Abstract
Does sodium dodecyl sulfate (SDS) denature proteins through electrostatic SDS-protein interactions? We show that a protein completely lacking charged side chains is unfolded by SDS in a manner similar to charged proteins, revealing that formal protein charges are not required for SDS-induced protein unfolding or binding.
Main Text
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) owes its huge success as an analytical tool in protein science to SDS’s ability to denature proteins (1). Early crystallographic studies with monomeric SDS highlighted electrostatic interactions between the sulfate headgroup and cationic protein side chains as well as hydrophobic interactions between the alkyl chains and protein (2). The relative importance of these two driving forces remains controversial (3, 4). Increasing chain length increases both surfactant self-association and the tendency to denature proteins (5, 6, 7). In contrast, nonionic surfactants largely do not interact with globular proteins (8, 9) and maintain the folded state of membrane proteins (10, 11). This highlights the denaturing role of SDS’s charged headgroup.
Ubiquitin-SDS studies (4) showed that positive protein residues were important for the interaction, not by acting as specific SDS binding partners but rather by providing a positive electrostatic potential to patches of the protein surface, promoting initial binding of SDS to predominantly hydrophobic amino acids in these regions. Neutralizing some of the positive charges in ubiquitin by acetylation slightly alters the amount of bound SDS before and after the unfolding of the protein but otherwise does not affect unfolding. Acetylating Lys residues in bovine carbonic anhydrase made the protein highly negative and dramatically increased SDS-induced unfolding half-lives from minutes to days (12), emphasizing the importance of charges on the kinetics in SDS unfolding.
A more radical approach to studying electrostatics in SDS denaturation is to entirely remove all charges from the protein, removing both attractive and repulsive electrostatic interactions between the protein and SDS. We recently developed a chargeless protein completely lacking ionizable side chains (13). We used the all-β 110-residue cellulose-binding domain (EXG:CBM) of a Cellulomonas fimi xylanase, which, in the wild-type state, only had 1 Lys, 1 Asp, 1 Arg, and 1 His, giving a total positive charge of 1–2 at a neutral pH. These charged side chains are replaced by neutral residues in the mutant EXG:CBMQQQW (i.e., mutations K28Q, D36Q, R68Q, and H90W), strongly diminishing the electrostatic surface potential. By comparing SDS denaturation of wild-type EXG:CBM and EXG:CBMQQQW, we identify charge-dependent steps of the SDS-induced unfolding mechanism.
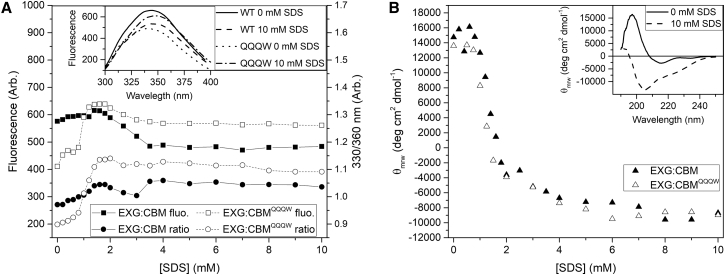
We monitored Trp fluorescence and far-ultraviolet circular dichroism (CD) signals for both EXG:CBM variants as a function of [SDS] (Fig. 1). Wild-type EXG:CBM has 5 Trp residues (positions 12, 17, 38, 54, and 72), of which Trp17, Trp54, and Trp72 are structured, aligned, and surface exposed, making up the cellulose-binding site, whereas Trp 12 and 38 are buried in the hydrophobic core. Trp12 is packed against Phe42, Phe44, Phe86, and Phe100, whereas Trp38 is flanked by several smaller nonaromatic residues (Val27, Val35, Leu40, Asn69, and Leu102). The mutant EXG:CBMQQQW contains an extra Trp residue at position 90, required to maintain stability when substituting His90 (13). Apart from minor effects of the extra buried Trp in EXG:CBMQQQW (we see a small reduction in overall Trp fluorescence in the native state but not in the SDS-denatured state, which may be caused by quenching due to residue 90’s spatial proximity to Trp12 or Trp38 in the crystal structure), the two variants follow the same pattern. Between 0 and 1.6 mM SDS, the fluorescence increases, followed by a drop between 2 and 4 mM, and finally a stable signal from 4 mM onward is obtained, coinciding with the onset of micelle formation (the critical micelle concentration (cmc) of SDS under these conditions is ∼4 mM). During the initial increase in intensity, we also observed a red shift in the fluorescence emission maximum (confer the 330:360 nm ratio; Fig. 1 A). By CD (Fig. 1 B); a baseline at 0–1 mM SDS is followed by a rapid change between 1 and 2 mM and a slower change from 2 to ∼4 mM SDS, after which the signal remains stable. The large CD spectral change (Fig. 1 B, inset) indicates conversion from mainly β-sheet to mainly α-helix in the SDS-EXG complex.
