Abstract
Immunoglobulin G3 (IgG3) has an uncertain role in the response to infection with and vaccination against human immunodeficiency virus (HIV). Here we describe a regulatory role for IgG3 in dampening the immune system–activating effects of chronic HIV viremia on B cells. Secreted IgG3 was bound to IgM-expressing B cells in vivo in HIV-infected chronically viremic individuals but not in early-viremic or aviremic individuals. Tissue-like memory (TLM) B cells, a population expanded by persistent HIV viremia, bound large amounts of IgG3. IgG3 induced clustering of B cell antigen receptors (BCRs) on the IgM+ B cells, which was mediated by direct interactions between soluble IgG3 and membrane IgM of the BCR (IgM-BCR). The inhibitory IgG receptor CD32b (FcγRIIb), complement component C1q and inflammatory biomarker CRP contributed to the binding of secreted IgG3 onto IgM-expressing B cells of HIV-infected individuals. Notably, IgG3-bound TLM B cells were refractory to IgM-BCR stimulation, thus demonstrating that IgG3 can regulate B cells during chronic activation of the immune system.
Several B cell abnormalities have been described in HIV infection since the virus was first identified in 19831, most notably in the memory compartment (reviewed in ref. 2). In contrast to healthy individuals, HIV-infected individuals show depletion of classic costimulatory receptor CD27–expressing resting memory (RM) B cells in most stages of infection, whereas nonconventional memory B cell populations are expanded, especially in HIV-viremic individuals3. These include tissue-like memory (TLM) B cells (CD21loCD27−), which exhibit increased expression of several inhibitory receptors and display features associated with exhaustion4, and activated memory (AM) B cells (CD21loCD27+), which are highly activated and are prone to extrinsic apoptosis5. The frequency of somatic hypermutation and capacity of derived antibodies to neutralize HIV are lower in TLM B cells than in RM B cells, suggestive of a defect in affinity maturation6. TLM B cells are not unique to HIV infection; similar B cell populations have been described in several infectious and non-infectious settings in which chronic activation of the immune system and inflammation are prevalent (reviewed in refs 7–11). Persistent stimulation, whether from viral infection12 or in models of aging and autoimmunity induced via Toll-like receptors13,14, has been associated with the expression, in B cells, of the transcription factor T-bet, a strong regulator of immunoglobulin class switching influenced by type 1 helper T cell responses15. In humans, IgG3 is most commonly associated with type 1 helper T cell–biased cytokines, as described in complement C3–deficient patients16, age-related effects of streptococcal infection17 and T-betexpressing B cells in HIV-infected individuals18. In almost all of those studies, B cells were shown to express several inhibitory markers, as well as the markers CD11c and CXCR3, which are uniquely expressed on TLM B cells in association with B cell exhaustion4.
HIV-induced hypergammaglobulinemia is dominated by IgG1, although serum concentrations of IgG3 are also elevated19. Several unique features make IgG3 an interesting candidate for further study. Among the IgG subclasses, IgG3 is the most flexible, due to its extended hinge region20, and IgG3 is the most polymorphic isotype21, which suggests that genetics might affect its function. IgG3 also has the highest affinity for C1q, the first component of the classical complement pathway22, which provides it with strong effector function that is, however, somewhat tempered by its relatively short half-life23. These properties of IgG3 might explain its proposed strong yet transient role in infection with and vaccination against HIV24–28. Here we describe a novel function for IgG3 as a regulator of TLM B cells in HIV-infected chronically viremic individuals.
Results
IgG3 bound to IgM+ B cells of HIV-viremic individuals.
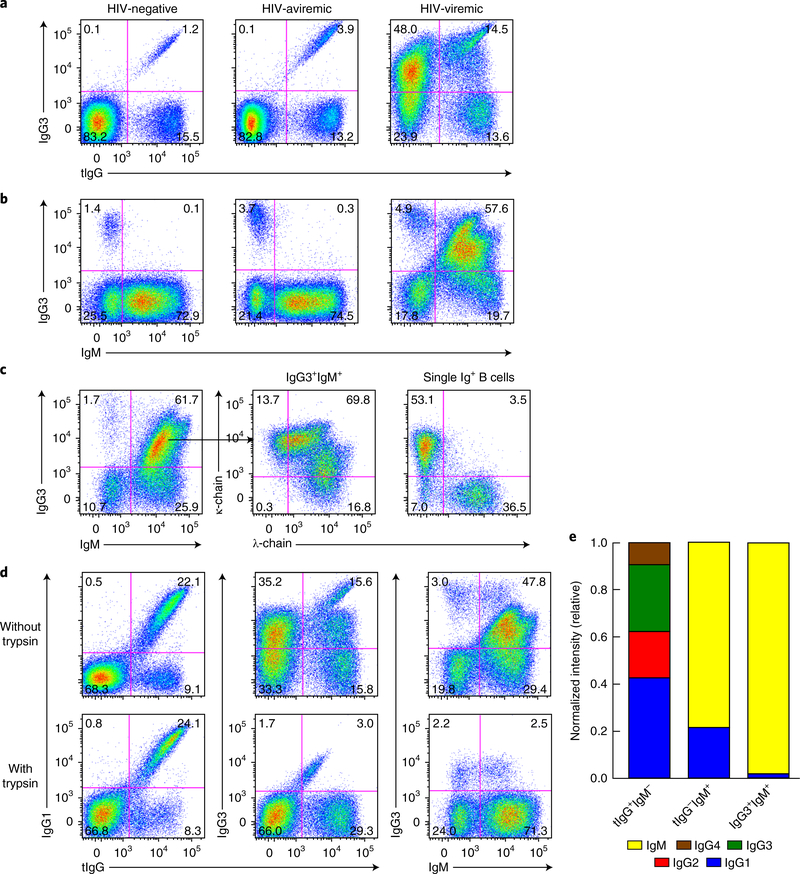
We evaluated the expression of total IgG (tIgG) and IgG3 on the surface of B cells of HIV-negative and HIV-infected individuals at various stages of disease. As expected for HIV-negative and HIV-aviremic individuals, a small yet clearly discernable fraction of tIgG+ B cells stained positively for the IgG3 isotype (Fig. 1a, diagonal pattern, top right quadrant). Unexpectedly, an unusually large proportion of B cells from HIV-viremic individuals were positive for IgG3, and most of these IgG3+ B cells were negative for tIgG (Fig. 1a). However, the same pan–IgG Fc–specific monoclonal antibody (mAb), clone G18–145, detected similar patterns of expression of IgG1 for all three groups of individuals investigated (Supplementary Fig. 1a). We determined that two other commercially available pan–IgG Fc–specific mAbs, clones ICO-97 and M1310G05, fully detected the IgG3 present on the surface of B cells of HIV-viremic individuals (Supplementary Fig. 1b). Furthermore, a large fraction of the IgG3+ B cells of HIV-viremic individuals, but not those of HIV-negative or HIV-aviremic individuals, were also positive for IgM (Fig. 1b). Collectively, these staining patterns suggested a unique association between IgG3 and IgM on B cells of HIV-viremic individuals in which the IgG3 was displayed in such a way as to inhibit recognition by certain pan-IgG mAbs.
Fig. 1 |. Soluble IgG3 binds to IgM-expressing B cells of HIV-viremic individuals in vivo.
a,b, Flow cytometry of B cells isolated from the peripheral blood of HIV-negative, HIV-aviremic and HIV-viremic individuals (above plots), CD20 gated (to exclude plasmablasts, which typically express CD19 but not CD20), and stained for IgG3 and tIgG (a) or IgM (b). Numbers in quadrants indicate percent cells in each throughout. c, Flow cytometry showing immunoglobulin light-chain expression (middle and right) on IgG3+IgM+ and combined single immunoglobulin-positive (Ig+) B cells of an HIV-viremic individual. d, Flow cytometry showing staining for tIgG, IgG1, IgG3 and IgM of B cells obtained from an HIV-viremic individual and treated with trypsin (bottom row) or not (top row) for 5 min at 37°C. e, Expression of immunoglobulin-encoding mRNA (key) by B cells sorted by the surface expression as tIgG+IgM−, tIgG−IgM+ or IgG3+IgM+ (horizontal axis); band intensity was normalized to that of control mRNA encoding β -actin. Data are representative of 83 experiments (HIV-negative), 17 experiments (HIV-aviremic) or 75 experiments (HIV-viremic) (a,b), 17 (c) or 13 (d) experiments, or three independent experiments (e).
The unusual IgG3+IgM+ B cells observed in HIV-viremic individuals could have arisen from cells expressing one immunoglobulin isotype and simultaneously binding a second soluble isotype. We addressed this possibility by staining for immunoglobulin light chains; expression patterns typically show mutual exclusion unless polyclonal soluble immunoglobulin is bound to the cells. Single IgG3+ and IgM+ B cells displayed an immunoglobulin lightchain pattern that was consistent with mutual exclusion; however, IgG3+IgM+ B cells revealed a mixed pattern (Fig. 1c), which suggested that a secreted polyclonal source of immunoglobulin was bound to this population. To distinguish between secreted forms of immunoglobulin and expressed forms of immunoglobulin, we treated B cells with trypsin, an enzyme to which most membrane forms of immunoglobulin are insensitive29 (data not shown). While the trypsin treatment had no effect on the cell-surface abundance of tIgG and IgG1, it greatly diminished IgG3 on IgG3+IgM+ B cells, with a concomitant increase in IgM-only B cells (Fig. 1d). To formally establish which immunoglobulin was expressed by IgG3+IgM+ B cells of HIV-viremic individuals, we performed mRNA analysis for genes encoding the constant regions of IgM and IgG subclasses on B cells that were stained for tIgG, IgG3 and IgM and sorted into different fractions. Single IgM-expressing B cells and IgG-expressing B cells expressed mRNA encoding IgM and mRNA encoding a mixed pattern of IgG subclasses, respectively, with IgG1 being dominant (Fig. 1e). In contrast, analysis of mRNA from IgG3+IgM+ B cells also revealed almost exclusive expression of mRNA encoding the immunoglobulin heavy-chain constantregion μ -chain (IGHM), similar to that of single IgM+ B cells (Fig. 1e). Collectively, these analyses revealed that IgG3+IgM+ B cells of HIV-viremic individuals were IgM-expressing B cells that bound soluble IgG3 in vivo.
