Abstract
Summary
Creating clear, visually pleasing 2D depictions of RNA and DNA strands and their interactions is important to facilitate and communicate insights related to nucleic acid structure. Here we present RiboSketch, a secondary structure image production application that enables the visualization of multistranded structures via layout algorithms, comprehensive editing capabilities, and a multitude of simulation modes. These interactive features allow RiboSketch to create publication quality diagrams for structures with a wide range of composition, size and complexity. The program may be run in any web browser without the need for installation, or as a standalone Java application.
Availability and implementation
1 Introduction
The base pairing and sequence information of RNA and DNA is frequently communicated via secondary structure diagrams, in which labeled nodes and lines represent nucleotides and their interactions. These images are valuable for determining and communicating structural hypotheses, conclusions and insights.
Due to the wide variety and complexity of RNA, layout algorithms cannot always generate a clean diagram or anticipate which structural features require emphasis in the context of a particular presentation. This creates the need for manual editing features in conjunction with automatic layout generation. Existing programs (Byun and Han, 2009; Darty et al., 2009; Elias and Hoksza, 2017; Hecker et al., 2013; Kerpedjiev et al., 2015; Wiese et al., 2005) provide varying levels of interactivity, but the ability to exactly position nucleotides and substructures is necessary to produce satisfactory images for the full range of RNA and DNA complexes. Because structures are often composed of multiple strands with interstrand, pseudoknot and non-canonical interactions, a program should both read input that accommodates such elements and facilitate precise editing. It is therefore desirable that a program offer complete control over the display of nucleotides and motifs of complex RNA and DNA, in combination with computational structure optimization features.
2 Main features
RiboSketch enables the manipulation of diverse structures through its interactive editing environment, accessible in both online and offline formats. Layouts can be generated automatically, manually (assisted by practical commands), by force simulation, or through any combination of methods.
2.1 Input and output
RiboSketch accepts many common secondary structure file formats, such as dot-bracket (Lorenz et al., 2011), MFold connect (Markham et al., 2005) and BPSEQ (Cannone et al., 2002). Many secondary structure file formats do not contain non-canonical base pairing information, so RiboSketch provides a general solution by enabling the addition of such interactions through direct input into a text field. Alternatively, RiboSketch can read a file format that does contain non-canonical base-pairs derived from the program DSSR (Lu et al., 2015). The state of the program may be saved and stored in a text file format to be later reloaded for continued work. RiboSketch can export rasterized or vector graphics images (PNG or SVG format respectively).
2.2 Depiction algorithms
Initial layouts are rendered using a modified version of the Shapiro/NAView approach (Bruccoleri et al., 1988; Shapiro et al., 1984), expanded to accommodate multiple strands and pseudoknots (the structure is first rendered based on a maximal subset of nested base pairs, after which the non-nested base pairs are added). For multistranded structures, a novel algorithm is applied in which strands are arranged to position related substructures close to one another and to minimize the number of pseudoknots, resulting in a more comprehensible image. RiboSketch can also create circular diagrams, in which nucleotides are positioned in a circle with base pairs drawn as chords.
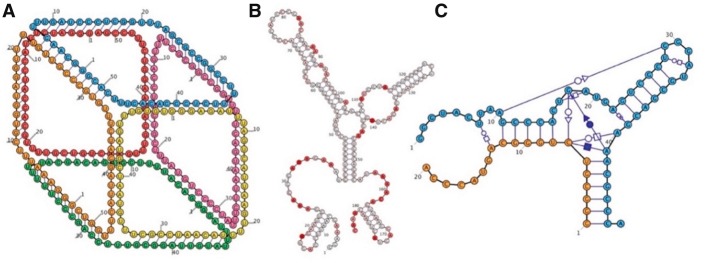
Customizable display properties include the size, color and labeling of nucleotides. RiboSketch provides an array of color schemes to highlight various structural properties. Additionally, the user may overlay chemical probing data or specify the coloring of individual nucleotides via the input of numerical values (see Fig. 1B). Uniquely, Ribosketch can create depth effects through its ‘bring to front’ and ‘send to back’ commands, allowing diagrams to imply 3D shape (see Fig. 1A). The user may also change the zoom factor. The force-directed-layout is optionally aided by an algorithm that suggests evenly spaced circular layouts of nucleotides in internal loops and multi-branch junctions.
Fig. 1.
