adversarial attacks – The shakiness of deep decisions
Several reports in the literature have shown that one can “fool” a deep ANN. Targeted manipulation of a very small number of pixels inside an image, which is called an adversarial attack, can mislead a heavy-duty deep network.[30] Apparently, some deep networks that act as a nonlinear and complex “lookup table” are prone to slipping into an adjacent cell of that (invisible) decision table that they implicitly store in their millions of weights. Such behavior is, of course, worrisome in the medical field. Does this imply that the minimal presence of noise or artifacts (e.g., tissue tears/folds, crushed cells, debris, and contamination[31,32]) in a deep ANN can mistakenly be diagnosed as cancer? The research community has still to find out how to create deep networks robust enough to avoid such mishaps. Perhaps, such uncertainty is a new manifestation of the old problem of “overfitting” in AI where a big solution swallows (i.e., memorizes) a small problem. Even so, verification of this new type of error is more difficult to deal with.
Challenge #9: Lack of transparency and interpretability
Deep ANNs have demonstrated several impressive success stories in object and scene recognition. However, they have not removed one of the major drawbacks of ANNs when used as classifiers, which is lack of interoperability. Some consider ANNs to embody a “black box” after they are trained.[33,34] Although researchers have started to investigate creative ways to explain the results of AI,[35] there is at present no established way to easily explain why a specific decision was made by a network when dealing with histopathology scans. In other words, the millions of multiplications and additions performed inside a deep ANN in order to provide an output (i.e., a decision) do not provide a verifiable path to understanding the rationale behind its decisions. This is generally unacceptable in the medical community, as physicians and other experts involved in the diagnostic field typically need to justify the underlying reasons for a specific decision. The pathway to a reliable diagnosis must be transparent and fully comprehensible. This is also important if a deep learning algorithm needs to be fixed (locked down) as well as obtains regulatory approval for its use in clinical practice.
Challenge #10: Realism of artificial intelligence
While there is currently much optimism that AI applied to pathology is going to soon deliver far-reaching benefits (e.g., increased efficiency such as automation, error reduction and greater diagnostic accuracy, and better patient safety), implementing these tools so that they function well in daily practice is going to be difficult to accomplish. Reports of AI failures in health care are not necessarily related to failed technology but rather difficulties deploying AI tools in practice.[36] Pathologists’ buy-in to employ these tools, irrespective of whether they intend to aid or replace them in practice, will depend on three key factors: (1) ease of use (e.g., uncomplicated preimaging demands, agnostic input, and generalizable, scalable, understandable output), (2) financial return on investment associated with using the app, and (3) trust (e.g., evidence of performance).
OPPORTUNITIES
Opportunity #1: Deep features – Pretraining is better
Transfer learning has gained much attention in recent years.[37] Customarily, one trains a deep network with a large set of images in a specific domain and uses the acquired knowledge in a different domain by either using the deep network as a feature extractor or by minimally re-training it with a (small) set of images in the new domain to fine-tune it for the new purpose.[38,39] Pretrained networks, hence, have clear potential for many domains including the medical field.[40] For instance, sentiment analysis in text documents in different domains can benefit from transfer learning, and features learned from a million natural images (animals, buildings, vehicles, etc.) may provide features for medical images. Moreover, some of the aforementioned challenges can be overcome if transfer learning is used instead of attempting to train a new network from scratch.