HIV disease conditions associated with IgG3+IgM+ B cells.
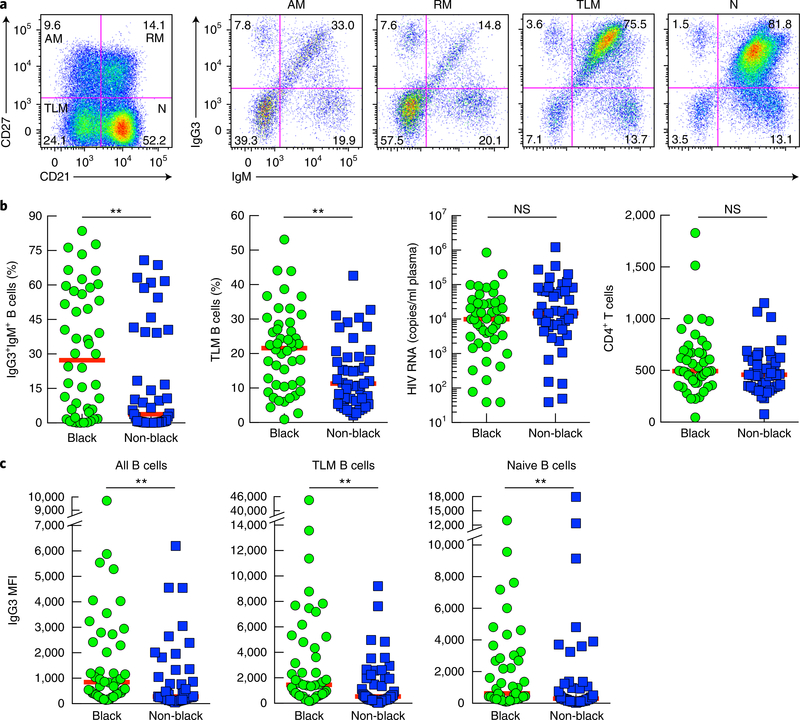
IgM-expressing B cells include naive B cells and unswitched memory B cells, the latter of which are present among several phenotypically distinct populations, including RM B cells, AM B cells and TLM B cells in HIV-viremic individuals2. Accordingly, we evaluated the distribution of IgG3+IgM+ B cells through the use of two markers that define these populations: CD21 and CD27. A high frequency of IgG3+IgM+ B cells was observed among TLM B cells, while the frequency of these cells was much lower among AM B cells and RM B cells (Fig. 2a). A high frequency of IgG3+IgM+ B cells was also observed among the naive B cells of HIV-viremic individuals (Fig. 2a). To understand the conditions that favor the presence of IgG3+IgM+ B cells, we performed extensive longitudinal and cross-sectional analyses. Longitudinally, IgG3+IgM+ B cells did not appear until the transition from early HIV viremia to chronic HIV viremia, and their frequency markedly diminished following sustained suppression of chronic viremia by anti-retroviral therapy (ART) (Supplementary Fig. 2a). For cross-sectional analyses, we studied 108 HIV-infected individuals categorized into three groups: early HIV viremia; chronic HIV viremia; and HIV aviremia by ART (Table 1). The frequency of IgG3+IgM+ B cell was significantly higher in the chronic HIV-viremic group than in the two other groups and was significantly higher in the two viremic groups than in the HIV-aviremic group (P < 0.05 and P < 0.0001 (two-tailed Mann-Whitney test after significance was obtained by Kruskal-Wallis analysis of variance (ANOVA) of the full set); Supplementary Fig. 2b). However, the frequency of IgG3+IgM+ B cells was not correlated with viral load, as viremia was significantly higher in the early stage of infection than in the chronic stage of infection (P < 0.01 and P < 0.0001 (tests as above); Table 1 and Supplementary Fig. 2b). The frequency of TLM B cells was significantly higher in the chronically infected HIV-viremic group than in the HIV-aviremic group, whereas CD4+ T cell counts did not significantly differ between the groups (P < 0.0001 (tests as above); Table 1 and Supplementary Fig. 2b).
Fig. 2 |. Enrichment for the IgG3+IgM+ profile among peripheral blood TLM B cells and naive B cells in chronic HIV viremia.
a, Flow cytometry of CD20-gated B cells of a chronically infected HIV-viremic individual, showing expression of CD27 and CD21 (far left), and the IgG3 and IgM profiles (middle and right) of the corresponding AM, RM, TLM and naive (N) B cell populations (above plots). Data are representative of 75 experiments. b, Frequency of IgG3+IgM+ B cells and TLM B cells in black and non-black subjects (horizontal axis), as measured by flow cytometry, as well as HIV viral load and CD4+ T-cell counts in those subjects. c, Mean fluorescence intensity (MFI) of IgG3 on all CD20-gated B cells and on TLM B cells and naive B cells from which IgG3-expressing B cells were excluded (Methods), for black and non-black subjects (horizontal axis), as measured by flow cytometry. Each symbol (b,c) represents an individual subject (n = 46 black subjects; n = 45 non-black subjects); red horizontal lines indicate the median. NS, not significant (P > 0.05); **P < 0.01 (two-tailed Mann-Whitney test).
Table 1 |.
Characteristics of HIV-infected individuals studied
| Early viremic | Chronic viremic | Aviremic | |
|---|---|---|---|
| n (total) | 16 | 75 | 17 |
| Age (years) | 32 (21–54) | 36 (20–66) | 35 (24–59) |
| Sex (F/M) | 0/16 | 13/62 | 5/12 |
| Race (black/non-black) | 7/9 | 39/36 | 4/11a |
| CD4+ T cell count(cells/μl) | 514 (330–693) | 482 (45–1828) | 544 (216–969) |
| HIV RNA (copies/ml) | 39,229 (39,229–81,957)**,**** | 7,100 (4,100–12,237)**** | <40 |
| Duration of ART (years) | NA | NA | 5.5(0.5–10.5)a |
Characteristics of study participants: age (in years) and T cell counts (cells per microliter of blood (cells/μl)) are presented as the median (with range in parentheses); sex is presented as the number of females versus the number of males (F/M); race is presented as the number of black subjects versus the number of non-black subjects (black/non-black); RNA (copies per milliliter of plasma (copies/ml)) is presented as the geometric mean (with the 95% confidence interval in parentheses). NA, not applicable.
P < 0.01
P < 0.0001
(two-tailed Mann-Whitney test after significance was obtained by Kruskal-Wallis ANOVA test of the full set; details, Supplementary Fig. 2b).
Among aviremic subjects, race was unknown for 2 of 17 subjects, and duration of ART was unknown for 1 of 17 subjects.
Among the 91 HIV-viremic individuals assessed, a wide spectrum of frequencies and patterns of IgG3+IgM+ B cells was noted independently of the phase of infection; the median frequency was 17% in the chronic HIV-viremia group, with a range 0.1% to 84% (Supplementary Fig. 2b). Among the demographics available, race had a strong influence: HIV-viremic individuals who were black (either African-American or immigrants from Africa) had a significantly higher frequency of IgG3+IgM+ B cells than that of non-black HIV-viremic individuals (Fig. 2b). All 91 HIV-viremic individuals were included in this black–versus–non-black comparison who otherwise did not significantly differ by phase of HIV viremia, plasma viral load, CD4+ T cell count, sex or age (Fig. 2b and Supplementary Table 1). However, the increased frequency of IgG3+IgM+ B cells among black HIV-viremic individuals was associated with a significantly higher frequency of TLM B cells than that of non-black individuals (Fig. 2b), an observation that also revealed a significant direct correlation between the frequency of IgG3+IgM+ B cells and that of TLM B cells when data points from all 91 HIV-viremic individuals were combined (data not shown; r = 0.4800 and P < 0.0001 (two-tailed Spearman’s rank correlation test)).
We extended the analysis of the frequency of IgG3+IgM+ B cells to include the intensity of IgG3 staining on IgM-expressing B cells. The intensity of IgG3 was significantly higher on IgG3+IgM+ total B cells as well as naive and TLM B cells of black individuals than on those of non-black individuals (Fig. 2c). The intensity of IgG3 was also significantly higher for the chronic HIV-viremia group than for the early-HIV-viremia or HIV-aviremia group (P < 0.05, P < 0.01, P ≤ 0.001 and P < 0.0001 (two-tailed Mann-Whitney test after significance was obtained by Kruskal-Wallis analysis of variance (ANOVA) of the full set); Supplementary Fig. 2c). Finally, the intensity of IgG3 was significantly higher on IgG3+IgM+ TLM B cells than on their naive B cell counterparts (P < 0.0001 (two-tailed Wilcoxon matched-pairs signed rank test); Supplementary Fig. 2d). Collectively, these observations suggested that IgG3+IgM+ B cells were most strongly associated with TLM B cells during chronic HIV viremia and were influenced by genetic background.
Co-localization of IgG3 and IgM on IgG3+IgM+ B cells.
