Abstract
Purpose of review:
Electronic health records (EHRs) contain valuable data for identifying health outcomes, but these data also present numerous challenges when creating computable phenotyping algorithms. Machine learning methods could help with some of these challenges. In this review, we discuss four common scenarios that researchers may find helpful for thinking critically about when and for what tasks machine learning may be used to identify health outcomes from EHR data.
Recent findings:
We first consider the conditions in which machine learning may be especially useful with respect to two dimensions of a health outcome: 1) the characteristics of its diagnostic criteria, and 2) the format in which its diagnostic data are usually stored within EHR systems. In the first dimension, we propose that for health outcomes with diagnostic criteria involving many clinical factors, vague definitions, or subjective interpretations, machine learning may be useful for modeling the complex diagnostic decision-making process from a vector of clinical inputs to identify individuals with the health outcome. In the second dimension, we propose that for health outcomes where diagnostic information is largely stored in unstructured formats such as free text or images, machine learning may be useful for extracting and structuring this information as part of a natural language processing system or an image recognition task. We then consider these two dimensions jointly to define four common scenarios of health outcomes. For each scenario, we discuss the potential uses for machine learning – first assuming accurate and complete EHR data and then relaxing these assumptions to accommodate the limitations of real-world EHR systems. We illustrate these four scenarios using concrete examples and describe how recent studies have used machine learning to identify these health outcomes from EHR data.
Summary:
Machine learning has great potential to improve the accuracy and efficiency of health outcome identification from EHR systems, especially under certain conditions. To promote the use of machine learning in EHR-based phenotyping tasks, future work should prioritize efforts to increase the transportability of machine learning algorithms for use in multi-site settings.
Keywords: electronic health records, machine learning, health outcomes, phenotyping, cohort identification
1. Introduction
Post-market evaluation of medical products is essential to ensure that patients are receiving safe and effective treatment under real-world conditions. Such evaluations are important because pre-market trials are often underpowered to detect rare adverse events, and their findings may not be generalizable to patterns of use in routine clinical practice among more diverse patient populations (1). Moreover, an increasing number of pre-market trials are using surrogate outcomes as endpoints, which often have questionable or weak associations with clinically meaningful health outcomes (2, 3).
The analysis of real-world data is therefore paramount to generating evidence about the real-world effects of medical products on health outcomes. Real-world data refers to “data obtained outside the context of randomized controlled trials generated during routine clinical practice” (4). Electronic health records (EHRs) and insurance claims are perhaps the most common sources of real-world data used for health outcomes research and pragmatic clinical trials (4, 5). Compared to claims data, EHR data may have greater utility for identifying health outcomes due to the presence of richer clinical information including vital signs, laboratory test results, progress notes, discharge summaries, and radiologic and pathologic images and reports, among other information (6). The creation of computer-processable algorithms to identify individuals with specific health conditions, diseases, or clinical events from electronic health data is often referred to as ‘computational phenotyping’ (7).
Researchers using EHR data to create electronic phenotyping algorithms often face challenges due in part to the 3Vs of big data: volume, variety, and velocity (8). In this review, we focus on some of the challenges that may arise due to the volume and variety of data in EHR systems. First, how does one efficiently query various types of EHR data to extract useful clinical information? Much of the data in EHR systems are stored in a variety of unstructured formats (eg, free text, images) that cannot be easily queried apart from manual inspection that is often time-consuming and labor-intensive (9, 10). Then, once extracted, how does one optimally use these large volumes of clinical data to accurately identify patients with the health outcome? This phenotyping task is especially challenging for conditions with diagnostic criteria that consider a multitude of factors with often vague definitions and subjective interpretations. In these cases, it may be difficult to devise an accurate phenotyping algorithm because a clinician’s diagnosis incorporates abstract elements of their judgment such as tacit knowledge, experience, and clinical gestalt (11).
The use of non-parametric modeling strategies from the machine learning literature could help address these challenges. Machine learning has been defined as “a program that learns to perform a task or make a decision automatically from data, rather than having the behavior explicitly programmed” (12). In practice, these programs or algorithms exist along a continuum, ranging from fully human-guided to fully machine-driven (12). Examples of successful machine learning applications based on non-EHR data include the use of support vector machines to identify flu-related tweets to discover influenza trends (13), the use of random forests and long short-term memory networks to detect credit card fraud (14), and the use of deep convolutional neural networks to recognize and classify objects in images (15), among many others.
Health outcomes research could benefit from the use of more flexible, data-driven machine learning methods, as many of the tasks being tackled using these approaches in other domains bear striking similarities to the challenges researchers face when using EHR data to identify health outcomes. Thus, the purpose of this review is to present a set of common scenarios that researchers may find helpful for thinking more critically about when and for what tasks machine learning may be useful for identifying health outcomes from EHR data. We begin by providing a brief overview of several machine learning techniques that have been commonly used to measure health outcomes from electronic health data. Next, we consider two dimensions of a health outcome and identify the conditions in each dimension where machine learning may be especially useful. We then consider these two dimensions jointly to create four common scenarios of health outcomes and discuss the potential uses for machine learning in each scenario – first assuming accurate and complete EHR data and then relaxing these assumptions to accommodate the limitations of real-world EHR systems. We illustrate these four scenarios using concrete examples and describe how recent studies have used machine learning to identify these health outcomes from EHR data. In all our discussions, we refer to machine learning as the use of algorithms that exist far along the machine-driven end of the ‘machine learning spectrum’ (12).
2. Overview of Common Machine Learning Techniques Used to Identify Health Outcomes
Machine learning applications in health care are usually supervised or unsupervised. Supervised learning uses labeled data (ie, data that has been tagged with respect to an outcome or response of interest) to train machines to predict these labels based on a set of inputs or predictors (16). In contrast, unsupervised learning neither requires labeled data nor predicts an outcome or response; rather, it uncovers patterns and relations in the data to cluster observations or variables into similar groups (16). Most existing machine learning applications in health care – including those that create electronic phenotyping algorithms – use supervised learning (17). In this section, we provide a brief overview of several machine learning techniques that have been commonly used to identify health outcomes from electronic health data, including supervised methods such as support vector machines and random forests, as well as neural networks and deep learning models that may be either supervised or unsupervised (17–19).