Figure 1.
Equilibrium fluorescence (A) and circular dichroism (CD) (B) as functions of [SDS]. (A) Samples were excited at 280 nm, and emission was measured at 360 nm. The right axis shows the 330:360 nm ratio. The inset shows the emission spectra at 0 and 10 mM SDS. (B) The CD signal at 200 nm in which the difference in ellipticity between the native and SDS-denatured state is greatest. The inset shows the far-ultraviolet CD spectrum of EXG:CBMQQQW in 0 and 10 mM SDS.
Thus, fluorescence and CD both indicate a major change in protein conformation between 1 and 2 mM, with minor additional changes between 2 and 4 mM. Despite the low number of charged residues in EXG:CBM and only the N- and C-termini of EXG:CBMQQQW, SDS remains a strong protein denaturant.
To investigate how a lack of charge affected SDS binding, we turned to isothermal calorimetry (ITC). Heat flow from a titration of SDS into protein solutions often reveals additional spectroscopically invisible protein-surfactant interaction phases. Further, varying [protein] provides the SDS/protein stoichiometry at each phase in the titration (8, 14).
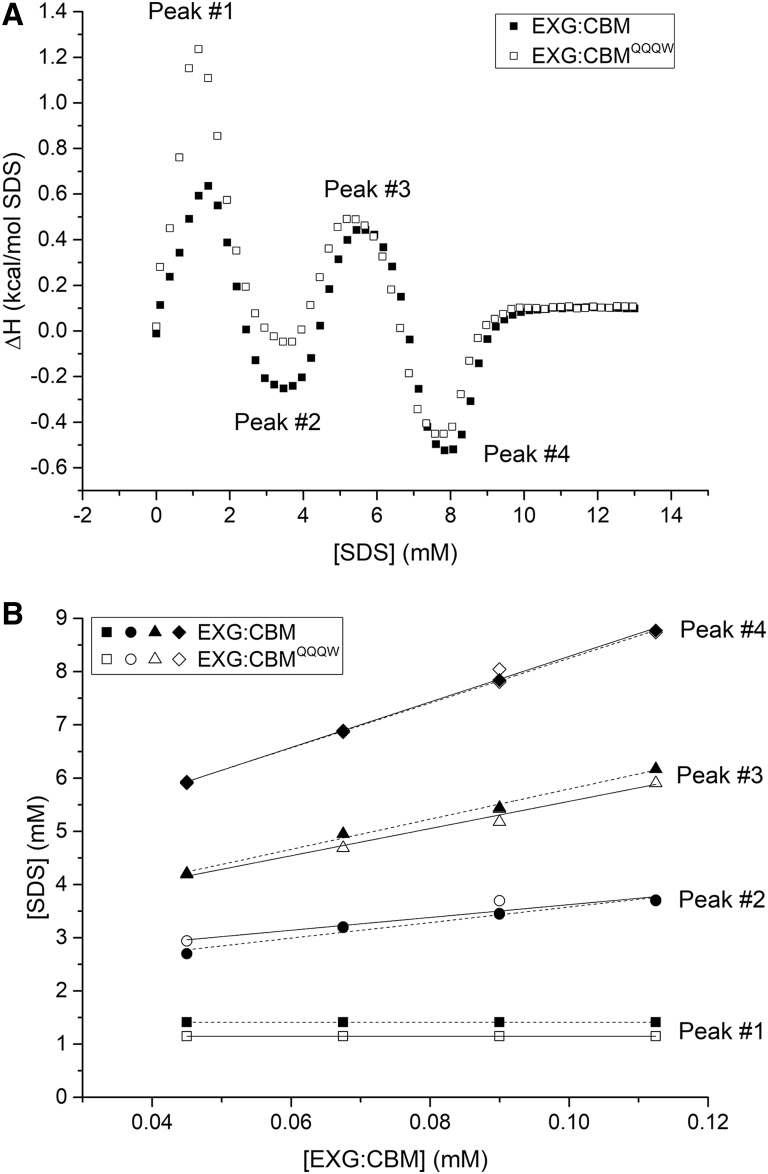
Remarkably, EXG:CBM and EXG:CBMQQQW showed nearly identical, complex titration behaviors with four distinctive peaks (two maxima and two minima) below or at the cmc (Fig. 2 A). There are minor variations in the magnitude of the enthalpic signal; this may be related to the removal of the charged side chains. The last three transitions depended linearly on [protein]; the amplitude but not the position of the first peak was influenced by [protein] (Fig. 2 