Abstract
We evaluated DNA binding of the B-HLH family members TCF4 and USF1 using protein binding microarrays (PBMs) containing double-stranded DNA probes with cytosine on both strands or 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC) on one DNA strand and cytosine on the second strand. TCF4 preferentially bound the E-Box motif (CAN∣NTG) with strongest binding to the 8-mer CAG∣GTGGT. 5mC uniformly decreases DNA binding of both TCF4 and USF1. The bulkier 5hmC also inhibited USF1 binding to DNA. In contrast, 5hmC dramatically enhanced TCF4 binding to E-Box motifs ACAT∣GTG and ACAC∣GTG, being better bound than any 8-mer containing cytosine. Examination of x-ray structures of the closely related TCF3 and USF1 bound to DNA suggests TCF3 can undergo a conformational shift to preferentially bind to 5hmC while the USF1 basic region is bulkier and rigid precluding a conformation shift to bind 5hmC. These results greatly expand the regulatory DNA sequence landscape bound by TCF4.
Cytosine methylation outside CG dinucleotide has recently been identified in stem cells and the brain. To explore potential changes in sequence-specific DNA binding of transcription factors, we use Agilent DNA microarrays and did the double stranding reaction with 5mC or 5hmC. Using this technical innovation we explored DNA binding specificity of two helix-loop-helix proteins. For USF1, these modifications inhibited binding, while for TCF4, new sequences were bound. This innovation of DNA microarray slides opens up new possibilities to explore how modification of DNA alters transcription factor binding to mediate changes of biological importance.
Introduction
Mammalian genomes have a bipartite structure, with 98% of the genome being depleted in CG dinucleotides and the cytosine on both DNA strands is methylated. The remaining 2% of the genome is CG dinucleotide rich regions, known as CG islands (CGIs) that are typically unmethylated and often have housekeeping gene regulatory functions 1. Two recent observations in mammals have expanded the possibilities for sequence-specific DNA binding of transcription factors (TFs). First, the TET family of dioxygenases can iteratively oxidize 5mC to 5hmC, then 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC) 2, resulting in five forms of cytosine in the genome whose abundance varies dramatically between cell types suggesting potential biological function 2–4. Several studies have recently reported that 5hmC is involved in gene activation in differentiating cells 5, 6. The presence of 5hmC at the boundaries of hypomethylated regions suggests the dynamic nature of these sharp boundaries 7.
The second observation is that 5mC can occur outside of CG dinucleotides, particularly in stem cells 8 and brain 9, 10, expanding the number of potentially modified cytosines from 42 million cytosines (two cytosines for the 21 million CG dinucleotides contained in the mouse genome) to 1.6 billion cytosines in the mouse genome. Methylation of cytosine not in CG dinucleotides, particularly in CA dinucleotides 9, 11, is the dominant form of methylation in mouse and human neurons, where it accounts for 53% of methylated cytosines 9. Recent studies show that 5hmC is abundant in the mammalian brain 9, 11, 12, and its levels are dynamically regulated during brain development, increasing from 0.2% in the fetal cortex to 0.87% in the adult cortex 9, indicating a likely biological role for 5hmC in normal neuronal development 11, 13. The effect of 5hmC, 5fC, and 5caC modifications within a CG dinucleotide on DNA binding of TFs has been investigated 14–17 for a few TFs binding a limited number of DNA sequences. However, little is known about how these cytosine modifications not is CG dinucleotides affect DNA binding of TFs.
There are over 60 members of the B-HLH family of transcription factors that dimerize as homodimers and heterodimers and bind to E-Box motifs (CAN∣NTG) 18, 19. Since each monomer of the B-HLH dimer binds a CAN∣ half-site, we place a vertical line at the center of the dyad for clarity. Many B-HLH proteins bind strongly to the CG dinucleotide containing palindromic E-box CAC∣GTG that is enriched in CGIs. Many of these proteins are involved in housekeeping functions, including ARNTL, BHLHE40, HEY2, MLXIP, MAX, and USF1. In contrast, some B-HLHs are involved in specifying cell identity and differentiation, like ASCL1, ATOH1, NEUROD1, NEUROG, TCF4 (E2A), TCF3 (E2–2, ITF2), and TCF12 (HEB) and bind to non-CG dinucleotide containing E-Box motifs like CAG∣GTG or CAG∣CTG that localize outside of CGIs 20. TCF4 binds E-Box motifs both as a homodimer and a heterodimer with various tissue specific B-HLH proteins to form transcriptional networks that regulate cellular differentiation of many cell types 21. Aberrant expression and/or mutations in TCF4 can cause abnormal brain development leading to neurodevelopmental disorders such as Pitt-Hopkins syndrome, schizophrenia, Fuchs’ corneal endothelial dystrophy, and primary sclerosing cholangitis 21, 22.
Since the E-Box motif contains a cytosine, we hypothesized that cytosine modifications may modulate B-HLH TF binding. To test this, we examined if 5mC and 5hmC changed the DNA binding of TCF4 and USF1, two B-HLH TFs. Using the Agilent HK Protein Binding Micoroarray (PBM) design 23, we performed DNA double-stranding reactions with cytosine, 5mC, or 5hmC, and examined the effect on DNA-binding of TCF4 and USF1. 5mC and 5hmC inhibits USF1 DNA-binding. 5mC inhibits TCF4 DNA binding. 5hmC in contrast enhances DNA-binding of TCF4 to E-box motifs containing a central CG dinucleotide (ACAC∣GTG). This suggesting that 5hmC can enhance TCF4 binding to E-Box motifs in CGIs to inhibit cell growth and facilitate cell differentiation.
