 Bio-layer interferometry is an excellent method to evaluate multivalent carbohydrate–protein interactions.
Bio-layer interferometry is an excellent method to evaluate multivalent carbohydrate–protein interactions.
Abstract
The study of complex multivalent carbohydrate–protein interactions remains highly complicated and sometimes rendered impossible due to aggregation problems. In this study, we demonstrate that bio-layer interferometry is an excellent complementary method to standard techniques such as SPR and ITC. Using tetra- and hexadecavalent GalNAc glycoconjugates and Helix pomatia agglutinin (HPA) as a model lectin, we were able to measure reliable kinetic and thermodynamic parameters of multivalent interactions going from the micro to the nanomolar range.
Introduction
Carbohydrate–lectin recognition plays central roles in a number of physiological and pathological processes such as cell adhesion, cancer metastasis or viral and bacterial infections.1–5 If monovalent interactions are typically in the millimolar range, high affinity can be achieved between multivalent ligands and lectins through complex binding mechanisms.6–8 The understanding of these interactions (also named the ‘cluster glycoside effect’) is of highest interest but still remains challenging because of the diversity of structural parameters that influence these processes. Therefore, complete investigation requires the utilization of analytical techniques such as enzyme-linked lectin assay (ELLA), hemagglutination inhibition assays (HIA), isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), atomic force microscopy (AFM), dynamic light scattering (DLS), frontal affinity chromatography (FAC)9 or nuclear magnetic resonance (NMR)10 all of which give complementary information. For example, SPR requires the immobilization of one partner (typically the protein) on a micro-fluidic device, whereas the second partner is injected in solution under continuous flow on the surface. It allows the real-time monitoring of the association and dissociation of the ligand, giving access to kinetic and thermodynamic parameters.9,11,12 ITC is also a physico-chemical technique of choice in this field.9,12–15 These experiments are performed in solution, do not require labelling or immobilization of any partner and offer the advantage of giving access to both thermodynamic parameters and stoichiometry of interaction which are essential to propose a binding mechanism. However, the main drawback of ITC relies on the fact that the formation of aggregates often observed with multivalent ligands may lead to uninterpretable data, thus precluding access to key binding parameters.