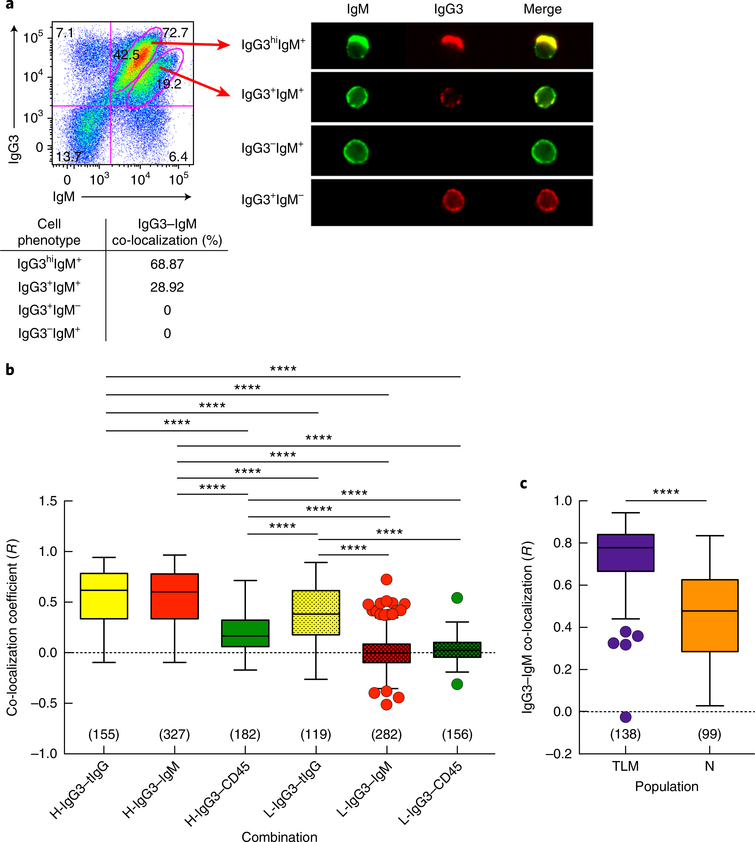
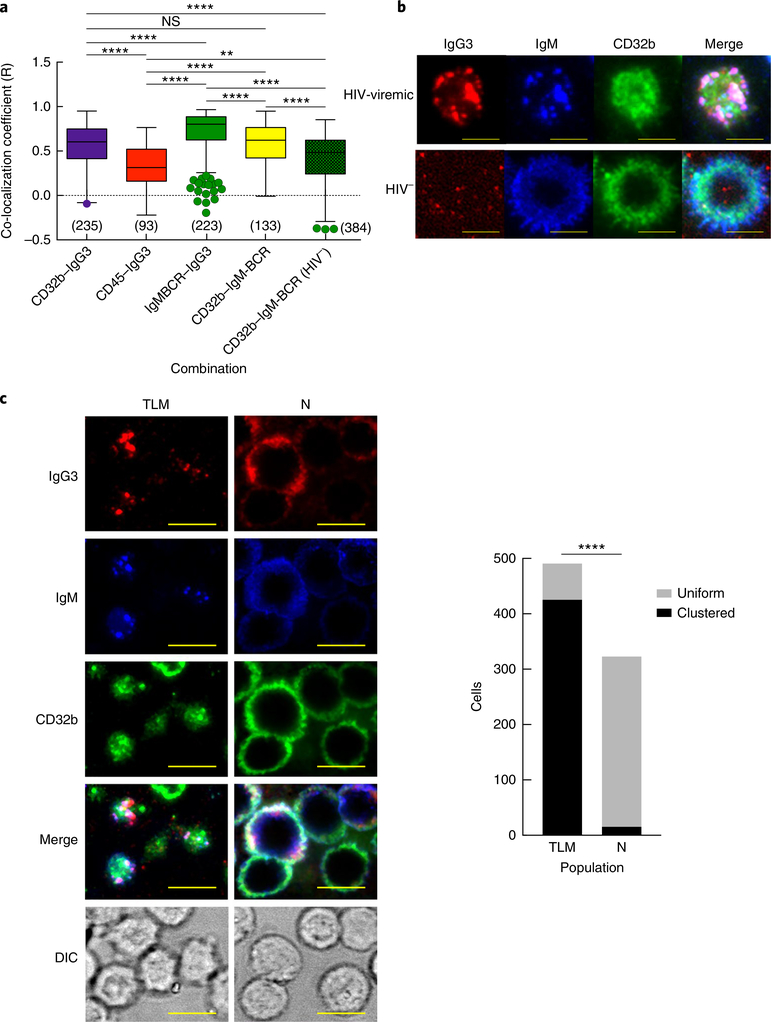
The presence of IgG3 on IgM-expressing B cells prompted us to consider IgG3–IgM co-localization. Using high-throughput imaging cytometry, we identified IgM+ B cells with varying intensities of IgG3, and we performed single-cell analysis of the gated populations to evaluate co-localization (Fig. 3a). There was a high frequency of IgG3–IgM co-localization among B cells that were IgG3hiIgM+, whereas the extent of such co-localization was lower for mid-intensity IgG3 on IgM+ B cells (Fig. 3a). We then used a high-resolution imaging technique, total internal reflection fluorescence (TIRF) microscopy, to determine the degree of IgG3–IgM co-localization on B cell surfaces and compared co-localization coefficients for HIV-infected individuals with high- or low-intensity IgG3 on IgM+ B cells. For high-intensity IgG3 on IgM+ B cells, the co-localization coefficient of IgG3–IgM was similar to that of IgG3–tIgG, which depicts maximum co-localization when two different antibodies (anti-IgG3 and anti-tIgG) are used to detect the same protein (IgG3) (Fig. 3b). For low-intensity IgG3 on IgM+ B cells, the co-localization coefficient of IgG3–IgM was significantly lower than that of IgG3–tIgG (Fig. 3b). The B cells were also stained for CD45, a pan–B cell marker that is expressed at a density similar to that of membrane IgM of the BCR (IgM-BCR) but is excluded from microclusters that form during BCR activation in response to antigen engagement30. The co-localization coefficient of IgG3–CD45 was significantly lower than that of IgG–tIgG for both high-intensity IgG3 and low-intensity IgG3 on IgM+ B cells and was significantly lower than the IgG3–IgM coefficient for high-intensity IgG3 on IgM+ B cells (Fig. 3b). Furthermore, the IgG3–IgM coefficient was significantly higher for TLM B cells than for naive B cells of individuals with high-intensity IgG3 on IgM+ B cells (Fig. 3c). Collectively, these findings strongly indicated that IgG3 and IgM were co-localized on IgG3+IgM+ B cells and that the interaction between IgG3 and IgM was strongest on TLM B cells.
Fig. 3 |. IgG3–IgM co-localization on IgG3+IgM+ B cells.
a, Imaging cytometry (top right) of CD20+ B cells from a chronically infected HIV-viremic individual, gated (left) on populations with different levels of IgG3 and IgM (arrows at left); below, frequency of IgG3–IgM co-localization, based on quantitative analysis of the relative intensity of two fluorescent images. b, Pearson’s co-localization coefficient (R) for various protein pairs (horizontal axis), as measured by TIRF microscopy of B cells from a chronically infected HIV-viremic individual with high-intensity IgG3 on IgM+ B cells (H-) and one with low-intensity IgG3 on IgM+ B cells (L-), stained for tIgG (with polyclonal anti-IgG), IgG3, IgM and CD45; R values were calculated pairwise for each marker and for each source of cells. c, R values for IgG3–IgM co-localization on the TLM B cells and naive B cells (horizontal axis) of a chronically infected HIV-viremic individual with high-intensity IgG3 on IgM+ B cells. Results in b,c are presented as ‘box and whiskers’ plots (middle line, mean; box limits, inter-quartile distance; extended lines, Tukey’s method); in parentheses (above horizontal axis), number of cells imaged for each analysis. ****P < 0.0001 (two-tailed unpaired Student’s t-test, after significance was obtained by one-way ANOVA of the full set (in b); only significant P values are shown in b). Data are representative of eight (a,b) or three (c) independent experiments.
Diminished BCR responses among IgG3+IgM+ TLM B cells.
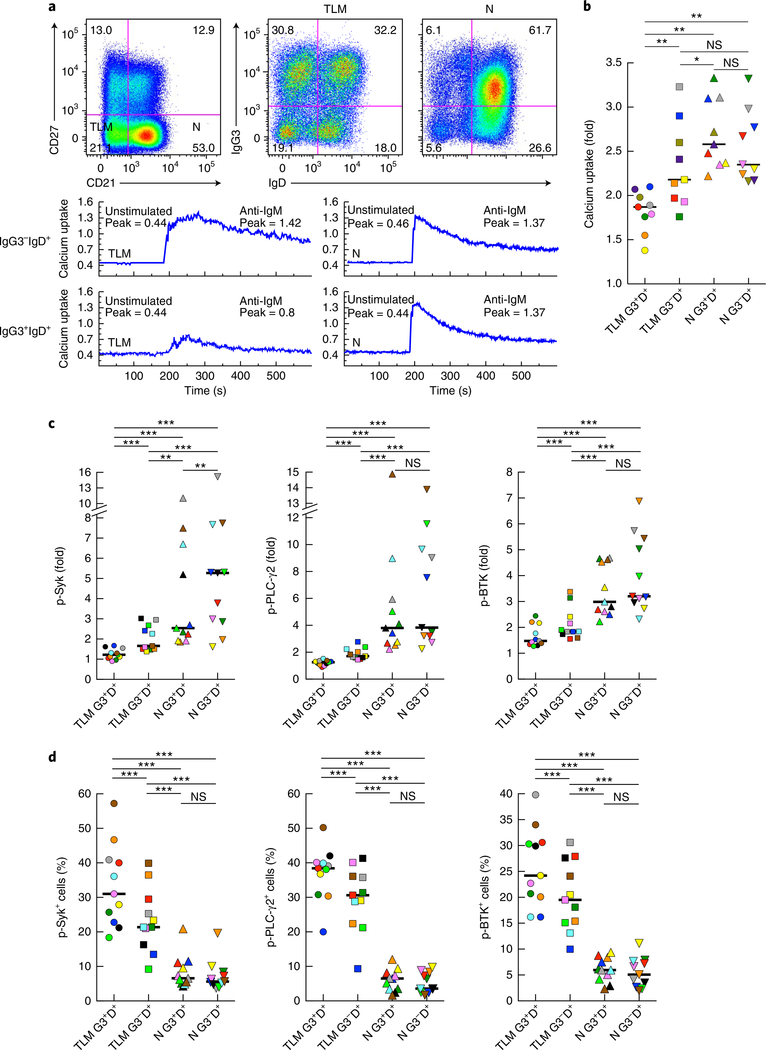
To evaluate whether in vivo binding of soluble IgG3 to IgM-expressing B cells affected BCR signaling ex vivo, we measured calcium mobilization following the crosslinking of IgM-BCR on B cells of HIV-viremic individuals with moderate- to high-intensity IgG3 on IgM+ B cells. IgD was used to identify IgM-expressing cells for these analyses. Calcium uptake following IgM-BCR crosslinking was equally substantial for naive B cells whether soluble IgG3 was bound to the cells or not (Fig. 4a,b). In contrast, while IgG3−IgD+ TLM B cells responded to IgM-BCR crosslinking with calcium fluxes that were not significantly different from those of their naive B cell counterparts, IgG3+IgD+ TLM B cells were distinctly and significantly unresponsive to IgM-BCR crosslinking (Fig. 4a,b).