Figure 1 illustrates the architecture of these different machine learning techniques. Support vector machines (Figure 1, panel A) classify observations by finding the optimal separating hyperplane between observations of different outcome classes in the covariate space (20). The optimal hyperplane is defined as the hyperplane that separates observations from different outcome classes with the maximum margin (ie, the largest distance between the hyperplane and the nearest observations on either side of it from different outcome classes, called the ‘support vectors’) (20). In contrast, random forests (21) (Figure 1, panel B) are rule-based ensemble learners that classify observations by averaging the predictions across a collection of decision trees. Each tree in the random forest is trained on a bootstrap sample of the data where at each node, the sample is split on the best (ie, most informative) predictor among a randomly selected subset of all candidate predictors (20).
Figure 1. Illustration of several machine learning techniques commonly used to identify health outcomes from electronic health data.
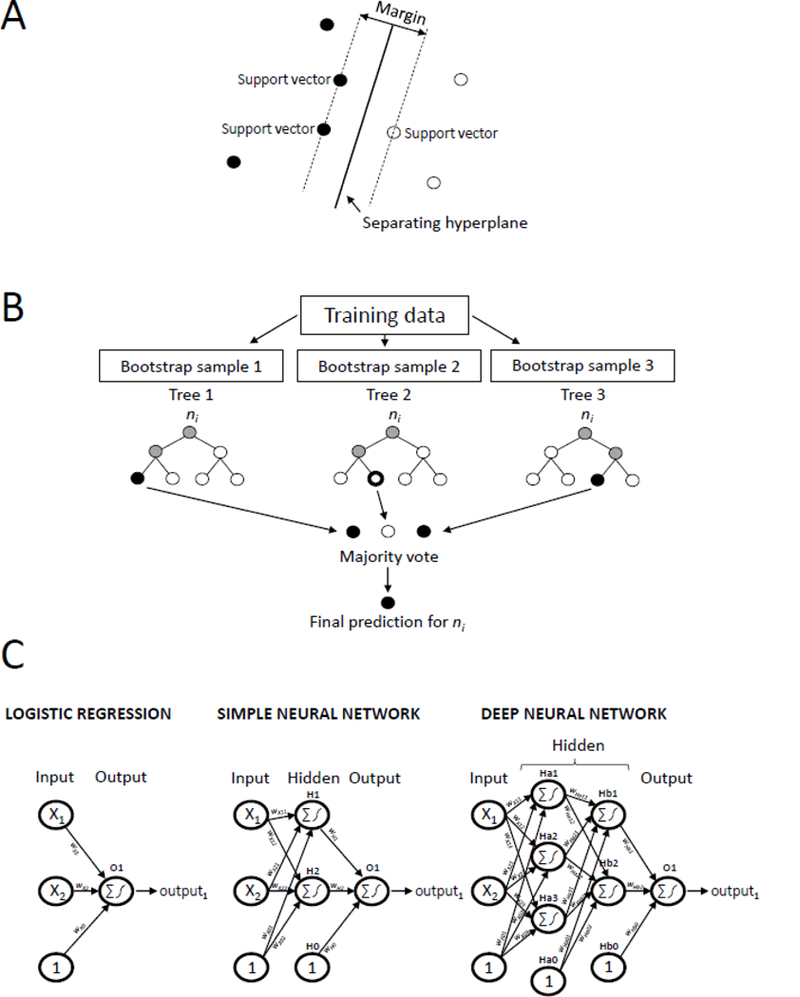
Panel A shows the architecture of a support vector machine, which classifies observations by finding the separating hyperplane with the largest margin between observations of different outcome classes on either side of it in the covariate space. Panel B shows the architecture of a random forest, which averages the predictions across a collection of decision trees, where each tree is trained on a bootstrap sample of the data. Panel C shows the similarities between the architectures of a logistic regression model (left), simple neural network (middle), and deep neural network (right). Nodes (represented by circles) are connected to subsequent nodes by a connection weight, w. Nodes in the input layer correspond to the variables presented to the model. Nodes with a ‘1’ represent bias terms. Nodes in the hidden and output layers take the weighted sum of its input nodes and pass this value through an activation function (eg, the logistic function) to produce its output. Logistic regression models do not contain any hidden layers, simple neural networks contain one hidden layer with any number of hidden nodes, while deep neural networks contain two or more hidden layers, each with any number of hidden nodes.
Neural networks and deep learning models have become increasingly popular in recent years, especially for diagnostic imaging applications and other prediction tasks (17). When used for supervised learning, these models can be viewed as increasingly complex extensions of the conventional regression framework (17). A conventional logistic regression model (Figure 1, panel C left) contains an input layer and an output layer, where the input layer contains a node for each predictor variable, and all input nodes are connected to the output node via a series of connection weights (or coefficients). The output node takes the weighted sum of the input nodes (ie, the sum of each variable multiplied by its corresponding connection weight) and passes this value through an activation function (in this case, the logistic function) to yield the final output from the model. Neural networks (Figure 1, panel C middle) expand on this framework by adding a hidden layer between the input and output layers, where the nodes in this hidden layer allow the neural network to automatically model more complex, non-linear relations between the predictor variables and the outcome (22). Deep learning models extend this framework even further, adding in multiple hidden layers between the input and output layers to capture even more intricacies in the data (17).
For all these algorithms, their architecture allows them to automatically model complex associations and interactions in the data with minimal assumptions and few a priori specifications from the investigator. However, these algorithms can also be challenging to interpret, may be prone to overfitting, and often require large amounts of training data to achieve adequate performance (12). Thus, when considering using machine learning to identify health outcomes from EHR data, researchers should think critically about when these methods are most warranted and for what tasks they should be used to both justify and optimize their use (23).
3. Setting the Stage: The Two Dimensions of a Health Outcome
In this section, we lay the groundwork for presenting the four scenarios by first considering two dimensions of a health outcome and identifying conditions in each dimension where machine learning may be most useful for computational phenotyping. In the first dimension, we consider the characteristics of the diagnostic criteria used to define the health outcome (Figure 2, Y-axis). In the second dimension, we consider the format in which diagnostic information about the health outcome is usually stored in EHR systems (Figure 2, X-axis).
Figure 2. Four common scenarios of health outcomes and their relation to the usefulness of machine learning for measuring health outcomes from electronic health record (EHR) data.