B). The calculated number of bound SDS molecules at each step did not differ significantly between the two EXG variants (Table 1). At saturation (peak no. 4 in the thermograms), both EXG variants bound ∼1.1 g SDS per gram of protein, only slightly lower than the ∼1.4 g/g seen for most globular proteins (15, 16). Other β-sheet proteins also bind a lower amount of SDS (∼1.2 g/g) (8), and the presence of a disulfide bond in EXG:CBM could further restrict complete exposure of the polypeptide chain to SDS micelles in the unfolded state.
Figure 2.
Titration of SDS into EXG:CBM and EXG:CBMQQQW followed by ITC. (A) Enthalpograms from 1 mg/mL protein samples (B) Peak positions (numbered as in A) as functions of [EXG:CBM] fitted to Eq. 1 (in the Supporting Materials and Methods).
Table 1.
Binding Stoichiometry at the Four ITC Peaks
| Protein | No. Transition Peak | [SDS]aq (mM)a | No. Bound SDSa |
|---|---|---|---|
| EXG:CBM | 1 | 1.4 | – |
| 2 | 2.1 ± 0.2 | 14.6 ± 2.0 | |
| 3 | 3.0 ± 0.1 | 28.4 ± 1.5 | |
| 4 | 4.04 ± 0.02 | 42.1 ± 0.2 | |
| EXG:CBMQQQW | 1 | 1.1 | – |
| 2 | 2.4 ± 0.2 | 12.0 ± 2.5 | |
| 3 | 3.0 ± 0.2 | 25.6 ± 2.0 | |
| 4 | 4.0 ± 0.2 | 42.8 ± 2.4 |
Parameters from fitting to Eq. 1 (in the Supporting Materials and Methods).
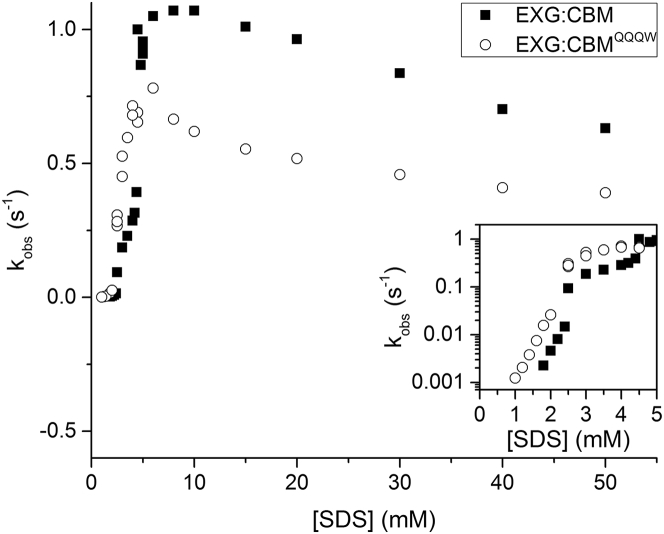
Although removal of positive charges from EXG:CBM to EXG:CBMQQQW did not affect the stoichiometry and number of steps in the protein-surfactant interaction, it could alter denaturation kinetics. Kinetic measurements (Fig. 3) were made using stopped-flow fluorescence (<2.5 mM SDS) and manual-mixing CD spectroscopy (>2.5 mM SDS).