Bio-Layer Interferometry (BLI) is a label-free method based on the real-time optical monitoring of biomolecular interactions.16–18 Briefly, experiments are performed in standard multi-well plates containing a solution of one partner in which a biosensor tip covalently functionalized with the second partner is immersed. Tips are composed of a biocompatible layer to minimize non-specific interactions with the sensor and a thin layer coated with reactive groups. Upon irradiation of the functionalized biosensor with a white light laser, the detection of interferometry variation occurring during association/dissociation steps allows for the determination of kinetic and thermodynamic parameters of the interaction, with comparable accuracy than other physico-chemical techniques.19,20 While being less sensitive to external factors such as aggregation and microfluidic troubles, BLI has never been used to study multivalent interactions. In this paper, we report the first evaluation of a series of multivalent glycoconjugates towards the hexameric αGalNAc-specific Helix pomatia agglutinin (HPA) used as a model lectin.
Results and discussion
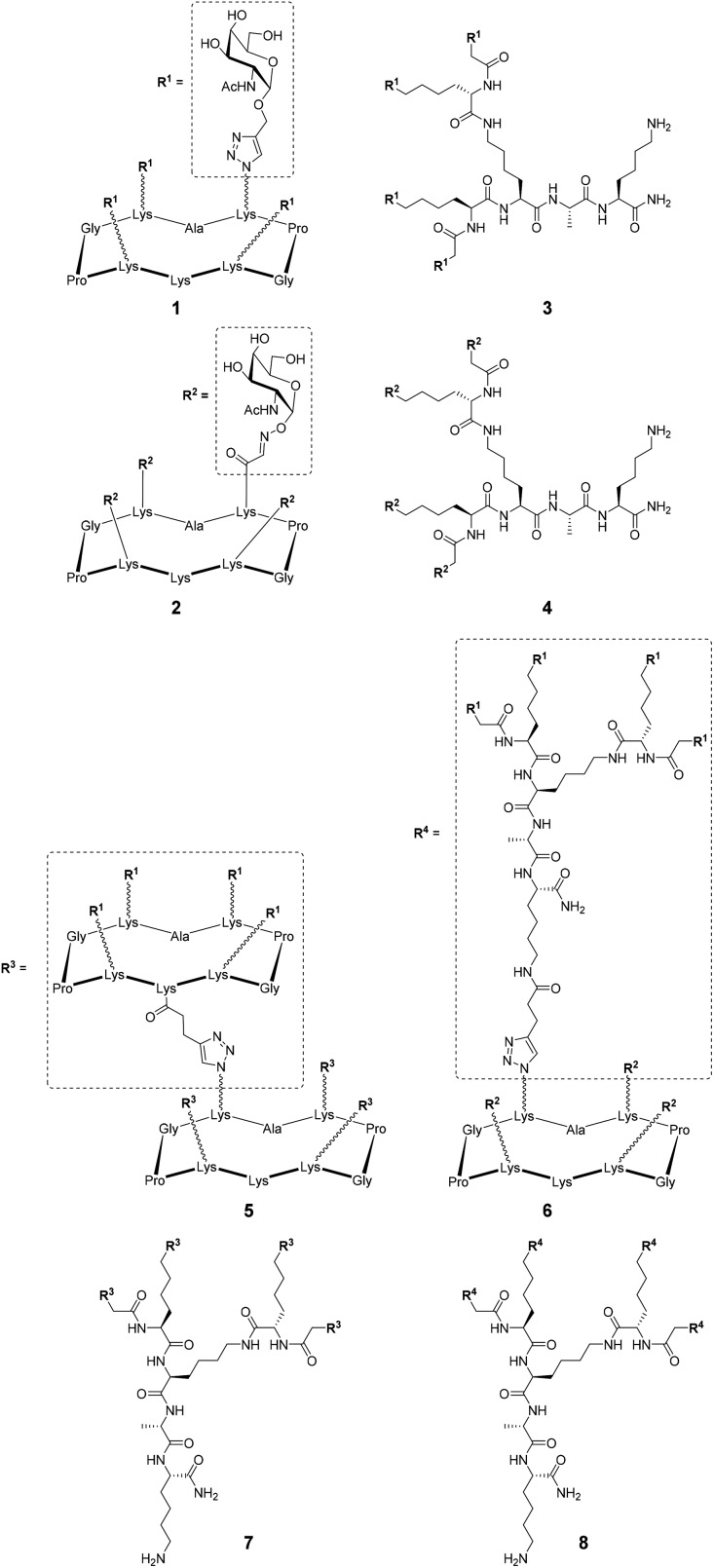
Several previously reported structures displaying 4 and 16 αGalNAc (Fig. 1)21–24 have been prepared by oxime ligation or copper-catalyzed azide–alkyne cycloaddition for this study. Recognition properties of some of these ligands for HPA have been studied by ELLA and microarray studies,25 however evaluation by ITC failed due to aggregation problems in particular for ligands with higher valency. To circumvent this issue and determine key binding parameters, BLI assays have been performed.
Fig. 1. Structures of tetravalent glycoclusters and hexadecavalent glycodendrimers.21–24 .
As BLI strongly depends on the thickness of the biolayer, the layer density and hydrophobic/-philic properties, two types of commercial sensors with different surface characteristics were used. The first one (SA) presents a 2D biolayer coated with streptavidin while the second one (SSA) is coated with a streptavidin-functionalized polymer giving a tridimensional biolayer. The first step of the experiment consisted of the immobilization of the biotinylated HPA by dipping the tip into a 1 μM lectin solution for 900 seconds, followed by a washing step (Fig. SI-1†). Lectin grafting was monitored by measuring the wavelength shift and a maximum rate of lectin immobilization was determined. After tip functionalization, the sensor was dipped successively into different concentrations of ligand solutions and then free buffer solution to measure the association and dissociation steps respectively.
In control experiments, the binding of HPA with a scaffold devoid of GalNAc was used as a negative control (see the ESI†) and no unspecific interaction with HPA was observed (Fig. SI-2†). In addition, a monovalent reference (i.e. GalNAc) has been evaluated, however no detectable signal could be measured with this compound with both the SA and SSA sensors (see Fig. SI-3 in the ESI†). This could be either due to the well-known weak affinity of lectin-monovalent ligand or to the molecular weight of the ligand which is too low to modify significantly the thickness of the biolayer and make the signal detectable. This lack of sensitivity clearly represents a limitation of BLI experiments for measuring biomolecular interactions.