Materials and methods
Cloning and expression of mouse TCF4 and USF1
Constructs containing the DNA binding domains (DBDs) and 50 flanking amino acids of mouse TCF4 and USF1 were obtained from Dr. Timothy Hughes, University of Toronto, Canada, as GST constructs cloned into the pET-GEXCT (C-terminal GST) vector 24. TCF4 and USF1 proteins were expressed using a PURExpress in vitro protein synthesis kit (NEB) as suggested by the manufacturer’s protocol. For each 25 µL of IVT reaction, 180 ng of plasmid containing TCF4 or USF1 tagged to GST was used for expression. Amino acid sequences for the B-HLH domains of TCF3, TCF4 and USF1 are shown with numbering for TCF3 and USF1 presented. The invariant glutamate is underlined.

Design of the 40,000 (40K) feature PBMs.
The 40K array design consists of a single-stranded 60-mer containing a variable probe sequence that is 35-bp long and a common 25-bp sequence near the glass surface which is complimentary to the primer sequence used in DNA double-stranding. The design of this 35-mer is based on deBruijn sequences, and each non-palindromic 8-mers occurs on 32 different probes in diverse flanking sequence contexts 23,25.
Double-stranding of the microarray with either cytosine, 5mC or 5hmC.
In order to analyze the effect of 5mC and 5hmC on DNA, we modified the double-stranding procedure described before 26 using either 5-methylcytosine (5mC, NEB) or 5-hydroxymethylcytosine (5hmC, Zymo Research). The resulting double-stranded DNA on the array will contain either cytosine on both strands or 5mC/5hmC on one strand and cytosine on the second strand. This results in a hemi-methylated or hemi-hydroxymethylated state. DNA double-stranding was performed as previously described 1, 27.
Protein binding reaction, image quantification and analyses of Z-scores.
Protein binding reactions, image quantification, and calculation of Z-scores were performed as described previously 27. The Z-scores for 8-mers were calculated by two different approaches: for each 8-mer, either the reverse complementary 8-mers was count as the same (32,896 8-mers, e.g. CCCCCCCC and GGGGGGGG were both count as CCCCCCCC) or not (65,536 8-mers, e.g. CCCCCCCCC and GGGGGGGG were two different 8-mers). For 32,896 8-mers, the Z-score was calculated from the average signal intensity across the 16 or 32 spots containing each 8-mer. For 65,536 8-mers, the Z-score was also calculated from the average signal intensity across spots containing each 8-mer, but at least 10 spots containing the 8-mer. The Z-score of 8-mer with less than 10 spots on the array was arbitrarily set as 0 to avoid noise. Z-scores for 8-mers used to describe datasets are from modified strand. We have deposited 3 replicates of TCF4 binding cytosine and 5hmC and 2 replicates to 5mC. For USF1, there are 2 replicates for cytosine 5mC, and 5hmC. The data are at ftp://helix.nih.gov/pcf/chuck/Array/TCF4_and_USF/.
Structural analysis.
Coordinates for crystal structure of TCF3 homodimer bound to E-box DNA were obtained from Dr. Ellenberger 28. The USF1 bound to DNA x-ray crystal structure identifier is PDB:1AN4 29. Molecular models were developed with the CHARMM software package 30. Alternate structures were obtained by energy minimization and molecular dynamics, allowing only the sidechains of the binding glutamic acid and arginine residues and the DNA modification groups to move. Figures were generated with the UCSF Chimera package 31.
Results
DNA double-stranding of the microarrays.
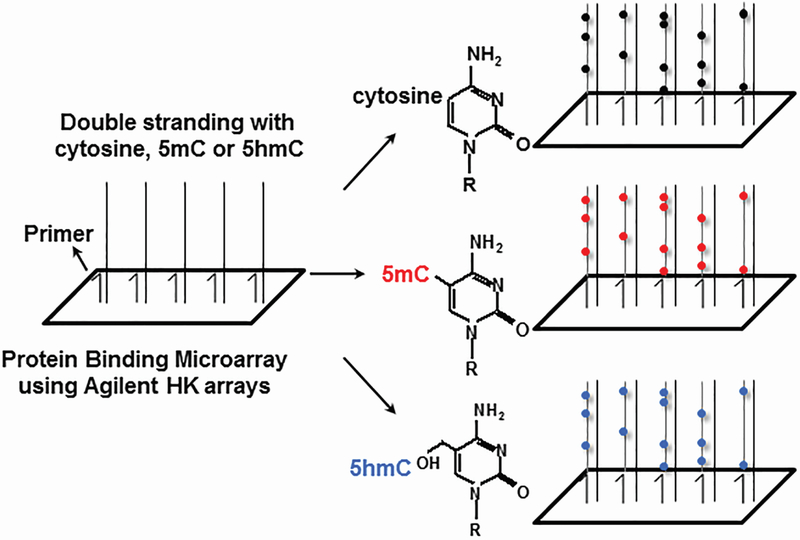
DNA polymerases can incorporate 5mC and 5hmC into DNA when double-stranding single-stranded DNA 32. We exploited this property to double-strand the single-stranded DNA on an Agilent microarray using 5mC or 5hmC (Figure 1). This generates double-stranded arrays with 5mC or 5hmC on one DNA strand, mimicking what occurs biologically in several cell types, including brain 9, 13.
Figure 1. Schematic of modified PBM double-stranding procedure.
Single stranded HK arrays were double-stranded with either cytosine (black spot), 5mC (red spot), or 5hmC (blue spot) using a common primer.
