SUMMARY
Mdn1 is an essential AAA (ATPase Associated with various Activities) protein that removes assembly factors from distinct precursors of the ribosomal 60S subunit. However, Mdn1’s large size (~5000aa) and its limited homology to other well-studied proteins have restricted our understanding of its remodeling functions. Here, we present structures for S. pombe Mdn1 in the presence of AMPPNP at up to ~4Å, or ATP plus Rbin-1, a chemical inhibitor, at ~8Å. These data reveal that Mdn1’s MIDAS domain is tethered to its ring-shaped AAA domain through an ~20nm long structured linker and a flexible ~500aa Asp/Glu-rich motif. We find that the MIDAS domain, which also binds other ribosome-assembly factors, docks onto the AAA ring in a nucleotide state-specific manner. Together, our findings reveal how conformational changes in the AAA ring can be directly transmitted to the MIDAS domain and thereby drive the targeted release of assembly factors from ribosomal 60S-subunit precursors.
INTRODUCTION
The biogenesis of ribosomes is a multi-step process that can establish the translational capacity of cells and plays important roles in cellular growth and survival (Kressler et al., 2012; Woolford and Baserga, 2013). In eukaryotes, ribosome biogenesis is initiated by transcription of ribosomal DNA (rDNA) in the nucleolus (Kressler et al., 2012; Woolford and Baserga, 2013). Association of ribosomal proteins and assembly factors with ribosomal RNAs (rRNAs) results in the formation of megadalton-sized complexes corresponding to precursors of the 40S (pre-40S) and 60S (pre-60S) ribosomal subunits (Kressler et al., 2012; Woolford and Baserga, 2013). Many of the key steps of this dynamic process depend on energy-consuming enzymes (Konikkat and Woolford, 2017; Kressler et al., 2012). In particular, Mdn1 (or Midasin; Rea1 in S. cerevisiae) is an essential ATPase required for the proper remodeling of pre-60S particles (Konikkat and Woolford, 2017; Kressler et al., 2012). Currently, we do not understand how this unusually large enzyme (~540 kDa), comprised of ~5000 amino acids (aa) in a single polypeptide, carries out its functions.
Mdn1 belongs to the AAA protein family (ATPase Associated with various Activities), whose members harness energy from ATP hydrolysis to perform mechanical work (Erzberger and Berger, 2006; Hanson and Whiteheart, 2005). While most AAA proteins function as hexameric rings assembled by identical AAA domains, Mdn1 has six non-equivalent AAA domains linked in a single polypeptide, an arrangement similar to that of the microtubule-based motor protein, dynein (Garbarino and Gibbons, 2002; Schmidt and Carter, 2016). However, the AAA domains in Mdn1 share <15% identity with those in dynein (for details, see Methods section). Further, the ~2000 aa in Mdn1 that separate its N-terminal AAA domains from the C-terminal MIDAS (Metal Ion Dependent Adhesion Site) domain do not share readily detectable homology with other known proteins. Therefore, it is difficult to develop reliable models for Mdn1 function based on our understanding of dynein or other AAA proteins.
Our current understanding of Mdn1 function is mainly based on characterization of Rea1, the Mdn1 ortholog in S. cerevisiae (Kressler et al., 2012). Rea1 has been proposed to remodel both nucleolar and nucleoplasmic pre-60S particles (Bassler et al., 2010; Ulbrich et al., 2009). In particular, yeast two-hybrid assays identified two assembly factors, Ytm1 and Rsa4, that interact with the C-terminal MIDAS domain in Rea1 (Ulbrich et al., 2009). Genetic disruption of the interaction between Rea1 and Ytm1, a nucleolar assembly factor, leads to retention of Ytm1 in nucleoplasmic pre-60S intermediates, suggesting that Rea1 activity is required for Ytm1 removal from the nucleolar pre-60S intermediates (Bassler et al., 2010). In addition, pulldown studies suggest that both Rea1 and Rsa4 are present in the Rix1 particle, a nucleoplasmic pre-60S intermediate. Furthermore, the Rea1-Rsa4 interaction is required for ATP-dependent removal of Rsa4 from the affinity-purified Rix1 particles (Matsuo et al., 2014; Nissan et al., 2004; Ulbrich et al., 2009). Experiments with S. pombe, using Rbin-1, a potent and selective chemical inhibitor of Mdn1, support these findings and also revealed that Mdn1 is likely involved in the proper formation of the nucleolar Nsa1 particle, a step that precedes the removal of Ytm1 from 60S precursors (Kawashima et al., 2016). However, it is unclear how Mdn1 binds to and selectively removes different assembly factors at distinct stages of ribosome biogenesis.
Negative-stain electron microscopy (EM) studies led to a model for how Rea1’s conformational changes during its ATPase cycle are coupled to its function (Ulbrich et al., 2009). Two-dimensional (2D) averages of S. cerevisiae Rea1 particles suggest that the MIDAS domain is connected to the AAA ‘ring’ through an elongated ‘tail’ that can have different orientations relative to the ring. Based on these data it was proposed that the tail domain can transmit force, similar to the lever-arm motion described for myosin (Spudich, 2001), from the AAA to the MIDAS domain (Bassler et al., 2010). An alternative hypothesis was also proposed that long-range structural communication may exist between the AAA and MIDAS domains and that conformational changes would be propagated across the entire tail (Ulbrich et al., 2009). However, no studies have directly observed the position of the substrate-binding MIDAS domain and linked the tail domain’s motion to specific nucleotide states. Higher resolution data are needed to determine if these proposed models explain Mdn1 function.
We report two single-particle cryo-EM maps for the full-length S. pombe Mdn1 that allowed us to build a pseudo-atomic model for the protein in the presence of AMPPNP, based on an ~4 Å resolution map for the AAA ring and an ~6 Å resolution map for the C-terminal tail, and a Cα-backbone model for Mdn1 in the presence of ATP plus the chemical inhibitor Rbin-1, based on an ~8 Å resolution map. Together, these data suggest that Mdn1’s MIDAS domain is tethered by a flexible ~500 aa Asp/Glu-rich motif to an ~20 nm long structured linker domain connected to the AAA ring. Importantly, the C-terminal MIDAS domain docks onto the AAA ring in the presence of ATP and the chemical inhibitor, but not in the presence of AMPPNP. Our findings, along with a previously determined cryo-EM map of the Rix1 particle, suggest how conformational changes in the AAA ring can be directly transmitted to the MIDAS domain. These conformational dynamics could allow Mdn1 to dissociate assembly factors, such as Rsa4, from precursors of the ribosomal 60S subunit.
RESULTS
Mdn1 Has a Characteristic AAA Ring and an Elongated Hook-shaped Tail
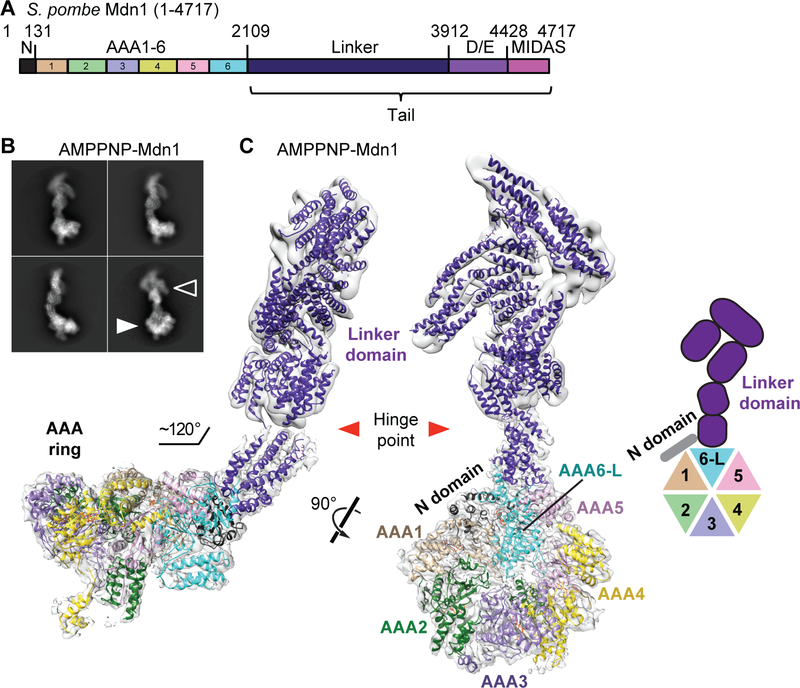
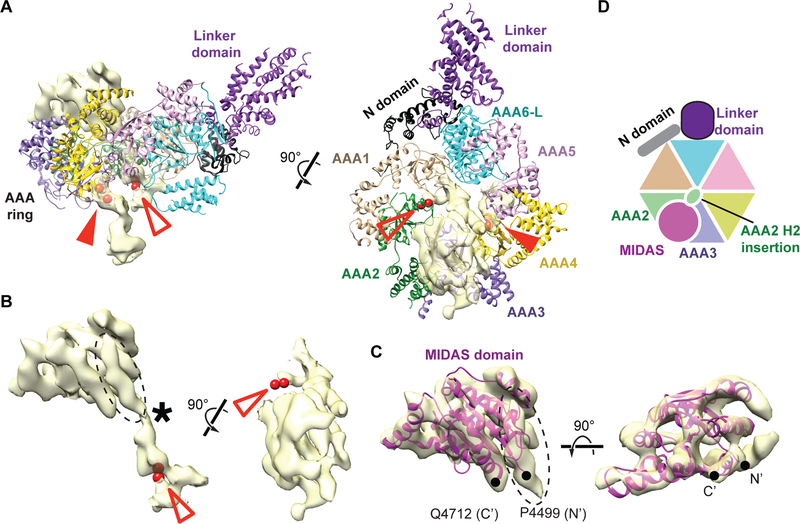
Alignment of primary sequences reveals that Mdn1 has a conserved domain organization across eukaryotes (Supplementary Data Item 1). At the N terminus, there is a small domain, named the N domain (~130 aa in S. pombe Mdn1), followed by six AAA domains, and a ‘tail’, which is comprised of the linker, Asp/Glu-rich (D/E-rich) and MIDAS domains (Figure 1A).
Figure 1. Overall Architecture of Full-length Mdn1 in the Presence of AMPPNP.
(A) Schematic for the domain organization of S. pombe Mdn1 (aa 1–4717). Residue numbers at selected domain boundaries are indicated. (B) Selected class averages of vitrified Mdn1 in the presence of AMPPNP (1 mM). The AAA ring (solid arrow) and the hook-shaped tail (open arrow) are indicated. (C) Two views of the Mdn1 density map (grey surface) with the model that was built into the map (ribbon representation, colored as in (A)) (hereafter, AMPPNP-Mdn1). The model and a schematic for the N, AAA and linker domains are shown. The hinge point (red arrowhead) in the tail region, where the density was split and processed separately, is also indicated in the model.
Proteins in the AAA family can couple ATP hydrolysis to large conformational changes (Erzberger and Berger, 2006; Hanson and Whiteheart, 2005). Therefore, full-length S. pombe Mdn1 was incubated with AMPPNP (1 mM), a nucleotide analog that inhibits the ATPase activity of recombinant Mdn1 (Kawashima et al., 2016), and analyzed using single-particle cryo-EM (Figures S1 and S2). Initial class averages indicated that Mdn1 adopts an elongated structure with a ‘ring’ (Figure 1B, solid arrowhead), an arrangement of AAA domains typical for proteins in the superfamily (Erzberger and Berger, 2006; Hanson and Whiteheart, 2005), connected to an elongated, hook-shaped ‘tail’ (Figure 1B, open arrowhead).
Three-dimensional (3D) classification of Mdn1 particles indicated that the ring and tail densities varied slightly in their relative orientations, likely due to flexibility at a single site in the tail, which we named ‘hinge point’ (Figure 1C, red arrowheads). Therefore, we masked the densities for the ring and tail regions, above and below the hinge point, and processed them separately. The final 3D maps were refined to an estimated resolution of ~4.0 Å for the ring region, and ~5.9 Å for the tail region (Figures 1C, S1C and S2A). At ~4.0 Å resolution, α-helices and individual β-strands can be resolved in Mdn1’s ring region (Figures 1C and S2C). We were also able to trace the backbone for the N-terminal portion, which included the N and AAA domains and the tail up to the hinge point (Figure 1C). A pseudo-atomic model for the AAA domains was built based on known AAA protein folds (Erzberger and Berger, 2006; See methods for details). The remaining tail density was modeled with polyalanine helices. Hereafter, we refer to this model as AMPPNP-Mdn1 (Movie S1).
The AAA ring of AMPPNP-Mdn1 has an ~140 Å outer diameter, ~30 Å inner diameter and a height of ~65 Å. The elongated tail region is composed of five bundles of α-helices. The tail first extends from the AAA ring for ~20 nm but then turns back to form a hook-like structure. The angle between the base of the tail and the plane of the AAA ring is ~120° (Figure 1C). Our model for the hook-shaped tail accounts for ~1400 aa, which includes ~50% of the C-terminal residues after the AAA domains (~2600 aa, Figure 1A), suggesting that a portion of the C-terminal tail domain may be disordered.