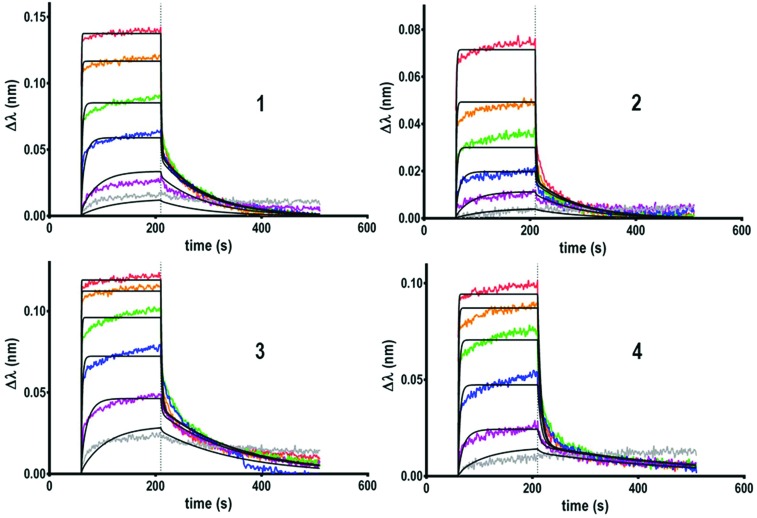
Next, the binding properties of the tetravalent cyclopeptide (1–2) and polylysine (3–4) compounds, either functionalized with GalNAc through a triazole linkage or oxime ether were evaluated. As shown in sensorgrams in Fig. 2, it was observed that the interaction between HPA and each tetravalent glycoconjugate is reversible. Indeed, during the dissociation step, the signal decreases down to the baseline, indicating a total lectin–ligand dissociation.
Fig. 2. Sensorgrams obtained for the tetravalent glycoconjugates 1–4.
Kinetic parameters of the interaction have been determined for all the tetravalent conjugates to evaluate whether the structure of the scaffold and/or the nature of the linker influence the lectin–ligand interaction. By fitting the sensorgrams with a heterogeneous model (2 : 1) compatible with the structural feature of HPA,26 association (ka or kon) and dissociation (kd or koff) constants have been measured and KD was calculated from the ratio of kd over ka. As summarized in Table 1, no significant differences have been observed between compounds 1, 2 and 4 which showed modest dissociation constants in the range of 20 μM, whereas a slightly better KD of 5 μM was observed for 3.
Table 1. Kinetic and thermodynamic parameters of the interaction between HPA and tetravalent glycoconjugates 1–4.
| Ligand | K D (μM) | k on (104 M–1 s–1) | k off (s–1) |
| 1 | 18.8 ± 3.8 | 2.5 ± 1.0 | 0.5 ± 0.2 |
| 2 | 26.3 ± 1.2 | 2.4 ± 1.9 | 0.6 ± 0.5 |
| 3 | 5.5 ± 0.9 | 15.2 ± 1.9 | 0.8 ± 0.2 |
| 4 | 22.1 ± 10.8 | 3.8 ± 3.2 | 0.5 ± 0.3 |
Although koff values are similar for all compounds, the association step seems more sensitive to the ligand structural feature. Higher kon values were indeed observed for conjugates 3–4 based on the more flexible polylysine scaffold than cyclopeptide-based compounds 1–2. The influence of the ligand structure is more significant when GalNAc is linked to the scaffold with a triazole spacer as found in 1 and 3. For this reason, we used the same linkage for the GalNAc functionalization of hexadecavalent glycodendrimers and evaluated the influence of the ligand valency. Four glycodendrimers (5–8) showing alternate combination of cyclopeptide and/or polylysine (Fig. 1) have thus been synthesized21–24 and evaluated by BLI (Fig. 3).
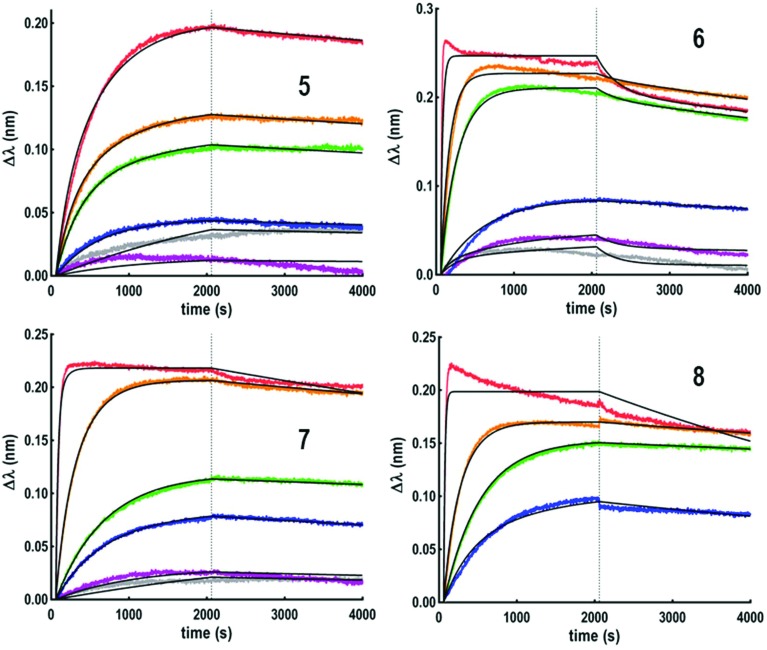
Fig. 3. Sensorgrams obtained for the hexadecavalent glycoconjugates 5–8.