We monitored the DNA double-stranding reactions with Cy3-dCTP (4%) and plotted the fluorescence intensities of the array spots after scanning at 570nm using an Agilent SureScan scanner (Figure 2). Overall, the fluorescence intensities from double-stranding with cytosine (Figure 2A) or 5hmC (Figure 2C) are similar and twice as high with 5mC (Figure 2B). We divided the fluorescence intensities of each feature by the number of cytosines in the 35-mer variable sequence that incorporates Cy3-dCTP. This analysis indicates that probes with more cytosines have more Cy3-dCTP signal (red lines in Figures 2 A, B and C). These results suggest that the DNA double-stranding reactions were successful when we used either cytosine, or 5mC, or 5hmC. Supplemental Figure S1 shows the occurrence of all 8-mers on the Watson and Crick strand. The vast majority of 8-mers (64,396) occur more than 10 times on each strand in the 40K array. 8-mers that occur less than 10 times (1,140 8-mers) on either the Watson or Crick strand were excluded from further analysis.
Figure 2. Double-stranding efficiency of the PBMs with cytosine, 5mC, and 5hmC.
Fluorescence intensities, from lowest to highest values, of the spiked Cy3-dCTP across 40k features of the HK array (blue), divided by the number of cytosines in the 35-mer variable sequence (red) for double-stranding with (A) 5mC and (B) 5hmC.
TCF4 binding to double-stranded DNA arrays containing cytosine, or 5mC, or 5hmC.
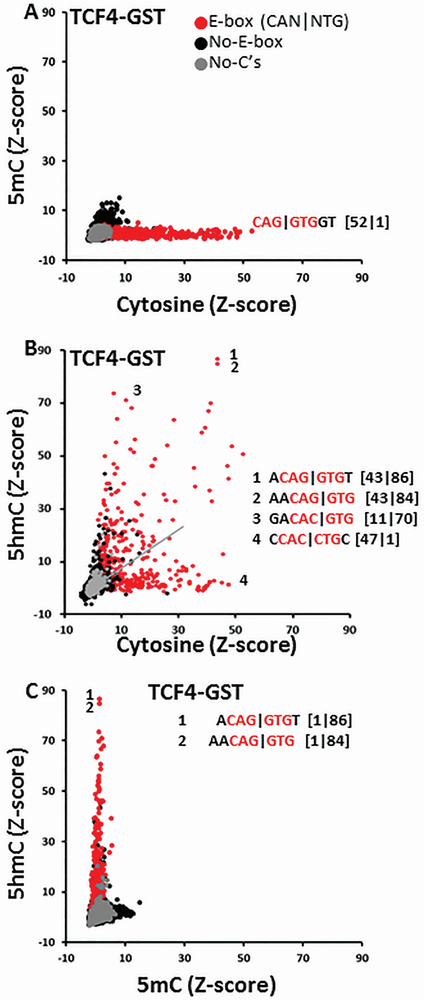
B-HLH dimers recognize the E-box motif (CAN∣NTG) 18, 19, 33. Figure 3A presents 8-mer Z-scores for the TCF4 homodimer binding to double-stranded DNA containing cytosine on both DNA strands (x-axis) compared to 5mC on one DNA strand and cytosine on the second DNA strand (y-axis). With cytosine, all of the well-bound 8-mers contain E-Boxes, with the non-CG dinucleotide 8-mer CAG∣GTGGT being the best bound with a Z-score of 52 (Figure 3A). The presence of 5mC on one DNA strand inhibited TCF4 binding with no 8-mers being well-bound. These data are reproducible (Supplemental Figure S2).
Figure 3. TCF4-GST B-HLH domain binding to DNA 8-mers containing cytosine, 5mC or 5hmC on one strand.
DNA 8-mers containing E-boxes are labeled as red spots, 8-mers with a cytosine are black, and 8-mers without a cytosine are grey. A. TCF4-GST binding to 8-mers containing cytosine (X-axis) or 5mC on one DNA strand (Y-axis). The Z-score values for cytosine and 5hmC are written in [x-axis : y-axis] format. B. TCF4-GST binding to 8-mers containing cytosine (X-axis) or 5hmC (Y-axis). C. TCF4-GST binding to 8-mers containing 5mC (X-axis) or 5hmC (Y-axis). 8-mers shown are from modified strand.
Figure 3B compares TCF4 binding to DNA with cytosine or 5hmC. 8-mers with no cytosines can be used to normalize Z-scores. These 8-mers are poorly bound. The slope of the line through 8-mers with no cytosines was 0.73. Some E-Boxes like GACAC∣GTG are only well bound to 8-mers that contain 5hmC while others like CCAC∣CTGC are only well-bound with cytosine. Other E-Boxes are well bound to DNA containing either cytosine or 5hmC (ACAG∣GTGT). Figure 3C compares the binding preferences of TCF4 to DNA containing either 5mC or 5hmC on one-strand.