(A) RiboSketch gives the user complete control over the positions of nucleotides, shown in this 6-stranded RNA cube (Halman et al., 2017). Nucleotide placement and choices of foreground/background layout creating a 3D perspective were performed interactively. (B) SHAPE data coloring of 5S rRNA, E.coli (Kladwang et al., 2014). (C) Depiction of Hammerhead Ribozyme including non-canonical interactions (Mir et al., 2016) (Color version of this figure is available at Bioinformatics online.)
2.3 Interactive editing
RiboSketch enables the user to modify the layout of secondary structure diagrams by clicking, dragging, rotating and utilizing a plethora of commands and features. Operations can be applied to the whole structure, individual nucleotides, or user-defined subsets. Bonds may be interactively added or deleted [in a manner similar to Forna (Kerpedjiev et al., 2015)], and the model dynamically updates. Distances between base-paired nucleotides and the lengths of backbones may be defined and modified. The editing process is made convenient through the ability to undo and redo actions, and to save and load the state of edited structures. By providing a high level of interactivity, the program is flexible and can process structures ranging from long ribosomal strands to synthetic nanoparticles.
2.4 Dynamic simulation
RiboSketch can subject structures to a force simulation in which backbone and base-pair interactions act as springs (similar to Kerpedjiev et al., 2015; Shabash and Wiese, 2017; Wiese et al., 2005), while nucleotides in close proximity undergo repulsion. Helices may be rigidified or left flexible. The user may turn this simulation on or off thus, switching between dynamic and static states to accelerate the editing process. This can be of particular use for processing structures that challenge algorithmic approaches, such as those with many pseudoknots.
2.5 Non-canonical interactions
RiboSketch can depict non-canonical interactions using the standard Leontis and Westhof symbol notation (Leontis and Westhof, 2001), as well as hybrid RNA-DNA interactions. Bases can interact with multiple partners (see Fig. 1C).
3 Implementation
The implementation for the standalone program utilizes the Java-based Processing environment (http://processing.org). In addition to a standalone Java application, we provide a web-based version that can be run within a web browser using JavaScript (without Java). The web version utilizes Processing.js (http://processingjs.org).
4 Conclusion
With precise editing features and algorithms to accommodate multiple strands and pseudoknots, RiboSketch is valuable for generating images of diverse RNA and DNA secondary structures.
Funding
This work was supported in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract No. HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Conflict of Interest: none declared.
References
- Bruccoleri R.E. et al. (1988) An improved algorithm for nucleic acid secondary structure display. Comput. Appl. Biosci. CABIOS, 4, 167–173. [DOI] [PubMed] [Google Scholar]
- Byun Y., Han K. (2009) PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics, 25, 1435–1437. [DOI] [PubMed] [Google Scholar]
- Cannone J.J. et al. (2002) The Comparative RNA Web (Crw) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K. et al. (2009) VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics, 25, 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias R., Hoksza D. (2017) TRAVeLer: a tool for template-based RNA secondary structure visualization. BMC Bioinformatics, 18, 487.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halman J.R. et al. (2017) Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res., 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker N. et al. (2013) RNA secondary structure diagrams for very large molecules: rNAfdl. Bioinformatics, 29, 2941. [DOI] [PubMed] [Google Scholar]
- Kerpedjiev P. et al. (2015) Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics, 31, 3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W. et al. (2014) Standardization of RNA chemical mapping experiments. Biochemistry, 53, 3063–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. (2001) Geometric nomenclature and classification of RNA base pairs. RNA, 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R. et al. (2011) ViennaRNA Package 2.0. Algorithms Mol. Biol., 6, 26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.-J. et al. (2015) DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res., 43, e142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N.R. et al. (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res., 33, W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A. et al. (2016) Two active site divalent ions in the crystal structure of the hammerhead ribozyme bound to a transition state analogue. Biochemistry, 55, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabash B., Wiese K.C. (2017) Numerical integration methods and layout improvements in the context of dynamic RNA visualization. BMC Bioinformatics, 18, 282.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B.A. et al. (1984) Generating non-overlapping displays of nucleic acid secondary structure. Nucleic Acids Res., 12, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese K.C. et al. (2005) JViz.Rna—a java tool for RNA secondary structure visualization. IEEE Trans. NanoBioscience, 4, 212–218. [DOI] [PubMed] [Google Scholar]