Figure 3.
Rate constants of unfolding in SDS obtained from kinetic studies with EXG:CBM and EXG:CBMQQQW. Kinetics below 2.5 mM SDS were measured with CD, and kinetics from 2.5 mM SDS and up were measured with stopped-flow fluorescence. The inset shows a zoom-in of the pre-cmc region. The cmc of SDS in our buffer system is ∼4mM.
The mutant showed similar kinetics with two distinctive unfolding modes. Up to ∼3 mM SDS, kinetics are slow and show a linear relationship between log (kunf) and [SDS]. Above the cmc, kinetics are faster but decrease with [SDS], suggesting inhibition by micellar SDS, as observed for other proteins (8, 17). The slow rate constants are comparable to the conventionally charged protein S6 unfolded at pH 8 (at which S6 has a small negative net charge) (18) and the β-sheet proteins TII27 and TNfn3 (8).
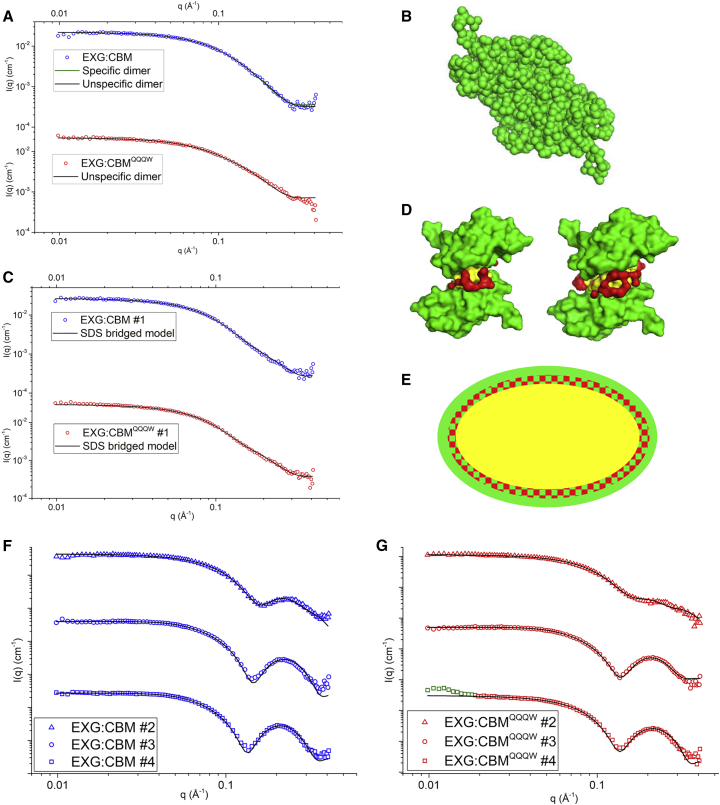
To investigate the structure of the protein-SDS complexes, we recorded small angle X-ray scattering (SAXS) data for both proteins alone and at [SDS] corresponding to the four ITC peak positions, which reflect characteristic transitions in the binding of SDS to protein (19, 20).