Secondary-structure prediction algorithms suggest that the ~500 aa-long D/E-rich domain, which follows the linker domain, likely contains only two α-helices (formed by ~30 aa) and no β-strands (Supplementary Data Item 1). To test if the D/E-rich domain is indeed not observed in the map, we generated and purified an Mdn1 construct that lacked the C-terminal D/E-rich and MIDAS domains (Mdn1-ΔC, aa 1–3911) (Figure S3A-C). We used single-particle cryo-EM to determine the structure of Mdn1-ΔC in the presence of AMPPNP. We obtained maps at a resolution of ~6.5 Å for the region including the AAA ring and ~7.4 Å for the rest of the protein (Figures S3D-H). The model for full-length Mdn1 could be placed into the density map of Mdn1-ΔC as a single rigid body without adjustment (Figure S3H). Together, these data indicate that the hook-shaped tail is formed by the linker domain alone (Figures 1C and S3H) and also confirm that the D/E-rich and MIDAS domains are not observed in the map for full-length Mdn1 in the presence of AMPPNP (Figure 1C). Further, these data also suggest that removal of the C-terminal D/E-rich and MIDAS domains does not affect the overall structure of the AAA and linker domains in the presence of the non-hydrolyzable ATP analog.
The Six AAA Domains in AMPPNP-Mdn1 are Asymmetrically Arranged
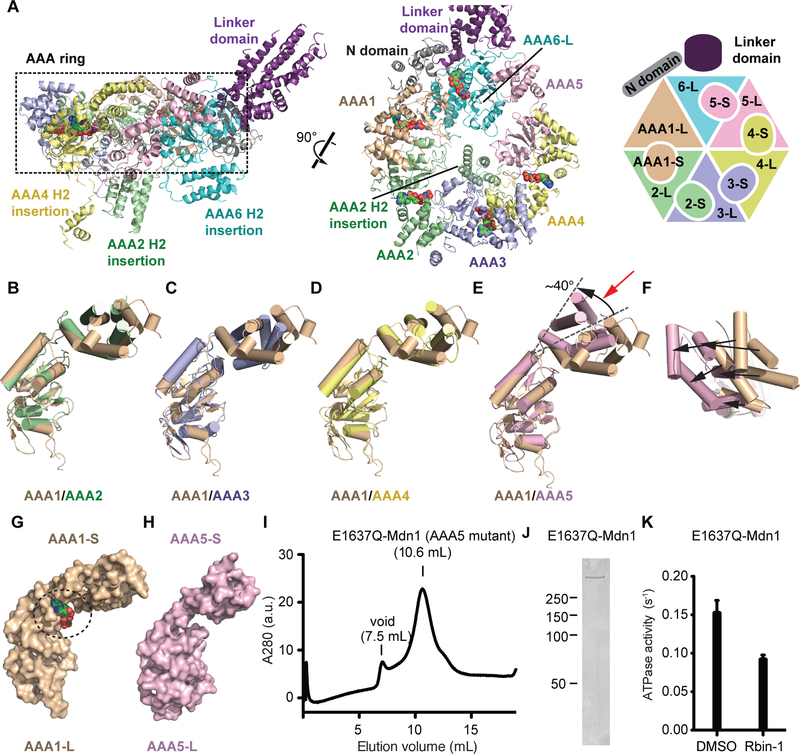
We next examined the organization of the six AAA domains in Mdn1. In general, a AAA domain is comprised of a large N-terminal subdomain (AAA-L) and a small C-terminal subdomain (AAA-S) (Figure S4A). Six AAA-L and five AAA-S subdomains could be readily identified in the cryo-EM map as they adopt characteristic folds (Figures 2A and S4C-4H) (Erzberger and Berger, 2006). In the orientation shown in the left panel of Figure 2A, the AAA-L domains are at the lower face of the ring, while the AAA-S domains lie above. The polypeptide chain following the C terminus of AAA6-L, which was expected to form the AAA6-S domain, lacks structural similarity to other AAA-S domains within Mdn1. We find that this region of the protein extends out of the ring and forms the base of the linker domain (Figure 2A). The N, AAA1-L and AAA6-L domains form a structural motif, which we name the ‘ring-linker junction’, that establishes the orientation of the hook-shaped linker domain relative to the AAA ring (Figures 2A and S4B).
Figure 2. Asymmetric Arrangement of Mdn1’s Six AAA Domains.
(A) Two views of the model for the six AAA domains and part of the linker domain, along with a schematic, are shown. The AAA ring core (dashed rectangle) is highlighted. (B-E) Alignment of the AAA2–5 domains with the AAA1 domain using their large subdomains. In (E), an ~40° difference between the orientations of the small subdomains in AAA5 and AAA1 is apparent. (F) AAA1 and AAA5 small subdomains viewed from the direction indicated by the red arrow in (E). Differences in the positions of the five equivalent α-helices (black arrows) are highlighted. (G and H) Surface representations of the AAA1 (G) and AAA5 (H) domains. In (G), the nucleotide (sphere representation) is also highlighted. (I) Size-exclusion chromatography profile for E1637Q-Mdn1 (AAA5 mutant) (elution volume: 10.6 mL, void 7.5 mL). (J) SDS-PAGE analysis of purified E1637Q-Mdn1 (Coomassie blue stain). (K) ATPase activities of Mdn1 in the presence of solvent control (DMSO) or Rbin-1 (1 μM, dissolved in DMSO) were measured using an NADH-coupled ATPase assay ([MgATP] = 1 mM).
Sequence alignments reveal that Mdn1 belongs to a subfamily of AAA proteins that possess insertions following helix H2 in the AAA-L subdomain (called H2 insertion, Figures S4A and Supplementary Data Item 1) (Erzberger and Berger, 2006). For three AAA-L subdomains (AAA1-L, AAA3-L, and AAA5-L), these insertions are <20 aa, while the other AAA-L subdomains (AAA2-L, AAA4-L and AAA6-L) have longer insertions (60–120 aa, Figures 2A and Supplementary Data Item 1). We find that these three longer H2 insertions project out from the same face of the AAA ring, opposite to that from which the tail extends (Figure 2A). The AAA2 H2 insertion (120 aa) forms a four-helix bundle that is proximal to the central hole of the AAA ring (Figure 2A). The AAA4 H2 insertion (60 aa) forms a bundle of three helices. Finally, the AAA6 H2 insertion (~100 aa) is also largely helical and adjoins the AAA6-L and AAA1-L domains (Figure 2A), possibly functioning as a bridging element.
We note that the AAA1 and AAA6 in S. pombe Mdn1 lack the “Asp-Glu” Walker B motif that is required for ATP hydrolysis in AAA proteins (Supplementary Data Item 1) (Erzberger and Berger, 2006), suggesting that ATPase activity in Mdn1 is restricted to its AAA2–5 ATPase sites. Consistent with this hypothesis, cell-based studies have shown that key residues in the Walker A, Walker B and arginine finger motifs in the AAA2–5 ATPase sites are all required for Mdn1 function (Kawashima et al., 2016). We next compared the relative orientations of the five AAA-L and associated AAA-S subdomains (Figures 2B-2E). AAA6 is not included as it lacks a typical AAA-S subdomain. The small and large subdomains in AAA2 to AAA4 show a similar arrangement as seen in AAA1 (Figures 2B-2D). In AAA5, however, the small subdomain is rotated by ~40° relative to its large subdomain, causing substantial displacements of the five α-helices in AAA5-S (Figures 2E and 2F). This difference in relative orientation results in an opening between the small and large subdomains in AAA5 (Figures 2G and2H).
Interestingly, we can assign densities corresponding to nucleotides in the AAA1-AAA4 and AAA6 ATPase sites but not in the AAA5 site (Figures S4C-S4H). As the cell-based studies indicate that ATP hydrolysis at the AAA5 ATPase site is needed for Mdn1 function, we expected that AAA5 contributes to Mdn1’s overall ATPase activity. To test this we expressed and purified a construct with a Walker B mutation in Mdn1’s AAA5 domain (E1637Q). Equivalent mutations have been used to disrupt ATP hydrolysis but not ATP binding in AAA proteins (Bhabha et al., 2014; Martin et al., 2005). The AAA5 mutant protein eluted from a size-exclusion column at 10.6 mL, similar to wild-type Mdn1 (elution volume: 10.6 mL) (Figure 2I and 2J). Negative-stain EM analysis revealed that the AAA5 mutant adopts an extended conformation, with the ring and tail regions appearing similar to those observed for wild-type Mdn1 (Figure S4I). We found the ATPase activity of the mutant construct is 0.15±0.02 ATP per second (Figure 2K), which is ~15% of what we measured under similar conditions for wild-type Mdn1 (Kawashima et al., 2016). Addition of Rbin-1 (1 μM) inhibited ~40% of the ATPase activity of the mutant, similar to what we observed for wild-type Mdn1 at this inhibitor concentration (Kawashima et al., 2016). These data are consistent with the observed ATPase activity being mainly due to the mutant Mdn1 construct. Together, our findings suggest that the AAA5 site contributes to Mdn1’s overall ATPase activity and that the ATPase sites are structurally asymmetric and likely function asynchronously.
Structure of Full-length Mdn1 in the presence of ATP and Rbin-1 Reveals Local Conformational Changes in the AAA Ring
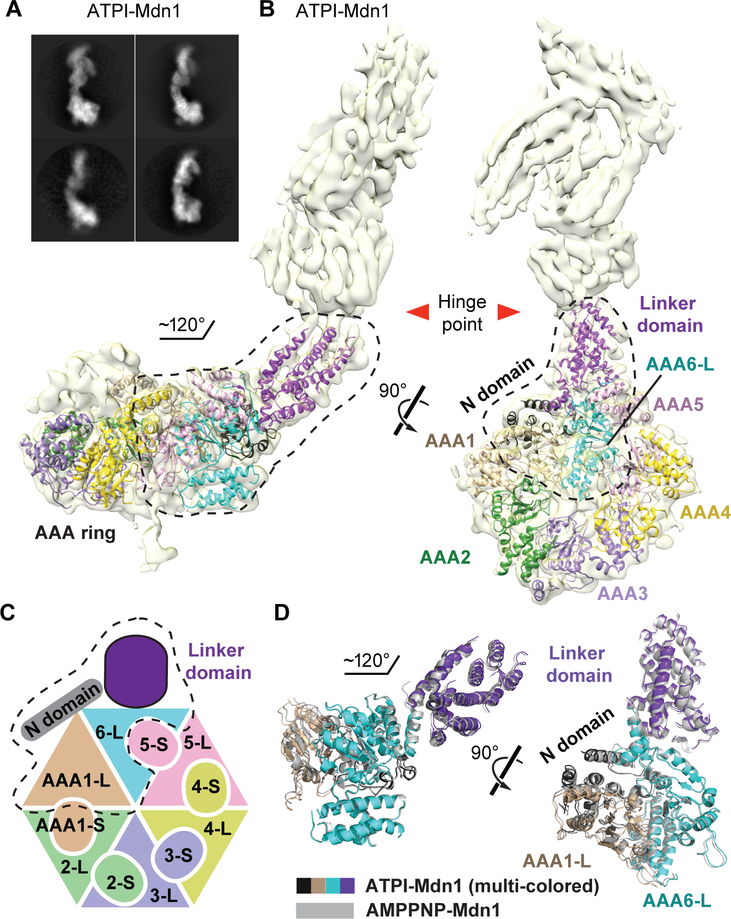
We next examined the potential conformational changes of Mdn1 during ATP hydrolysis by determining the cryo-EM map of Mdn1 in the presence of ATP (1 mM) and Rbin-1 (1 μM) (Kawashima et al., 2016). This cell-permeable chemical inhibitor can potentially trap Mdn1 in a conformational state distinct from that of AMPPNP-Mdn1. Class averages revealed an elongated overall structure similar to that of AMPPNP-Mdn1 (Figure 3A). 3D classification of the entire molecule yielded a class with well-defined density for both the ring and tail regions. The final density map was refined to a resolution of ~7.7 Å (Figures 3B and S5, hereafter named ATPI-Mdn1), with α-helices and β-sheets, but not individual β-strands, readily distinguishable.
Figure 3. Overall Architecture of Mdn1 in the Presence of ATP and Rbin-1.
(A) Selected class averages of vitrified Mdn1 in the presence of ATP (1 mM) and Rbin-1 (1 μM), a chemical inhibitor of Mdn1 (hereafter, ATPI-Mdn1). (B) Two views of the density map (yellow surface) with a Cα-backbone model for the region of Mdn1 N-terminal of the hinge point (ribbon colored as in Figures 1A and3C). This model does not include the H2 insertions of AAA2 and AAA4. The ring-linker junction, comprised of N-domain, AAA1-L, AAA6-L and linker domain, is outlined (black dashed lines). (C) Schematic highlighting the ring-linker junction (dashed line) is shown. (D) Two views of the overlay of the ring-linker junctions in AMPPNP-Mdn1 (grey ribbon) and ATPI-Mdn1 (multi-colored ribbon, colored as in (C)).
We found that the maps revealed similar overall organizations for ATPI- and AMPPNP-Mdn1 (Figures 3A-3C). For the N-terminal region of ATPI-Mdn1, up to the hinge point in the linker domain, we could build a backbone model using the AMPPNP-Mdn1 model (Figure 3B and Movie S2). Nucleotides, Rbin-1 and the H2 insertions of AAA2 and AAA4 could not be readily assigned. The poly-alanine model for the entire hook-shaped tail from AMPPNP-Mdn1 could be placed into the ATPI-Mdn1 density as a single rigid body without further adjustment (Figure S6A). Interestingly, ATPI-Mdn1 also has its tail oriented ~120° relative to the AAA ring (Figure 3B), as observed in AMPPNP-Mdn1 (Figure 1C). Overlaying the ‘ring-linker junction’ from these two structures also did not reveal substantial differences (Figure 3D).