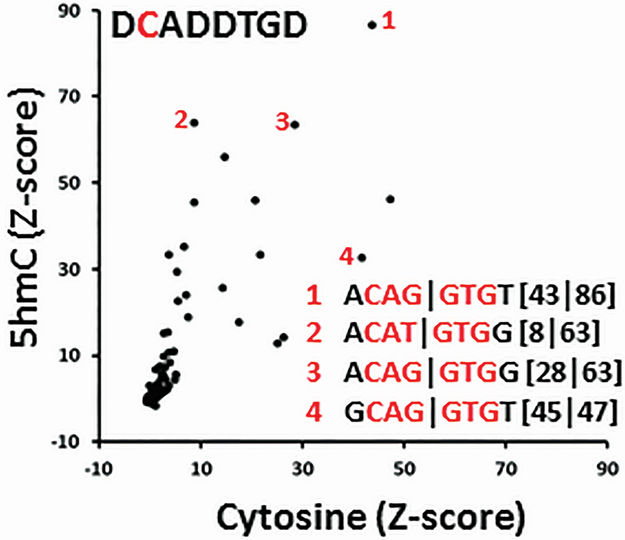
We next examined E-Box 8-mers containing only one cytosine, the cytosine in the E-Box CAN∣NTG. 5hmC has little effect on DNA binding for some 8-mers (GCAG∣GTGT), but drastically increases DNA binding for others (ACAT∣GTGG) (Figure 4). When we examined 8-mers with one cytosine that is not the canonical cytosine in the E-box motif, DNA binding is very poor and thus the contribution of the 5mC and 5hmC at different positions in the TFBS could not be evaluated.
Figure 4. Effect of 5hmC on TCF4-GST binding to E-Box 8-mers containing only one cytosine.
E-boxes containing DCAD∣DTGD were well bound by both cytosine and 5hmC. Four E-box motifs are labeled to highlight the differences in binding with cytosine and 5hmC. D is the IUPAC DNA code for A, T, or G.
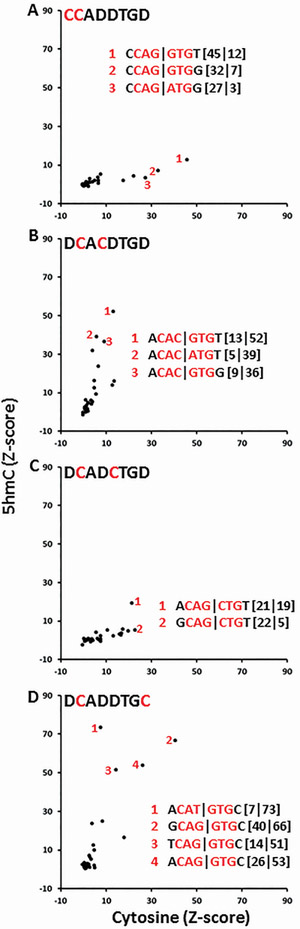
An alternative method to evaluate the contribution of cytosine, 5mC, and 5hmC at different positions in the E-Box is to examine E-Boxes with two cytosines, the cytosine in the canonical E-Box (CAN∣NTG) and a second cytosine elsewhere in the motif. CCAD∣DTGD (Figure 5A) and DCAD∣CTGD (Figure 5C) (where D is A, T, or G) are well bound only with cytosine, suggesting these two positions in the E-Box motif are better bound when they contain cytosine compared to 5hmC. 5hmC enhanced binding to DCAC∣DTGD with Z-scores increasing from 13 to 52 (Figure 5B). These results are more dramatic than for E-Boxes containing one cytosine DCAD∣DTGD suggesting that 5hmC contributes more than cytosine at both positions in the TFBS to TCF4 binding. DCAD∣DTGC is the most variable, with some E-Boxes increasing binding; for example the Z-score for ACAT∣GTGC (with 5hmC) increases from 7 to 73 while others are well bound with either cytosine or 5hmC (Figure 5D).
Figure 5. Effect of two cytosines within E-box 8-mers on TCF4-GST binding to DNA with cytosine or 5hmC.
A. CCAD∣DTGD is only bound with cytosine. B. DCAC∣DTGD is only bound with 5hmC. C. DCAD∣CTGD is only bound by cytosine. D. DCAD∣DTGC is the most variable, with 5hmC generally favoring DNA binding.
TCF4 binding to the four trimer half sites: effect of cytosine or 5hmC.
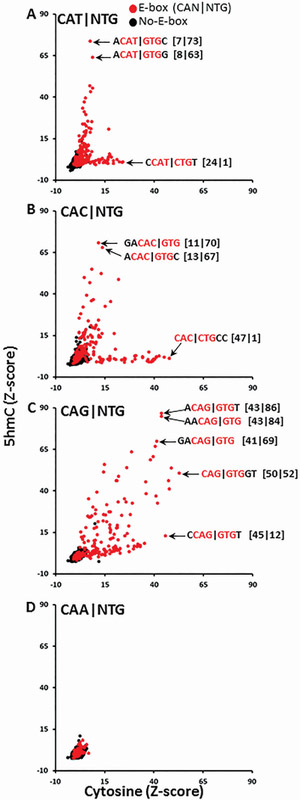
An alternative analysis is to examine how 5hmC affects TCF4 binding to the four possible E-Box half sites: CAT∣NTG, CAC∣NTG, CAG∣NTG, and CAA∣NTG (Figure 6). For CAT∣NTG and CAC∣NTG, some 8-mers are well bound with either cytosine or 5hmC, but not both. Closer examination indicates that 5hmC enhances the binding of TCF4 when the 4th position is guanine (G) (e.g., CAC∣GTG or CAT∣GTG), but inhibits binding if the 4th position is 5hmC (CAT∣CTG and CAC∣CTG) (Figure 6A, 6B). The increase in binding with 5hmC and guanine in the 4th position indicates that several E-box motifs with a central CG dinucleotide that are well-bound only with 5hmC (Supplemental Figure S3). CAG∣NTG has more variability. Some 8-mers are bound only with cytosines; others only with 5hmC, and some are well bound with either cytosine or 5hmC (Figure 6C). CAA∣NTG is not well bound with either cytosine or 5hmC (Figure 6D), in agreement with previous results 19.