Fig. 4 |. IgG3+IgM+ B cells have reduced responses to BCR stimulation.
a, Flow cytometry (top) of CD20-gated B cells of a chronically infected HIV-viremic individual, showing expression of CD27 and CD21 (far left) and the IgG3 and IgD profiles of the corresponding TLM B cell and naive B cell populations (middle and right); below, calcium uptake by IgG3−IgD+ and IgG3+IgD+ (far left margin) TLM B cells (left) and naive B cells (right) before (Unstimulated) and after (Anti-IgM) 2 min of incubation with F(ab’)2 anti-IgM; the peak (number in plots) is the ratio of bound fluorescent Ca2+ indicator dye Indo-1 to unbound Indo-1. Data are representative of nine experiments. b, Calcium uptake by IgG3+IgD+ TLM B cells (TLM G3+D+), IgG3−IgD+ TLM B cells (TLM G3−D+), IgG3+IgD+ naive B cells (N G3+D+) and IgG3−IgD+ naive B cells (N G3−D+) from chronically infected HIV-viremic individuals with moderate- to high-intensity IgG3 on IgM+ B cells, presented as the peak ratio (as in a) after incubation with anti-IgM relative to that ratio in unstimulated cells (‘fold’ values). c,d, Quantification of phosphorylated (p-) Syk, PLC-γ2 and Btk (measured by flow cytometry) in B cell populations as in b (horizontal axis) after 2 min of incubation with F(ab’)2 anti-IgM (c) or in the absence of such incubation with anti-IgM (d); results in c after incubation with anti-IgM are presented relative to baseline (‘fold’ values). Each symbol (b–d) represents an individual subject (n = 9 (b) or n = 11 (c,d)); small horizontal lines indicate the median. *P < 0.05, **P < 0.01 and *** P ≤ 0.001 (two-tailed Wilcoxon matched-pairs signed rank test after significance was obtained by Friedman ANOVA of the full set).
In addition, we evaluated the phosphorylation of downstream intermediates associated with signal transduction through the BCR. One of the earliest mediators of BCR signaling is the kinase Syk, which is also involved in formation of BCR signaling microclusters30. Two other BCR signaling mediators were also evaluated: Bruton’s tyrosine kinase (Btk), and its downstream target, phospholipase PLC-γ2. The induced phosphorylation of all three proteins followed a pattern that was largely similar to that observed for calcium mobilization (Fig. 4c). The induced phosphorylation of Syk, Btk and PLC-γ2 following IgM-BCR crosslinking was significantly lower in IgG3+IgD+ TLM B cells than in IgG3−IgD+ TLM B cells, as well as both naive B cell populations. Activation-induced phosphorylation of these proteins was also lower in the IgG3−IgD+ TLM B cells than in the naive B cell populations, and induced phosphorylation of Syk was lower in IgG3+IgD+ naive B cells than in IgG3−IgD+ naive B cells (Fig. 4c). One possible explanation for the deficiency in the induced phosphorylation of Syk, Btk and PLC-γ2 in TLM B cells, and in particular in IgG3+IgD+ TLM B cells, was that their baseline content of phosphorylated Syk, Btk and PLC-γ2 was greater than that of the naive B cell populations (Fig. 4d). Collectively, these data suggested that while IgG3 bound to both naive B cells and TLM B cells, functionally, the IgM-BCR of TLM B cells was more adversely affected.
Transfer of IgG3 to healthy donor B cells.
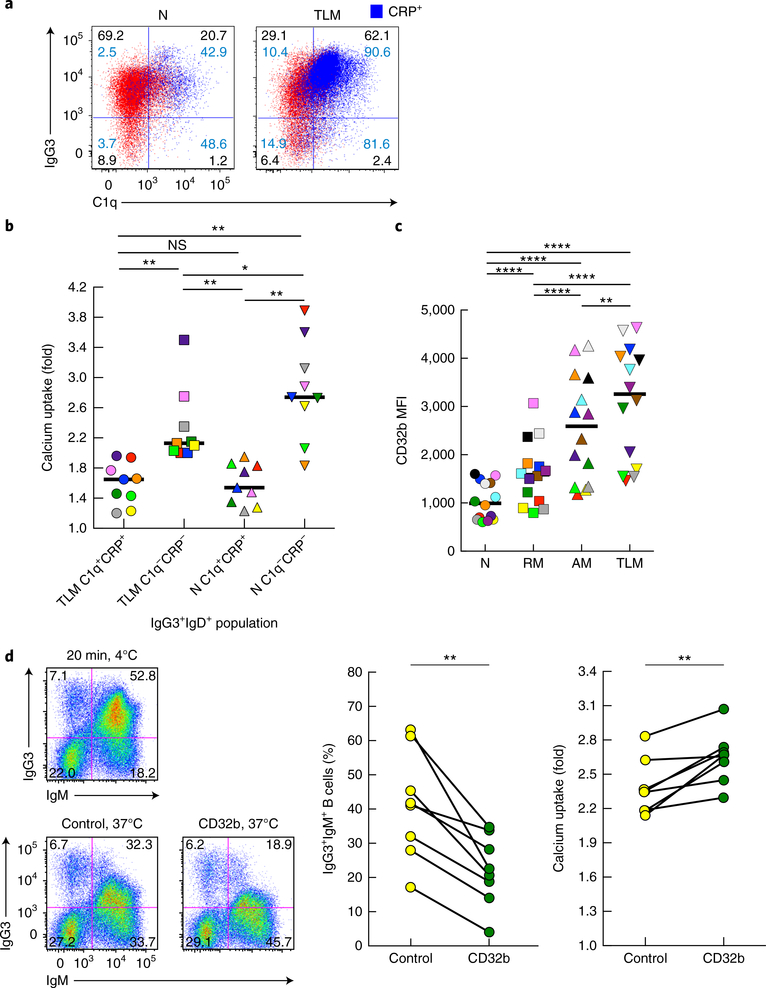
Next we considered whether soluble IgG3 isolated from the serum of HIV-infected individuals with IgG3+IgM+ B cells was transferable to B cells of individuals who did not have IgG3+IgM+ B cells (called ‘unaffected B cells’ here). Accordingly, we performed comparisons within the cohort of HIV-viremic individuals by pairing serum from those with the highest intensity of IgG3 on their IgM-expressing B cells with serum from those with the lowest such intensity. Polyethylene glycol (PEG) precipitation (PEG-IgG3) of serum from individuals with high-intensity IgG3 on IgM+ B cells led to binding of IgG3 to unaffected B cells, but such precipitation of serum from those with low-intensity IgG3 on IgM+ B cells did not (Supplementary Fig. 3a). When IgG3 was enriched from serum through the use of an agarose-bead IgG3-specific approach, binding of IgG3 from individuals with high-intensity IgG3 on IgM+ B cells to unaffected B cells was also observed, but at lower and more variable frequencies (Supplementary Fig. 3a). Of note, none of these conditions led to the binding of IgG3 by B cells when the source of serum was HIV-negative individuals (data not shown). The recipient cells themselves also contributed to this pattern for the binding of IgG3 by B cells in that TLM B cells, found at a low yet discernable frequency in the peripheral blood of HIV-negative individuals, typically bound PEG-IgG3 at the highest frequency (Supplementary Fig. 3b). Notably, binding of PEG-IgG3 also attenuated IgM-BCR signaling (Supplementary Fig. 3c), with effects similar to those described for IgG3+IgM+ B cells of HIV-viremic individuals.
We considered other serum-related factors that might contribute to varying intensities of IgG3 on IgM+ B cells. Factors related to HIV infection, including the abundance of HIV RNA and HIV-specific antibody reactivity in PEG-IgG3 did not differ by the intensity status of IgG3 on IgM+ B cells (data not shown). However, when the serum concentrations of IgG subclasses were measured, significantly greater amounts of IgG3 and IgG1, but not of IgG2 or IgG4, were observed for individuals with high-intensity IgG3 on IgM+ B cells than for those with low-intensity IgG3 on IgM+ B cells (P < 0.001 and P < 0.0001 (two-tailed Mann-Whitney test); Supplementary Fig. 4a). Notably, the serum concentration of IgG2 in healthy adults is generally higher than that of IgG3, an observation that held for individuals with low-intensity IgG3 on IgM+ B cells but not for those with high-intensity IgG3 on IgM+ B cells, as indicated by the median ratio of IgG3 to IgG2 (0.3 and 2.5, respectively; Supplementary Fig. 4a). Those differences were also reflected in the amount of IgG3 in the PEG preparations; however, even when an equal amount of IgG3 in PEG-IgG3 was added, IgG3 from individuals with low-intensity IgG3 on IgM+ B cells remained largely non-transferable (data not shown).