It was initially observed that the dissociation of interaction in buffer solution was not possible with these compounds, thus leading to unexploitable data. To circumvent this issue, a regeneration step was included, by dipping the tip in a 1 M GalNAc solution for 30 seconds. However, this protocol was excluded due to the experiment time (superior to 7 hours) leading to a partial evaporation of the ligand solutions. To shorten the regeneration step, 6 sensors in parallel were used which gave the possibility to extend the association and dissociation times to 2000 seconds. Using this experimental protocol, the thermodynamic constants indicated in Fig. 4 could be determined.
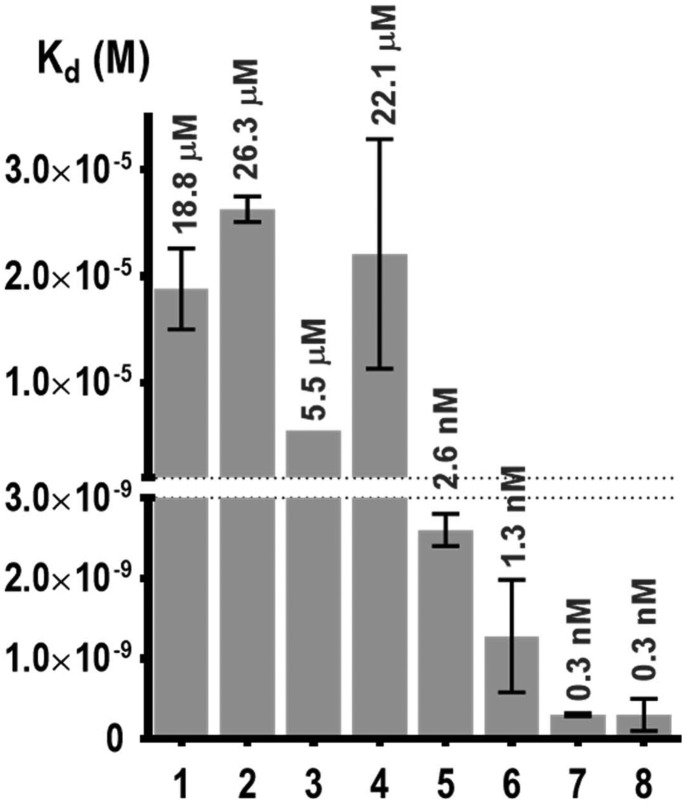
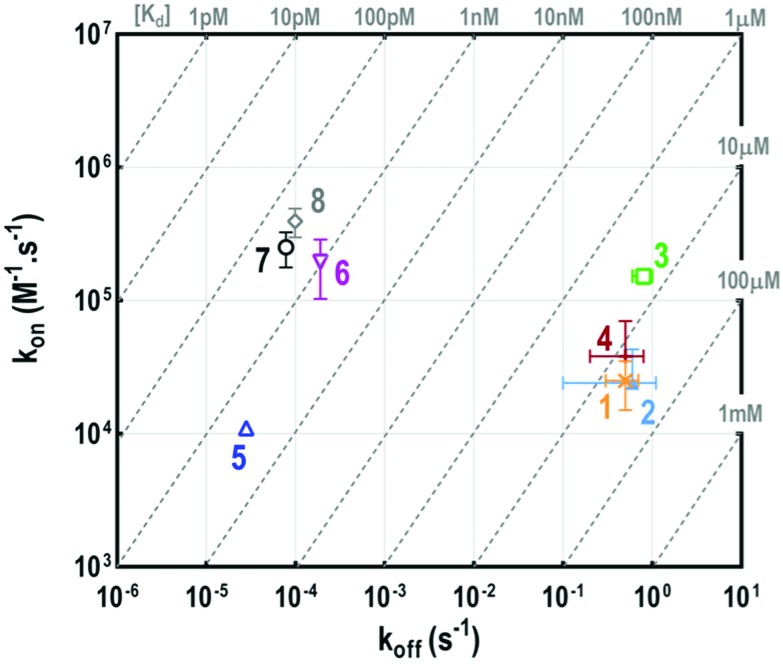
Fig. 4. Thermodynamic dissociation constants determined for the multivalent glycoconjugates 1–8 and HPA interactions.