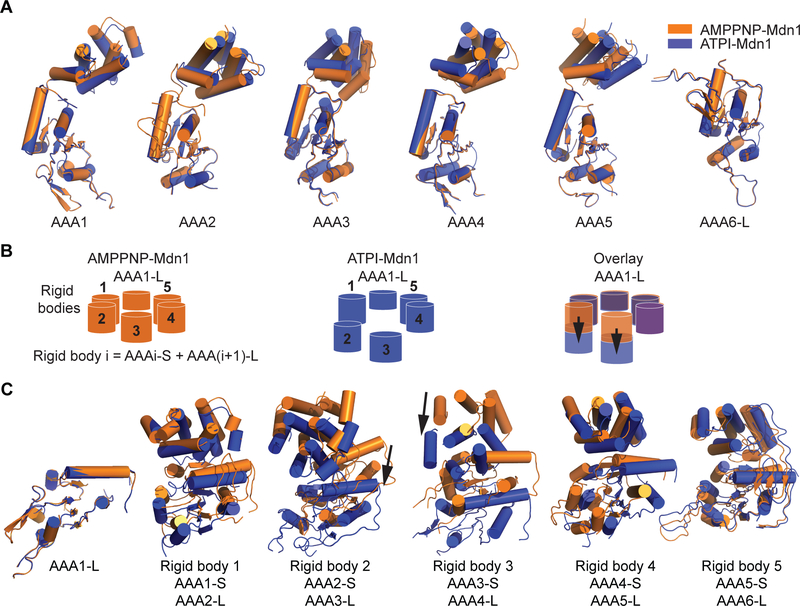
Pairwise comparisons of individual AAA domains reveal that the conformations of AAA1 and AAA6 are similar between AMPPNP- and ATPI-Mdn1 (Figure 4A). This may also contribute to the similar orientation of the tail relative to the AAA ring. In contrast, substantial differences are found across the AAA2-AAA5 domains in these two structures (Figure 4A). To examine these changes, we focused on the structural units called ‘rigid bodies’ (‘rigid body i’ is defined as the combination of the AAAi-S and AAA(i+1)-L subdomains; as shown in Figure S4A), which during conformational changes in AAA proteins can move as single structural units (Figure S6B) (Glynn et al., 2009; Matyskiela et al., 2013). Rigid bodies 1, 4 and 5 rotate slightly but are essentially on the same plane as the AAA1-L subdomain and occupy similar positions in AMPPNP- and ATPI-Mdn1 (Figures 4B, 4C and Movie S3). In contrast, large displacements (>15 Å) of rigid bodies 2 and 3 between the two states are observed. (Figures 4B and 4C). Together, these data suggest that local conformational changes of Mdn1’s AAA ring are linked to ATPase activity at a subset of sites.
Figure 4. Comparisons of AAA Domains in AMPPNP-Mdn1 and ATPI-Mdn1.
(A) Alignment of the individual AAA domains (AAA1-AAA5 and AAA6-L) in AMPPNP-Mdn1 (orange ribbon) and ATPI-Mdn1 (blue ribbon). The models were aligned using the large subdomains. (B) Schematics of the five rigid bodies (cylinders) and AAA1L (shorter cylinder) in AMPPNP-Mdn1 (orange) and ATPI-Mdn1 (blue) and an overlay of them aligned using AAA1-L. The displacements of rigid bodies 2 and 3 (black arrows) are also indicated in the overlay. (C) Arrangement of individual rigid bodies in AMPPNP-Mdn1 (orange ribbon) and ATPI-Mdn1 (blue ribbon) when the two models are aligned using their AAA1-L subdomain. The displacements of rigid bodies 2 and 3 (black arrows) are indicated.
Mdn1 Undergoes Long-range Intramolecular Dynamics
We next examined the densities in the ATPI-Mdn1 map that could not be readily assigned using the AMPPNP-Mdn1 model (Figure 5A). First, on the face of the AAA ring, opposite to the one from which the tail extends, there is unassigned density that adjoins AAA4-L (Figure 5A). Further, this density is directly connected to the residues flanking the AAA4 H2 insertion (Figure 5A, filled red arrowheads) that initially could not be modeled. We find that the three-helix bundle modeled for the AAA4 H2 insertion in the AMPPNP-Mdn1 structure fits into this density (Figure S7A). The relative orientation of the AAA4-L subdomain and its H2 insertion differs only slightly between the AMPPNP- and ATPI-Mdn1 structures (Figure S7A and S7B), suggesting that the AAA4 H2 insertion moves together with the AAA4-L subdomain during ATP hydrolysis.
Figure 5. Occlusion of the Central Hole of the AAA Ring and Docking of the C-terminal MIDAS Domain onto the N-terminal AAA Ring in ATPI-Mdn1.
(A) Two views of the unassigned density (yellow surface) and the initial ATPI-Mdn1 model (ribbon representation colored as in Figure 1A). The red spheres indicate the residues flanking the H2 insertions of AAA4 (filled red arrowheads) and AAA2 (open red arrowheads). (B) Two views of the unassigned densities (yellow surface) that extend from the vicinity of the residues flanking the AAA2 H2 insertion (red spheres, views as in (A)). A constriction in this density (asterisk) is also highlighted. (C) Two views of the homology model for the MIDAS domain (magenta ribbon, aa 4499–4712) that was fit into the density shown in (B) (yellow surface). Only the density above the constriction in (B) is shown. The N-terminal end (Pro-4499) and C-terminal end (Gln-4712) of the model are labeled. The N-terminal end of the model (N’) connects to an unassigned density, whose location is indicated by dashed circles in (B) and (C). (D) Schematic of ATPI-Mdn1 model highlighting the docking of MIDAS domain onto the AAA2 and AAA3 domains is shown.
An additional density that extends from the same face of the AAA ring as the AAA4 H2 insertion was assigned to the AAA2 H2 insertion, as it connects to residues flanking the H2 insertion of AAA2 (Figures 5A and 5B, open red arrowheads). This density extends into the central hole of the AAA ring. The additional unassigned density that passes through the ring is not likely to be part of the AAA2 H2 insertion, as it is separated from the density representing the H2 insertion by a constriction (Figure 5B, asterisk). In AMPPNP-Mdn1, the AAA2 H2 insertion forms a four-helix bundle and covers only part of the central hole (Figure 2A). If the four-helix bundle maintained this same position in ATPI-Mdn1, it would clash with the AAA4 H2 insertion density (Figure S7C). This result suggests that displacement of the AAA4-L domain during the ATPase cycle leads to the reorganization of the AAA2 H2 insertion and occlusion of the central hole in the AAA ring, reminiscent of substrate binding in AAA unfoldases (Monroe et al., 2017; Puchades et al., 2017; Ripstein et al., 2017).
In AAA unfoldases, conserved aromatic residues in an ‘Ar-Φ-Gly’ tripeptide (Ar: aromatic residue; Φ: hydrophobic residue, named ‘pore loop’) interact with substrates inside the central hole of the AAA ring (Monroe et al., 2017; Puchades et al., 2017). Interestingly, Mdn1’s AAA3 domain possesses a conserved ‘Tyr-Ile/Leu-Gly’ motif. The other AAA domains in Mdn1 lack this motif but are enriched in glycine, aromatic and hydrophobic residues at corresponding sites near the beginning of the H2 helix (Supplementary Data Item 1). All of these aromatic residues lie proximal to the central hole in AMPPNP-Mdn1 and only Tyr-198 in AAA1 could be modeled in our ATPI-Mdn1 map (Figures S7D and S7E). Tyr-198 in the AAA1 domain makes direct contact with the density inside the central hole that we assigned as the AAA2 H2 insertion (Figure S7E), suggesting that this residue may contribute to interactions with and stabilization of the motif that occludes the central hole.
Finally, the unassigned density on the same face as the hook-shaped tail positioned above rigid bodies 2 and 3 is sufficiently resolved to reveal well-defined secondary-structure elements (Figure 5B). These structural motifs were similar to those observed in MIDAS domains of other unrelated proteins (Song et al., 2005). Indeed, a homology model for the MIDAS domain, which consists of a central β-sheet surrounded by seven α-helices, could be unambiguously fit into this density (Figure 5C). Together, these data indicate that, when the central hole is blocked, the C-terminal MIDAS domain, located ~2000 aa from the N-terminal AAA domains, docks onto the AAA ring (Figure 5D and Movie S2).
ATPI-Mdn1 Can be Docked into Cryo-EM Maps of the Rix1 Particle
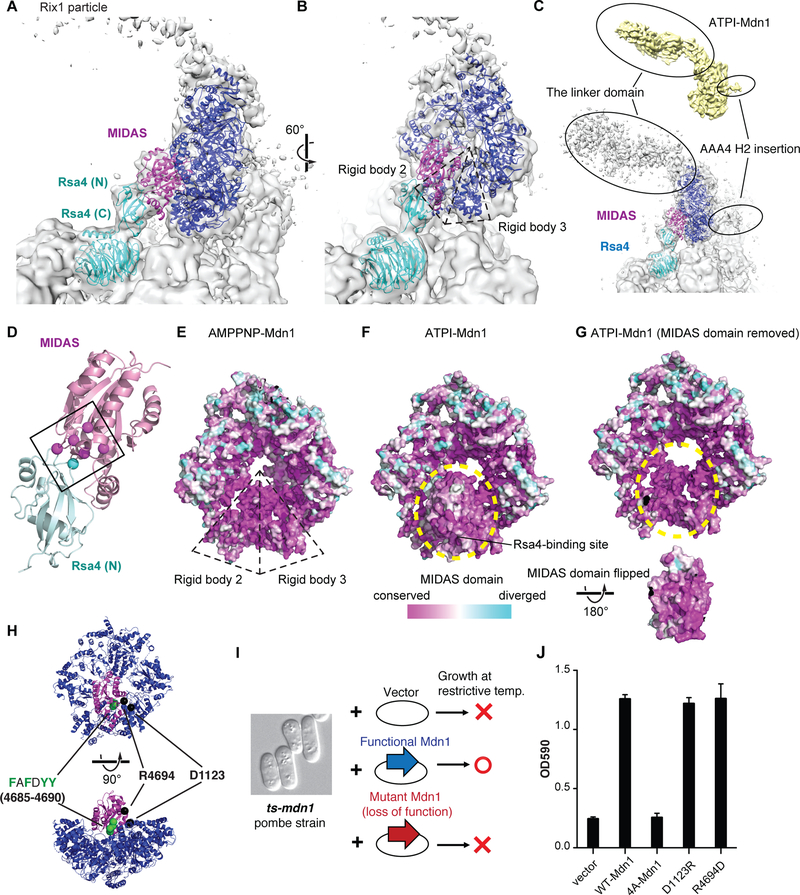
Mdn1 is required for the removal of assembly factors, including the removal of Rsa4 from the Rix1 particle, a precursor of the 60S subunit (Kressler et al., 2012; Matsuo et al., 2014; Ulbrich et al., 2009). Recent studies have characterized the structures of the S. cerevisiae Rix1 and Nog2 particles (Barrio-Garcia et al., 2016; Wu et al., 2016). These structures indicate that Rea1, the S. cerevisiae Mdn1 ortholog, binds the core of the pre-60S particle through a limited portion of the AAA ring’s outer edge. In the map of the Rix1 particle the tail of Rea1, for which only weak density is observed, extends away from the core of the pre-60S particle (Barrio-Garcia et al., 2016). We find that our ATPI-Mdn1 model fits well into the Rea1 density in this map (Figures 6A-6C and Movies S4 and S5). Remarkably, the ATPI-Mdn1 MIDAS domain occupies density in the Rix1-particle map that was previously unassigned (Barrio-Garcia et al., 2016; Wu et al., 2016). Importantly, this density which we assign to the MIDAS domain, is adjacent to the density corresponding to the ribosome assembly factor Rsa4 in the Rix1-particle model (Barrio-Garcia et al., 2016; Wu et al., 2016). In this arrangement, residues that have been shown to be critical for Mdn1-Rsa4 interaction localize to the interface between the MIDAS and Rsa4 domains (Figure 6D) (Ulbrich et al., 2009). When contoured at a lower threshold level, the Rix1-particle map also reveals densities that correspond to the linker domain and the AAA4 H2 insertion in our ATPI-Mdn1 structure (Figure 6C and Movie S5). These analyses reveal that our ATPI-Mdn1 structure depicts a conformation of Mdn1 in the Rix1 particle, in which the MIDAS domain can directly interact with Rsa4. This model suggests inhibition of Mdn1 by Rbin-1 would not preclude the assembly of the Rix1 particle but could block the release of Rsa4, which is consistent with our findings that acute inhibition of Mdn1 by Rbins can result in an enrichment of Rsa4 on Rix1 particles purified from fission yeast cells (Kawashima et al., 2016).
Figure 6. Docking ATPI-Mdn1 into the Rix1 particle Cryo-EM Map and testing the model using a cell-based assay.