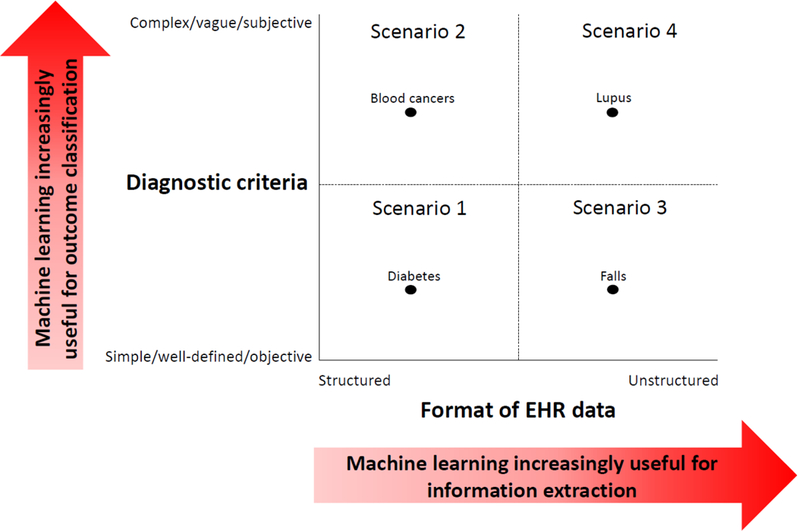
These scenarios arise by jointly considering two dimensions of a health outcome and the conditions under which machine learning may be useful in each dimension. The first dimension (Y-axis) considers several characteristics related to the diagnostic criteria for a given health outcome: 1) the number of clinical parameters involved, which may vary from few (simple) to many (complex), 2) the clarity of the outcome definition, which may vary from having clear boundaries (well-defined) to fuzzy boundaries (vague), and 3) the degree to which clinicians draw upon their personal interpretations during the diagnostic process, which may vary from small (objective) to large (subjective). For health outcomes with more complex, vague, or subjective diagnostic criteria, machine learning may be increasingly useful for predicting the presence of the health outcome based on a vector of clinical inputs (‘outcome classification’). The second dimension (X-axis) considers the format in which important diagnostic information about the health outcome is stored within EHR systems (which may vary from structured to unstructured). For health outcomes where diagnostic information is stored largely in unstructured formats, machine learning may be increasingly useful for facilitating the extraction of important clinical information in a more structured form (‘information extraction’). We dichotomize these two dimensions to create four common scenarios of health outcomes, illustrated by the four quadrants. The black dots represent examples of health conditions that fall within each quadrant.
3.1. Diagnostic Criteria Characteristics
The ability to identify patients with a given health outcome may be more straightforward or challenging depending on several factors related to its diagnostic criteria: i) the number of clinical parameters involved, ii) the clarity of the outcome definition, and iii) the degree to which clinicians draw upon their personal interpretations during the diagnostic process. Given these factors, diagnostic criteria can be characterized as simple or complex (ie, involving few or many clinical parameters), well-defined or vague (ie, involving clear or fuzzy boundaries), and objective or subjective (ie, involving little or much personal interpretation from clinicians). Table 1 gives examples to illustrate each of these concepts. When health outcomes have diagnostic criteria that are simple, well-defined, and objective, the task of classifying patients is usually straightforward once the necessary clinical parameters have been collected or observed. One would also expect little variability in human determination of these outcomes. In contrast, when health outcomes have more complex, vague, or subjective diagnostic criteria, the task of classifying patients is considerably more difficult because it is not easy to predict how clinicians interpret the information to arrive at a final diagnosis. Moreover, it is likely that greater variation exists between clinicians in their determination of these health outcomes. In such cases, machine learning algorithms – when trained on expert-labeled data – could be useful for handling large numbers of clinical variables and detecting intricate associations among them to produce highly accurate and individualized predictions and even reduce classification variability among clinicians with varying levels and degrees of expertise (24). We refer to this task of predicting the presence of a health outcome based on a vector of clinical inputs or parameters as ‘outcome classification’.
Table 1.
Impact of diagnostic criteria characteristics on the ability to identify health outcomes from electronic health record (EHR) data
| Diagnostic criteria | Ability to identify health outcomes from EHR data |
|
|---|---|---|
| Easier | More difficult | |
| Number of clinical parameters involved | Few parameters (simple) Example: Hypertension Diagnosis usually based on two or more blood pressure readings obtained on separate occasions (65). |
Many parameters (complex) Example: Acute leukemia Diagnosis usually based on multiple laboratory test results and other clinical factors (66). |
| Clarity of outcome definition | Clear boundaries (well-defined) Example: Falls “Inadvertently coming to rest on the ground, floor or other lower level, excluding intentional change in position to rest in furniture, wall or other objects” – World Health Organization (67) |
Fuzzy boundaries (vague) Example: Pain “An unpleasant sensory and emotional experience associated with actual or potential tissue damage, or described in terms of such damage” – International Association for the Study of Pain(68), also adopted by the World Health Organization (69) |
| Degree of personal interpretation required from clinicians | Little personal interpretation required (objective) Example: Obesity Diagnosis based on having a body mass index (BMI) of ≥30 kg/m2 (70). |
Much personal interpretation required (subjective) Example: Systemic lupus erythematous Diagnosis largely based on clinical judgement after assessing clinical manifestations, laboratory findings, and serological and histological tests (35, 71). |
3.2. Format of the EHR Data
Clinical data generated during the diagnostic process can be stored in either structured or unstructured formats within EHR systems. Coded billing data such as diagnostic and procedure codes, certain laboratory test results, and some vital signs are stored in structured formats that allow clinical information to be easily extracted from the EHR (6, 9). Other types of EHR data, including admission and discharge summaries, progress notes, and pathology or radiology test results are stored as unstructured text or images (6) and account for up to 80% of data in EHR systems (25). The ability to extract useful clinical information from these rich but unstructured data types is a challenge because the same retrieval methods for structured EHR data cannot be used and manual review is not feasible on a large scale (9). Once again, machine learning algorithms could help – this time by facilitating the extraction and structuring of useful clinical information from unstructured EHR data as part of a natural language processing (NLP) system or image recognition task, a task we refer to as ‘information extraction’. In fact, modern deep learning methods are revolutionizing the way computers analyze human language, producing impressive results on a variety of NLP tasks in many domains (26).
4. The Four Scenarios
In this section, we jointly consider these two dimensions of a health outcome and discuss their combined impact on determining when and for what tasks machine learning may be most useful for computational phenotyping. To simplify the discussion, we dichotomize each dimension to create a 2×2 grid of their combined values, yielding four common scenarios of health outcomes (Figure 2). We define diagnostic criteria as the set of clinical findings normally used to diagnose patients with a given health outcome and assume that these criteria contribute toward a ‘true’ phenotyping mechanism (ie, with the greatest accuracy). In the first part of our discussion, we assume an ideal EHR system with accurate and complete data capture (ie, no medical care has occurred outside the EHR system, all diagnostic tests required to identify the outcome have been performed, and all test results have been accurately measured and stored in their usual format within the system). In the next section, we relax these assumptions and extend our discussion to accommodate the more realistic setting where some of these assumptions are not met.