To characterize constituents within PEG-IgG3 that were responsible for the binding of IgG3 to B cells of HIV-negative individuals, we used size-exclusion chromatography (SEC) to generate aggregated and non-aggregated fractions. As a polymeric control with binding properties similar to those of immunocomplexes, we used heat-aggregated IgG3; as expected, the unfractionated form and first elution (void volume; SEC-1) bound unaffected B cells (Supplementary Fig. 4b). When PEG precipitates derived from HIV-viremic individuals were fractionated by SEC, binding of IgG3 to unaffected B cells was restricted to the second retarded fraction (SEC-2; Supplementary Fig. 4c), consistent with a monomeric state. Immunoblot analysis revealed that IgG3 was present in unfractionated PEG and SEC-2, while it was largely absent from SEC-1 (Supplementary Fig. 5a). Furthermore, substantial amounts of C1q, known to have a strong affinity for IgG322, were detected in unfractionated PEG and SEC-1, as expected, given its size of ~450 kDa (Supplementary Fig. 5a). However, small amounts of C1q were also present in SEC-2 (Supplementary Fig. 5a). Given the known interactions among the inflammatory biomarker CRP, C1q and IgG receptors (FcγRs)31–33, as well as the size compatibility of CRP and SEC-2, we considered the involvement of CRP in the binding of PEG-IgG3 and its SEC fractions to unaffected B cells. CRP was consistently observed in unfractionated PEG-IgG3 and SEC-2 but was absent from SEC-1 (Supplementary Fig. 5a). Furthermore, depletion of C1q or CRP or both led to a significant reduction in the binding of PEG-IgG3 to unaffected B cells (P < 0.05 (two-tailed Wilcoxon matched-pairs signed rank test, after significance was obtained by Friedman ANOVA of the full set; Supplementary Fig. 5b). We also considered other factors that could promote the binding of soluble IgG3 to IgM-expressing B cells. CD32b (the low-affinity inhibitory FcγR for IgG that contains one cytoplasmic immunoreceptor tyrosine-based inhibitory motif) is expressed on most human B cells and has been shown to efficiently bind CRP33, as well as aggregated IgG, with the highest affinity for IgG334. Preincubation of unaffected B cells with ligand-blocking anti-CD32b led to a significant reduction in the binding of PEG-IgG3 (P < 0.05 (test as above without Friedman ANOVA); Supplementary Fig. 5c). Interactions among immunoglobulins, C1q and FcRs are modulated by carbohydrate moieties35. Full deglycosylation of PEG-IgG3 via the glycosidase PNGase F largely abolished the binding of IgG3 to unaffected B cells, whereas the more IgG-restricted endoglycosidase EndoS236 also caused a significant, albeit more variable, reduction in binding (P < 0.05 (tests as above); Supplementary Fig. 5d). Collectively, these data suggested that non-aggregated IgG3 in the serum of HIV-infected individuals with IgG3+IgM+ B cells was able to bind to unaffected B cells through interactions that depended on glycans and associations with C1q, CRP and CD32b.
Mechanism for the binding of IgG3 to IgM-expressing B cells.
We considered the involvement of C1q and CRP in the binding of IgG3 to B cells in vivo. C1q and CRP were consistently present along with IgG3 on IgG3+IgM+ TLM B cells and, to a lesser extent, naive B cells of HIV-viremic individuals (Fig. 5a). We considered how the presence of C1q and CRP on IgG3+IgM+ B cells of HIV-viremic individuals affected IgM-BCR signaling. Calcium uptake following stimulation with anti-IgM was lowest for CRP+C1q+IgG3+IgD+ TLM B cells and naive B cells and was significantly lower than that for their CRP−C1q− counterparts, whereas the responses of CRP−C1q− TLM B cells were significantly lower than those of their respective naive B cell counterparts, but the responses of CRP+C1q+ TLM B cells were not (Fig. 5b). Therefore, when C1q and CRP were present, both IgG3+IgM+ TLM B cells and IgG3+IgM+ naive B cells displayed diminished BCR signaling; however, TLM B cells exhibited additional functional deficiencies.
Fig. 5 |. Role of C1q, CRP and CD32b in IgG3 binding and signaling by IgM-BCR B cells.
a, Flow cytometry of CD20-gated B cells from a chronically infected HIV-viremic individual with high-intensity IgG3 on IgM+ B cells, showing binding of C1q and IgG3 on naive B cells and TLM B cells (blue, CRP overlay). Numbers in blue indicate percent CRP among cells in that quadrant. Data are representative of 11 experiments. b, Calcium uptake by C1q+CRP+and C1q−CRP− populations (horizontal axis) of IgG3+IgD+ TLM B cells and naive B cells from chronically infected HIV-viremic individuals (n = 9) with moderate- to high-intensity IgG3 on IgM+ B cells (presented as in Fig. 4b). c, Mean fluorescence intensity of CD32b (evaluated by flow cytometry) on B cells isolated from HIV-viremic individuals (n = 14) and identified as naive, RM, AM or TLM B cells (horizontal axis) by their expression of CD21 and CD27. d, Flow cytometry (left half) showing the expression of IgG3 and IgM by CD20-gated B cells obtained from a chronically infected HIV-viremic individual with high-intensity IgG3 on IgM+ B cells and maintained for 20 min at 4 °C (top) or treated at 37 °C with control goat IgG (bottom left) or goat anti-CD32b (bottom right); middle and right, frequency of IgG3+IgM+ B cells (middle right) and calcium uptake (far right) among B cells obtained from chronically infected HIV-viremic individuals (n = 8 subjects (middle right) or n = 7 subjects (far right)) with moderate- to high-intensity IgG3 on IgM+ B cells and treated with antibody as at bottom left (horizontal axis); results for calcium uptake presented as in b. Each symbol (b–d) represents an individual subject (identified by color in b,c); small horizontal lines (b,c) indicate the median; diagonal lines (d) connect values for the same subject. *P < 0.05, **P < 0.01 and ****P ≤ 0.0001 (two-tailed Wilcoxon matched-pairs signed rank test, after significance was obtained by Friedman ANOVA of the full set (in b,c)).
Given the role of CD32b in the transfer of serum-derived IgG3 to unaffected B cells, we considered its role in the binding of IgG3 to B cells in vivo. Consistent with published findings showing increased expression of multiple inhibitory receptors on TLM B cells4, expression of CD32b was higher on TLM B cells than on other B cell populations (Fig. 5c). Treatment of IgG3+IgM+ B cells from HIV-viremic individuals with anti-CD32b caused a significantly greater decrease in bound IgG3 than did treatment with a control antibody (Fig. 5d). This decrease in bound IgG3 was accompanied by a modest yet significant increase in calcium uptake following IgM-BCR stimulation (Fig. 5d). Treatment with anti-CD32b affected both naive B cells and TLM B cells (data not shown).
To evaluate the proximity of CD32b to IgG3 and IgM, we performed TIRF microscopy of IgG3+IgM+ B cells and unaffected B cells. On IgG3+IgM+ B cells, the co-localization coefficient was highest for IgG3–IgM and lowest for IgG3–CD45 (Fig. 6a). The co-localization coefficient of CD32b–IgG3 was similar to that of CD32b–IgM, and while both values were significantly higher than that of CD32b–IgM on unaffected B cells, they were significantly lower than the co-localization coefficient of IgG3–IgM (Fig. 6a). Among B cells of individuals with high-intensity IgG3 on IgM+ B cells, TIRF images revealed the presence of highly clustered IgM-BCR containing both IgG3 and CD32b. However, while the IgG3 patterns almost completely overlapped those of IgM-BCR, consistent with their high co-localization coefficient, CD32b presented a mixed pattern of high-intensity clustering with IgG3 and IgM-BCR and areas of evenly distributed discrete punctae (Fig. 6b). These patterns of polarization and clustering were more strongly associated with IgG3+IgM+ TLM B cells than with naive B cells (Fig. 6c), and they were not observed on unaffected B cells (Fig. 6b).
Fig. 6 |. Co-localization of CD32b with IgG3 and IgM on IgG3+IgM+ B cells.
a, Pearson’s co-localization coefficient (R) for various protein pairs (horizontal axis), as measured by TIRF microscopy of B cells from a chronically infected HIV-viremic individual with high-intensity IgG3 on IgM+ B cells and an HIV-negative individual (HIV−), stained for CD32b, IgG3, IgM and CD45 (presented as in Fig. 3b). **P < 0.01 and ****P < 0.0001 (two-tailed unpaired Student’s t-test after significance was obtained by one-way ANOVA of the full set). b, TIRF microscopy of B cells from an HIV-viremic individual (top) and an HIV-negative individual (bottom), stained as in a (above images). Scale bars, 5 μm. c, TIRF microscopy (left) of contact areas and differential interference contrast (DIC) of B cells isolated from a chronically infected HIV-viremic individual whose IgG3+IgM+ profile was associated predominantly with TLM B cells (left column) and another whose IgG3+IgM+ profile was associated predominantly with naive B cells (right column); right, quantification of B cells with a uniform or clustered distribution of IgM (key) for each individual identified by their predominant B cell population (horizontal axis). Scale bars (left), 9 μm. ****P < 0.0001 (two-tailed Fisher’s exact test). Data are representative of four (a,b) or three (c) independent experiments.
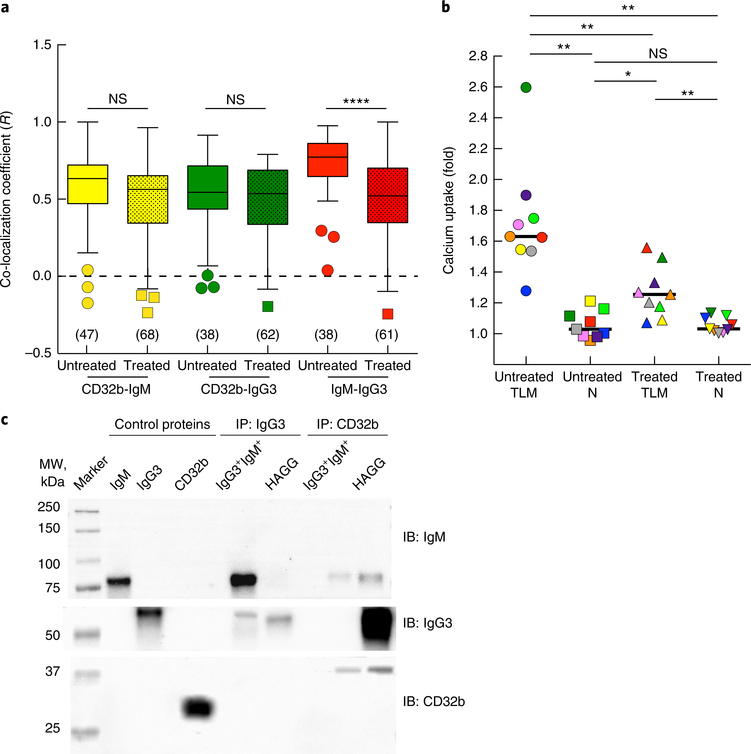
The stronger association between IgG3 and IgM-BCR on IgG3+IgM+ B cells than between IgG3 and CD32b on such cells was further investigated. First, treatment of IgG3+IgM+ B cells with exogenous IgG3 led to a significant reduction in IgG3–IgM co-localization (Fig. 7a) and a strong concomitant reduction in the inhibitory effect of IgG3 on calcium uptake following IgM-BCR stimulation of TLM B cells but not after such stimulation of naive B cells (Fig. 7b). We considered the possibility that IgG3 interacted directly with IgM-BCR. B cells isolated from HIV-viremic individuals with a high frequency of IgG3+IgM+ B cells were compared with HIV-negative B cells incubated with heat-aggregated IgG3, for which CD32b is the known binding receptor34. When cell lysates were immunoprecipitated with either anti-IgG3 or anti-CD32b, IgG3 specifically co-precipitated IgM from IgG3+IgM+ B cells but not from heat-aggregated IgG3+ B cells and, conversely, CD32b co-precipitated IgG3 from heat-aggregated IgG3+ B cells but not from IgG3+IgM+ B cells (Fig. 7c). After treatment with anti-CD32b, small amounts of IgM were detected in lysates of both cell types, whereas IgG3 was detected in heat-aggregated IgG3+ B cells but not in IgG3+IgM+ B cells. Collectively, these observations indicated that while CD32b was involved in the binding of IgG3 to IgM-expressing B cells and in modulating IgM-BCR responses, we found evidence of direct interactions between IgG3 and IgM that also affected IgM-BCR responses.