SDS-free data could not be fitted with the EXG NMR monomer structure alone (21). Using an unspecific dimerization structure factor in the modeling, we obtained good fits, which yielded aggregation factors of 1.64 and 1.45 for EXG:CBM and EXG:CBMQQQW, respectively, indicating a certain level of dimerization. For more specific analysis, we generated specific dimer Protein Data Bank (PDB) files from the monomer PDB file and fitted a linear combination of the monomer and dimer to the SAXS data. For wild-type EXG:CBM, a very good fit was obtained using 27.5% monomer and 72.5% dimer (Fig. 4). Sedimentation equilibrium experiments have previously shown that 40% of EXG:CBM is a dimer at 1.1 mg/mL (21), consistent with most of the protein being in the dimer form at the 2 mg/mL used in the SAXS experiment. EXG:CBMQQQW did not fit well as a linear combination of monomer and any of the specific dimers we managed to generate, despite being well fitted by the unspecific dimer model.
Figure 4.
(A) SAXS data of SDS-free EXG:CBM fitted to the indicated models. (B) The structure of the specific dimer model. (C) SAXS data of EXG:CBM at the first ITC peak fitted to the SDS bridged model, shown in (D) for EXG:CBM (left) and EXG:CBMQQQW (right),. Green: protein, yellow: surfactant alkyl chains, red: surfactant headgroups. (E) A schematic presentation of the core-shell model used for fitting the data in (F) and (G). The hydrocarbon core is shown in yellow, and the shell with headgroups and protein is shown in checkered red/green and green. (F and G) SAXS data corresponding to peaks no. 2–4 fitted by the core-shell model with the resulting parameters shown in Table 2 Green points in (G) were omitted in the fitting. To see this figure in color, go online.
SAXS data of the two EXG:CBM proteins corresponding to the first ITC peak fitted almost equally well to a model of two EXG:CBM proteins bridged by 4.37 and 6.60 SDS molecules for EXG:CBM and EXG:CBMQQQW, respectively (Fig. 4). Fitting to a more conventional model of protein-decorated SDS micelles (9, 22) was unsuccessful for both SDS-protein complexes at peak no. 1, whereas spectra recorded at [SDS] corresponding to ITC binding peak positions no. 2–4 fitted very well (Fig. 4; χ2 values: 1.87–3.94). This model provides the aggregation number for SDS in the micelle (Nagg), the amount of protein per complex (MProt), and the ellipticity parameter ε (ε = 1 is a sphere, ε < 1 is an oblate ellipse, and ε > 1 is a prolate ellipse; Table 2). Binding point no. 2 gave an oblate micelle, whereas binding points no. 3–4 gave prolate micelles. Unsurprisingly, higher [SDS] increased Nagg, with very similar values for EXG:CBM and EXG:CBMQQQW; these values were comparable to, although slightly larger than, the binding numbers derived from ITC. We calculated that the number of protein molecules per complex decreased from ∼1.8 to ∼1.2 as [SDS] increased. Thus, at lower [SDS], a larger proportion of complexes contains at least two protein molecules.
Table 2.
Parameters from Protein-SDS Core-Shell Model Fitting of ITC Peaks No. 2–4
| ITC Peak | EXG:CBM |
EXG:CBMQQQW |
||||
|---|---|---|---|---|---|---|
| No. 2 | No. 3 | No. 4 | No. 2 | No. 3 | No. 4 | |
| Dhead (Å)a | 9.24 ± 0.07 | 7.16 ± 0.05 | 6.82 ± 0.05 | 8.66 ± 0.09 | 9.62 v 0.04 | 6.28 ± 0.14 |
| Rcore (Å)a | 16.78 ± 0.14 | 12.09 ± 0.06 | 13.02 ± 0.05 | 19.08 ± 0.18 | 12.24 ± 0.07 | 12.92 ± 0.09 |
| εb | 0.40 ± 0.01 | 1.88 ± 0.03 | 1.79 ± 0.03 | 0.25 ± 0.01 | 1.56 ± 0.03 | 1.92 ± 0.06 |
| Naggb | 22.4 | 39.5 | 47.0 | 20.5 | 35.0 | 49.2 |
| MProt (kDa)b | 18.6 | 16.8 | 13.4 | 20.5 | 16.2 | 14.1 |
| No. protein/complex | 1.7 | 1.5 | 1.2 | 1.8 | 1.5 | 1.3 |
| χ2 | 3.07 | 3.11 | 2.41 | 2.56 | 1.87 | 3.94 |
Rcore is the radius of the micelle, whereas Dhead is the thickness of the layer around it in the core-shell model, which includes headgroup and protein.