4.1. Scenario 1
Scenario 1 (Figure 2, lower left quadrant) includes health outcomes that have simple, well-defined, and objective diagnostic criteria based on the results of diagnostic tests and procedures stored in structured EHR fields. In an ideal EHR system with accurate and complete data capture, machine learning would generally not be needed to identify patients with these health outcomes because one would be able to easily extract the relevant clinical information from the EHR system and use it to classify patients according to simple methods (eg, logic rules or scoring systems) that reflect the clearly-defined and objective diagnostic criteria for the health outcome.
Obesity, hypertension, and diabetes are all examples of health outcomes in this scenario. Because all the clinical measurements needed to diagnose patients with these conditions (height, weight, vital signs, and laboratory tests results) are typically captured in structured fields within an EHR database, one would have little reason to use machine learning to identify these patients from an ideal EHR system. Indeed, Makam et al. (27) devised an accurate case-finding algorithm for diabetes that achieved a sensitivity of 97% and specificity of 90% using only data from structured EHR fields and an expert-derived scoring system that did not involve the use of any machine learning or statistical modeling methods.
4.2. Scenario 2
Scenario 2 (Figure 2, upper left quadrant) includes health outcomes that are diagnosed based on the results of multiple diagnostic tests and procedures that are typically stored in structured EHR fields. However, the exact interpretation of each test result and the interplay between them in identifying patients with the health outcome is not highly explicable, often requiring a clinician’s judgment to make the final diagnosis. For such health outcomes, machine learning could be useful for modeling the complex diagnostic decision-making process to predict the presence of the health outcome based on a battery of test results (ie, outcome classification). On the other hand, machine learning would generally not be needed to extract these test results from an ideal EHR system because they already exist in a machine-readable format.
For example, hematologic diseases are diagnosed largely based on the results of laboratory blood tests (28). To make a diagnosis, clinicians must be able to recognize patterns of disease amongst a myriad of blood test results to arrive at the most likely diagnosis (29). Given this complex diagnostic decision-making process, machine learning could be useful for systematically detecting disease-specific patterns from blood test results to improve the detection of hematologic diseases, especially diseases that lack widely accepted diagnostic criteria, such as basophilic leukemias (30). Gunčar et al. (28) built random forests to perform differential diagnosis of hematologic diseases based on a wide array of structured blood test results (181 attributes from 179 different blood tests). Among 43 possible hematological disease categories, their final random forest correctly identified the true disease category – as recorded among patients’ admission and discharge diagnoses in the hospital system – with an overall accuracy (ie, percent correct classification) of 0.60, which was on par with the average accuracy of six hematology specialists (0.62) and better than the average accuracy of eight non-hematology internal medicine specialists (0.26). When the top five (rather than top one) most likely disease categories predicted by the random forest and the hematology specialists were compared to the true disease category, the accuracy of the random forest increased to 0.90 while the accuracy of the hematology specialists increased to only 0.77. Moreover, the authors created a random forest using only 61 blood parameters that achieved similar performance as the random forest using the full set of 181 parameters. These findings suggest that for health outcomes involving many clinical parameters with complex interactions and lacking well-defined, objective diagnostic criteria, machine learning methods could be useful not only for facilitating accurate disease classification, but also identifying important disease markers to reduce unnecessary testing.
4.3. Scenario 3
Scenario 3 (Figure 2, lower right quadrant) includes health outcomes that have clear and objective diagnostic criteria based on only a few clinical measurements, but these measurements are typically captured in unstructured formats within EHR systems. For such health outcomes, machine learning could be useful for facilitating the extraction of useful clinical information from unstructured EHR data into a more structured form (for example, as part of a machine learning-based NLP system applied to clinical free text). However, once the necessary clinical measurements have been extracted from the EHR system, machine learning would generally not be needed to predict the presence of the health outcome based on these measurements because their role in the diagnostic process is already clearly defined.
For example, falls are unambiguous clinical events that are poorly captured in structured diagnostic codes (31), but may be better documented within clinical notes. McCart et al. (32) used machine learning to classify ambulatory care clinical documents as being fall-related or not. For this NLP task, the authors represented the text in each document using the classic ‘bag of words’ technique to transform the document into a structured, machine-readable format, which was then used as input to either a logistic regression model or support vector machine. This process of extracting terms from unstructured text to create features or inputs for shallow machine learning models such as support vector machines or logistic regression is a popular approach that has been used for decades to tackle NLP problems (26, 33). When the authors assessed the performance of their algorithms on test sets from four different medical sites, they found that the support vector machine (trained using a cost-sensitive classification approach (34) to maximize sensitivity) consistently achieved the best discrimination, with an area under the receiver operating characteristic curve (AUC) ranging from 0.95 to 0.98 across the four sites. This support vector machine also had excellent sensitivity (ranging from 0.89 to 0.93) and specificity (ranging from 0.88 to 0.94).
4.4. Scenario 4
Scenario 4 (Figure 2, upper right quadrant) includes health outcomes that have more vague and subjective diagnostic criteria based on a variety of clinical measurements largely captured in unstructured EHR data. These health outcomes are arguably among the most challenging to identify from EHRs using traditional phenotyping approaches because: i) useful diagnostic information about the outcome cannot be easily extracted from a typical EHR system, and ii) the role of each clinical measurement in the diagnostic decision-making process is not well-described. In these cases, machine learning could be useful for both information extraction and outcome classification tasks.
For example, systemic lupus erythematosus (SLE) is an autoimmune disease whose diagnosis relies largely on clinical judgment (35). Patients with SLE display a wide range of signs and symptoms that would likely only be documented in clinical notes, such as malar or discoid rashes, photosensitivity, oral ulcers, or non-scarring alopecia, among others (36). Turner et al. (37) used an NLP system involving machine learning to identify patients with SLE from outpatient clinical notes. Similar to the previous example for falls (32), the authors extracted medical concepts from clinical notes and used them as inputs to one of four classifiers: a shallow neural network, random forest, naïve Bayes, or support vector machine. The authors extracted medical terms from the text using the clinical Text Analysis and Knowledge Extraction System (cTAKES) (38) – a pipeline commonly used for medical NLP tasks whereby words in the text are mapped to standardized concepts from the Unified Medical Language System (UMLS). The authors found that the random forest, shallow neural network, and support vector machine using standardized UMLS concepts as inputs all had excellent performance with accuracies ranging from 0.91 to 0.95 and AUCs ranging from 0.97 to 0.99 when compared to the classifications determined by trained rheumatologists as the gold standard.