Fig. 7 |. Role of IgG3 in binding and regulating IgM-BCR.
a, Pearson’s co-localization coefficient (R) for various protein pairs (below plot), as measured by TIRF microscopy of cells obtained from a chronically infected HIV-viremic individual with high-intensity IgG3I on IgM+ B cells, then left untreated or treated with irrelevant IgG3 mAb (horizontal axis) and stained for CD32b, IgG3 and IgM (presented as in Fig. 3b). ****P < 0.0001 (two-tailed unpaired Student’s t-test after significance was obtained by one-way ANOVA of the full set). b, Calcium uptake by TLM B cells and naive B cells (below plot) obtained from chronically infected HIV-viremic individuals with medium- to high-intensity IgG3 on IgM+B cells, then left untreated or stimulated with anti-IgM (horizontal axis); results for IgG3− B cells are presented relative to those for IgG3+ B cells (‘fold’ values). Each symbol represents an individual subject (n = 9; identified by color); small horizontal lines indicate the median. *P < 0.05 and **P < 0.01 (two-tailed Wilcoxon matched-pairs signed rank test after significance was obtained by Friedman ANOVA of the full set). c, Immunoblot analysis (IB) of control proteins or cell lysates that were immunoprecipitated (IP) with anti-IgG3 or anti-CD32b (above blot) and probed with anti-IgM (15 min), anti-IgG3 (3 min) or anti-CD32b (10 min) (right margin; film exposure time (after enhanced chemiluminescence) in parentheses); lysates were prepared from B cells of a chronically infected HIV-viremic individual with an IgG3+IgM+ profile (IgG3+IgM+) or from B cells obtained from a healthy donor and treated with heat-aggregated IgG3 (HAGG) (above blot). Data are from one experiment representative of five individuals with medium-intensity IgG3 on IgM+ B cells and from one experiment representative of three independent experiments with HAGG.
Discussion
Chronic HIV viremia leads to numerous B cell perturbations2,11,37,38. Here we extended those observations by identifying IgG3 as a regulator of IgM-expressing naive and TLM B cells. The highest frequencies and intensities of IgG3 on IgM+ B cells were observed among TLM B cells of chronically infected HIV-viremic black individuals. High-throughput and high-resolution imaging revealed substantial IgG3–IgM co-localization, especially among TLM B cells, and IgM-BCR polarization was similar to what has been described for antigen-activated B cells39. We also demonstrated that the close proximity of IgG3 and IgM on IgG3+IgM+ B cells was the result of direct IgG3–IgM-BCR interactions and that CD32b, C1q and CRP contributed to the binding of IgG3 by IgM-expressing B cells and diminished IgM-BCR signaling among both TLM B cells and a small proportion of naive B cells. CD32b, C1q and CRP also contributed to non-aggregated glycosylation-dependent transfer of IgG3 derived from the serum of individuals with IgG3+IgM+ B cells to unaffected B cells.
What are the conditions in the setting of HIV infection that promote the binding of non-aggregated IgG3 to B cells during chronic viremia through interactions with IgM-BCR and involvement of CD32b, CRP and C1q? For TLM B cells, it is conceivable that IgG3-induced clustering of IgM-BCRs changes the local microenvironment of the plasma membrane to enable other proteins, including inhibitory receptors such as CD32b, to be in sufficiently close proximity to impede BCR responses. That is consistent with the observations that the addition of exogenous IgG3 both disrupted the co-localization of IgG3 with IgM-BCR and reversed the inhibitory effect of the endogenous IgG3, thus demonstrating its regulatory effect. In addition, CD32b, which becomes selectively enriched within microclusters following activation40, might also co-aggregate and contribute to the inhibitory effect on IgM-BCR. However, CD32b has also been shown to influence the BCR even in the absence of clustering41, which might explain the influence of CD32b on naive B cells, despite lower levels of IgG3 and less colocalization with IgM-BCR than that of TLM B cells. In addition, CRP, which has been shown to interact with and activate FcγRs, including CD32b31, can also associate with C1q, which in turn can interact with the Fc regions of IgG3 and IgM42. Such interactions are probably in a delicate balance, given overlapping binding sites for FcγR within the Fc region of IgG and CRP31 or C1q43. While the effects of the interacting elements were strongest on TLM B cells, it is conceivable that they extended to a lesser degree on naive B cells. This proposed distinction is consistent with the cumulative findings of lower intensities of IgG3, C1q, CRP and CD32b, less IgG3–IgM co-localization, and diminished effects of bound IgG3 on BCR signaling on IgM-expressing naive B cells than on TLM B cells.
The frequency and intensity of IgG3 on IgM-expressing B cells were clearly influenced by a number of factors, best described as a spectrum of biochemical, temporal, genetic and, possibly, antigen-driven effects. Even among TLM B cells, for which the intensity of surface-bound IgG3 and polarization with IgM-BCR were strongest, not all clusters contained CD32b, consistent with evidence showing that IgG3 directly interacts with IgM-BCR rather than with its FcγR. Genetic background might help explain why race was a determining factor in the frequency, intensity and (possibly) lability of IgG3+IgM+ B cells. There is a high degree of polymorphism in the constant regions of IgG3 that might, along with polymorphisms in CD32b, influence associations with C1q and CRP44. In addition, polymorphisms in CD32b, which have been shown to influence the mobility and activation of CD32b45, have been associated with susceptibility to autoimmune and infectious diseases46,47. Other factors might have modulated IgG3–IgM-BCR activities and warrant further investigation; these include several potential receptors for C1q expressed on B cells48, and our finding of increased binding of soluble IgM on TLM B cells due to expression of the IgM Fc receptor TOSO, as well as the presence of IgG3 on non-B cells from the peripheral blood of individuals with an IgG3+IgM+ B-cell profile (L.K. et al., data not shown).
Finally, while serologic concentrations of IgG3 were clearly a factor in distinguishing individuals with low-intensity IgG3 on IgM+ B cells from those with high-intensity IgG3 on IgM+ B cells and might help to explain the paucity of IgG3+IgM+ B cells in early HIV viremia, the nature of the interactions and possibly specificity of IgG3 for IgM-BCR remains unclear. The partial occlusion of the Fc region of IgG3 on IgG3+IgM+ B cells and the low yet detectable binding of HIV antigen to bound IgG3 would suggest that the interactions occur outside the antigen-binding site. While many modes of interaction are possible49, the precise mode involved here might be difficult to address without identifying the IgG3-producing B cells. While we found no evidence of greater HIV reactivity among the IgG3 of individuals with IgG3+IgM+ B cells than among other IgG isotypes or HIV-viremic individuals without this profile, IgG3 usage is common among antibodies that are specific for the membrane-proximal external region of the HIV envelope protein subunit gp41, and a majority of these IgG3 antibodies are autoreactive and/or polyreactive, reacting with host cell membrane proteins50. Thus, IgG3 that binds to the IgM-BCR might arise in the setting of persistent HIV viremia and act as a sensor of chronic activation.
Methods
Study subjects and ethics approval.
HIV-infected individuals provided written informed consent for and were enrolled under NIAID Institutional Review Board-approved protocol 02-I-0202. HIV-negative individuals were enrolled under NIH Clinical Center Institutional Review Board-approved protocol 99-CC-0168. We complied with all relevant ethical regulations. Leukapheresis products were obtained from HIV-infected and HIV-negative individuals. Three groups of HIV infected individuals were studied (Table 1 and Supplementary Table 1): early infected HIV-viremic individuals, defined as having been infected within 6 months and not receiving ART; chronically infected HIV-viremic individuals, defined as having been infected at least 6 months and not receiving ART; HIV-infected individuals on ART with suppressed HIV plasma viremia (< 40 or < 50 copies of HIV RNA per ml). Protocol participation was contingent on HIV-infected study subjects’ maintaining a primary care physician, and they were encouraged to initiate ART, per current HIV treatment guidelines of the US Department of Health and Human Services (http://aidsinfo.nih.gov/contentfiles/lvguidelines/AdultandAdolescentGL.pdf).
B cell isolation.
Peripheral blood mononuclear cells (PBMCs) were obtained by density-gradient centrifugation and either were used fresh or were from cryopreserved samples. Mature (CD10−) B cells were isolated from PBMCs by negative magnetic bead–based selection using a B cell enrichment cocktail (StemCell Technologies) that was supplemented with tetrameric mAb to CD10 (clone FR4D11; StemCell Technologies).
Flow and imaging cytometry analyses.