Opportunity #2: Handcrafted features – Do not forget computer vision
The success of deep learning in developing some AI algorithms has pushed many computer vision schemes aside, among others the role of incorporating handcrafted features. Many well-established feature extraction methods such as local binary patterns,[41] and more recently encoded local projections[42] have demonstrated to be at least on par with deep features and even better in some cases. Their behavior can be fully understood, their results can be interpreted (at least by humans), and their extraction does not need excessive computational resources for learning. Whereas deep learning and other AI algorithms are quite exotic technologies for the pathology community, using “projections” and other conventional technologies may be more in alignment with the knowledge of many medical professionals. Many computer vision methods that utilize handcrafted features (e.g., nuclear size and gland shape) can be much more easily employed in digital pathology to deliver high identification accuracies.[43,44,45,46]
Opportunity #3: Generative frameworks: Learning to see and not judge
Most successful AI techniques belong to the class of discriminative models, methods that can classify data into different groups, but most commonly into two groups (e.g., malignant versus benign findings). Discriminative models are subject to most of the challenges we have already listed, most notably that their development needs labeled data. Generative models, in contrast, focus on learning to (re) produce data without making any decisions.[47,48] Naïve Bayes, restricted Boltzmann machines, and generative adversarial networks are examples of generative methods. If an algorithm can generate image data, it must have understood the image to be able to generate it. In general, generative algorithms learn joint probability (the statistics that characterize the image features) and guess the label (say what is in the image), whereas discriminative models directly estimate the class label. Deep generative models have been used, among others, for interstitial lung disease classification and for functional magnetic resonance imaging analysis.[48]
Opportunity #4: Unsupervised learning: When we do not need labels
Prior success stories for deep solutions have led to overuse of supervised algorithms. These are algorithms that need labeled data, images in which regions of interest are manually delineated by human experts. Unsupervised learning, however, has been a pillar of AI for decades that has been almost shoved into oblivion in recent years perhaps because of the impressive success of supervised AI methods.[49,50] We need to rediscover the potential of unsupervised algorithms, such as self-organizing maps[51] and hierarchical clustering,[52] and adequately integrate them into the workflow of routine pathology practice. Since labeling images is not part of the daily routine of pathologists, extracting features without supervision (i.e., labeled data) may be very valuable.[53]
Opportunity #5: Virtual peer review – Placing the pathologist in the center
Putting all of the challenges and opportunities together, it is obvious that the pathologist should be central to both algorithm development and execution; we need pathologists for the former to validate algorithm performance, while the latter will serve the pathologist with some extracted knowledge. Instead of making decisions on behalf of pathologists, smart algorithms could rather provide reliable information extracted from proven diagnosed cases in an archive when they are (anatomically/pathologically) similar to the relevant characteristics of the patient being examined. The task of finding similar cases (already diagnosed and treated by other colleagues) can be performed using the laboratory's archive, a regional archive, or even a national or global repository of vetted diagnosed cases. For instance, if a patient has a biopsy, then the diagnosis can be compared to a prior specimen for quality assurance purposes (e.g., comparing a cervix biopsy histologic diagnosis to a recent Pap test interpretation for real-time cytologic-histopathologic correlation). With AI, it may be more palatable to let ultimate decision-making reside with pathologists and in so doing provide them with as much extrinsic meaningful knowledge as possible to assist in this process. With respect to the problem of interobserver variability,[54,55] accessing image data to facilitate consensus would be beneficial.[56,57]
Opportunity #6: Automation
AI software tools, if exploited and implemented well, have the possibility of handling laborious and mundane tasks (e.g., counting mitoses and screening for easily identifiable cancer types) and simplifying complex tasks (e.g., triaging biopsies that need urgent attention and ordering appropriate stains upfront when indicated). For instance, it has been recently demonstrated for breast cancer that image retrieval for “malignant regions” that “can be easily recognized by pathologists” can be performed by AI methods with a sensitivity above 92%.[58] This can certainly contribute to reducing the workload of pathologists and assist with case triage.