Turner et al. (37) also explored the use of a more contemporary machine learning-based NLP approach to identify patients with SLE from clinical notes. This approach involved using the increasingly popular Word2Vec system (39) to create features from words in the text using word embeddings. Word embeddings or semantic vectors are numerical vectors trained by unsupervised neural networks whereby semantically similar words often end up having vectors with a smaller distance between them (40). These word embeddings were then used with Bayesian inversion methods (41) to classify documents. The Word2Vec inversion method had an accuracy of 0.90 and an AUC of 0.91, which was on par with the performance of International Classification of Diseases, Ninth Revision (ICD-9) billing codes. Although the Word2Vec inversion approach did not perform as well as the other more traditional machine learning-based NLP approaches also used in this study (described in the previous paragraph), the authors hypothesized that the performance of this newer approach would improve with more training data. In fact, the use of word embeddings as features in machine learning classifiers is a growing trend in the field of NLP (26) and has been shown to produce state-of-the-art results in other medical (40) and non-medical (42) NLP tasks. Indeed, Gehrmann et al. (33) recently used word embeddings trained by Word2Vec as inputs to deep learning convolutional neural networks to identify 10 different phenotypes from discharge summaries, including advanced lung disease, metastatic cancer, psychiatric disorders, and chronic neurologic dystrophies, among others. For nearly all phenotypes, the authors found that this approach outperformed concept extraction-based methods that used the cTAKES pipeline to create inputs to shallow machine learning classifiers.
Scenario 4 may also include health outcomes that are diagnosed largely based on clinicians’ impressions from medical images. During the diagnostic process for such health outcomes, two distinct types of unstructured documents are usually produced: the raw radiologic or pathologic image and the clinician’s report summarizing his or her impressions based on the image. Machine learning could be useful for not only extracting details about a clinician’s impression from the diagnostic report (ie, as part of an NLP task), but also predicting the presence of the health outcome directly from features in the medical image itself (ie, as an image recognition task). In fact, an increasing number of studies are using deep learning models for a variety of medical-related image classification tasks, yielding impressive results (10, 43–46). For example, Rajpurkar et al. (45) trained a 121-layer convolutional neural network called CheXNet to detect pneumonia from chest X-ray images. When the classifications from CheXNet and each of four radiologists were compared to each other as ‘ground truth’ (eg, CheXNet compared to the classifications from the four radiologists or radiologist 1 compared to the classifications from CheXNet and the other three radiologists), CheXNet outperformed three of the four radiologists with an F1 score (representing the harmonic average of sensitivity and positive predictive value) of 0.44 versus an average F1 score of 0.39 across the four radiologists. Given that the identification of pneumonia and other pathologies from medical images can be a subjective and time-consuming task (47, 48), these findings suggest that deep learning models – when trained on expert-labeled data – could help automate this diagnostic process at the level of domain experts and even reduce the level of inter-rater variability that may occur when using classifications from clinicians with varying levels and areas of expertise.
5. Relaxing the Assumptions
Unlike the ideal setting in the previous section where we assumed accurate and complete data capture in the EHR system, real-world EHR systems typically contain data that can be inaccurate (eg, due to measurement error or erroneous information) and incomplete (eg, due to missing documentation or medical encounters occurring outside the EHR system) (49, 50). Thus, even for simple, well-defined, and objective health outcomes like hypertension, an electronic phenotyping algorithm based solely on the presence of diagnostic codes or test results in the EHR system may not be sufficient. Moreover, if treatment can modify the levels of important diagnostic markers (eg, hypertension medications for lowering blood pressure), then patients undergoing treatment for their condition could still be misclassified based on traditional diagnostic parameters (eg, blood pressure readings), even if accurate and complete data on them are available in the EHR system.
To address these issues, most researchers include additional information besides the usual diagnostic markers in their phenotyping algorithms, such as information on medication use, procedures performed, patient demographics, and even genomic factors (51, 52). As a result, the location of a health outcome on the two-dimensional “grid of scenarios” (Figure 2) may shift from its theoretical position (ie, assuming accurate and complete data capture in the EHR system) upward, to where machine learning is more useful for performing outcome classification, particularly when associations between the health outcome and the additional variables in the phenotyping algorithm are complex – for example, when standard treatments for a health outcome may be used for multiple conditions (5) (eg, beta-blockers used to treat not only hypertension, but also anxiety and thyrotoxicosis (53)).
Researchers may also compensate for the limitations of real-world EHR data by measuring a single concept in the phenotyping algorithm using multiple data types (51). For example, investigators may additionally search the clinical notes for concepts such as medication use that are normally captured in structured EHR fields (51, 54, 55). As a result, the location of a health outcome may shift from its theoretical location on the “grid of scenarios” (Figure 2) rightward, to where machine learning is more useful for performing information extraction, particularly when much of the data being queried are unstructured.
In practice, researchers will often use a combination of these strategies to create the most accurate phenotyping algorithm possible (ie, include more clinical variables in the phenotyping algorithm and query more diverse types of EHR data to measure these variables). To help assess the potential benefits that machine learning may afford for a particular health outcome, researchers may find it helpful to go through the following mental exercise: 1) identify the closest quadrant on the “grid of scenarios” that represents the health outcome of interest under ideal conditions (ie, assuming accurate and complete capture of diagnostic information in the EHR system), 2) identify all clinical measurements and data types from the EHR – beyond the usual diagnostic markers and data types – that one can access and would like to include in the phenotyping algorithm, and 3) consider by how much the inclusion of these additional measurements and data types moves the health outcome from its theoretical location on the grid upward and rightward, to where machine learning is more useful for outcome classification and information extraction tasks, respectively. After completing this mental exercise, investigators may realize that even for health outcomes that normally belong in Scenario 1 (where machine learning would not generally be required under ideal conditions), machine learning may still offer some benefits for identifying these health outcomes from real-world EHR systems. Indeed, Teixeria et al. (55) created an electronic phenotyping algorithm for hypertension that considered not only diagnostic codes for hypertension and blood pressure measurements, but also anti-hypertensive medications and other vital signs. To measure these concepts, the authors not only used information from structured fields in the EHR, but also used NLP systems to search for this information in narrative clinical documents. The authors found that while the use of blood pressure cutoffs alone could identify hypertensive individuals with good discrimination (AUC of 0.85), the consideration of more clinical variables increased the AUC to 0.96 when using standard logistic regression methods, which increased slightly further to 0.98 when using a random forest model.