(A and B) Two views of the ATPI-Mdn1 model and the Rsa4 crystal structure (cyan, C. thermophilum, PDB ID: 4WJS) placed into the density map of the Rix1 particle (EMDB ID: 3199). For clarity, only the AAA ring (blue) and MIDAS domain (magenta) of ATPI-Mdn1 are shown. Details of the model fitting are described in the Methods section. Rigid bodies 2 and 3 of Mdn1’s AAA ring (dashed triangles) are indicated in (B). (C) A zoomed-out view of (A) and the map of ATPI-Mdn1 (yellow surface). A lower threshold compared to (A) was used to contour the map of the Rix1 particle. The densities in the Rix1 particle that likely correspond to the linker domain and the AAA4 H2 insertion are highlighted (black ovals). (D) The model for the MIDAS domain (magenta) and the N-terminal ubiquitin-like domain of Rsa4 (cyan) shown in the same orientation as in (B). The Cα carbons of residues (spheres) implicated in the interaction between the MIDAS domain (Asp-4511, Ser-4513, Ser-4515, Thr-4582 and Asp-4619 in S. pombe Mdn1, also labeled in Supplementary Data Item 1) and Rsa4 (Glu-117 in C. thermophilum Rsa4, equivalent to Glu-114 in S. cerevisiae Rsa4) are highlighted. (E-G) Residue conservation mapped onto the surfaces of (D) AMPPNP-Mdn1, (E) ATPI-Mdn1, and (F) ATPI-Mdn1 without the MIDAS domain and the surface of the MIDAS domain that interacts with the AAA ring. Rigid bodies 2 and 3 of Mdn1’s AAA ring (dashed triangles) are indicated in (E). The region where the MIDAS domain docks onto the AAA ring are highlighted (yellow dashed ovals) in (F) and (G). (H) Two orthogonal views of the AAA (blue) and MIDAS (magenta) domains in the ATPI-Mdn1 model. The Cα atoms of the aromatic residues of the FAFDYY sequence (aa 4685–4690, green spheres) and the Cα atoms of residues D1123 and R4694 (black spheres) are indicated. (I) Schematic of the yeast complementation assay using a ts-mdn1 strain. (J) The average OD590 values of exponentially growing cell cultures of ts-mdn1 strains overexpressing wild-type (WT) Mdn1 or the indicated mutants (4A-Mdn1: aa 4685–4690, ‘FAFDYY’ to ‘AAADAA’, D1123R-Mdn1 and R4694D-Mdn1) of full-length Mdn1 (n=3, mean ± SD). The data for the vector control are also shown.
Interestingly, our model also reveals that ATPI-Mdn1 makes direct contact with the core of the Rix1 particle through its rigid bodies 2 and 3 (Figures 6A and 6B), which we find undergo substantial displacements relative to their positions in AMPPNP-Mdn1 to create a binding site for the MIDAS domain. On the other hand, the AMPPNP-Mdn1 model could not be docked into the Rix1-particle map. In particular, there is a mismatch between AMPPNP-Mdn1’s AAA ring and the density proximal to the core of the Rix1 particle (Figure S7F), suggesting that Mdn1 associates with the Rix1 particle in a nucleotide state-specific manner.
We then analyzed the conservation of the primary sequence among Mdn1 orthologs from yeast to human and mapped the conservation scores of individual residues onto the surface models of both AMPPNP- and ATPI-Mdn1 using ConSurf (Figures 6E-6G) (Ashkenazy et al., 2016). Interestingly, AMPPNP-Mdn1 has a conserved surface above rigid bodies 2 and 3 in the AAA ring (Figure 6E), which is covered by the MIDAS domain in ATPI-Mdn1 (yellow dashed circle in Figures 6F). The surface of the MIDAS domain is also conserved, including the region facing the AAA ring (Figure 6G) and the solvent-exposed Rsa4-binding site (Figure 6F). The conservation level of the outer circumference of the AAA ring is variable, with higher conservation scores for residues near rigid bodies 2 and 3 that are in direct contact with the core of the Rix1 particle (Figures 6A-C, 6E and 6F).
To test the functional significance of the residues at the interface between the MIDAS domain and the AAA ring we used fission yeast genetics. In particular, we used a ‘ts-mdn1 complementation assay’, in which different Mdn1 mutants are overexpressed in a temperature-sensitive mdn1 mutant strain and the growth of cells at the restrictive temperature is measured (Figure 6H-J) (Kawashima et al., 2016). We focused on a conserved loop in the MIDAS domain that is enriched in aromatic residues and is buried when the MIDAS domain docks onto the AAA ring. We generated a mutant construct (aa 4685–4690: FAFDYY to AAADAA, hereafter, named 4A-Mdn1) with the goal to disrupt this interaction (Figure 6H). As controls, we also tested mutant constructs in which residues at the edge of the same docking interface were mutated to reverse their charges (D1123R-Mdn1 and R4694D-Mdn1). The cell-based assay indicated that 4A-Mdn1 cannot complement Mdn1 function in cells at the restrictive temperature, while the D1123R and R4694D mutants can (Figure 6J). We also generated recombinant wild-type MIDAS (aa 4381–4717) and mutant MIDAS (aa 4381–4717, FAFDYY to AAADAA, named 4A-MIDAS hereafter) domain constructs and found that both proteins eluted from a size-exclusion column at similar volumes (9.9 and 10.0 mL) (Figure S7G), suggesting that mutations in the aromatic loop in the MIDAS domain do not affect its overall folding. Together, these data are consistent with our model that the intramolecular docking of MIDAS on the AAA ring is required for Mdn1 function.
DISCUSSION
Our single-particle cryo-EM studies reveal two distinct conformations of Mdn1. These structural models, along with biochemical and cell-based analyses, suggest how conformational changes in Mdn1 contribute to ribosome biogenesis.
Insights into the Conformational Dynamics of Mdn1’s C-terminal Tail
Our studies shed light on the conformational dynamics of Mdn1’s C-terminal tail, comprised of the linker, D/E-rich and MIDAS domains. The linker domain adopts similar conformations in all three of our Mdn1 density maps (Figures 1C, 3B and S3H) and also has a similar orientation relative to the AAA ring in Rea1 in the published map (~10 Å local resolution) of the budding yeast Rix1 particle (Figure 6C). In our datasets, different 3D classes reveal a hinge point in the linker domain that results in small changes in orientation (<10°, Figure S1B and S5B). We did not see any 3D classes in which the linker bends substantially. Moreover, for Mdn1’s D/E-rich domain, the repulsion between the abundant negatively charged residues (182 Asp/Glu out of 516 aa, ~35%) would disfavor compact and well-defined folds, likely resulting in a flexible polypeptide. We propose that Mdn1’s linker domain serves as a structural extension from the AAA ring, similar to a fishing pole, to which the MIDAS domain is tethered through the D/E-rich motif, effectively a fishing line. The flexibility of the D/E-rich domain allows intramolecular docking of the MIDAS domain on the AAA ring in Mdn1. We propose that conformational changes of the AAA domain can be directly transmitted to Mdn1’s MIDAS domain. Our data are inconsistent with a potential lever-arm mechanism for transmitting forces across Mdn1’s C-terminal tail, similar to motor proteins like myosins or dynein (Bhabha et al., 2014; Spudich, 2001).
Analysis of the sequences of Mdn1 in different eukaryotes reveals different levels of conservation of its individual domains. In particular, comparisons of corresponding domains within H. sapiens and S. pombe Mdn1 indicate that the linker and D/E-rich domains (~20% identity) are less conserved compared to the AAA and MIDAS domains (~40% identity). Furthermore, H. sapiens Mdn1 has ~20–30% longer linker and D/E-rich domains compared to S. pombe Mdn1, while the lengths of the AAA and MIDAS domains are very similar (<2% difference). On the other hand, a much smaller Mdn1 ortholog is found in the eukaryotic parasite E. caniculi (2832 aa), in which the primary sequence suggests that the lengths of the linker and D/E-rich domains are reduced by ~60%, compared to S. pombe Mdn1, while the lengths of the AAA and MIDAS domains are reduced by only ~20% (Garbarino and Gibbons, 2002). These comparisons suggest that the linker and D/E-rich domains in Mdn1 are much more prone to genetic drift and tolerant to reductions in length, while there are more stringent constraints on the residues in the AAA and MIDAS domains, which are involved in specific intramolecular or intermolecular interactions critical for Mdn1 function (Figures 6). Our analysis also reveals that the length of the linker domain correlates with that of the D/E-rich domain, indicating that a longer linker domain may be compensated by a longer D/E-rich domain serving as a tether, and suggests that our proposed intramolecular docking mechanism for Mdn1 is likely conserved across eukaryotes.
Mdn1 Remodels Precursors of the 60S Ribosome
Our structures also suggest a simple model for how nucleotide-dependent conformational changes of Mdn1 contribute to ribosome biogenesis. In solution, Mdn1 could bind ATP and adopt conformations similar to that of AMPPNP-Mdn1. In this state, the MIDAS domain would sample different positions but not stably dock onto the AAA ring. ATP hydrolysis within the AAA2-AAA5 domains would drive displacements of rigid bodies 2 and 3 (Figure 7A). These movements would displace AAA4-L and its H2 insertion, resulting in a steric clash with Mdn1’s AAA2 H2 insertion, causing its reorganization such that it inserts into the central hole of the AAA ring. Together, these changes would create a site for the C-terminal MIDAS domain to dock onto the AAA ring (Figure 7A). This conformation is capable of stabilizing an interaction network formed by the core of the Rix1 particle, Rsa4 and Mdn1’s MIDAS domain and rigid bodies 2 and 3 (Figure 7B). It is also possible that Rsa4 and the pre-60S particle core can stimulate Mdn1’s activity and catalyze these conformational transitions. As the final step, additional ATP binding, hydrolysis or nucleotide release could reorganize these dynamic regions in Mdn1 and promote the breakdown of the interaction network in the Rix1 particle to release Rsa4 and Mdn1 (Figure 7B).
Figure 7. Model for the Function of Mdn1 During Ribosome Biogenesis.
(A) Schematics of two distinct Mdn1 conformations observed in this study. The linker, D/E-rich and MIDAS domains are indicated. Individual AAA domains are colored as in Figure 1A. The magenta arrows indicate the undefined location of the MIDAS domain in AMPPNP-Mdn1. Additional ATP binding and hydrolysis by Mdn1 can lead to additional conformational changes (dashed arrow). (B) Mdn1 adopts a conformation similar to ATPI-Mdn1 in the Rix1 particle. Additional ATP binding and hydrolysis can lead to local conformational changes in the AAA ring and displace the MIDAS domain to release Rsa4 from the Rix1 particle.
Our proposed model is consistent with previous biochemical assays showing that addition of ATP, but not AMPPNP, to affinity-purified Rix1 particle triggers the release of Mdn1 and Rsa4. It has also been shown that a Walker A lysine-to-alanine substitution at the AAA3 ATPase site does not affect the association of Mdn1 with the Rix1 particle but blocks the release of Mdn1/Rsa4 upon ATP addition (Barrio-Garcia et al., 2016). Interestingly, our data show that Mdn1 bound to the Rix1 particle adopts a conformation corresponding to ATPI-Mdn1, which has a wider opening in AAA3 compared to what is observed in AMPPNP-Mdn1 (Figure 4A), consistent with AAA3 being in the apo state. In addition, our data suggest that rigid bodies 2 and 3 in Mdn1, which flank the AAA3 ATPase site, would likely be displaced in the ATPase cycle. We speculate that the lack of ATP binding or hydrolysis in the AAA3 ATPase site of the AAA3 Walker A mutant restricts conformational changes of rigid bodies 2 and 3 needed for the release of Rsa4. In addition, we can use our structural model for Mdn1 to map the single-point mutations that alter the potency of Rbin-1 in fission yeast cells (Figure S7H) (Kawashima et al., 2016). Interestingly, seven different resistance-conferring mutations and one sensitivity-conferring mutation all lie in the AAA3-S and AAA4-L subdomains, suggesting that Rbin-1 directly or allosterically affects the conformational transitions of rigid body 3 in Mdn1.
Previous studies have revealed that the H2 insertion of AAA2 in Mdn1 interacts with the C-terminal sequence of Rix1 in a Rix1-Ipi1-Ipi3 complex and this interaction is required for the recruitment of Mdn1 to the Rix1 particle (Barrio-Garcia et al., 2016). Currently, it is not known whether this interaction exists in the Rix1 particle or is only established before the assembly of the Rix1 particle. Our structures reveal the dynamic nature of this H2 insertion in Mdn1 and suggest that its displacement could be coupled to its binding state with the C-terminal sequence of Rix1.
Studies in S. cerevisiae indicate that in addition to remodeling the Rix1 particle, Mdn1 also releases Ytm1 from nucleolar pre-60S intermediates. This remodeling step requires the interaction between Mdn1’s MIDAS domain and the N-terminal domain of Ytm1 (Bassler et al., 2010; Kater et al., 2017). A recent cryo-EM structure of the Nsa1 particle reveals the position of Ytm1 on the pre-ribosomes (Kater et al., 2017). Alignment of the Nsa1 particle to the Rix1 particle, using common motifs, indicates that Ytm1 binds to the core of the pre-60S particle at essentially the opposite side from where Rsa4 binds (separation ~25 nm) (Barrio-Garcia et al., 2016; Kater et al., 2017). However, Rea1 and the N-terminal domain of Ytm1 are not resolved in the Nsa1 particle (Kater et al., 2017). Therefore, at this stage, we do not know the precise interactions needed to remodel the Nsa1 particle or other nucleolar pre-ribosomes. It is likely that the MIDAS domain would dock on the AAA ring and then interact with the N-terminal domain of Ytm1, similar to the AAA ring-MIDAS-Rsa4 configuration in the Rix1 particle. This would suggest that Mdn1’s AAA ring has to be recruited proximal to Ytm1 in the nucleolar pre-60S particle. Further studies are needed to test this hypothesis.