Opportunity #7: Re-birth of the hematoxylin and eosin image
In recent years, we have witnessed an increase in molecular testing, sometimes in lieu of tissue morphologic evaluation. However, by grinding up tissue for such analyses, we risk losing valuable insight into histopathology (e.g., host stromal reaction to cancer[59]) and spatial relations (e.g., tumor microenvironment, immune response to neoplasia, and rejection in transplantation). With the advent of computational pathology,[60,61] especially when combined with emerging technologies (e.g., multiplexing and three-dimensional imaging), we have the ability to more deeply analyze individual pixels of pathology images to unlock diagnostic, theranostic, and potentially untapped prognostic information. Moreover, most AI approaches to mitosis detection, segmentation, nucleus classification, and predicting Gleason scores have been using H and E-stained images.[62]
Opportunity #8: Making data science accessible to pathologists
AI has the potential to favorably modify the pathologist's role in medicine. Despite the perceived threat of AI, it is plausible that AI tools that generate and/or analyze big image data will be a boon to pathologists by increasing their value, efficiency, accuracy, and personal satisfaction.[63]
SUMMARY
The accelerated adoption of digital pathology in clinical practice has ushered in new horizons for both computer vision and AI.[64,65] Because of recent success stories in image recognition for nonmedical applications, many researchers and entrepreneurs are convinced that AI in general and deep learning in particular may be able to assist with many tasks in digital pathology. However, there are no commercial AI-driven software tools available just yet. Hence, pathologist buy-in from the outset (i.e., even when developing algorithms) is critical to make sure that these eagerly anticipated software packages fill germane gaps without disrupting clinical workflow. What parts of the clinical workflow and which human tasks can be improved or may be even replaced by AI algorithms remains to be seen. The adoption of AI in pathology is certainly not going to be as straightforward as the current enthusiasm appears to suggest. Regulatory approval of AI tools is likely to significantly promote their adoption in clinical practice. The US Food and Drug Administration (FDA) has already approved AI apps in other fields such as ophthalmology and radiology.[66,67] Such FDA approval provides reassurance to both clinicians and patients that an AI app is trustworthy for clinical use. For clinicians, this implies less personal liability when using this tool, greater chance of receiving reimbursement, and in pathology less of a burden on the laboratory for self-validation. However, for developers of deep learning algorithms destined to be submitted for regulatory clearance, greater documentation of their model and technical decisions is required. Furthermore, commercialization implications such as scaling and deployment of their tool will need to be taken into consideration. These factors, in turn, can drive up the cost of algorithm development. Unfortunately, the AI community has experienced several major setbacks in the past when promised performances could not be delivered leading to very pessimistic views on AI.[68,69] The danger of overselling AI is still omnipresent. Nonetheless, there is clear potential for breakthrough with AI in medical imaging, particularly in digital pathology, if we suitably manage the strengths and pitfalls.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2018/9/1/38/245402
REFERENCES
- 1.Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantanowitz L, Parwani AV. pathology. ASCP Press; 2017. p. 304. ISBN: 978-08189-6104. [Google Scholar]
- 3.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–93.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma G, Carter A. Artificial intelligence and the pathologist: Future frenemies? Arch Pathol Lab Med. 2017;141:622–3. doi: 10.5858/arpa.2016-0593-ED. [DOI] [PubMed] [Google Scholar]
- 5.Holzinger A, Malle B, Kieseberg P, Roth PM, Müller H, Reihs R, et al. Towards the augmented pathologist: Challenges of explainable-ai in digital pathology. arXiv Preprint arXiv: 1712.06657. 2017 [Google Scholar]
- 6.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong ST. Is pathology prepared for the adoption of artificial intelligence? Cancer Cytopathol. 2018;126:373–5. doi: 10.1002/cncy.21994. [DOI] [PubMed] [Google Scholar]