6. Discussion
In this paper, we presented four common scenarios that researchers may find helpful for thinking critically about when and how machine learning may be used to identify health outcomes from EHR data. We identified these scenarios by jointly considering two dimensions of a health outcome and the conditions under which machine learning may be most useful. We discussed specific tasks for which machine learning might be used to help identify health outcomes in each scenario – first under ideal conditions of accurate and complete data capture in EHR systems and then under more realistic conditions. We also gave examples of health outcomes in each scenario and described how recent studies have used machine learning to identify these outcomes from EHR data. These studies all produced encouraging results, demonstrating the potential for machine learning techniques to improve the ability to accurately and efficiently identify health outcomes from EHR data. Although our discussions focused on health outcomes defined by a binary state (ie, presence or absence of the outcome), the concepts we discussed can be extended to include health outcomes defined by changes in disease state (ie, improvement or worsening).
6.1. Additional Considerations
The scenarios discussed in this paper provide a good starting point for thinking about the complexities of health outcomes and EHR data and how they may relate to the phenotyping process. Besides the volume and variety of data in EHR systems, researchers may also consider the impact of other data complexities such as the velocity at which data is being generated and the veracity of different data types in the EHR system (23).
Researchers may also want to consider other issues surrounding the use of machine learning for electronic phenotyping, such as the transparency and transportability of these algorithms and the training sizes required for their adequate performance. We discuss each of these issues briefly in the sections below.
6.1.1. Transparency
The lack of transparency associated with sophisticated machine learning algorithms like deep learning models may create barriers to their use in phenotyping tasks, particularly when the stakes are high and trust in the algorithm is needed by its end-users. For instance, clinicians want algorithms to support or enhance their medical knowledge rather than simply dictate their decision making, and regulatory bodies need algorithms to be interpretable for accountability purposes because their classifications could have drastic financial or legal implications (33). Thus, efforts to increase the interpretability of such ‘black box’ models are of utmost importance.
Two of the studies previously mentioned in this paper (33, 45) used recently developed methods to interpret the predictions from their deep learning models, yielding impressive results. Gehrmann et al. (33) calculated a modified version of saliency (56), which they termed ‘phrase-saliency’, to identify the most important phrases from clinical text used by their convolutional neural networks for prediction. The authors found that these salient phrases were rated by clinicians as being more specific and relevant to the phenotype of interest than the most important features identified using a more classical concept extraction-based NLP approach. Rajpurkar et al. (45) used class activation mappings (57) to produce heatmaps showing the most important areas of chest X-ray images used by their convolutional neural network for predicting chest pathologies, which agreed with the interpretations from radiologists. Such efforts to improve the transparency and interpretability of complex machine learning models build up the trust and confidence of clinicians and other end-users in these methods, thus promoting their use.
6.1.2. Transportability
Due to privacy concerns and administrative hurdles, many electronic phenotyping studies occur in single-site settings (9, 52, 58). Given the large amount of time and resources usually required to develop phenotyping algorithms, there is growing interest in sharing these algorithms amongst researchers and health care systems to increase their scalability (59). Indeed, initiatives like the Phenotype Knowledgebase (PheKB) (59) – an online environment designed to support researchers in building, sharing, and validating electronic phenotyping algorithms – demonstrate the efforts being made toward this end. Currently, however, few phenotyping algorithms – especially those involving machine learning – have been adapted for use in multiple settings (9).
To create phenotyping algorithms that are scalable, these algorithms must first be externally validated to assess their transportability, and then if necessary, tweaked to accommodate site-specific idiosyncrasies. This ‘validation-adaptation’ process can be extremely time-consuming and labor-intensive, especially for phenotyping algorithms involving NLP systems (58). For phenotyping algorithms that incorporate machine learning, it is especially important to externally validate these algorithms before applying them in different settings because these flexible methods are prone to overfitting (60). However, if further tweaking is required (eg, to model associations differently or recognize new abbreviations in clinical notes), machine learning algorithms may in fact require less human effort to retrain than algorithms relying more heavily on manual engineering. For example, NLP systems that use deep learning methods generally do not use any manually-created input and can be easily retrained to new data (33). When deep learning models are used to identify health outcomes from medical images, these models could potentially be even more transportable than when they are used for NLP tasks due to the lower amount of between-site variability in medical images compared to clinical narratives.
6.1.3. Training Size
Machine learning algorithms, especially deep learning models, require large amounts of annotated training data to achieve optimal performance (61). Indeed, the previously mentioned CheXNet algorithm (45) achieved its expert-level performance after being trained on over 100,000 labeled images. However, many researchers may not have access to such large amounts of training data due to lack of time or resources to annotate the data (62) or because the EHR system may simply not contain sufficient numbers of cases (for example, when identifying rare health outcomes). Furthermore, pooling labeled data across sites may often not be a viable option due to privacy concerns and administrative obstacles, as previously mentioned (52). However, innovative strategies such as data augmentation of images (63) and active learning (sequentially selecting the most informative examples for training) (64) may help reduce the number of training examples required to achieve adequate performance. For example, Chen et al. (62) used active learning with a support vector machine to create an electronic phenotyping algorithm for rheumatoid arthritis and found that active learning reduced the number of labeled samples required to achieve an AUC of 0.95 by 68%.
7. Conclusions
Modern EHR systems contain vast amounts and varieties of clinical data that could be valuable for identifying patients with different health outcomes. However, researchers face various challenges when using these data to create patient phenotyping algorithms. Machine learning methods may be useful for addressing some of these challenges. In this review, we presented a set of common scenarios that researchers may find helpful for thinking critically about when and for what tasks machine learning methods may be used to identify health outcomes from EHR data. We gave examples of health outcomes in each scenario and described recent studies that used machine learning to identify these health outcomes from EHR data, all of which produced encouraging results.
To promote the widespread and successful use of machine learning methods for EHR-based phenotyping tasks, future work should prioritize efforts to increase the transportability of machine learning algorithms for use in multiple settings. Efforts to address issues around the transparency and training sizes required for adequate performance of machine learning algorithms could also help encourage the use of these methods in electronic phenotyping tasks.
Acknowledgments:
We thank Dr. Lisa Herrinton from Kaiser Permanente Division of Research, Northern California for reviewing this paper and providing valuable feedback. We also thank Jacqueline Cellini from the Countway Library of Medicine for her help in identifying references for this review paper. Dr. Wong and Dr. Murray Horwitz are supported by the Thomas O. Pyle Fellowship from Harvard Medical School & Harvard Pilgrim Health Care Institute. Dr. Li is partially supported by the Agency for Healthcare Research and Quality (R01HS022728, R01HS025375, and R01HS024264). Dr. Toh is partially supported by the National Institute of Biomedical Imaging and Bioengineering (U01EB023683).