Figure 6. Effect of CAT, CAC, CAG or CAA in the E-box motif (CAN∣NTG) on binding of TCF4-GST to 5hmC and Cytosine.
A. E-box half site CAT∣NTG motifs binding DNA containing cytosine or 5hmC. ACAT∣GTG is only well bound with 5hmC, while CAT∣CTG is only bound with cytosine. B. E-box half site CAC∣NTG motifs binding DNA containing cytosine or 5hmC. Similar to CAT∣NTG, E-box motifs containing ACAC∣GTG preferentially bind DNA containing 5hmC, while CAC∣CTG preferentially binds DNA containing cytosine. C. E-box half site CAG∣NTG motifs binding DNA containing cytosine or 5hmC produces a more complex pattern. D. E-box motifs containing CAA∣NTG with cytosine or 5hmC are not well bound by TCF4-GST. DNA 8-mers containing E-box are red spots, 8-mers with a cytosine are black.
Complementary 8-mers and 5hmC.
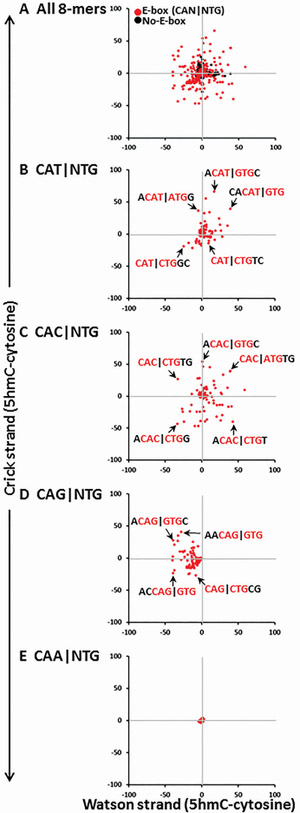
An intriguing trait of the asymmetric modification of cytosines is that modifications of complementary 8-mers may have different effects on binding of a TF. We next examined how 5hmC affects TCF4 binding to complementary 8-mers. Figure 7A shows the difference in binding (i.e. difference in Z-score) between 5hmC and cytosine for the Watson strand and Crick strand. There is a lot of variability with 8-mers in all four quadrants. We next plotted the four E-Box half motifs. CAT∣NTG E-Boxes tend to be better bound with 5hmC on either strand Figure 7B. CAC∣NTG shows the most variability with 8-mers in all four quadrants (Figure 7C). CAG∣NTG tends be poorly bound with 5hmC, but the complement is good (Figure 7D). CAA∣NTG is poorly bound in both cases (Figure 7E).
Figure 7. TCF4-GST binding to complementary DNA 8-mers containing cytosine or 5hmC on one strand.
A) The difference in Z-scores (5hmC-cytosine) for 8-mers from Watson-strand plotted against the difference in Z-scores for the complimentary Crick strand. 8-mers shown are from the Watson strand. Red spots contain the E-Box CAN∣NTG, black spots are 8-mers with a cytosine. B) E-Box CAT∣NTG. C) E-Box CAC∣NTG. D) E-Box CAG∣NTG. E) E-Box CAA∣NTG.
USF1 binding to double-stranded DNA arrays containing cytosine, 5mC, or 5hmC.
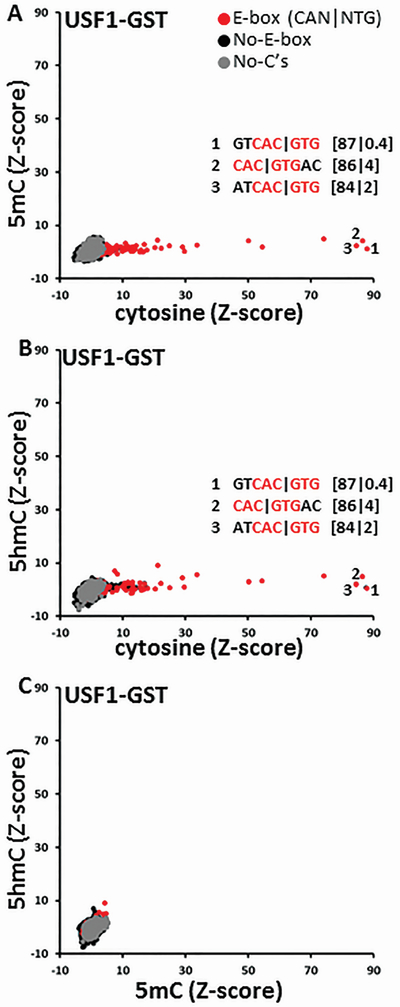
We next examined the DNA binding specificity of USF1, a B-HLH protein involved in housekeeping functions that preferentially binds the CG dinucleotide containing E-Box CAC∣GTG 20 to evaluate if 5hmC also increased binding. Figure 8A presents 8-mer Z-scores for the USF1 homodimer binding to double-stranded DNA containing cytosine on both strands or DNA with one strand containing 5mC. With cytosine, all of the well-bound 8-mers contain the E-Box with the GTCAC∣GTG 8-mer being the best bound with a Z-score of 87 (Figure 8A, x-axis). 5mC on one DNA strand inhibited binding of USF1, similar to TCF4. However, unlike TCF4, the presence of 5hmC inhibited USF1 binding (Figure 8B, y-axis, Figure 8C, Supplemental Figure S4 A-J).
Figure 8. USF1-GST B-HLH domain binding to DNA 8-mers containing cytosine, 5mC or 5hmC on one strand. A.