- 8.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A. 2018;115:E2970–E2979. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson S, Azizpour H, Smith K, Hartman J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl Res. 2018;194:19–35. doi: 10.1016/j.trsl.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, et al. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18:281. doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camelyon. 2016. [Last accessed on 2018 Apr 17]. Available from: https://www.camelyon16.grand-challenge.org/
- 14.Babaie M, Kalra S, Sriram A, Mitcheltree C, Zhu S, Khatami A, Rahnamayan S, Tizhoosh HR. Classification and retrieval of digital pathology scans: A new dataset. CVMI Workshop@ CVPR. 2017 [Google Scholar]
- 15.Ahmad J, Muhammad K, Baik SW. Data augmentation-assisted deep learning of hand-drawn partially colored sketches for visual search. PLoS One. 2017;12:e0183838. doi: 10.1371/journal.pone.0183838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, Yener B. Histopathological image analysis: A review. IEEE Rev Biomed Eng. 2009;2:147. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 18.Pena GP, Andrade-Filho JS. How does a pathologist make a diagnosis? Arch Pathol Lab Med. 2009;133:124–32. doi: 10.5858/133.1.124. [DOI] [PubMed] [Google Scholar]
- 19.Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313:1122–32. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH, et al. Patch-based convolutional neural network for whole slide tissue image classification. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016. 2016:2424–33. doi: 10.1109/CVPR.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turing A. Computing Machinery and intelligence. Mind. 1950;59:433–60. [Google Scholar]
- 22.Warwick K, Shah H. Passing the turing test does not mean the end of humanity. Cognit Comput. 2016;8:409–19. doi: 10.1007/s12559-015-9372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. Malaysia: Pearson Education Limited. 2016 [Google Scholar]
- 24.Goertzel B. Vol. 2. New York: Springer; 2007. Artificial general intelligence. In: Pennachin C, editor. Artificial General Intelligence. [Google Scholar]
- 25.Everitt T, Goertzel B, Potapov A. Heidelberg: Springer; 2017. Artificial general intelligence. Lecture Notes in Artificial Intelligence. [Google Scholar]
- 26.Chen XW, Lin X. Big data deep learning: Challenges and perspectives. IEEE Access. 2014;2:514–25. [Google Scholar]
- 27.Coates A, Huval B, Wang T, Wu D, Catanzaro B, Andrew N. Deep learning with COTS HPC systems. Int Conf Mach Learn 2013. :1337–45. [Google Scholar]
- 28.Andrade G, Ferreira R, Teodoro G, Rocha L, Saltz JH, Kurc T. Efficient execution of microscopy image analysis on CPU, GPU, and MIC equipped cluster systems. Computer Architecture and High Performance Computing (SBAC-PAD). 2014 IEEE 26th International Symposium on. IEEE. NIH Public Access. 2014:89. doi: 10.1109/SBAC-PAD.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos V, Sastre F, Yagües M, Bellver M, Giró-i-Nieto X, Torres J. Distributed training strategies for a computer vision deep learning algorithm on a distributed GPU cluster. Procedia Comput Sci. 2017;108:315–24. [Google Scholar]
- 30.Athalye A, Sutskever I. Synthesizing robust adversarial examples. arXiv preprint arXiv: 1707. 2017 [Google Scholar]
- 31.Rastogi V, Puri N, Arora S, Kaur G, Yadav L, Sharma R, et al. Artefacts: A diagnostic dilemma – A review. J Clin Diagn Res. 2013;7:2408–13. doi: 10.7860/JCDR/2013/6170.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee S. Artefacts in histopathology. J Oral Maxillofac Pathol. 2014;18:S111–6. doi: 10.4103/0973-029X.141346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight W. The dark secret at the heart of AI. [Last accessed on 2018 Sep 22];Technol Rev. 2017 120:54–63. Available from: http://www.technologyreview.com . [Google Scholar]
- 34.Pande V. Artificial intelligence's ‘black box’ is nothing to fear. The New York Times. 2018 [Google Scholar]
- 35.Montavon G, Samek W, Müller KR. Methods for interpreting and understanding deep neural networks. Digit Signal Proc. 2018;73:1–15. [Google Scholar]
- 36.Freedman DH. A reality check for IBM's AI ambitions. Technol Rev. 2017. [Last accessed on 2018 Sep 22]. Available from: http://www.technologyreview.com .
- 37.Bengio Y. Deep learning of representations for unsupervised and transfer learning. Proceedings of ICML Workshop on Unsupervised and Transfer Learning. Edinburgh, Scotlan. 2012;27:17–36. [Google Scholar]
- 38.Bar Y, Diamant I, Wolf L, Greenspan H. Deep learning with non-medical training used for chest pathology identification. Medical Imaging 2015: Computer-Aided Diagnosis. International Society for Optics and Photonics. 2015;9414:94140V. [Google Scholar]
- 39.Kieffer B, Babaie M, Kalra S, Tizhoosh HR. Convolutional neural networks for histopathology image classification: Training vs. using pre-trained networks. arXiv preprint arXiv: 1710.05726. 2017 [Google Scholar]
- 40.Chen H, Cui S, Li S. Application of transfer learning approaches in multimodal wearable human activity recognition. arXiv Preprint arXiv: 1707.02412. 2017 [Google Scholar]
- 41.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell. 2002;24:971–87. [Google Scholar]