Footnotes
Compliance with Ethical Standards:
Conflict of Interest: The authors have no conflicts of interest to disclose.
Human and Animal Rights and Informed Consent:This article does not contain any studies with human or animal subjects performed by the authors.
References
Papers of particular interest, published recently have been highlighted as:
● Of importance
●● Of major importance
- 1.Singh S, Loke YK. Drug safety assessment in clinical trials: methodological challenges and opportunities. Trials. 2012;13:138–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp R, Prasad V. Surrogate endpoints in oncology: when are they acceptable for regulatory and clinical decisions, and are they currently overused? BMC Medicine. 2017;15:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Agostino RB. Debate: The slippery slope of surrogate outcomes. Current Controlled Trials in Cardiovascular Medicine. 2000;1(2):76–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger ML, Sox H, Willke RJ, Brixner DL, Eichler HG, Goettsch W, et al. Good practices for real-world data studies of treatment and/or comparative effectiveness: Recommendations from the joint ISPOR-ISPE Special Task Force on real-world evidence in health care decision making. Pharmacoepidemiology and drug safety. 2017;26(9):1033–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.● Lanes S, Brown JS, Haynes K, Pollack MF, Walker AM. Identifying health outcomes in healthcare databases. Pharmacoepidemiology and drug safety. 2015;24(10):1009–16.Discusses important methodological issues for researchers to consider when identifying health outcomes from both claims and EHR databases.
- 6.Denny JC. Chapter 13: Mining Electronic Health Records in the Genomics Era. PLoS Computational Biology. 2012;8(12):e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richesson RL, Smerek MM, Blake Cameron C. A Framework to Support the Sharing and Reuse of Computable Phenotype Definitions Across Health Care Delivery and Clinical Research Applications. eGEMs. 2016;4(3):1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onukwugha E Big Data and Its Role in Health Economics and Outcomes Research: A Collection of Perspectives on Data Sources, Measurement, and Analysis. PharmacoEconomics. 2016;34:91–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.● Ford E, Carroll JA, Smith HE, Scott D, Cassell JA. Extracting information from the text of electronic medical records to improve case detection: a systematic review. Journal of the American Medical Informatics Association : JAMIA. 2016;23(5):1007–15.Reviews the methods and findings from previously published studies using information from free text in electronic medical records for patient phenotyping.
- 10.Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLOS ONE. 2017;12(6):e0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kienle GS, Kiene H. Clinical judgement and the medical profession. Journal of Evaluation in Clinical Practice. 2011;17(4):621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.●● Beam AL, Kohane IS. Big Data and Machine Learning in Health Care. Jama. 2018;319(13):1317–8.Provides an excellent non-technical overview of machine learning and big data and gives reasonable expectations for their roles in health care.
- 13.Alessa A, Faezipour M. A review of influenza detection and prediction through social networking sites. Theoretical Biology & Medical Modelling. 2018;15:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurgovsky J, Granitzer M, Ziegler K, Calabretto S, Portier P-E, He-Guelton L, et al. Sequence classification for credit-card fraud detection. Expert Systems with Applications. 2018;100:234–45. [Google Scholar]
- 15.Krizhevsky A, Sutskever I, Hinton GE ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. [Google Scholar]
- 16.James G, Witten D, Hastie T, Tibshirani R An Introduction to Statistical Learning: with Applications in R: Springer Publishing Company, Incorporated; 2014. 430 p. [Google Scholar]
- 17.●● Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, et al. Artificial intelligence in healthcare: past, present and future. Stroke and Vascular Neurology. 2017;2(4):230–43.Reviews the current state of machine learning applications in health care from PubMed with respect to different types of data used, areas of disease focus, and techniques used.
- 18.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436. [DOI] [PubMed] [Google Scholar]
- 19.Resta M, Sonnessa M, Tànfani E, Testi A Unsupervised neural networks for clustering emergent patient flows. Operations Research for Health Care. 2018;18:41–51. [Google Scholar]
- 20.Hastie T, Tibshirani R, Friedman J The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition: Springer; New York; 2009. [Google Scholar]
- 21.Breiman L Random Forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 22.Tu JV. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. Journal of Clinical Epidemiology. 1996;49(11):1225–31. [DOI] [PubMed] [Google Scholar]
- 23.●● Raghupathi W, Raghupathi V Big data analytics in healthcare: promise and potential. Health Information Science and Systems. 2014;2:3.Provides a broad overview of Big Data analytics and discusses important issues for consideration.
- 24.Darcy AM, Louie AK, Roberts L Machine learning and the profession of medicine. Jama. 2016;315(6):551–2. [DOI] [PubMed] [Google Scholar]
- 25.Murdoch TB, Detsky AS The inevitable application of big data to health care. Jama. 2013;309(13):1351–2. [DOI] [PubMed] [Google Scholar]
- 26.Young T, Hazarika D, Poria S, Cambria E Recent Trends in Deep Learning Based Natural Language Processing. CoRR. 2017;abs/1708.02709. [Google Scholar]
- 27.Makam AN, Nguyen OK, Moore B, Ma Y, Amarasingham R Identifying patients with diabetes and the earliest date of diagnosis in real time: an electronic health record case-finding algorithm. BMC Medical Informatics and Decision Making. 2013;13:81–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunčar G, Kukar M, Notar M, Brvar M, Černelč P, Notar M, et al. An application of machine learning to haematological diagnosis. Scientific Reports. 2018;8(1):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wians FH. Clinical Laboratory Tests: Which, Why, and What Do The Results Mean? Laboratory Medicine. 2009;40(2):105–13. [Google Scholar]
- 30.Valent P, Sotlar K, Blatt K, Hartmann K, Reiter A, Sadovnik I, et al. Proposed diagnostic criteria and classification of basophilic leukemias and related disorders. Leukemia. 2017;31:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters TM, Chandler AM, Mion LC, Daniels MJ, Kessler LA, Miller ST, et al. Use of ICD-9-CM Codes to Identify Inpatient Fall-Related Injuries. Journal of the American Geriatrics Society. 2013;61(12):10.1111/jgs.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCart JA, Berndt DJ, Jarman J, Finch DK, Luther SL Finding falls in ambulatory care clinical documents using statistical text mining. Journal of the American Medical Informatics Association : JAMIA. 2013;20(5):906–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.●● Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One. 2018;13(2):e0192360.Demonstrates the superiority of modern deep learning models over classical concept extraction based methods for performing NLP on unstructured clinical text for a variety of phenotyping tasks.