USF1-GST binding to 8-mers containing cytosine (X-axis) or 5mC (Y-axis). B. USF1-GST binding to 8-mers containing cytosine (X-axis) or 5hmC (Y-axis). C. USF1-GST binding to 8-mers containing 5mC (X-axis) or 5hmC (Y-axis). DNA 8-mers containing E-boxes are labeled as red spots, 8-mers with a cytosine are black, and 8-mers without a cytosine are grey.
Structural analysis of TCF3 homodimers binding to 5mC or 5hmC in the E-box.
To understand the structural effect of 5mC and 5hmC on the DNA binding of TCF4, we compared the amino acid sequence of the DNA binding region of TCF4 and USF1. There are many differences in the amino acids near the invariant glutamic acid that interacts with the CA dinucleotide in the E-Box, suggesting it may be possible to map amino acids that contribute to the differential interaction with 5hmC. Since the TCF4 crystal structure in not available, we instead examined the X-ray crystal structure of the closely related TCF3 homodimer bound to the E-Box motif CAC∣CTG (Ellenberger et al., 1994: coordinates obtained from the authors) 28 (see amino acid sequence alignment in Materials and methods).
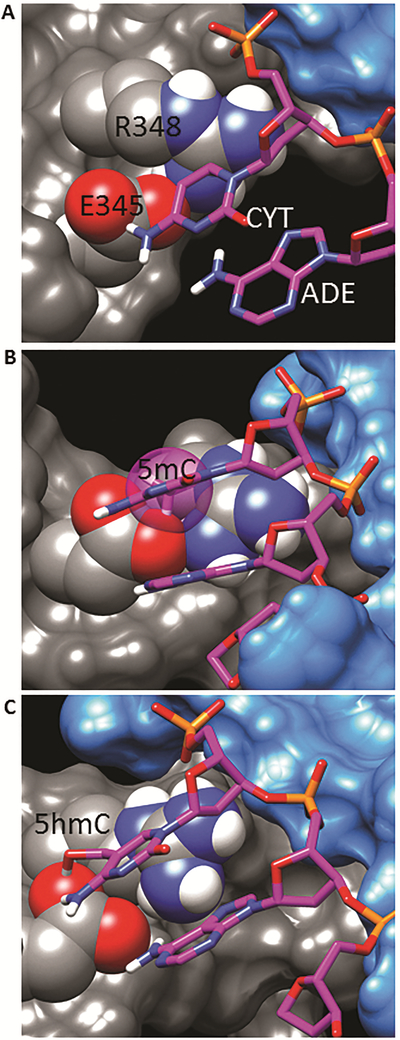
Figure 9A shows the invariant glutamic acid, E345 (TCF3 numbering) forming hydrogen bonds to the NH2 groups of both the cytosine and adenine in the CA dinucleotide of the E-box motif. This interaction captures the propensity for B-HLH proteins to bind the E-Box CAN∣NTG. The complex is further stabilized by salt-bridges between the conserved R348 side-chain with both E345 and the 5’-phosphate of the same cytosine.
Figure 9. Structural modeling of TCF3 with cytosine, 5mC and 5hmC.
Crystal structure of TCF3 (E47) homodimer bound to E-box DNA (Ellenberger et al., 1994). One protein monomer is represented as a grey surface, and the other monomer as a blue surface. Highlighted amino acid side-chains are shown as van der Waals spheres, and DNA is shown as sticks. Atom color code: protein carbon – grey, DNA carbon – magenta, oxygen – red, nitrogen – blue, phosphorous – yellow, hydrogen – white. A) Invariant glutamic acid interacting with the CA dinucleotide. Focus on the interface of the protein with E-box DNA bases. B) Steric clash of 5mC modification with E345 and R348. The added methyl carbon is shown as a transparent VDW sphere. C) Alternate structure with 5hmC modification.
Figure 9B is the same structure with an additional methyl group to produce 5mC. The carbon of this added methyl group is represented as a transparent sphere to illustrate the steric clash with both E345 and R348. The destabilization of the structure that this would cause is likely responsible for the observed decrease in binding affinity of TCF4 with 5mC containing E-box DNA. A larger, steric destabilization would occur for a 5hmC modification, since it has an additional hydroxyl group. Though 5hmC is larger than 5mC, it enhances TCF4 binding suggesting the protein and DNA form an alternate conformation.
To investigate what this alternative structure might be, molecular dynamic simulations were performed on the 5hmC-modified model. This was done very conservatively, allowing only the sidechains of E345 and R348, and the added 5hmC group to move. Consistent with the experimental data, many steric and energetically feasible structures were found containing small conformational changes of the two amino acid sidechains. As an example, one of the more stabilized structures is shown in Figure 9C. While a hydrogen bond is lost between the carboxylate group of E345 and the NH2 group of the cytosine, this is compensated for by a new hydrogen bond between E345 and 5hmC. This structure also maintains the stabilizing E345 hydrogen bond with the NH2 of the adenine base, and the R348 salt bridges with E345 and the 5’-phosphate group. Finally, it is concluded that the reason that a similar, stabilized complex does not form with the 5mC modification is that it lacks the added hydroxyl group of the 5hmC moiety to form compensating hydrogen bonds.
Structural analysis of USF1 homodimers binding to 5mC or 5hmC in the E-box.