Ellipticity parameter, aggregation number, and amount of protein per micelle, as described in the text.
In summary, at sufficiently high SDS stoichiometry, EXG:CBM forms the same conventional core-shell complex as other (charged) proteins; thus, charge is not essential to guide SDS-protein interactions leading to this structure. At low stoichiometry, EXG:CBM has a propensity to form dimers. SDS is known to induce dimers of ACBP (Acyl CoA Binding Protein) (20) and higher-order complexes with α-synuclein (19) and other proteins, and for EXG:CBM, this occurs through shared small SDS aggregates. The role of SDS in providing a “bridging” structure between two separate protein molecules at low SDS stoichiometry is novel, but it could be related to the tendency of EXG:CBM to form dimers also in the absence of SDS. The number of SDS molecules involved is too small to form a micelle, so SDS might connect hydrophobic regions of the protein (the SAXS structure is too low resolution to provide details on this).
Altogether, we only found minor differences between the SDS-induced unfolding of EXG:CBM and the EXG:CBMQQQW. This suggests that the few formal charges that distinguish the two proteins do not have any major impact on the unfolding mechanism in SDS. This could reflect that the electrostatic surface potential is not affected to a great extent by this difference or that charges simply do not play a big part in the unfolding after all. The latter is supported by the fact that SDS does unfold EXG:CBMQQQW, even well below the cmc. Further, the results from EXG:CBMQQQW do not deviate considerably from previous results for many other more highly charged proteins, questioning electrostatics’ role in direct interactions between protein and SDS.
Nevertheless, the anionic headgroup is obviously still needed for denaturation because its replacement by nonionic groups such as sugars completely removes denaturation potency as well as strongly reducing binding to the protein. These two observations may be reconciled by a simple model in which hydrophobic interactions allow SDS to bind strongly to the protein, whereas repulsion between the sulfate headgroups requires the protein to expand by denaturation. Also, the SDS headgroup is sterically small compared to that of nonionic surfactants, giving the protein easier access to SDS micelles’ amphiphilic interface between core and headgroup shell. This interface might denature the protein. In nonionic micelles, this access is screened by the larger headgroups. Another effect of the negatively charged headgroup of SDS could be to keep the SDS from forming micelles, increasing the cmc and making monomeric surfactants available for protein interactions. This is in line with the recent findings that SDS-denatured proteins can refold when nonionic surfactants are added to the mixture (9), implying that nonionic surfactants outcompete proteins in binding to SDS.
Author Contributions
C.H., J.R.W., and D.E.O. conceived experiments. C.H. and H.V.S. performed experiments. C.H., J.S.P., and D.E.O. analyzed data. C.H., D.E.O., and J.S.P. wrote the manuscript.
Acknowledgments
This research was supported by the Department of Biology, University of Copenhagen.
Editor: David Eliezer.
Footnotes
Supporting Materials and Methods are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31211-6.