- 42.Tizhoosh H, Babaie M. Representing medical images with encoded local projections. IEEE Trans Biomed Eng. 2018;65:2267–77. doi: 10.1109/TBME.2018.2791567. [DOI] [PubMed] [Google Scholar]
- 43.Alhindi TJ, Kalra S, Ng KH, Afrin A, Tizhoosh HR. Comparing LBP, HOG and deep features for classification of histopathology images. arXiv preprint arXiv: 1805.05837. 2018 [Google Scholar]
- 44.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016;33:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, et al. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging (Bellingham) 2014;1:034003. doi: 10.1117/1.JMI.1.3.034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susskind J, Mnih V, Hinton G. On deep generative models with applications to recognition. Computer Vision and Pattern Recognition (CVPR), 2011 IEEE Conference. IEEE. 2011 [Google Scholar]
- 48.Salakhutdinov R. Learning deep generative models. Ann Rev Stat Appl. 2015;2:361–85. [Google Scholar]
- 49.Fergus R, Perona P, Zisserman A. Proceedings. 2003 IEEE Computer Society Conference. Vol. 2. Madison WI, USA: IEEE; 2003. Object class recognition by unsupervised scale-invariant learning. Computer Vision and Pattern Recognition, 2003. [Google Scholar]
- 50.Lee H, Grosse R, Ranganath R, Ng AY. Montreal, Quebec, Canada: ACM; 2009. Convolutional deep belief networks for scalable unsupervised learning of hierarchical representations. Proceedings of the 26th Annual International Conference on Machine Learning; pp. 609–16. [Google Scholar]
- 51.Kohonen T. The self-organizing map. Proc IEEE. 1990;78:1464–80. [Google Scholar]
- 52.Murtagh F, Contreras P. Algorithms for hierarchical clustering: An overview. Wiley Interdiscip Rev. 2012;2:86–97. [Google Scholar]
- 53.Arevalo J, Cruz-Roa A, Arias V, Romero E, González FA. An unsupervised feature learning framework for basal cell carcinoma image analysis. Artif Intell Med. 2015;64:131–45. doi: 10.1016/j.artmed.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 54.Lawton TJ, Acs G, Argani P, Farshid G, Gilcrease M, Goldstein N, et al. Interobserver variability by pathologists in the distinction between cellular fibroadenomas and phyllodes tumors. Int J Surg Pathol. 2014;22:695–8. doi: 10.1177/1066896914548763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson SR, Rao P, Hes O, Epstein JI, Smith SC, Picken MM, et al. Challenges in pathologic staging of renal cell carcinoma: A study of interobserver variability among urologic pathologists. Am J Surg Pathol. 2018;42:1253–61. doi: 10.1097/PAS.0000000000001087. [DOI] [PubMed] [Google Scholar]
- 56.Mazzanti M, Shirka E, Gjergo H, Hasimi E. Imaging, health record, and artificial intelligence: Hype or hope? Curr Cardiol Rep. 2018;20:48. doi: 10.1007/s11886-018-0990-y. [DOI] [PubMed] [Google Scholar]
- 57.Tizhoosh HR, Czarnota GJ. Fast barcode retrieval for consensus contouring. arXiv preprint arXiv: 1709.10197. 2017 [Google Scholar]
- 58.Zheng Y, Jiang Z, Zhang H, Xie F, Ma Y, Shi H, et al. Histopathological whole slide image analysis using context-based CBIR. IEEE Trans Med Imaging. 2018;37:1641–52. doi: 10.1109/TMI.2018.2796130. [DOI] [PubMed] [Google Scholar]
- 59.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3:108–13. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 60.van Laak J, Rajpoot N, Vossen D. The Promise of Computational pathology: Part 1. The pathologist. 2018a. [Last accessed on 2018 Sep 22]. Available from: https://thepathologist.com .
- 61.van Laak J, Rajpoot N, Vossen D. The promise of computational pathology: Part 2. The pathologist. 2018b. [Last accessed on 2018 Sep 22]. Available from: https://thepathologist.com .
- 62.Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Jha S, Topol EJ. Adapting to artificial intelligence: Radiologists and pathologists as information specialists. JAMA. 2016;316:2353–4. doi: 10.1001/jama.2016.17438. [DOI] [PubMed] [Google Scholar]
- 64.Fornell D. How Artificial Intelligence Will Change Medical Imaging. Imaging Technology News. 2017. [Last accessed on 2018 Sep 22]. Available from: https://www.itnonline.com .
- 65.Granter SR, Beck AH, Papke DJ., Jr AlphaGo, deep learning, and the future of the human microscopist. Arch Pathol Lab Med. 2017;141:619–21. doi: 10.5858/arpa.2016-0471-ED. [DOI] [PubMed] [Google Scholar]
- 66.FDA News Release. FDA Permits Marketing of Artificial Intelligence-Based Device to detect Certain Diabetes-Related Eye Problems. FDA. 2018a. [Last accessed on 2018 Apr 11]. Available from: http://www.fda.gov .
- 67.FDA News Release. FDA Permits Marketing of Artificial Intelligence Algorithm for Aiding Providers in Detecting Wrist Fractures. FDFA. 2018b. [Last accessed on 2018 May 24]. Available from: http://www.fda.gov .
- 68.Dreyfus H. Cambridge, MA, USA: MIT Press; 1972. What Computer Still Can't Do: A Critique of Artificial Reason. [Google Scholar]
- 69.Dreyfus H, Dreyfus SE, Athanasiou T. Cambridge, MA, USA: MIT Press; 1972. What Computer Still Can't Do: A Critique of Artificial Reason. [Google Scholar]