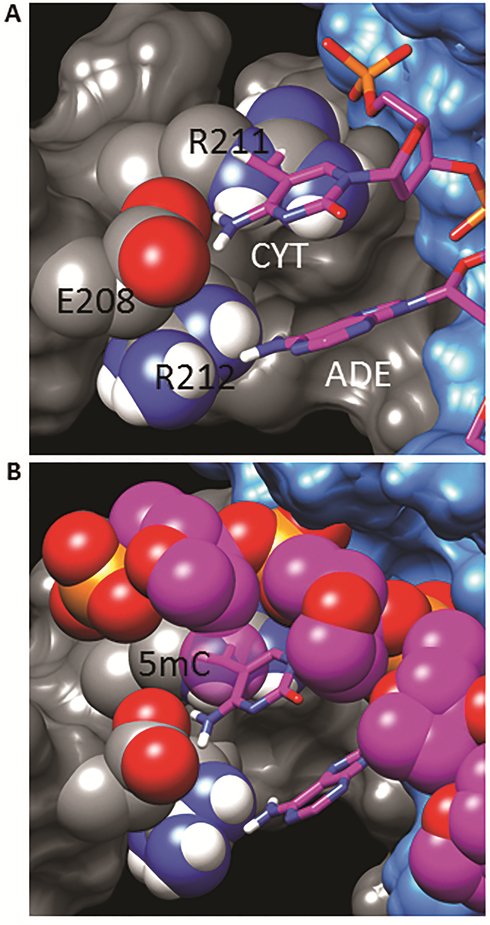
A similar structure analysis for the USF1 homodimer binding to the double-stranded E-box DNA motif CAC∣GTG 29 (PDB:1AN4) was performed. Figure 10A shows the interface of the protein and nucleotide bases with the conserved glutamic acid E208 (USF1 numbering) sidechain hydrogen bonding to the NH2 group of the first cytosine of the E-box motif, and the sidechain of R211 interacting with both E208 and the 5’-phosphate of the same cytosine. However, the conformations of the two complexes (TCF3 and USF1) differ such that an added 5mC group no longer overlaps with the glutamic acid, but forms a greater steric conflict with the arginine sidechain. As shown in Figure 10A, the added methyl group is half buried in the R211 guanidinium group. Molecular dynamics simulations were able to identify alternate conformations to relieve the steric conflict (not shown), the E208 and R211 sidechains were significantly more distorted than for the TCF3 complex model. An additional conflict is shown in Figure 10B, where the added 5mC methyl group also overlaps with the C2’ carbon of the deoxyribose group of the previous nucleotide on the 5’ side. Thus, not only does the modification sterically clash to a greater extent with USF1 than TCF3, but it also prohibits the conformation of the DNA preferentially bound by the protein. This observation is also true for the 5hmC modification, since both 5mC and 5hmC moieties have a bulky carbon at the same position. Another factor that would restrict the ability of USF1 to bind the modified DNA is that the position of the E208 sidechain is conformationally constrained by the bulky sidechain of R212 (Figure 10A). The analogous position in TCF3 and TCF4 is a smaller valine residue. The qualitatively greater steric conflicts in the USF1 structure are consistent with the experimental findings that this protein is unable to form a stable complex with either 5mC or 5hmC containing E-box motif.
Figure 10. Structural modeling of USF1 with 5mC.
Crystal structure of USF homodimer bound to E-box DNA (Ferre-D’Amare et al., 1994). Representations are similar to Figure 9. A) Shown is the interface of the protein with the E-box DNA bases, illustrating 5mC modification overlap with R211. B) Alternate view showing steric conflict of 5mC modification and deoxyribose of previous nucleotide. The 5mC methyl carbon and sugar-phosphate backbone are shown as VDW spheres.
This structural analysis only focused on the most obvious, steric and hydrogen bonding effects of cytosine modification on protein binding at the primary site of interaction of the protein with the E-box bases (i.e., the CA dinucleotide). More complex methods are required to explain the subtler experimental results presented here.
Discussion
5mC and 5hmC can occur outside of CG dinucleotides, particularly in stem cells 8 and brain 10, expanding the landscape of sequence-specific DNA binding of TFs. We examined how double-stranded DNA containing 5mC or 5hmC on one DNA strand changed DNA binding of TCF4 and USF1, two members of the B-HLH domain protein family. 5mC on one strand inhibits DNA binding of both TCF4 and USF1. 5hmC eliminates USF1 binding but dramatically enhances TCF4 binding to the E-Box motifs ACAT∣GTG and ACAC∣GTG sequences, which are better bound than any 8-mer with cytosine.
The biological importance of binding DNA containing 5hmC outside of CG dinucleotides is difficult to evaluate. TCF4 ChIP-seq can evaluate if 5hmC containing E-Box motifs are bound in vivo 34, 35. Cytosine modifications outside of CG dinucleotides are rare and never become prominent in a population of cells making it very difficult to biochemically examine them. Potentially, biological samples will be discovered or created where 5hmC not in CG dinucleotides is prominent in a population of cells. A potential method to evaluate TCF4 binding to E-Box motifs containing 5hmC is genetic, it may be possible to design alleles that do not bind to unmodified DNA but still bind to 5hmC containing DNAs. If these alleles have biological activity, it suggests that 5hmC binding is biological important.
The amino acid sequence around the invariant glutamate that interact with the cytosine in the E-Box CAN∣NTG is different between TCF4 and USF1, suggesting that a structural understanding of TCF4 binding to 5hmC might be possible. USF1 has four bulky arginines following the glutamic acid (ERRRR) while TCF4 has only two (ERLRV) suggesting that the TCF4 structure may be more amenable to conformational changes when it preferentially binds 5hmC. This conformational flexibility is seen in the two forms of the TCF3-DNA complex in the X-ray structure 28.