- 34.MetaCost Domingos P.: a general method for making classifiers cost-sensitive. Proceedings of the fifth ACM SIGKDD international conference on Knowledge discovery and data mining; San Diego, California, USA. 312220: ACM; 1999. p. 155–64. [Google Scholar]
- 35.Larosa M, Iaccarino L, Gatto M, Punzi L, Doria A Advances in the diagnosis and classification of systemic lupus erythematosus. Expert review of clinical immunology. 2016;12(12):1309–20. [DOI] [PubMed] [Google Scholar]
- 36.Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis and rheumatism. 2012;64(8):2677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner CA, Jacobs AD, Marques CK, Oates JC, Kamen DL, Anderson PE, et al. Word2Vec inversion and traditional text classifiers for phenotyping lupus. BMC Medical Informatics and Decision Making. 2017;17:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.● Mikolov T, Chen K, Corrado G, Dean J Efficient Estimation of Word Representations in Vector Space. CoRR. 2013;abs/1301.3781.Describes Word2Vec - an increasingly popular method for automatically engineering features from free text using machine learning to represent words in NLP tasks.
- 40.Luo Y, Cheng Y, Uzuner O, Szolovits P, Starren J Segment convolutional neural networks (Seg-CNNs) for classifying relations in clinical notes. Journal of the American Medical Informatics Association : JAMIA. 2018;25(1):93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taddy M Document Classification by Inversion of Distributed Language Representations. CoRR. 2015;abs/1504.07295. [Google Scholar]
- 42.Rudkowsky E, Haselmayer M, Wastian M, Jenny M, Emrich Š, Sedlmair M More than Bags of Words: Sentiment Analysis with Word Embeddings. Communication Methods and Measures. 2018;12(2–3):140–57. [Google Scholar]
- 43.Pak M, Kim S, editors. A review of deep learning in image recognition. 2017 4th International Conference on Computer Applications and Information Processing Technology (CAIPT); 2017 8–10 Aug 2017. [Google Scholar]
- 44.Litjens G, Sanchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. CoRR. 2017;abs/1711.05225. [Google Scholar]
- 46.Ciompi F, Chung K, van Riel SJ, Setio AAA, Gerke PK, Jacobs C, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Scientific Reports. 2017;7:46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brady AP. Error and discrepancy in radiology: inevitable or avoidable? Insights into Imaging. 2017;8(1):171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman MI, Lee EY, Bixby S, Diperna S, Hellinger, Markowitz R, et al. Variability in the interpretation of chest radiographs for the diagnosis of pneumonia in children. Journal of hospital medicine. 2012;7(4):294–8. [DOI] [PubMed] [Google Scholar]
- 49.Bowman S. Impact of Electronic Health Record Systems on Information Integrity: Quality and Safety Implications. Perspectives in Health Information Management. 2013;10(Fall):1c. [PMC free article] [PubMed] [Google Scholar]
- 50.Lin KJ, Glynn RJ, Singer DE, Murphy SN, Lii J, Schneeweiss S Out-of-system Care and Recording of Patient Characteristics Critical for Comparative Effectiveness Research. Epidemiology (Cambridge, Mass). 2018;29(3):356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. Journal of the American Medical Informatics Association : JAMIA. 2016;23(e1):e20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.● Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2014;21(2):221–30.Reviews different approaches, including machine learning methods, used in the recent literature to identify patients with a common phenotype from EHR data.
- 53.Jackson RE, Bellamy MC Antihypertensive drugs. BJA Education. 2015;15(6):280–5. [Google Scholar]
- 54.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis care & research. 2010;62(8):1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teixeira PL, Wei WQ, Cronin RM, Mo H, VanHouten JP, Carroll RJ, et al. Evaluating electronic health record data sources and algorithmic approaches to identify hypertensive individuals. Journal of the American Medical Informatics Association : JAMIA. 2017;24(1):162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Chen X, Hovy E, Jurafsky D, editors. Visualizing and Understanding Neural Models in NLP2016: Association for Computational Linguistics. [Google Scholar]
- 57.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A, editors. Learning Deep Features for Discriminative Localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 27–30 June 2016. [Google Scholar]
- 58.Carrell DS, Schoen RE, Leffler DA, Morris M, Rose S, Baer A, et al. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. Journal of the American Medical Informatics Association. 2017;24(5):986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. Journal of the American Medical Informatics Association : JAMIA. 2016;23(6):1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster KR, Koprowski R, Skufca JD. Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomedical engineering online. 2014;13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asperti A, Mastronardo C, editors. The Effectiveness of Data Augmentation for Detection of Gastrointestinal Diseases from Endoscopical Images. BIOIMAGING; 2018.
- 62.Chen Y, Carroll RJ, Hinz ERM, Shah A, Eyler AE, Denny JC, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e253–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong SC, Gatt A, Stamatescu V, McDonnell MD, editors. Understanding Data Augmentation for Classification: When to Warp? 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA); 2016 Nov. 30 2016-Dec. 2 2016. [Google Scholar]
- 64.Lewis DD, Gale WA A sequential algorithm for training text classifiers. Proceedings of the 17th annual international ACM SIGIR conference on Research and development in information retrieval; Dublin, Ireland. 188495: Springer-Verlag New York, Inc.; 1994. p. 3–12. [Google Scholar]
- 65.Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Dennison Himmelfarb C, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Journal of the American College of Cardiology. 2018;71(19):e127–e248. [DOI] [PubMed] [Google Scholar]
- 66.Arber DA, Borowitz MJ, Cessna M, Etzell J, Foucar K, Hasserjian RP, et al. Initial Diagnostic Workup of Acute Leukemia: Guideline From the College of American Pathologists and the American Society of Hematology. Archives of Pathology & Laboratory Medicine. 2017;141(10):1342–93. [DOI] [PubMed] [Google Scholar]
- 67.World Health Organization. WHO Global Report on Falls Prevention in Older Age. 2007.
- 68.Loeser JD, Treede R-D The Kyoto protocol of IASP Basic Pain Terminology. PAIN®. 2008;137(3):473–7. [DOI] [PubMed] [Google Scholar]
- 69.World Health Organization. WHO guidelines on the pharmacological treatment of persisting pain in children with medical illnesses. Genva, Switzerland; 2012. [PubMed] [Google Scholar]
- 70.Frantzides CT, Luu MB. BMJ Best Practice: Obesity in adults. November 2017. [Available from: http://bestpractice.bmj.com/topics/en-us/211.
- 71.Thong B, Olsen NJ. Systemic lupus erythematosus diagnosis and management. Rheumatology. 2017;56(suppl_1):i3–i13. [DOI] [PubMed] [Google Scholar]