Comparisons of Mdn1 to dynein and other AAA unfoldases
While the overall conformational dynamics of Mdn1 can be considered to be unlike what has been described for dynein, some common principles are apparent. Dynein, like Mdn1, has a pseudo-hexameric AAA ring and >10 nm protrusions extending from the ring, all within a single polypeptide (Schmidt and Carter, 2016). The ATPase activity of AAA1 in dynein is thought to drive local conformational changes of the AAA ring, and to create two alternative intramolecular binding sites for its cargo-binding domain (Schmidt and Carter, 2016). Our structural data reveal that, similar to dynein, Mdn1 also couples nucleotide hydrolysis at a subset of ATPase sites to create an intramolecular docking site for its MIDAS domain.
Studies of AAA proteins have revealed functional coupling between different AAA ATPase sites (Bhabha et al., 2014; DeWitt et al., 2015). We find that a mutation in AAA5 reduces the ATPase activity of Mdn1 to ~15% of the wild-type Mdn1. As Rbin-1 inhibits up to ~40% of the wild-type and the AAA5 mutant Mdn1’s ATPase activities, it is likely that Rbin-1 blocks the activity at AAA sites other than AAA5. Therefore, we suggest that AAA5 alone does not contribute to ~85% of Mdn1’s overall ATPase activity. We propose that similar to dynein’s AAA3 domain, which can allosterically regulate the ATPase activity at AAA1 (Bhabha et al., 2014; DeWitt et al., 2015), the nucleotide states of Mdn1’s AAA5 domain modulate the ATPase activity at other AAA sites.
Mdn1 also shares similarities with AAA unfoldases, such as Yme1 and Vps4. These unfoldases translocate different protein substrates through the central hole of the AAA ring to unfold them for further processing (e.g., proteolysis) (Monroe et al., 2017; Puchades et al., 2017). In the case of Mdn1, the central hole in its AAA ring is also occupied by a polypeptide whose insertion appears to be nucleotide state-dependent. However, unique to Mdn1, the inserted sequence does not derive from a different substrate protein, but is a segment of the same polypeptide. We posit that the occlusion of the central hole by the H2 insertion of AAA2 serves an auto-regulatory function. Together, our data reveal how conformational dynamics required for the functions of different AAA proteins can be combined in Mdn1, an unusually large protein, to remodel ribosome precursors.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tarun M. Kapoor (kapoor@rockefeller.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Insect cell culture
SF9 insect cells were cultured in Gibco sf-900 II SFM medium (Thermo Fisher 10902088) supplemented with 10% FBS (Thermo Fisher 10082139) and 1% Antibiotic-Antimycotic (Thermo Fisher 15240062). High Five insect cells were cultured in Express Five SFM (Thermo Fisher 10486025) supplemented with 1% Antibiotic-Antimycotic. Insect cells were maintained at 27 °C.
Yeast cell culture
The S. pombe strain SAK2189 (h+ leu1 mdn1-ts26<<hygr) (Kawashima et al. 2016) was transformed with wild-type or mutant pREP1NTAPΔNdeI-mdn1 plasmid. The cells were grown in minimum medium (EMM) supplemented with Leu at 25 °C (permissive temperature) or 36 °C (restrictive temperature).
Bacterial culture
The E. coli cells (BL21(DE3) chemically competent E. coli, Thermo Fisher C60600) used for the MIDAS domain overexpression were cultured in LB BROTH MILLER medium (FORMEDIUM LMM0102) at 37 °C to O.D ~ 0.8 before induction (at 18 °C for 18 hours) using 1 mM IPTG (Gold Biotechnology I2481C100).
METHOD DETAILS
Protein sequence alignment and comparison.
The sequences of Mdn1 orthologs (S. pombe Mdn1, Uniprot: O94248; S. cerevisiae Mdn1, Uniprot: Q12019; B. taurus Mdn1, Uniprot: E1BC24; H. sapiens Mdn1, Uniprot: Q9NU22) were aligned in ClustalW, which was also used to align and analyze the sequences of the AAA domains from S. pombe Mdn1 (Uniprot: O94248, aa 1–2196) and S. pombe dynein (Uniprot: O13290, aa 1852–4196).
Protein expression and purification.
Full-length S. pombe Mdn1 and Mdn1-ΔC were cloned into the pFastBac HTC vector (Thermo Fisher 10584027). Quick-change mutagenesis was used to generate E1637Q-Mdn1 (AAA5 Walker B mutant) using PfuUltra II polymerase (Agilent 600670). Mdn1 proteins were expressed in insect cells (High Five Cells) and purified as previously reported (Kawashima et al., 2016). Briefly, we used the Bac-to-Bac system (Thermo Fisher) to generate recombinant baculoviruses. High Five cells (Thermo Fisher B85502) were grown to ~2.5 million cells/mL in Express Five SFM (Thermo Fisher 10486025) and then infected (1:50 dilution of P2 virus). The cells were cultured in suspension at 27 °C and harvested at 48 hr after infection. All of the following steps were carried out on ice or at 4 °C. Cells were lysed by sonication in an equal volume of lysis buffer (50 mM Tris [pH 7.5], 400 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 200 μM ATP, 3 U/mL benzonase, 1× Roche complete protease inhibitor without EDTA, 10% glycerol). The homogenized lysate was then centrifuged at 60,000 rpm for 1 hr. The supernatant was incubated with Ni-NTA beads (QIAGEN) for 40 min. The beads were extensively washed using wash buffer (50 mM Tris [pH 7.5], 400 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 200 μM ATP, 10% glycerol). The protein was then eluted using high-imidazole buffer (20 mM Tris [pH 7.5], 120 mM NaCl, 300 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 200 μM ATP, 10% glycerol). The eluted fractions were filtered and loaded onto a Mono Q column 5/50 GL (GE Healthcare Life Sciences). The eluted fractions were collected and analyzed by SDS-PAGE. The relevant fractions were pooled and then concentrated using Amicon Ultra-4 Centrifugal Filter Units. The concentrated sample was then loaded onto a Superose 6 10/300 GL column (GE Healthcare Life Sciences) using the FPLC SEC buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 1mM EGTA, 50 μM ATP and 1 mM DTT). Typical yield of these preparations is ~ 0.1 mg/3 L culture. Freshly purified Mdn1 was used immediately for EM studies.
Recombinant wild-type MIDAS domain (aa 4381–4717) or 4A-MIDAS domain (aa 4685–4690: FAFDYY to AAADAA) were cloned into pET28 vector and overexpressed in E. coli. The cells were lysed by French press in lysis buffer (20 mL lysis buffer per 1 L culture; 50 mM Tris [pH 7.5], 150 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 3 U/mL benzonase, 1× Roche complete protease inhibitor without EDTA and 10% glycerol). All of the following steps were carried out on ice or at 4 °C. The lysate was centrifuged at 55,000 rpm for 30 min. The supernatant was incubated with Ni-NTA beads (QIAGEN) for 40 min. The beads were washed using wash buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 20 mM imidazole, 1 mM MgCl2, 5 mM 2-mercaptoethanol and 10% glycerol) and then eluted with 300 mM imidazole in the washing buffer. The eluted protein was concentrated using Amicon Ultra-4 Centrifugal Filter Units (Millipore UFC805008) and loaded on a Superdex 75 16/60 column (GE Healthcare Life Sciences) in size-exclusion buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM MgCl2 and 2 mM 2-mercaptoethanol). The eluted fractions were analyzed by SDS-PAGE and fractions corresponding to expected molecular weight (~42 kDa) were concentrated and loaded on a Superdex 75 10/300 column (GE Healthcare Life Sciences) using the same size-exclusion buffer.
NADH-coupled steady-state ATPase assay.
For each steady-state ATPase reaction, the final volume was 25 μL. The final concentration of E1637Q-Mdn1 (AAA5 Walker B mutant) was 40nM. Sodium sulfate, NADH (Sigma N7410), phosphoenol pyruvic acid monopotassium salt (Sigma P7127), D-lactic dehyrogenase (Sigma L3888) and pyruvate kinase (ammonium sulfate suspension, Sigma P1506) were added to final concentrations of 2 mM, 200 μM, 1.0 mM, 30 U/mL and 30 U/mL, respectively. The presence of 2 mM sodium sulfate enhances inhibition of Mdn1’s ATPase activity by Rbins. 1 μl DMSO or compound dissolved in DMSO was added and mixed. 5 μl 5 mM MgATP (pH = 7.0), was added to make the final total volume to be 25 μl and the final concentration of MgATP to be 1 mM. Time course of fluorescence (440 nm) decrease due to NADH oxidation was measured using a Synergy NEO Microplate Reader. The fluorescence values were plotted against time and fit by linear regression. The slopes of these lines were used to calculate the ATPase rate.
Negative-Stain EM sample preparation and processing.
The grids for negative staining were prepared by following a published protocol (Ohi et al., 2004). Freshly purified E1637Q-Mdn1 (AAA5 mutant) was mixed with 1 mM ATP and diluted to ~0.01 mg/mL before applying to the grid. The grid was stained with 0.7% uranyl formate (PFALTZ&BAUER, Inc. U01000). The images were then collected at room temperature with a Philips CM10 electron microscope equipped with a tungsten filament operating at 100 kV. All images were recorded on an AMT XR16L-ActiveVu charge-coupled device camera (Woburn, MA, USA) using a defocus of approximately −1.5 μm and a nominal magnification of 52000×.
Cryo-EM sample preparation and data collection.
To prepare cryo-EM grids of full-length Mdn1 with ATP and Rbin-1, the purified protein was concentrated to ~0.15 mg/mL using centrifugal filters (MW cutoff: 50 kDa; Millipore) and mixed with 10× (ATP+Rbin-1) stock solution (final concentration: 1 mM ATP, 1 μM Rbin-1, 2 mM Na2SO4 and 0.2% DMSO). Samples were applied to Quantifoil R1.2/1.3 400 mesh Cu holey carbon grids (Quantifoil) covered with a homemade thin carbon film, and then blotted and frozen in liquid ethane with a Cryoplunge 3 system (Gatan). Cryo-EM data were collected on a 200-kV Talos Arctica electron microscope (Thermo Fisher Scientific) equipped with a K2 Summit detector (Gatan) at a nominal magnification of 28,000× in super-resolution counting mode. After binning over 2 × 2 pixels, the calibrated pixel size was 1.5 Å on the specimen level. Exposures of 15 s were dose-fractionated into 50 frames with a dose rate of 10 electrons pixel−1 s−1, resulting in a total dose of 66.7 electrons Å−2. Defocus values ranging from −2.0 to −4.0 μm were used.
To prepare cryo-EM grids of full-length Mdn1 with AMPPNP and Mdn1-ΔC with AMPPNP, the purified protein was concentrated to ~0.15 mg/mL using centrifugal filters (MW cutoff: 50 kDa; Millipore) and mixed with 10× AMPPNP stock solution (final concentration: 1 mM AMPPNP). To minimize the potential hydrolysis of AMPPNP, AMPPNP was added to Mdn1 immediately before the grids were frozen (<10 min). Samples were applied to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil), blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data were collected on a 300-kV Titan Krios electron microscope (Thermo Fisher Scientific) equipped with a K2 Summit detector at a nominal magnification of 22,500× in super-resolution counting mode. After binning over 2 × 2 pixels, the calibrated pixel size was 1.3 Å on the specimen level. Exposures of 15 s were dose-fractionated into 50 frames with a dose rate of 10 electrons pixel−1 s−1, resulting in a total dose of 88.8 electrons Å−2. Defocus values ranging from −1.4 to −3.0 μm were used. Cryo-EM data collection statistics can be found in Table S1.
Cryo-EM data processing.
All movie frames were corrected with a gain reference collected in the same EM session, and specimen movement was corrected using MotionCor2 with dose weighting (Zheng et al., 2017). Images showing substantial ice contamination, abnormal background, thick ice or low contrast were discarded. The contrast transfer function (CTF) was estimated using CTFFIND4 (Rohou and Grigorieff, 2015). Approximately 10,000 particles were manually picked, subjected to 2D classification, and the resulting averages were used as templates for automatic particle picking using either RELION 1.4 for the pilot dataset of ATPI-Mdn1 only (Scheres, 2012) or Gautomatch for all other datasets (http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/). In particular, six representative 2D averages were rotated every 30° over 360° to generate seventy-two 2D views and then used as templates in Gautomatch. Particles selected for all micrographs were visually inspected, and poorly selected particles were manually adjusted or discarded. The final datasets for ATPI-Mdn1, AMPPNP-Mdn1 and AMPPNP-Mdn1-ΔC contained 247,841, 398,829, and 217,779 particles, respectively.