This experiment first indicated that increasing valency drastically influences the lectin–ligand interaction to reach a nanomolar range, allowing gains of affinity from 7000 to 18 000 compared to tetravalent structures. The strongest interaction with HPA was obtained for a scaffold containing polylysine as the central core, with the dissociation constant being 0.3 nM for both 7 and 8. In the case of hexadecavalent structures with cyclopeptide as the central core, the dissociation constant is slightly better for 6 displaying polylysine at the periphery (1.3 nM) compared to compound 5 entirely based on cyclopeptide (2.6 nM). These observations are in agreement with previous microarray studies (KD = 12–92 nM for 5–8). Association and dissociation kinetic constants were reported on the isoaffinity graph presented in Fig. 5, and in Table SI-2 in the ESI,† to compare all ligands. Interestingly, as observed for tetravalent structures, the more flexible hexadecavalent ligands showed higher kon than the more rigid structure.
Fig. 5. Isoaffinity graph plotted from the HPA–glycoconjugate 1–8 interaction.
Dendrimer 5 indeed displays the slowest association step compared to the other hexadecavalent structures. By contrast compounds 8 and 7 showed higher dissociation constants due to a faster association step, with respective association rates 25 and 40 times higher than for 5.
Conclusion
To conclude, we demonstrated that the interaction between a lectin and multivalent glycoconjugates can be evaluated by BLI as an alternative to ITC which led to aggregation problems. We were able to measure affinities for hexadecavalent ligands in the nanomolar range. These results are in good agreement with previous studies, thus confirming the reliability of BLI for studying multivalent interactions. Moreover, we could highlight significant differences in kinetic association constants depending on the ligand flexibility. Besides determining both kinetic and thermodynamic parameters, BLI experiments offer the advantages that they can be performed fast, can be operated at low cost and require a lower quantity of both lectin and ligand compared to other analytical techniques. While low affinity ligands could not be evaluated, we believe that BLI represents an excellent complementary study to SPR and ITC to gather binding parameters and understand multivalent interactions in deeper details. This technology is currently used in our laboratory to screen and optimize multivalent ligands against bacterial lectins for the development of antiadhesive agents.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors thanks the CNRS, the Université Grenoble Alpes, the Institut de Chimie Moléculaire de Grenoble (ICMG, FR 2607). This work was supported by the French ANR project Glyco@Alps (ANR-15-IDEX-02) and Labex ARCANE (ANR-11-LABX-003). O. R. acknowledges the European Research Council Consolidator Grant “LEGO” (647938) for D. G. and E. L.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ob01664j
References
- Lis H., Sharon N. Chem. Rev. 1998;98:637. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- Sharon N., in Toward Anti-Adhesion Therapy for Microbial Diseases, ed. I. Kahane and I. Ofek, Springer US, 1996, pp. 1–8. [DOI] [PubMed] [Google Scholar]
- Imberty A., Varrot A. Curr. Opin. Struct. Biol. 2008;18:567. doi: 10.1016/j.sbi.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Boukerb A. M., Rousset A., Galanos N., Méar J. B., Thépaut M., Grandjean T., Gillon E., Cecioni S., Abderrahmen C., Faure K., Redelberger D., Kipnis E., Dessein R., Havet S., Darblade B., Matthews S. E., De Bentzmann S., Guéry B., Cournoyer B., Imberty A., Vidal S. J. Med. Chem. 2014;57:10275. doi: 10.1021/jm500038p. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Jiménez-Barbero J., Casnati A., De Castro C., Darbre T., Fieschi F., Finne J., Funken H., Jaeger K.-E., Lahmann M., Lindhorst T. K., Marradi M., Messner P., Molinaro A., Murphy P., Nativi C., Oscarson S., Penadés S., Peri F., Pieters R. J., Renaudet O., Reymond J.-L., Richichi B., Rojo J., Sansone F., Schäffer C., Turnbull W. B., Velasco-Torrijos T., Vidal S., Vincent S., Wennekes T., Zuilhof H., Imberty A. Chem. Soc. Rev. 2013;42:4709. doi: 10.1039/c2cs35408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. C., Lee R. T. Acc. Chem. Res. 1995;28:321. [Google Scholar]
