Abstract
Summary
A database of curated genomic variants with clinically supported drug therapies and other oncological annotations is described. The accompanying web portal provides a search engine with two modes: one that allows users to query gene, cancer type, variant type or position for druggable mutations, and another to search for and to visualize, on three-dimensional protein structures, putative druggable sites that cluster with known druggable mutations.
Availability and implementation
1 Introduction
The number of drug therapies is ever-growing, thanks to ongoing research and clinical trials to develop and test treatment strategies (Siu et al., 2015). Drug therapies are designed to target various components of the biological machinery, including specific genomic variants in the patient’s DNA. Examination of approved ‘precision-medicine’ solutions may directly suggest putative treatments, e.g. a genomic variant that acts as a biomarker of response to a given drug across histological types of cancer. In this context, methods that open up new directions for investigating personalized therapies would be valuable.
We have developed the DEPO database and web portal to allow users to query a curated database of clinically relevant genomic variants and their respective drug therapies using standard gene names and, optionally, a cancer type, variant type or variant position. The DEPO portal supports basic queries, extensible to include all cancer types, and is unique in predicting putative druggable mutations based on protein structure based mutational clusters using the HotSpot3D software suite (Niu et al., 2016). In addition, it incorporates the JSmol 3D molecular viewer for visualizing these potentially druggable sites.
2 Materials and methods
2.1 Database overview
DEPO’s database consists of observed and validated, druggable genomic events: single nucleotide polymorphisms (missense, frameshift and nonsense mutation SNPs), in-frame insertions and deletions, fusions, copy number alterations (amplification and loss) and expression changes. It draws data from the Cancer Biomarkers oncology database within Cancer Genome Interpreter (Tamborero et al., 2018), which is curated and maintained by expert oncologists. (This database is licensed under a Creative Commons Public Domain Dedication.) Additional variant/drug interactions are manually curated by reviewing peer-reviewed literature and conference abstracts, the NCCN Biomarkers Compendium (nccn.org), MyCancerGenome (Swanton, 2012), CancerDR (Kumar et al., 2013), SageDB (Dienstmann et al., 2015) (synapse.org/#! Synapse: syn2370773/wiki/62707), the Personalized Cancer Therapy website (Johnson et al., 2015) (pct.mdanderson.org), and cross-referencing all data against primary sources. DEPO’s variant/drug entries are therefore paired with annotations of potential interest to oncologists and use standards established by the Clinical Genome Resource (ClinGen) (Ritter et al., 2016) for clinically relevant cancer variants. HotSpot3D (Niu et al., 2016) is a structure-based tool that maps mutations onto three-dimensional protein structural models and performs computational discovery of clusters of spatially proximal mutations. The DEPO portal enables search and visualization of residue sites identified by HotSpot3D as proximal to curated druggable mutations.
DEPO is updated monthly by downloading flat files from each of the external databases and standardizing the information using several Python scripts in conjunction with manual curation. Perl scripts then prepare the data for MySQL table insertions, which are carried out by PHP scripts. HotSpot3D predictions are updated for every major release of publically available mutation sets and integrated into the portal.
2.2 Web portal architecture
The portal is built using HTML and CSS and is based on the Bootstrap (getbootstrap.com) front-end framework. AJAX (jquery.com) is used to launch server-side PHP scripts for performing queries on a backend MySQL relational database containing DEPO and HotSpot3D data and for updating the search results section accordingly. The DataTables jQuery plug-in (www.datatables.net) provides styling for displaying and further interrogating search results. The JSmol 3D structure viewer (wiki.jmol.org/index.php/JSmol) is used for visualizing DEPO mutations in HotSpot3D clusters. Finally, the Chart.js (www.chartsjs.org) JavaScript graphics library is used to display DEPO summary graphs.
3 Results
Therapeutic data in DEPO are served through an accompanying web portal (see Fig. 1), through which users can perform queries based on two entities, druggable alterations and HotSpot3D cluster information. We describe these briefly. The Search tab allows users to query druggable variants by gene and provides additional menus to restrict searches by cancer type, variant type (copy number alteration, expression outlier, gene fusion and mutation), or variant protein position. The option to leave cancer type unspecified is a design feature intended to reveal the repurposing potential of drug therapies across cancer types. The portal only requires position information when querying variants, thereby providing wildcard functionality for alternative alleles. A special ‘intramolecular cluster’ variant type allows users to enter a gene name and protein position to search for all DEPO mutational sites identified by HotSpot3D as being in close proximity to that position. These query positions are predicted to be putative functional mutations and therefore potential candidates for druggability. (Validation of such candidates requires experimental confirmation that is beyond the scope of DEPO.) Clicking the ‘View 3D’ web button opens the JSmol molecular viewer in a panel for interactive visualization of the mutation clusters.
Fig. 1.