For ATPI-Mdn1, an initial pilot dataset containing 80,152 particles (box size: 256 × 256 pixels) was subjected to 2D classification using RELION 1.4 (Figure S5). Classes that produced poor averages were discarded. Averages showing representative views were used to generate an initial density map with the VIPER command implemented in SPARX (Hohn et al., 2007). The particles after 2D clean-up were then subjected to 3D classification into eight classes using the VIPER map as a reference. Subsequent refinement and post-processing of the best class yielded a map at ~17.5 Å resolution (FSC = 0.143). A mask big enough to cover all 3D classes was then generated and used to process the complete dataset. For the complete dataset, all particles (247,841 particles) were subjected to 2D classification in RELION 2.0 and poor averages were discarded (Kimanius et al., 2016). The remaining 210,668 particles were refined to the final map from the pilot dataset filtered to 25 Å resolution with the mask. A subsequent 3D classification into eight classes without alignment yielded one class (containing 54,557 particles) that showed better-resolved structural features. Refinement of this class yielded the final density map at a resolution of ~7.7 Å (FSC = 0.143, Figure S5D). Local-resolution map of ATPI-Mdn1 was calculated by ResMap in RELION 2.0 (Figure S5E). These local-resolution maps represent relative differences in resolution across the maps but the absolute values may not be exact.
For AMPPNP-Mdn1, an initial pilot dataset containing 60,632 particles (box size: 300 × 300 pixels) was subjected to 2D classification using RELION 1.4 (Figure S1B). Classes that produced poor averages were discarded. Subsequent 3D classification into five classes was carried out in RELION 2.0, using the ATPI-Mdn1 map filtered to 25 Å as the initial reference. The resulting class averages differed slightly in the angle between the ring region (up to the hinge in the tail) and the rest of the tail. The class average showing the clearest structural features was further refined, resulting in a density map at ~8.0 Å resolution (FSC = 0.143). This map was used to generate generous masks for the entire protein, the ring region, and the tail region. The full dataset containing 398,829 particles was processed using RELION 2.0 (Figure S1C). All particles after 2D clean-up (313,336 particles) were first refined against the final map obtained from the pilot dataset filtered to 25 Å resolution using the mask for the entire protein. Then, 3D classifications into six classes for the ring region and eight classes for the tail region were performed separately, using the local search option and the respective masks. In both cases, the best-resolved class accounts for ~30% of the total number of particles. Refinement of the best class for the ring region (containing 87,580 particles) produced a map at ~4.4 Å resolution, and refinement of the best class for the tail region (containing 94,649 particles) yielded a map at ~5.3 Å resolution (FSC = 0.143). We observed that the FSC curve for the tail region approached the 0.143 level (at ~5.9 Å) but then increased abruptly again before crossing the 0.143 level at a later point (at ~5.3 Å) (Figure S2A). This map was low-pass filtered to 5.9 Å and used in figures. The map of the ring region after post-processing was used as the reference for refinement of all particles after 2D clean-up (313,336 particles) in FREALIGN (Grigorieff, 2016), yielding the final map at a resolution of ~4.0 Å (FSC = 0.143) (Figure S2A). Local-resolution maps of the ring and tail regions of AMPPNP-Mdn1 were calculated by ResMap in RELION 2.0 (Figures S2B and S2F). These local-resolution maps represent relative differences in resolution across the maps but the absolute values may not be exact.
The AMPPNP-Mdn1-ΔC dataset was processed similarly as the full dataset of AMPPNP-Mdn1 in RELION 2.0 (Figure S3). All particles after autopicking (217,779 particles, box size: 300 × 300 pixels) were subjected to 2D classification. Classes generating poor averages were discarded, and the remaining 175,881 particles were refined to the AMPPNP-Mdn1 map filtered to 25 Å resolution using the mask for the entire protein. 3D classifications into five classes using the local search option were carried out separately for the ring and tail regions with the respective masks. Refinement of the best class for the ring region (containing 26,164 particles) produced a map at ~6.1 Å resolution, and refinement of the best class for the tail region (containing 22,921 particles) yielded a map at ~7.4 Å resolution (FSC = 0.143) (Figure S3F). We observed the FSC curve for the ring region approached the 0.143 level (at ~6.5 Å) but then increased abruptly again before crossing the 0.143 level at a later point (at ~6.1 Å) (Figure S3F). This map was low-pass filtered to 6.5 Å and used in figures. Local-resolution maps of the ring and tail regions of AMPPNP-Mdn1-ΔC were calculated by ResMap in RELION 2.0 (Figure S3G). These local-resolution maps represent relative differences in resolution across the maps but the absolute values may not be exact.
Model building and refinement.
The structural model for AMPPNP-Mdn1 was built in three steps: first, the AAA domains were built without the flexible loops and H2 insertions; then, the linker domain, and finally the N domain, subset of the loops where densities can be seen and H2 insertions were built. Phyre2 was used to calculate initial homology models of the AAA domains (Kelley et al., 2015). Flexible loops as well as the H2 insertions were removed from the homology models, and the AAA domains were split into their large and small subdomains. These subdomains were fit into the ring region of the ~4.0-Å resolution AMPPNP-Mdn1 map using Chimera (Pettersen et al., 2004). Since most side chains were not resolved in the cryo-EM density map, we removed all side chains beyond the β-carbons using the phenix.pdbtools command in PHENIX (Adams et al., 2010). The β-carbons and sequence information was preserved to constrain further refinement using the phenix.real_space_refine command in PHENIX with morphing, simulated annealing and secondary structure restrains.
For the density of the hook-shaped tail in AMPPNP-Mdn1, α-helical segments were first identified using the phenix.find_helices_strands command in PHENIX. Polyalanine α-helices were built into the density, which accounted for most, if not all, of the tail density. Guided by the secondary structure predictions from SPIDER2 (Yang et al., 2017) and PSIPRED (Buchan et al., 2013), we were able to trace the backbone and to establish the connectivity of the α-helices from the C-terminal end of the AAA6-L domain up to the hinge. Due to the ambiguity in the helical registry, we only built a polyalanine model for the traced backbone. For the cryo-EM map of the rest of the tail, i.e., the C-terminal density after the hinge, which was masked and refined separately and resolved at ~5.9 Å, we used the identified polyalanine helices without connecting loops to present the overall architecture of the helical bundles (as shown in Figure 1C).
For the N domain, loops and H2 insertions, we first combined the models of the AAA domains (the backbone model with β-carbons) and the linker domain up to the hinge (the polyalanine model) and then focused on unassigned densities for the ring region. Ab initio model building for the N domain, loops and H2 insertions was carried out in COOT (Emsley et al., 2010), guided by secondary-structure predictions. For regions in which bulky side chains were resolved as bumps (as shown in Figure S2C), we used these residues as anchor points during model building. Further refinement using the phenix.real_space_refine command in PHENIX with secondary structure restrains and manual adjustments in COOT were performed iteratively, yielding our final model for the N-terminal density. The refinement statistics are summarized in Supplementary Table S1. For model validation, the final model for the N-terminal region (up to the hinge) was refined against one of the independent half maps (work) of the corresponding ~4.0 Å resolution map. FSC curves were then calculated between the refined model and half map 1 (work), half map 2 (free) as well as the combined map (Figure S2E).
To interpret the map for ATPI-Mdn1, the AMPPNP-Mdn1 model was divided into the N domain, the individual AAA subdomains and the linker domain before the hinge. Using Chimera, the models for these domains were individually fit as rigid bodies into the ATPI-Mdn1 density map. The entire AAA6-L subdomain, including its H2 insertion, and the H2 insertion of AAA4 from AMPPNP-Mdn1 fit well into the ATPI-Mdn1 map. The model for the H2 insertion of AAA2 in AMPPNP-Mdn1 could not be readily fit and was therefore not modeled in the ATPI-Mdn1 map. The models for the different domains were then combined, and rigid-body refinement was performed using the phenix.real_space_refine command in PHENIX with secondary structure restraints.
Models for the MIDAS domain were built using RosettaCM (Song et al., 2013). Homology modeling was carried out using seven templates (PDB ID: 1AUQ, 1IJB, 1MF7, 1SHU, 4HQO, 5HGJ, and 5HJ2) identified by hhsearch (Soding, 2005). For each template, a “partial thread” was generated, in which residue identities were changed to the target sequence but the backbone was kept fixed. Residues not present in each template were not modeled. To determine the overall orientation of these models within the target density in the ATPI-Mdn1 map, a rotational search was carried out in Rosetta around this density region, using a modified version of a previously developed fragment-docking method (Wang et al., 2015). For each of the seven templates, the rotational search identified a single orientation with much better density agreement than any other. Rosetta-based homology modeling was then carried out, combining pieces from the seven homologous structures and refining the models with the Rosetta all-atom energy function that was augmented with a term assessing the agreement of the model to the density. In total, 1500 models were generated, and the models with the lowest overall energy were selected. The five lowest-energy models were quite well converged (inter-structure root-mean-square deviations < 1.5 Å), giving fairly high confidence in the final models.
Fitting of the ATPI-Mdn1 model into the Rix1-particle map.
As shown in Figures 6A-C, the model for ATPI-Mdn1 and the crystal structure of Rsa4 (cyan, C. thermophilum, PDB ID: 4WJS) were docked into the published density map of the Rix1 particle (gray surface, EMDB ID: 3199). The docking of Rsa4 was guided by the reported model for the Rix1 particle (PDB ID: 5FL8) that was based on the same map (EMDB ID: 3199). A slight adjustment was necessary to prevent inappropriate fitting of the Rsa4 structure into a previously unassigned density, which we assigned, based on our model in the current work, as the MIDAS domain of Mdn1.
Calculation of surface conservation with the ConSurf server.
The Clustal Omega server was used to align the Mdn1 orthologs from sixteen species (Schizosaccharomyces pombe Mdn1, Uniprot ID: O94248; Saccharomyces cerevisiae Mdn1, Uniprot ID: Q12019; Candida albicans Mdn1, Uniprot ID: A0A1D8PL61; Komagataella phaffii Mdn1, Uniprot ID: F2QMK3; Arabidopsis thaliana Mdn1, Uniprot ID: A0A1P8AUY4; Dictyostelium discoideum Mdn1, Uniprot ID: Q869L3; Caenorhabditis elegans Mdn1, NCBI accession: NP_001263748.1; Drosophila melanogaster Mdn1, NCBI accession: NP_001097279.2; Danio rerio Mdn1, NCBI accession: NP_003200751.2; Xenopus tropicalis Mdn1, NCBI accession: NP_002940362.2; Anolis carolinensis Mdn1, Uniprot ID: G1KPU9; Equus caballus Mdn1, Uniprot ID: F6XJW2; Bos taurus Mdn1, Uniprot ID: E1BC24; Canis lupus Mdn1, Uniprot ID: J9NZY7; Mus musculus Mdn1, Uniprot ID: A2ANY6; Homo sapiens Mdn1, Uniprot ID: Q9NU22). The alignment was then analyzed with the ConSurf server to determine the conservation values for the individual residues (Ashkenazy et al., 2016). The output PDB file, which contains the conservation scores as B-factor values, was then displayed in PYMOL.
Yeast complementation assay.
The yeast complementation assays were performed based on a previously described protocol (Kawashima et al., 2016). Mutations were introduced into N (1–8528 bp) or C (8529–14154 bp) fragments of the mdn1 gene using PrimeSTAR Mutagenesis Basal Kit (TAKARA). N and C fragments were ligated to generate the full-length mdn1 mutant genes, which were cloned into pREP1NTAPΔNdeI vector. The SAK2189 (h+ leu1 mdn1-ts26<<hygr) strain was transformed with wild-type or mutant pREP1NTAPΔNdeI-mdn1 plasmid, and grown on EMM-Leu plate containing 30 μM thiamine at 25 °C. Colony PCR and DNA sequencing was carried out to confirm that transformants harbored the correct plasmid DNA. 5 mL EMM-Leu pre-cultures containing 30 μM thiamine were incubated overnight at 25 °C till the OD (590 nm) was ~0.5. Cells were washed with doubly distilled water to remove thiamine, followed by resuspension in thiamine-free EMM-Leu medium to OD (590 nm) = 0.01 and the volume adjusted to 2 mL. These cultures were then incubated for 24 h at 36 °C and OD (590 nm) values were measured.
QUANTIFICATION AND STATISTICAL ANALYSIS.
The number of replicates of biochemical assays are indicated in corresponding figure legends. Cryo-EM data collection and refinement statistics can be found in Supplementary Table S1. Details of cryo-EM data analyses are provided in METHOD DETAILS.
DATA AND SOFTWARE AVAILABILITY.
The accession numbers for the model of ring region of AMPPNP-Mdn1 and the fitted model for ATPI-Mdn1 reported in this paper are PDB: 6EDO and 6EES, respectively. The accession numbers for the corresponding cryo-EM maps of the ring region of AMPPNP-Mdn1, the tail region of AMPPNP-Mdn1, the ring region of AMPPNP-Mdn1-ΔC, the tail region of AMPPNP-Mdn1-ΔC, and ATPI-Mdn1 reported in this paper are EMDB: EMD-9032, EMD-9033, EMD- 9034, EMD-9035, and EMD-9036, respectively.
Supplementary Material
(A) A cryo-EM image of AMPPNP-Mdn1 processed with MotionCor2. Some Mdn1 particles are marked (white circles). Scale bar: 20 nm. (B) The workflow used to process the pilot dataset. 3D classification revealed structural variability due to movement around a hinge point (red arrows). After refinement of the class showing the most structural detail, the final map was used to generate masks for the entire elongated molecule, as well as for the AAA ring region (N-terminal of the hinge point) and tail region alone (C-terminal of the hinge point). The density map was also used as the initial reference for processing the complete dataset. (C) The workflow used to process the complete dataset. The angular distributions of the particles in the final reconstructions are shown next to the corresponding maps. Note: the resolution of the final reconstruction is further discussed in Figure S2A.