All antibodies (and some other reagents) are identified in Supplementary Table 2. Multicolor flow and imaging cytometry were performed using monoclonal or polyclonal antibodies to the following: CD20-APC-H7 (clone 2H7), tIgG-PE-Cy7 (clone G18–145), IgD-APC-H7 (clone IA6–2), CD32-PE (clone FLI8.26), kappa-PE (clone TB28–2), lambda-BV421(clone JDC-12), and CD27-BUV395 (clone L128) (all from BD Biosciences); tIgG-BV421 (clone M1310G05), CD3-BV650 (clone OKT3), CD21-FITC (clone BU32), IgM-BV510 or -BV570 (clone MHM-88), IgD-PE-Cy7 or BV605 (clone IA6–2), CD27-PerCP-Cy5.5 (clone O323) and CD21-PerCP-Cy5.5 (clone BU32) (all from BioLegend); C1q-biotin (clone JL-1), streptavidin-Pacific Green, streptavidin-PE (ThermoFisher); IgG1-PE (clone HP6001), and IgG3-Alexa Fluor 647 (clone HP6050) (all from Southern Biotech); CRP-FITC (Cat# #ab34659; Abcam); and tIgG Alexa Fluor 405 (clone ICO-97; Novus). Flow cytometry was performed on an LSRFortessa (BD Biosciences), with data analyses performed using FlowJo software (TreeStar). In preliminary analyses, the CD32b-specific clone ch8B5N297Q-Alexa Fluor 448 (Macrogenics) was used to verify that the pan-CD32 clone FLI8.26 could be used to evaluate CD32b expression on B cells. To distinguish between secreted forms of immunoglobulin and expressed forms of immunoglobulin, B cells from HIV-viremic individuals were treated with 0.25% trypsin (Lonza) in PBS for 5 min at 37 °C, followed by staining for flow cytometry. To determine mean fluorescent intensity (MFI) for cell-bound IgG3, IgG3-expressing B cells were excluded by gating on tIgG+IgG3+ B cells that were making clear diagonal pattern, as illustrated (Supplementary Fig. 1c). IgG3–IgM co-localization Fig. 1c). IgG3–IgM co-localization was analyzed using an ImageStream Mark II Imaging Cytometer and IDEAS Software version 6.2 (EMD Millipore). Fluorescent images were collected at × 60 magnification.
Total internal reflection fluorescence (TIRF) microscopy.
For co-localization analyses, TIRF microscopy was carried out as described previously with modifications51. In brief, B cells were placed in poly-d-lysine–coated chambers and were stained with monoclonal or polyclonal antibodies to the following: IgG3-Alexa Fluor 647 (flow cytometry mAb); IgM-DyLight 549 (Cat# 109–507-043) or tIgG-DyLight 549 (Cat# 109–505-008) (both from Jackson ImmunoResearch Laboratories); and CD32-FITC (clone FLI8.26) or CD45-FITC (clone H130) (both from BD Biosciences). After cells were fixed with 4% paraformaldehyde, TIRF was performed using an Olympus IX-81 inverted microscope equipped with a 100 × objective lens (Olympus), a TIR controlling system, EMCCD camera, and 488-, 561- and 640-nm laser lines controlled by Metamorph software (Molecular Devices). Pearson’s co-localization coefficient (R) between any two molecules was calculated by intensity correlation analysis from single cell regions of interest after background subtraction, image filtering and masking of background noise using a MATLAB program (MathWorks). For classification of clustered distribution versus uniform distribution of IgM on cell surfaces, contact areas of B cells stained for CD32b, IgM and IgG3 were analyzed by Image J software. In brief, after the background area on TIRF images was established using the automatic threshold function, clustered areas within positive signal pixels were defined using the particle analysis function. Particle sizes over 0.5 μm2 were used to define IgM clustering: cluster-positive cells were counted as clustered cells, and cells showing homogeneous circular distribution of IgM with no distinct clusters were counted as uniform.
Isotype-specific PCR.
For analysis of immunoglobulin-encoding mRNA, tIgG+IgM−, tIgG−IgM+ and IgG3+IgM+ B cells from HIV-viremic individuals with high-intensity IgG3 on IgM+ B cells were sorted into Trizol LS (Sigma) using a FACSAria II (BD Biosciences). Total RNA was isolated, was concentrated using the RNeasy Micro Kit (Qiagen) and was reverse-transcribed using oligo-dT (Integrated DNA Technologies) and SuperScript II (ThermoFisher). Isotype-specific PCR was performed using a common, sense primer in the framework 3 region and unique antisense primers specific for each isotype, as previously described52. After visualization of PCR products by agarose-gel electrophoresis, relative isotype expression was calculated by quantification of band intensities using Image Lab Software (Bio-Rad).
Phosphorylation assay.
B cells stained with flow cytometry mAbs to CD20, CD27, CD21, IgG3, and IgD (identified above) were stimulated with 10 μg/ml goat F(ab’)2 anti-IgM (Jackson ImmunoResearch Laboratories; Cat# 109–006-129) at 37 °C for 2 min. For the detection of phosphorylated signaling intermediates, cells were fixed and permeabilized using BD Cytofix and Phosflow Perm/Wash buffers (BD Biosciences) and were stained separately with PE-conjugated mAbs to Syk phosphorylated at Tyr348 (clone I120–722), Btk phosphorylated at Tyr223 (clone N35–86) and PLC-γ2 phosphorylated at Tyr759 (clone K86–689.37) (all from BD Biosciences). Flow cytometry was performed on an LSRFortessa.
Calcium influx.
B cells were incubated with 0.2 μM Indo-1 AM dye (Life Technologies) containing 2% Pluronic F-127 (Life Technologies) for 20 min at 37 °C. Indo-1 AM–loaded B cells were labeled with flow-cytometry antibodies to CD20, CD27, CD21, IgG3, IgD, C1q and CRP (identified above). Changes in fluorescence intensity were monitored on an LSRFortessa flow cytometer. Readings were recorded for 3–4 min to establish a baseline measurement, followed by BCR stimulation induced by anti-IgM (identified above). IgD (antibody identified above) was used to identify IgM-expressing B cells, due to the interference between the anti-IgM used to stimulate B cells and the mAb used to detect IgM expression (both antibodies identified above), and because both IgD and IgM were expressed on the vast majority of cells involved in these analyses (data not shown). The intracellular calcium concentration was quantified by calculation of the ratio of Indo-1 AM emission at 405 nm to that at 475 nm (bound/unbound).
Serum analyses.
The concentration of IgG subclasses in serum was measured by multiplex cytometric bead array (CBA) using a FACSArray instrument (BD Biosciences), with the exception that IgG1 was calculated by subtraction of the concentrations of IgG2–4 from tIgG. The CBA analyses were conducted according to the manufacturer’s instructions. Data were analyzed using FCAP Array Software (version 1.01; BD Biosciences), which converts sample mean fluorescent intensity values into a concentration using the standard curve.
Isolation of IgG3 from serum.
IgG3 was isolated from serum using two methods: polyethylene glycol (PEG) and agarose-bead-enrichment. For PEG precipitation, serum from HIV-viremic individuals with high- or low-intensity IgG3 on IgM+ B cells was mixed with an equal volume of 7% PEG 8000 (Sigma) and sodium borate buffer (0.1 M Na2B4O, 0.075 M NaCl), pH 8.4, as described previously53. PEG-precipitate concentrations of IgG subclasses were measured by multiplex human isotyping panel using a Bio-Plex 200 instrument (Bio-Rad). Assays were performed according to the manufacturer’s instructions. Agarose-bead IgG3 enrichment was performed using CaptureSelect IgG3 Affinity Matrix (ThermoFisher), according to the manufacturer’s instructions.
Size-exclusion chromatography (SEC).
SEC was performed on an ÄKTA prime plus liquid chromatography system using a Superdex200 10/300GL (GE Healthcare) with a mobile phase of Tris buffer (0.1 M Tris and 50 mM NaCl), pH 8, at a flow rate of 0.5 ml/min. Absorbance at 280 nm was used to detect proteins. The injection amount was 250 μ g protein in a volume of 250 μ l. The elution volume for fraction SEC-1 was at 7–10 ml, and the elution volume for fraction SEC-2 was at 11–15 ml.
N-glycosylation analyses.
Glycans were removed from PEG-IgG3 using enzymatic digestion with peptide–N-glycosidase F (PNGase F, New England Biolabs) and endoglycosidase S2 (EndoS2, Genovis), according to the manufacturer’s instructions. In brief, 20 μ g of PEG-IgG3 was incubated for 30 min at 37 °C with or without 500 units of PNGase F or 40 units of EndoS2. To determine the effect of glycan removal, binding of enzyme-treated PEG-IgG3 to the B cells from HIV-negative individuals was compared with that of untreated PEG-IgG3.
Immunoprecipitation.
B cells (5 × 106) were lysed in ice-cold lysis buffer (PBS/1% Triton X-100 supplemented with protease inhibitor cocktail (Roche)) for 1 h at 4 °C with mixing and lysates were incubated overnight with mAbs to IgG3 (Southern Biotech; clone HP6050, biotinylated) or CD32 (BioRad; clone AT10, biotinylated), followed by incubation with Pierce Streptavidin magnetic beads (ThermoFisher) for 1 h at 4 °C with mixing. Co-immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblot analysis. Control human proteins consisted of monoclonal recombinant IgG3 (Bio-Rad; clone ABD18705), recombinant CD32b/c (R&D Systems; Cat# 1875-CD-050), and plasma-derived IgM (Abcam; Cat# ab91117).
SDS-PAGE and immunoblot analysis.
Protein purity and identity were evaluated by SDS-PAGE on a 10% Tris-Glycine gel (Bio-Rad). Separation was performed using a Mini-PROTEAN Electrophoresis System (Bio-Rad) at constant voltage of 90 V for 40 min, followed by 140 V for 45 min and transfer to a PVDF membrane (Bio-Rad). The membrane was blocked with 4% milk powder in 0.1% Tween in PBS for 1 h at 20 °C. Membranes were sequentially incubated with goat anti-IgM-HRP (Southern Biotech; Cat# 2020–05) or primary antibody rabbit anti-C1q (Novus; Cat# H00000712-D01P), mouse anti-CRP (R&D Systems; clone 232026), rabbit anti-CD32 (GeneTex; Cat# GTX133371) or rabbit anti- IgG3 (Abcam; clone EPR4419), followed by the secondary antibody, goat anti-rabbit-HRP (Bio-Rad; Cat# 1662408EDU) or goat anti-mouse-HRP (Bio-Rad; Cat# 1000450), diluted in 4% milk powder in 0.1% Tween in PBS.