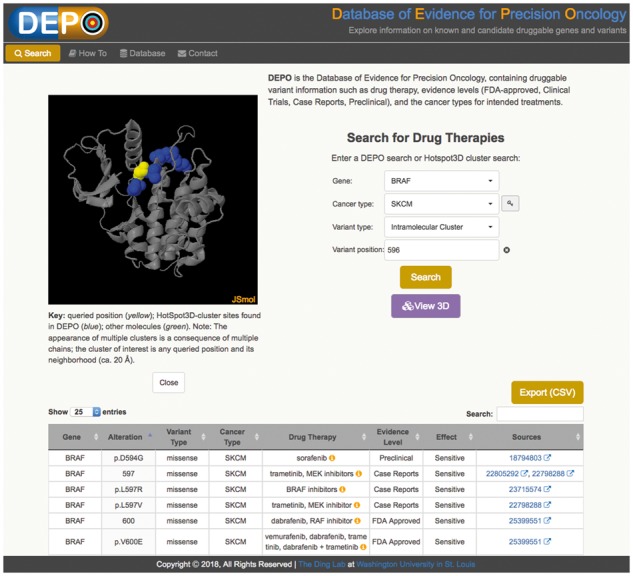
Screenshot of DEPO portal. Example showing a search for drug therapies pertaining to the BRAF gene in skin cutaneous melanoma (SKCM). JSmol provides visualization of protein residue locations with known drug activity as reported in the DEPO database (blue) and of the query site (yellow), which HotSpot3D has predicted to be putative candidate site with functional significance. The table below lists the curated drug information for the mutationally clustered sites (blue), i.e. drug therapy, drug class, evidence level, drug effect and hyperlinks to literature sources
The search results table lists drug therapies (with drug class information obtainable via mouse-over on the informational icon), evidence levels, drug effect and literature sources that match the user query (i.e. gene name and any of the following: variant type, cancer type, variant position). Here, drug effect describes whether a variant correlates with increased sensitivity or resistance of a tumor to a drug. Evidence level describes the quality of data supporting a given variant/drug entry (i.e. preclinical, case report, clinical trial and FDA-approved). Drug class is determined using a look-up table generated manually from DrugBank/NIHClasses. Literature sources involving PubMed IDs are hyperlinked to the corresponding articles or are otherwise briefly cited in plain text format. Dynamic refinement of search results may be conducted by typing text into the result table’s search box. Users can easily generate a comma-delimited text file for exporting the search results. The How To tab lists several examples of search types and provides user interactivity features. Finally, the Database tab generates an on-the-fly summary of DEPO variant classes (copy number variation, gene fusion, expression outlier and mutations) and druggable mutations by gene and evidence level (FDA approved, clinical trials, case reports and preclinical) with mouse-over display of the count of each item.
4 Conclusion
DEPO, a curated database and web portal for precision oncology, has been created to facilitate searches for drug therapies clinically associated with genetic biomarkers. Given that regulatory approval for drug therapies typically proceeds incrementally for each variant and cancer type combination, DEPO is implemented to promote insight into potential drug repurposing by allowing queries across multiple cancer types. Furthermore, although other independently developed online resources, such as CIViC (Griffith et al., 2017), also present clinically relevant cancer variants, DEPO is unique in both identifying mutations that potentially affect drug binding affinity and response based on their proximity to other mutations having druggability information and in providing visualization of these 3D mutational clusters within a common interface.
Acknowledgements
L.D. designed and supervised research. R.J.M, A.D.S. and W.W. developed the portal. S.S., R.J.M., A.D.S., L.B.W. and M.A.W. developed the portal content. S.Q.S. and P.B. constructed DEPO. R.J.M. and S.S. wrote the manuscript. S.S., R.J.M., S.Q.S. and L.D. revised the manuscript.
Funding
This work was supported by the National Cancer Institute Grants R01CA178383, R01CA180006, 5U24CA10972-02 and 5U24CA211006-02 to L.D. and National Human Genome Research Institute Grants U01HG006517 and 1R01HG009711-01 to L.D.
Conflict of Interest: none declared.
References
- Dienstmann R. et al. (2015) Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov., 5, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M. et al. (2017) CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet., 49, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. et al. (2015) The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discov. Today, 20, 1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. et al. (2013) CancerDR: cancer drug resistance database. Sci. Rep., 3, 1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B. et al. (2016) Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet., 48, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter D.I. et al. (2016) Somatic cancer variant curation and harmonization through consensus minimum variant level data. Genome Med., 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu L.L. et al. (2015) Next-generation sequencing to guide clinical trials. Clin. Cancer Res., 21, 4536–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C. (2012) My Cancer Genome: a unified genomics and clinical trial portal. Lancet Oncol., 13, 668–669.22748256 [Google Scholar]
- Tamborero D. et al. (2018) Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med., 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]