The fitting of the model into the Rix1 particle map (grey surface, EMDB ID: 3199) contoured at a threshold at which the density extending from the ring region of Rea1 is observed. The models for AAA core (blue ribbon), the linker (purple ribbon), the MIDAS (magenta ribbon) domains of Mdn1 and the Rsa4 (cyan ribbon) are shown.
(A) Two views of the ATPI-Mdn1 map (yellow surface) with the polyalanine-helix model for the linker domain (purple ribbon representation) generated based on the AMPPNP-Mdn1 map. A density not accounted for by the polyalanine-helix model is highlighted (dashed circle). (B) Individual rigid bodies and AAA1-L in AMPPNP-Mdn1 (orange cartoon) and ATPI-Mdn1 (blue cartoon) are aligned based on the large subdomains. Note that no substantial differences in the conformations of individual rigid bodies are found when comparing AMPPNP-Mdn1 and ATPI-Mdn1. Note that the rigid bodies 2 and 3 are displaced when the two models are aligned using AAA1-L as shown in Figure 4C.
(A) The model for the AAA4 H2 insertion generated based on the AMPPNP-Mdn1 map (blue ribbon) was fit into the ATPI-Mdn1 map (yellow surface). (B) Alignment of the models of AAA4-L with its H2 insertion from the structures of AMPPNP-Mdn1 (orange ribbon) and ATPI-Mdn1 (blue ribbon). (C) Potential steric clashes between the H2 insertions of AAA2 and AAA4 in ATPI-Mdn1 if the AAA2 H2 insertion adopts the same conformation as in AMPPNP-Mdn1. The model for AAA2-L with its H2 insertion from AMPPNP-Mdn1 (orange ribbon) was aligned to the model from ATPI-Mdn1 based on the AAA2-L subdomain. The model for the AAA4 H2 insertion from the ATPI-Mdn1 structure is also shown (blue). (D) Two views of the AAA ring in the model for AMPPNP-Mdn1 (orange ribbon), with AAA2 H2 insertion (green ribbon) highlighted. Conserved aromatic residues (black spheres) near the beginning of the H2 helix in Mdn1's AAA domains (also labeled in Data S1). All of these conserved residues are close to the central hole of the AAA ring. (E) Model for the AAA ring (blue ribbon) of ATPI-Mdn1 and density representing the AAA2 H2 insertion and the MIDAS domain (yellow surface). The conserved Tyr-198 residue is in direct contact with the density assigned as the AAA2 H2 insertion (as shown in Figure 5B, density below the constriction indicated by the asterisk). (F) Docking of the AMPPNP- and ATPI-Mdn1 models generated in this work into the density map of the Rix1 particle (EMDB ID: 3199). (G) Overlay of the size-exclusion chromatography profiles for the wild-type MIDAS domain (aa 4381–4717) and 4A-MIDAS domain (aa 4381–4717, loop 4685–4690 mutation: FAFDYY to AAADAA). The elution volumes of both proteins are indicated for comparison. The SDS-PAGE analysis (Coomassie blue stain) of WT-MIDAS and 4A-MIDAS are shown in the inset. The major bands match the expected molecular weight of the MIDAS domain. (H) The α carbons of residues that can suppress or sensitize the activity of Rbin-1 in cells are highlighted (green and red spheres, respectively) in the ATPI-Mdn1 model. Rigid bodies 2 and 3 are outlined (dashed triangles).
The sequences were aligned and displayed in Jalview (Waterhouse et al., 2009). Domain boundaries and selected structural and catalytic motifs are marked. The numbers following the Walker A, Walker B and arginine finger motifs indicate the AAA domain in which they are located. The aromatic-rich motifs (Ar motifs) are also shown. Secondary-structure elements in S. pombe Mdn1 were predicted using SPIDER2 (Yang et al., 2017) and the potential α-helices (black cylinders) and β-strands (grey arrows) are indicated above the S. pombe sequence. Residues that are important for the interaction between the MIDAS domain and Rsa4 in S. cerevisiae are boxed and labeled using red dots above the sequence (Asp-4511, Ser-4513, Ser-4515, Thr-4582 and Asp-4619 in S. pombe Mdn1) (Ulbrich et al., 2009).
TABLE S1. Cryo-EM Data Collection and Refinement Statistics, Related to Star Methods Section.
The fitting of the AMPPNP-Mdn1 model for the ring region (blue ribbon, comprised of the N and AAA domains) and tail region (purple ribbon, the linker domain) into the AMPPNP-Mdn1 density (grey surface) is shown.
(A) Gold-standard Fourier Shell Correlation (FSC) curves calculated between independently refined half maps (each half map was reconstructed using randomly selected 50% of the particles) for the N- and C-terminal regions of AMPPNP-Mdn1 relative to the hinge point. For the map of the C-terminal density, the resolution was estimated as ~5.9 Å, more conservatively than the value suggested by the FSC = 0.143 criterion (~5.3 Å). (B) Local-resolution map of the N-terminal density that includes the AAA ring calculated by ResMap in RELION 2.0. (C) Representative EM densities are shown (grey mesh) for the map of the N-terminal AMPPNP-Mdn1 region, together with the built model (green ribbons). The β-carbons are shown for selected side chains. (D) The densities corresponding to the nucleotides in AAA4 and AAA6 of AMPPNP-Mdn1 are shown (grey mesh), together with the modeled AMPPNP molecules (sticks). The β-carbons of the key lysine residues in the Walker A motifs in AAA4 of Mdn1 (K1199) and AAA1 of dynein (K1882) are shown. The alpha-carbons of residues in the N loops that are important for the interaction with the adenine ring of the nucleotides, are also shown. For comparison, the box to the right shows ADP bound to AAA1 in D. discoideum dynein (PDB: 3VKG) (Kon et al., 2012). (E) Validation of the model for the N-terminal ring region of AMPPNP-Mdn1 (as also shown in Figures 1C and 2A) by cross-validation FSC curves: “work” (blue curve), refined model versus half map 1 used for refinement (work map); “free” (red curve), refined model versus half map 2 that was not used for refinement (free map); “combined” (black), refined model versus the combined final map. The similarity of the “work” and “free” curves suggests no substantial over-fitting. (F) Local-resolution map of the density of the C-terminal tail region of AMPPNP-Mdn1 calculated by ResMap in RELION 2.0.
The fitting of the ATPI-Mdn1 model for the ring region (blue ribbon, comprised of the N and AAA domains), tail region (purple ribbon, the linker domain) and MIDAS domain (magenta ribbon) into the ATPI-Mdn1 density (yellow surface) is shown.
(A) Schematic of the Mdn1-ΔC construct that lacks the C-terminal D/E-rich and MIDAS domains (aa 1–3911). The D/E-rich domain starts at aa 3912. (B) Size-exclusion chromatography profile for Mdn1-ΔC (elution volume: 11.5 mL). The elution volumes of the void peak (7.5 mL) and full-length Mdn1 (Mdn1-FL, 10.6 mL) are also indicated for comparison. (C) Peak fractions from the size-exclusion chromatography of Mdn1-ΔC and Mdn1-FL were analyzed by SDS-PAGE (Coomassie blue stain). (D) A cryo-EM image of AMPPNP-Mdn1-ΔC processed with MotionCor2. Some Mdn1-ΔC particles are marked (white circles). Contaminant particles are also marked (red circles). Scale bar: 20 nm. (E) Workflow used to process the dataset. Classes representing contaminant particles (red circles) were removed from the dataset. The angular distributions of the particles in the final reconstructions are shown next to the corresponding maps. Note: the resolution of the final reconstructions is further discussed in (F). (F) Gold-standard FSC curve calculated between independently refined half maps for the N- and C-terminal regions of AMPPNP-Mdn1-ΔC. For the map of the N-terminal density, the resolution was estimated as ~6.5 Å, more conservatively than the value suggested by the FSC = 0.143 criterion (~6.1 Å). (G) Local-resolution maps for the N- and C-terminal regions of Mdn1-ΔC calculated by ResMap in RELION 2.0. (H) Two views of the EM density map of Mdn1-ΔC (yellow surface). The Mdn1 model for full-length Mdn1 (ribbons, colored blue and purple for the AAA ring and hook-shaped tail regions, respectively) could be placed into the Mdn1-ΔC map without further refinement.
The linear morph of the core AAA domain between the models for AMPPNP-Mdn1 and ATPI-Mdn1 is shown. The AAA1L (green ribbon), which was used to align those two models, and rigid bodies 2 and 3 (blue ribbon), which undergo substantial displacements, are highlighted.
(A) Schematic of secondary-structure elements in two sequential AAA domains and the arrangement of their subdomains. α-Helices (grey cylinders) and β-strands (yellow arrows) are shown. The N and C termini of the first AAA domain (solid red linkers) and those of the second AAA domain (faded red linkers) are shown. The dashed box indicates rigid body i, which is formed by subdomains AAAi-S and AAA(i+1)-L. Helices 0 (H0), 1 (H1) and 2 (H2) are the first three helices in the large subdomain. The H2 insertions, which are present in all six AAA-L domains of Mdn1 but have different lengths (15–120 aa), are shown as green rectangles. The aromatic motif at the beginning of H2 helix is also labeled (blue bar). These motifs are also labeled in the alignment of Mdn1 orthologs' sequences in Data S1. (B) Two views of the 'ring-linker junction' motif that connects the tail to the AAA ring in AMPPNP-Mdn1 are shown (ribbon representation). The N, AAA1-L, and AAA6-L domains and part of the linker domain are colored as in Figure 2A. (C-H) The density map of each AAA domain (grey mesh) is shown with the built model (ribbon representation). The nucleotides (stick representation) bound between the large and small subdomains of AAA1–4 and next to the AAA6-L subdomain are shown. All nucleotide densities are modeled as AMPPNP molecules. The models are colored as in Figure 1A. Note that AAA5 shows no density for a nucleotide. (I) Negative-stain EM image of Mdn1 carrying the E1637Q mutation in the Walker B motif of AAA5. Scale bar: 20 nm.
The fitting of the ATPI-Mdn1 model into the Rix1 particle map (grey surface, EMDB ID: 3199) contoured at a threshold at which the map density matches the size of the model for AAA core (blue ribbon) with the docked MIDAS domain (magenta ribbon) of Mdn1. The models for the linker domain of Mdn1 (purple ribbon) and Rsa4 (cyan ribbon) are also shown.
(A) A cryo-EM image of ATPI-Mdn1 processed with MotionCor2. Some Mdn1 particles are marked with white circles. Scale bar: 20 nm. (B) The workflow used to process the pilot dataset. (C) The workflow used to process the complete dataset. The angular distribution of the particles in the final reconstruction is shown next to the final map. (D) Gold-standard FSC curve calculated between independently refined half maps for ATPI-Mdn1. The resolution was estimated as ~7.7 Å (FSC = 0.143). (E) Local-resolution map calculated by ResMap in RELION 2.0.
ACKNOWLEDGEMENTS.
We thank Mark Ebrahim and Johanna Sotiris at the Evelyn Gruss Lipper Cryo-EM Resource Center at Rockefeller University for assistance in data collection. We thank Zongli Li and Melissa Chambers for help with early negative-stain EM work. We thank Yixiao Zhang and Hyojin Kim from the Walz laboratory for advice on sample preparation and data processing. We also thank Nathan J. Harper for help in preparing MIDAS domain constructs. This work is supported by JSPS KAKENHI Grant Number JP18K19138 (to S.A.K.), the Robertson Foundation (to T.M.K.) and NIH/NIGMS GM98579 (to T.M.K.). Z.C. was also supported by the Tri-institutional Program in Chemical Biology and the Rockefeller University Graduate Program.
Footnotes
DECLARATION OF INTERESTS.