- Mammen M., Choi S.-K., Whitesides G. M. Angew. Chem., Int. Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lundquist J. J., Toone E. J. Chem. Rev. 2002;102:555. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- Cecioni S., Imberty A., Vidal S. Chem. Rev. 2015;115:525. doi: 10.1021/cr500303t. [DOI] [PubMed] [Google Scholar]
- Ardá A., Jiménez-Barbero J. Chem. Commun. 2018;54:4761. doi: 10.1039/c8cc01444b. [DOI] [PubMed] [Google Scholar]
- Sattin S., Daghetti A., Thépaut M., Berzi A., Sanchez-Navarro M., Tabarani G., Rojo J., Fieschi F., Clerici M., Bernardi A. ACS Chem. Biol. 2010;5:301. doi: 10.1021/cb900216e. [DOI] [PubMed] [Google Scholar]
- Cecioni S., Praly J.-P., Matthews S. E., Wimmerova M., Imberty A., Vidal S. Chem. – Eur. J. 2012;18:6250. doi: 10.1002/chem.201200010. [DOI] [PubMed] [Google Scholar]
- Turnbull W. B., Precious B. L., Homans S. W. J. Am. Chem. Soc. 2004;126:1047. doi: 10.1021/ja0378207. [DOI] [PubMed] [Google Scholar]
- Vincent S. P., Buffet K., Nierengarten I., Imberty A., Nierengarten J.-F. Chem. – Eur. J. 2016;22:88. doi: 10.1002/chem.201504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyard D., Baldoneschi V., Varrot A., Fiore M., Imberty A., Richichi B., Renaudet O., Nativi C. Bioconjugate Chem. 2018;29:83. doi: 10.1021/acs.bioconjchem.7b00616. [DOI] [PubMed] [Google Scholar]
- Yang L., Connaris H., Potter J. A., Taylor G. L. BMC Struct. Biol. 2015;15:1. doi: 10.1186/s12900-015-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeneni S., Ji Y., Watson D. C., Young N. M., Woods R. J. Front. Immunol. 2014;5:1. doi: 10.3389/fimmu.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjuler C.C. T.T., Maolanon N. N., Sauer J., Stougaard J., Thygesen M. B., Jensen K. J. Nat. Protoc. 2017;12:2411. doi: 10.1038/nprot.2017.109. [DOI] [PubMed] [Google Scholar]
- Pioszak A. A., Parker N. R., Gardella T. J., Xu H. E. J. Biol. Chem. 2009;284:28382. doi: 10.1074/jbc.M109.022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauschopf U., Lilie H., Honold K., Wozny M., Reusch D., Esswein A., Schäfer W., Rücknagel K. P., Rudolph R. Biochemistry. 2000;39:8878. doi: 10.1021/bi0001426. [DOI] [PubMed] [Google Scholar]
- Bossu I., Šulc M., Křenek K., Dufour E., Garcia J., Berthet N., Dumy P., Křen V., Renaudet O. Org. Biomol. Chem. 2011;9:1948. doi: 10.1039/c0ob00772b. [DOI] [PubMed] [Google Scholar]
- Berthet N., Thomas B., Bossu I., Dufour E., Gillon E., Garcia J., Spinelli N., Imberty A., Dumy P., Renaudet O. Bioconjugate Chem. 2013;24:1598. doi: 10.1021/bc400239m. [DOI] [PubMed] [Google Scholar]
- Thomas B., Pifferi C., Daskhan G. C., Fiore M., Berthet N., Renaudet O. Org. Biomol. Chem. 2015;13:11529. doi: 10.1039/c5ob01870f. [DOI] [PubMed] [Google Scholar]
- Pifferi C., Thomas B., Goyard D., Berthet N., Renaudet O. Chem. – Eur. J. 2017;23:16283. doi: 10.1002/chem.201702708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang A., Laigre E., Goyard D., Defrancq E., Vinet F., Dumy P., Renaudet O. Org. Biomol. Chem. 2017;15:5135. doi: 10.1039/c7ob00889a. [DOI] [PubMed] [Google Scholar]
- Sanchez J. F., Lescar J., Chazalet V., Audfray A., Gagnon J., Alvarez R., Breton C., Imberty A., Mitchell E. P. J. Biol. Chem. 2006;281:20171. doi: 10.1074/jbc.M603452200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.