Supporting Material
References
- 1.Otzen D. Protein-surfactant interactions: a tale of many states. Biochim. Biophys. Acta. 2011;1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Yonath A., Podjarny A., Traub W. Crystallographic studies of protein denaturation and renaturation. 2. Sodium dodecyl sulfate induced structural changes in triclinic lysozyme. Biochemistry. 1977;16:1418–1424. doi: 10.1021/bi00626a028. [DOI] [PubMed] [Google Scholar]
- 3.Otzen D.E. Proteins in a brave new surfactant world. Curr Opin Colloid In. 2015;20:161–169. [Google Scholar]
- 4.Shaw B.F., Schneider G.F., Whitesides G.M. Complexes of native ubiquitin and dodecyl sulfate illustrate the nature of hydrophobic and electrostatic interactions in the binding of proteins and surfactants. J. Am. Chem. Soc. 2011;133:17681–17695. doi: 10.1021/ja205735q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw B.F., Schneider G.F., Whitesides G.M. Effect of surfactant hydrophobicity on the pathway for unfolding of ubiquitin. J. Am. Chem. Soc. 2012;134:18739–18745. doi: 10.1021/ja3079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ospinal-Jiménez M., Pozzo D.C. Structural analysis of protein complexes with sodium alkyl sulfates by small-angle scattering and polyacrylamide gel electrophoresis. Langmuir. 2011;27:928–935. doi: 10.1021/la103495k. [DOI] [PubMed] [Google Scholar]
- 7.Andersen K.K., Otzen D.E. How chain length and charge affect surfactant denaturation of acyl coenzyme A binding protein (ACBP) J. Phys. Chem. B. 2009;113:13942–13952. doi: 10.1021/jp905553h. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen M.M., Andersen K.K., Otzen D.E. Unfolding of beta-sheet proteins in SDS. Biophys. J. 2007;92:3674–3685. doi: 10.1529/biophysj.106.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaspersen J.D., Søndergaard A., Pedersen J.S. Refolding of SDS-unfolded proteins by nonionic surfactants. Biophys. J. 2017;112:1609–1620. doi: 10.1016/j.bpj.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau F.W., Bowie J.U. A method for assessing the stability of a membrane protein. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 11.Otzen D.E. Folding of DsbB in mixed micelles: a kinetic analysis of the stability of a bacterial membrane protein. J. Mol. Biol. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]
- 12.Gitlin I., Gudiksen K.L., Whitesides G.M. Peracetylated bovine carbonic anhydrase (BCA-Ac18) is kinetically more stable than native BCA to sodium dodecyl sulfate. J. Phys. Chem. B. 2006;110:2372–2377. doi: 10.1021/jp055699f. [DOI] [PubMed] [Google Scholar]
- 13.Højgaard C., Kofoed C., Winther J.R. A soluble, folded protein without charged amino acid residues. Biochemistry. 2016;55:3949–3956. doi: 10.1021/acs.biochem.6b00269. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen A.D., Arleth L., Westh P. Interactions of Humicola insolens cutinase with an anionic surfactant studied by small-angle neutron scattering and isothermal titration calorimetry. Langmuir. 2005;21:4299–4307. doi: 10.1021/la047299+. [DOI] [PubMed] [Google Scholar]
- 15.Pitt-Rivers R., Impiombato F.S. The binding of sodium dodecyl sulphate to various proteins. Biochem. J. 1968;109:825–830. doi: 10.1042/bj1090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds J.A., Tanford C. Binding of dodecyl sulfate to proteins at high binding ratios. Possible implications for the state of proteins in biological membranes. Proc. Natl. Acad. Sci. USA. 1970;66:1002–1007. doi: 10.1073/pnas.66.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otzen D.E. Protein unfolding in detergents: effect of micelle structure, ionic strength, pH, and temperature. Biophys. J. 2002;83:2219–2230. doi: 10.1016/S0006-3495(02)73982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otzen D.E., Nesgaard L.W., Sehgal P. Aggregation of S6 in a quasi-native state by sub-micellar SDS. Biochim. Biophys. Acta. 2008;1784:400–414. doi: 10.1016/j.bbapap.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Giehm L., Oliveira C.L., Otzen D.E. SDS-induced fibrillation of α-synuclein: an alternative fibrillation pathway. J. Mol. Biol. 2010;401:115–133. doi: 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- 20.Andersen K.K., Oliveira C.L., Otzen D. The role of decorated SDS micelles in sub-CMC protein denaturation and association. J. Mol. Biol. 2009;391:207–226. doi: 10.1016/j.jmb.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Xu G.Y., Ong E., Harvey T.S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 22.Mortensen H.G., Madsen J.K., Pedersen J.S. Myoglobin and α-Lactalbumin Form Smaller Complexes with the Biosurfactant Rhamnolipid Than with SDS. Biophys. J. 2017;113:2621–2633. doi: 10.1016/j.bpj.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.