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic acids research 44, W344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Bassler J, Beckmann R, and Hurt E (2016). Architecture of the Rix1-Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nature structural & molecular biology 23, 37–44. [DOI] [PubMed] [Google Scholar]
- Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, and Hurt E (2010). The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Molecular cell 38, 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Cheng HC, Zhang N, Moeller A, Liao M, Speir JA, Cheng Y, and Vale RD (2014). Allosteric communication in the dynein motor domain. Cell 159, 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, and Jones DT (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic acids research 41, W349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, and Yildiz A (2015). The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nature structural & molecular biology 22, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta crystallographica Section D, Biological crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, and Berger JM (2006). Evolutionary relationships and structural mechanisms of AAA+ proteins. Annual review of biophysics and biomolecular structure 35, 93–114. [DOI] [PubMed] [Google Scholar]
- Garbarino JE, and Gibbons IR (2002). Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC genomics 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, and Sauer RT (2009). Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2016). Frealign: an exploratory tool for single-particle cryo-EM. Methods in enzymology 579, 191–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, and Whiteheart SW (2005). AAA+ proteins: have engine, will work. Nature reviews Molecular cell biology 6, 519–529. [DOI] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, and Ludtke SJ (2007). SPARX, a new environment for cryo-EM image processing. Journal of structural biology 157, 47–55. [DOI] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio-Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I, et al. (2017). Visualizing the assembly pathway of nucleolar pre-60S ribosomes. Cell 171, 1599–1610e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Chen Z, Aoi Y, Patgiri A, Kobayashi Y, Nurse P, and Kapoor TM (2016). Potent, reversible, and specific chemical inhibitors of eukaryotic ribosome biogenesis. Cell 167, 512–524e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, and Kurisu G (2012). The 2.8 Å crystal structure of the dynein motor domain. Nature 484, 345–350. [DOI] [PubMed] [Google Scholar]
- Konikkat S, and Woolford JL Jr. (2017). Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. The Biochemical journal 474, 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bergler H, and Bassler J (2012). The power of AAA-ATPases on the road of pre-60S ribosome maturation molecular machines that strip pre-ribosomal particles. Biochimica et biophysica acta 1823, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, and Sauer RT (2005). Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, and Hurt E (2014). Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Lander GC, and Martin A (2013). Conformational switching of the 26S proteasome enables substrate degradation. Nature structural & molecular biology 20, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe N, Han H, Shen PS, Sundquist WI, and Hill CP (2017). Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MP, Berger F, Rao L, Brenner S, Cho C, and Gennerich A (2015). Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proceedings of the National Academy of Sciences of the United States of America 112, 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, and Hurt E (2004). A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Molecular cell 15, 295–301. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, and Walz T (2004). Negative staining and image classification – powerful tools in modern electron microscopy. Biological procedures online 6, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera – a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, Glynn SE, and Lander GC (2017). Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, and Rubinstein JL (2017). Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, and Carter AP (2016). Structure and mechanism of the dynein motor ATPase. Biopolymers 105, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J (2005). Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960. [DOI] [PubMed] [Google Scholar]
- Song G, Yang Y, Liu JH, Casasnovas JM, Shimaoka M, Springer TA, and Wang JH (2005). An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin αLβ2. Proceedings of the National Academy of Sciences of the United States of America 102, 3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, and Baker D (2013). High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA (2001). The myosin swinging cross-bridge model. Nature reviews Molecular cell biology 2, 387–392. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, and Hurt E (2009). Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138, 911–922. [DOI] [PubMed] [Google Scholar]
- Wang RY, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, and DiMaio F (2015). De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature methods 12, 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr., and Baserga SJ (2013). Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. (2016). Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, Sattar A, and Zhou Y (2017). SPIDER2: a package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. Methods in molecular biology 1484, 55–63. [DOI] [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, and Penczek PA (2012). Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A cryo-EM image of AMPPNP-Mdn1 processed with MotionCor2. Some Mdn1 particles are marked (white circles). Scale bar: 20 nm. (B) The workflow used to process the pilot dataset. 3D classification revealed structural variability due to movement around a hinge point (red arrows). After refinement of the class showing the most structural detail, the final map was used to generate masks for the entire elongated molecule, as well as for the AAA ring region (N-terminal of the hinge point) and tail region alone (C-terminal of the hinge point). The density map was also used as the initial reference for processing the complete dataset. (C) The workflow used to process the complete dataset. The angular distributions of the particles in the final reconstructions are shown next to the corresponding maps. Note: the resolution of the final reconstruction is further discussed in Figure S2A.
The fitting of the model into the Rix1 particle map (grey surface, EMDB ID: 3199) contoured at a threshold at which the density extending from the ring region of Rea1 is observed. The models for AAA core (blue ribbon), the linker (purple ribbon), the MIDAS (magenta ribbon) domains of Mdn1 and the Rsa4 (cyan ribbon) are shown.
(A) Two views of the ATPI-Mdn1 map (yellow surface) with the polyalanine-helix model for the linker domain (purple ribbon representation) generated based on the AMPPNP-Mdn1 map. A density not accounted for by the polyalanine-helix model is highlighted (dashed circle). (B) Individual rigid bodies and AAA1-L in AMPPNP-Mdn1 (orange cartoon) and ATPI-Mdn1 (blue cartoon) are aligned based on the large subdomains. Note that no substantial differences in the conformations of individual rigid bodies are found when comparing AMPPNP-Mdn1 and ATPI-Mdn1. Note that the rigid bodies 2 and 3 are displaced when the two models are aligned using AAA1-L as shown in Figure 4C.
(A) The model for the AAA4 H2 insertion generated based on the AMPPNP-Mdn1 map (blue ribbon) was fit into the ATPI-Mdn1 map (yellow surface). (B) Alignment of the models of AAA4-L with its H2 insertion from the structures of AMPPNP-Mdn1 (orange ribbon) and ATPI-Mdn1 (blue ribbon). (C) Potential steric clashes between the H2 insertions of AAA2 and AAA4 in ATPI-Mdn1 if the AAA2 H2 insertion adopts the same conformation as in AMPPNP-Mdn1. The model for AAA2-L with its H2 insertion from AMPPNP-Mdn1 (orange ribbon) was aligned to the model from ATPI-Mdn1 based on the AAA2-L subdomain. The model for the AAA4 H2 insertion from the ATPI-Mdn1 structure is also shown (blue). (D) Two views of the AAA ring in the model for AMPPNP-Mdn1 (orange ribbon), with AAA2 H2 insertion (green ribbon) highlighted. Conserved aromatic residues (black spheres) near the beginning of the H2 helix in Mdn1's AAA domains (also labeled in Data S1). All of these conserved residues are close to the central hole of the AAA ring. (E) Model for the AAA ring (blue ribbon) of ATPI-Mdn1 and density representing the AAA2 H2 insertion and the MIDAS domain (yellow surface). The conserved Tyr-198 residue is in direct contact with the density assigned as the AAA2 H2 insertion (as shown in Figure 5B, density below the constriction indicated by the asterisk). (F) Docking of the AMPPNP- and ATPI-Mdn1 models generated in this work into the density map of the Rix1 particle (EMDB ID: 3199). (G) Overlay of the size-exclusion chromatography profiles for the wild-type MIDAS domain (aa 4381–4717) and 4A-MIDAS domain (aa 4381–4717, loop 4685–4690 mutation: FAFDYY to AAADAA). The elution volumes of both proteins are indicated for comparison. The SDS-PAGE analysis (Coomassie blue stain) of WT-MIDAS and 4A-MIDAS are shown in the inset. The major bands match the expected molecular weight of the MIDAS domain. (H) The α carbons of residues that can suppress or sensitize the activity of Rbin-1 in cells are highlighted (green and red spheres, respectively) in the ATPI-Mdn1 model. Rigid bodies 2 and 3 are outlined (dashed triangles).
The sequences were aligned and displayed in Jalview (Waterhouse et al., 2009). Domain boundaries and selected structural and catalytic motifs are marked. The numbers following the Walker A, Walker B and arginine finger motifs indicate the AAA domain in which they are located. The aromatic-rich motifs (Ar motifs) are also shown. Secondary-structure elements in S. pombe Mdn1 were predicted using SPIDER2 (Yang et al., 2017) and the potential α-helices (black cylinders) and β-strands (grey arrows) are indicated above the S. pombe sequence. Residues that are important for the interaction between the MIDAS domain and Rsa4 in S. cerevisiae are boxed and labeled using red dots above the sequence (Asp-4511, Ser-4513, Ser-4515, Thr-4582 and Asp-4619 in S. pombe Mdn1) (Ulbrich et al., 2009).
TABLE S1. Cryo-EM Data Collection and Refinement Statistics, Related to Star Methods Section.
The fitting of the AMPPNP-Mdn1 model for the ring region (blue ribbon, comprised of the N and AAA domains) and tail region (purple ribbon, the linker domain) into the AMPPNP-Mdn1 density (grey surface) is shown.
(A) Gold-standard Fourier Shell Correlation (FSC) curves calculated between independently refined half maps (each half map was reconstructed using randomly selected 50% of the particles) for the N- and C-terminal regions of AMPPNP-Mdn1 relative to the hinge point. For the map of the C-terminal density, the resolution was estimated as ~5.9 Å, more conservatively than the value suggested by the FSC = 0.143 criterion (~5.3 Å). (B) Local-resolution map of the N-terminal density that includes the AAA ring calculated by ResMap in RELION 2.0. (C) Representative EM densities are shown (grey mesh) for the map of the N-terminal AMPPNP-Mdn1 region, together with the built model (green ribbons). The β-carbons are shown for selected side chains. (D) The densities corresponding to the nucleotides in AAA4 and AAA6 of AMPPNP-Mdn1 are shown (grey mesh), together with the modeled AMPPNP molecules (sticks). The β-carbons of the key lysine residues in the Walker A motifs in AAA4 of Mdn1 (K1199) and AAA1 of dynein (K1882) are shown. The alpha-carbons of residues in the N loops that are important for the interaction with the adenine ring of the nucleotides, are also shown. For comparison, the box to the right shows ADP bound to AAA1 in D. discoideum dynein (PDB: 3VKG) (Kon et al., 2012). (E) Validation of the model for the N-terminal ring region of AMPPNP-Mdn1 (as also shown in Figures 1C and 2A) by cross-validation FSC curves: “work” (blue curve), refined model versus half map 1 used for refinement (work map); “free” (red curve), refined model versus half map 2 that was not used for refinement (free map); “combined” (black), refined model versus the combined final map. The similarity of the “work” and “free” curves suggests no substantial over-fitting. (F) Local-resolution map of the density of the C-terminal tail region of AMPPNP-Mdn1 calculated by ResMap in RELION 2.0.
The fitting of the ATPI-Mdn1 model for the ring region (blue ribbon, comprised of the N and AAA domains), tail region (purple ribbon, the linker domain) and MIDAS domain (magenta ribbon) into the ATPI-Mdn1 density (yellow surface) is shown.
(A) Schematic of the Mdn1-ΔC construct that lacks the C-terminal D/E-rich and MIDAS domains (aa 1–3911). The D/E-rich domain starts at aa 3912. (B) Size-exclusion chromatography profile for Mdn1-ΔC (elution volume: 11.5 mL). The elution volumes of the void peak (7.5 mL) and full-length Mdn1 (Mdn1-FL, 10.6 mL) are also indicated for comparison. (C) Peak fractions from the size-exclusion chromatography of Mdn1-ΔC and Mdn1-FL were analyzed by SDS-PAGE (Coomassie blue stain). (D) A cryo-EM image of AMPPNP-Mdn1-ΔC processed with MotionCor2. Some Mdn1-ΔC particles are marked (white circles). Contaminant particles are also marked (red circles). Scale bar: 20 nm. (E) Workflow used to process the dataset. Classes representing contaminant particles (red circles) were removed from the dataset. The angular distributions of the particles in the final reconstructions are shown next to the corresponding maps. Note: the resolution of the final reconstructions is further discussed in (F). (F) Gold-standard FSC curve calculated between independently refined half maps for the N- and C-terminal regions of AMPPNP-Mdn1-ΔC. For the map of the N-terminal density, the resolution was estimated as ~6.5 Å, more conservatively than the value suggested by the FSC = 0.143 criterion (~6.1 Å). (G) Local-resolution maps for the N- and C-terminal regions of Mdn1-ΔC calculated by ResMap in RELION 2.0. (H) Two views of the EM density map of Mdn1-ΔC (yellow surface). The Mdn1 model for full-length Mdn1 (ribbons, colored blue and purple for the AAA ring and hook-shaped tail regions, respectively) could be placed into the Mdn1-ΔC map without further refinement.
The linear morph of the core AAA domain between the models for AMPPNP-Mdn1 and ATPI-Mdn1 is shown. The AAA1L (green ribbon), which was used to align those two models, and rigid bodies 2 and 3 (blue ribbon), which undergo substantial displacements, are highlighted.
(A) Schematic of secondary-structure elements in two sequential AAA domains and the arrangement of their subdomains. α-Helices (grey cylinders) and β-strands (yellow arrows) are shown. The N and C termini of the first AAA domain (solid red linkers) and those of the second AAA domain (faded red linkers) are shown. The dashed box indicates rigid body i, which is formed by subdomains AAAi-S and AAA(i+1)-L. Helices 0 (H0), 1 (H1) and 2 (H2) are the first three helices in the large subdomain. The H2 insertions, which are present in all six AAA-L domains of Mdn1 but have different lengths (15–120 aa), are shown as green rectangles. The aromatic motif at the beginning of H2 helix is also labeled (blue bar). These motifs are also labeled in the alignment of Mdn1 orthologs' sequences in Data S1. (B) Two views of the 'ring-linker junction' motif that connects the tail to the AAA ring in AMPPNP-Mdn1 are shown (ribbon representation). The N, AAA1-L, and AAA6-L domains and part of the linker domain are colored as in Figure 2A. (C-H) The density map of each AAA domain (grey mesh) is shown with the built model (ribbon representation). The nucleotides (stick representation) bound between the large and small subdomains of AAA1–4 and next to the AAA6-L subdomain are shown. All nucleotide densities are modeled as AMPPNP molecules. The models are colored as in Figure 1A. Note that AAA5 shows no density for a nucleotide. (I) Negative-stain EM image of Mdn1 carrying the E1637Q mutation in the Walker B motif of AAA5. Scale bar: 20 nm.
The fitting of the ATPI-Mdn1 model into the Rix1 particle map (grey surface, EMDB ID: 3199) contoured at a threshold at which the map density matches the size of the model for AAA core (blue ribbon) with the docked MIDAS domain (magenta ribbon) of Mdn1. The models for the linker domain of Mdn1 (purple ribbon) and Rsa4 (cyan ribbon) are also shown.
(A) A cryo-EM image of ATPI-Mdn1 processed with MotionCor2. Some Mdn1 particles are marked with white circles. Scale bar: 20 nm. (B) The workflow used to process the pilot dataset. (C) The workflow used to process the complete dataset. The angular distribution of the particles in the final reconstruction is shown next to the final map. (D) Gold-standard FSC curve calculated between independently refined half maps for ATPI-Mdn1. The resolution was estimated as ~7.7 Å (FSC = 0.143). (E) Local-resolution map calculated by ResMap in RELION 2.0.