Some B-HLH proteins preferentially bind to ACAC∣GTG motif with unmodified cytosine including Bhlhe41, Clock, Hey2, and Npas2 20. It will be interesting to determine if 5hmC inhibits DNA binding as occurs with USF1. This could act potentially as a switch with one protein binding with cytosine and TCF4 binding when the motif contains 5hmC. This manuscript has presented a new method to examine how 5mC and 5hmC affects DNA binding of TF. This method can be expanded to additional modified bases like 5fC, 5caC, and N6-methyladenine 36. This expanded DNA sequence landscape shows dramatic biochemical changes in DNA binding that may be biologically important.
Conclusion
In summary, we developed a new protein binding microarray method, in which single-stranded oligonucleotide arrays were double-stranded with either 5-methyl cytosine or 5-hydroxymethyl cytosine. The modified double-stranding procedure creates asymmetric distribution of cytosine mimicking what occurs in mammalian stem cells and brain tissues. Using this modified arrays we examined the DNA binding of two B-HLH proteins: TCF4 and USF1. DNA binding of both proteins was inhibited by 5mC. 5hmC increased DNA binding of TCF4 to E-box motifs ACAC∣GTG and ACAT∣GTG. 5hmC inhibited DNA binding of USF1. These highlight the utility of the modified protein binding microarray method to examine how modified cytosines alter the DNA binding of sequence-specific TFs.
Supplementary Material
Acknowledgements
We thank Dr. Tom Ellenberger for providing the crystal structure co-ordinates of TCF3 homodimer.
Funding
This work is supported by the intramural research project of National Cancer Institute, NIH, Bethesda, USA.
References
- 1.He X, Tillo D, Vierstra J, Syed KS, Deng C, Ray GJ, Stamatoyannopoulos J, FitzGerald PC and Vinson C, Genome biology and evolution, 2015, 7, 3155–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C and Zhang Y, Science, 2011, 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastor WA, Aravind L and Rao A, Nature reviews. Molecular cell biology, 2013, 14, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H and Zhang Y, Cell, 2014, 156, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubeler D, Nature, 2015, 517, 321–326. [DOI] [PubMed] [Google Scholar]
- 6.Ko M, An J and Rao A, Current opinion in cell biology, 2015, 37, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S and Rao A, Nature, 2011, 473, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B and Ecker JR, Nature, 2009, 462, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM and Ecker JR, Science, 2013, 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, Lin S, Lin Y, Jung I, Schmitt AD, Selvaraj S, Ren B, Sejnowski TJ, Wang W and Ecker JR, Nature, 2015, 523, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, Yu M, Liu X, Zhu P, Li X, Hou Y, Guo H, Wu X, He C, Li R, Tang F and Qiao J, Genome biology, 2014, 15, R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriaucionis S and Heintz N, Science, 2009, 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y and Ecker JR, Annual review of genomics and human genetics, 2015, 16, 55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T and Vermeulen M, Cell, 2013, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 15.Golla JP, Zhao J, Mann IK, Sayeed SK, Mandal A, Rose RB and Vinson C, Biochemical and biophysical research communications, 2014, 449, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayeed SK, Zhao J, Sathyanarayana BK, Golla JP and Vinson C, Biochimica et biophysica acta, 2015, 1849, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto H, Zhang X, Vertino PM and Cheng X, The Journal of biological chemistry, 2015, 290, 20723–20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR and Stuiver MH, Biochimica et biophysica acta, 1994, 1218, 129–135. [DOI] [PubMed] [Google Scholar]
- 19.De Masi F, Grove CA, Vedenko A, Alibes A, Gisselbrecht SS, Serrano L, Bulyk ML and Walhout AJ, Nucleic acids research, 2011, 39, 4553–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, Zheng H, Goity A, van Bakel H, Lozano JC, Galli M, Lewsey MG, Huang E, Mukherjee T, Chen X, Reece-Hoyes JS, Govindarajan S, Shaulsky G, Walhout AJ, Bouget FY, Ratsch G, Larrondo LF, Ecker JR and Hughes TR, Cell, 2014, 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E and Blake DJ, Trends in molecular medicine, 2014, 20, 322–331. [DOI] [PubMed] [Google Scholar]
- 22.Sweatt JD, Experimental & molecular medicine, 2013, 45, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML and Hughes TR, Cell, 2008, 133, 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharrocks AD, Gene, 1994, 138, 105–108. [DOI] [PubMed] [Google Scholar]
- 25.Philippakis AA, Qureshi AM, Berger MF and Bulyk ML, Journal of computational biology : a journal of computational molecular cell biology, 2008, 15, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR and Bulyk ML, Science, 2009, 324, 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR and Vinson C, Genome research, 2013, DOI: gr.146654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellenberger T, Fass D, Arnaud M and Harrison SC, Genes & development, 1994, 8, 970–980. [DOI] [PubMed] [Google Scholar]
- 29.Ferre-D’Amare AR, Pognonec P, Roeder RG and Burley SK, The EMBO journal, 1994, 13, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM and Karplus M, Journal of computational chemistry, 2009, 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE, Journal of computational chemistry, 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 32.Chen CC, Wang KY and Shen CK, The Journal of biological chemistry, 2012, 287, 33116–33121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markus M, Du Z and Benezra R, The Journal of biological chemistry, 2002, 277, 6469–6477. [DOI] [PubMed] [Google Scholar]
- 34.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B and He C, Cell, 2012, 149, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X, Li X, Yuan B, Ke Y, He C, Zhou L, Liu J and Ci W, Cell research, 2016, 26, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, Li C, Liu B, Luo Y, Zhu Y, Zhang N, He S, He C, Wang H and Chen D, Cell, 2015, 161, 893–906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.