In vitro binding assay.
B cells (5 × 105) of HIV-negative individuals were incubated for 20 min at 37 °C with 5 μ g of heat-aggregated IgG3 (Sigma; Cat# I5654) or (Bio-Rad; clone ABD18705), PEG-IgG3, fractions SEC-1 and SEC-2 or agarose-bead-enriched IgG3, isolated from the serum of HIV-viremic individuals. For blocking experiments, B cells of HIV-negative individuals were pre-treated with 10 μ g/ml control goat IgG (Cat# AB-108-C) or goat anti-CD32 (Cat# AF1330) (both from R&D Systems) for 30 min at 4 °C and then were treated with PEG-IgG3. To define the association of CD32b with bound IgG3, B cells from HIV-viremic individuals with a strong IgG3+IgM+ pattern were treated with 10 μg/ml control goat IgG or goat anti-CD32 (both identified above) for 10–20 min at 37 °C. To evaluate disruption of bound IgG3, 5 × 105 B cells from HIV-viremic individuals with a strong IgG3+IgM+ pattern were treated with 5 μ g of IgG3 (Bio-Rad; clone ABD18705) for 15 min at 37 °C. To define involvement of C1q and CRP in the binding of IgG3 to the IgM-expressing B cells, 40 μg of PEG-IgG3 was incubated for 60 min at 4 °C with sheep anti-rabbit IgG M-280 Dynabeads (ThermoFisher) pre-conjugated to 2 μ g control rabbit IgG (R&D Systems; Cat# AB-105-C), rabbit anti-C1q (Dako; Cat# A0136) or rabbit anti-CRP (Abcam; clone Y284) or combinations thereof, according to the manufacturer’s instructions. Depletion was also performed on serum before PEG precipitation or after PEG-IgG3 fractionation. PEG-IgG3 depleted of C1q or CRP or both, or control IgG–treated PEG-IgG3, was incubated with B cells as described above. The cells were stained and flow cytometry was performed on an LSRFortessa.
HIV specificity assays.
B cells and PEG precipitate were evaluated for HIV specificity by ELISA and flow cytometry, respectively, using soluble HIV YU2-derived gp140 trimers, as previously described6.
Statistical analyses.
Data were analyzed with Prism version 7 (Graphpad Software). P values <0.05 were considered significant. The test used for each experiment and P value ranges are indicated in figure legends.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank the study volunteers for their participation in this study; C. Rehm and S. Jones for specimen procurement; A.S. Fauci for discussions and suggestions. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing interests.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41590-018-0180-5.
Data availability.
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary information is available for this paper at https://doi.org/10.1038/s41590-018-0180-5.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lane HC et al. Abnormalities of B-cell activation and immunoregulation in patients with the acquired immunodeficiency syndrome. N. Engl. J. Med 309, 453–458 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Moir S & Fauci AS B-cell responses to HIV infection. Immunol. Rev 275, 33–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moir S et al. B cells in early and chronic HIV infection: evidence for preservation of immune function associated with early initiation of antiretroviral therapy. Blood 116, 5571–5579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moir S et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med 205, 1797–1805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho J et al. Two overrepresented B cell populations in HIV-infected individuals undergo apoptosis by different mechanisms. Proc. Natl. Acad. Sci. USA 103, 19436–19441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meffre E et al. Maturational characteristics of HIV-specific antibodies in viremic individuals. JCI Insight 1, e84610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karnell JL et al. Role of CD11c+ T-bet+ B cells in human health and disease. Cell. Immunol 321, 40–45 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Naradikian MS, Hao Y & Cancro MP Age-associated B cells: key mediators of both protective and autoreactive humoral responses. Immunol. Rev 269, 118–129 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Portugal S, Obeng-Adjei N, Moir S, Crompton PD & Pierce SK Atypical memory B cells in human chronic infectious diseases: An interim report. Cell. Immunol 321, 18–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pupovac A & Good-Jacobson KL An antigen to remember: regulation of B cell memory in health and disease. Curr. Opin. Immunol 45, 89–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knox JJ, Kaplan DE & Betts MR T-bet-expressing B cells during HIV and HCV infections. Cell. Immunol 321, 26–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnett BE et al. Cutting edge: B cell-intrinsic T-bet expression is required to control chronic viral infection. J. Immunol 197, 1017–1022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubtsov AV et al. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood 118, 1305–1315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubtsova K et al. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J. Clin. Invest 127, 1392–1404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng SL, Szabo SJ & Glimcher LH T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. USA 99, 5545–5550 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pekkarinen PT et al. Dysregulation of adaptive immune responses in complement C3-deficient patients. Eur. J. Immunol 45, 915–921 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Mortensen R et al. Adaptive immunity against Streptococcus pyogenes in adults involves increased IFN-γ and IgG3 responses compared with children. J. Immunol 195, 1657–1664 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Knox JJ et al. T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight 2, e92943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckner CM et al. Characterization of plasmablasts in the blood of HIV-infected viremic individuals: evidence for nonspecific immune activation. J. Virol 87, 5800–5811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux KH, Strelets L & Michaelsen TE Flexibility of human IgG subclasses. J. Immunol 159, 3372–3382 (1997). [PubMed] [Google Scholar]
- 21.Lefranc MP & Lefranc G Human Gm, Km, and Am allotypes and their molecular characterization: a remarkable demonstration of polymorphism. Methods Mol. Biol 882, 635–680 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Michaelsen TE, Sandlie I, Bratlie DB, Sandin RH & Ihle O Structural difference in the complement activation site of human IgG1 and IgG3. Scand. J. Immunol 70, 553–564 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Morell A, Terry WD & Waldmann TA Metabolic properties of IgG subclasses in man. J. Clin. Invest 49, 673–680 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dugast AS et al. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur. J. Immunol 44, 2925–2937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung AW et al. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163, 988–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharf O et al. Immunoglobulin G3 from polyclonal human immunodeficiency virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J. Virol 75, 6558–6565 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yates NL et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci. Transl. Med 6, 228ra39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yates NL et al. Multiple HIV-1-specific IgG3 responses decline during acute HIV-1: implications for detection of incident HIV infection. AIDS 25, 2089–2097 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wesemann DR et al. Immature B cells preferentially switch to IgE with increased direct Sμ to Sε recombination. J. Exp. Med 208, 2733–2746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Depoil D et al. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat. Immunol 9, 63–72 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Lu J et al. Structural recognition and functional activation of FcγR by innate pentraxins. Nature 456, 989–992 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bang R et al. Analysis of binding sites in human C-reactive protein for FcγRI, FcγRIIA, and C1q by site-directed mutagenesis. J. Biol. Chem 280, 25095–25102 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Bharadwaj D, Stein MP, Volzer M, Mold C & Du Clos TW The major receptor for C-reactive protein on leukocytes is Fcgγ receptor II. J. Exp. Med 190, 585–590 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruhns P et al. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Seeling M, Brückner C & Nimmerjahn F Differential antibody glycosylation in autoimmunity: sweet biomarker or modulator of disease activity? Nat. Rev. Rheumatol 13, 621–630 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Sjögren J et al. EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyses N-linked glycans on IgG and α 1-acid glycoprotein. Biochem. J 455, 107–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noto A & Pantaleo G B-cell abnormalities and impact on antibody response in HIV infection. Curr. Opin. HIV AIDS 12, 203–208 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Pallikkuth S, de Armas L, Rinaldi S & Pahwa S T follicular helper Cells and B cell dysfunction in aging and HIV-1 infection. Front. Immunol 8, 1380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce SK & Liu W The tipping points in the initiation of B cell signalling: how small changes make big differences. Nat. Rev. Immunol 10, 767–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W, Won Sohn H, Tolar P, Meckel T & Pierce SK Antigen-induced oligomerization of the B cell receptor is an early target of FcγRIIB inhibition. J. Immunol 184, 1977–1989 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davey AM & Pierce SK Intrinsic differences in the initiation of B cell receptor signaling favor responses of human IgG+ memory B cells over IgM+ naive B cells. J. Immunol 188, 3332–3341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sörman A, Zhang L, Ding Z & Heyman B How antibodies use complement to regulate antibody responses. Mol. Immunol 61, 79–88 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Ugurlar D et al. Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 359, 794–797 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Hogarth PM & Pietersz GA Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov 11, 311–331 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Xu L et al. Impairment on the lateral mobility induced by structural changes underlies the functional deficiency of the lupus-associated polymorphism FcγRIIB-T232. J. Exp. Med 213, 2707–2727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clatworthy MR et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcγRIIb reduce susceptibility to malaria. Proc. Natl. Acad. Sci. USA 104, 7169–7174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willcocks LC et al. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 107, 7881–7885 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son M, Diamond B & Santiago-Schwarz F Fundamental role of C1q in autoimmunity and inflammation. Immunol. Res 63, 101–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nezlin R, & Ghetie V Interactions of immunoglobulins outside the antigen-combining site. Adv. Immunol 82, 155–215 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Molinos-Albert LM, Clotet B, Blanco J & Carrillo J Immunologic insights on the membrane proximal external region: a major human immunodeficiency virus type-1 vaccine target. Front. Immunol 8, 1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohn HW, Krueger PD, Davis RS & Pierce SK FcRL4 acts as an adaptive to innate molecular switch dampening BCR signaling and enhancing TLR signaling. Blood 118, 6332–6341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerutti A et al. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center and plasmacytoid phenotypic differentiation in a human monoclonal IgM+IgD+ B cell line. J. Immunol 160, 2145–2157 (1998). [PMC free article] [PubMed] [Google Scholar]
- 53.Brunner M & Sigal LH Immune complexes from serum of patients with Lyme disease contain Borrelia burgdorferi antigen and antigen-specific antibodies: potential use for improved testing. J. Infect. Dis 182, 534–539 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.