Abstract
Background
The roots and rhizomes of Cimicifuga yunnanensis Hsiao are commonly used as anti-inflammatory, antipyretic, and analgesic remedies and detoxification agents in traditional Chinese medicine (TCM). Although C. yunnanensis has been considered as supplementary medicine for several disorders, the antitumor effect of this herb and its key components has not been explored.
Materials and methods
The rhizomes of C. yunnanensis were isolated by chromatographic techniques. Structures of isolated compounds were identified based on spectroscopic methods and comparison with published data. The in vitro anticancer activities of purified components were also performed by MTT experiments. The in vivo anticancer activities were examined by subcutaneous tumor model or a breast cancer liver metastasis model.
Results
In this study, we aimed to identify and characterize the effective antitumor components from the rhizomes of C. yunnanensis. By bioassay-guided fractionation techniques and chemical characterization, 12 cycloartane triterpenes and four chromones were isolated, among them, 11 compounds were identified in this genus at first. The identified two compounds showed dramatic inhibitory activities against breast cancer cells: compound 4 (23-epi-26-deoxyactein) and compound 13 (cimigenol). Then, we examined the antitumor effect of these two selective candidate chemicals on triple-negative breast cancer (TNBC) cells in vivo and found that they could reduce tumor growth in subcutaneous tumor model or breast cancer liver metastasis model.
Conclusion
These results suggested that the selective compounds isolated from C. yunnanensis Hsiao could be the promising new agents for TNBC treatment.
Keywords: Cimicifuga, Cimicifuga yunnanensis, cycloartane triterpenoids, anticancer activity, triple-negative breast cancer, breast cancer liver metastasis
Introduction
Cimicifuga is a genus belonging to the Ranunculaceae family, which composes of 18 species in the whole world and eight species in the People’s Republic of China.1 The rhizomes of Cimicifuga dahurica (Turcz) Maxim, Cimicifuga heracleifolia Kom, and Cimicifuga foetida Linnaeus are called “Shengma”, in traditional Chinese medicine (TCM), which have been used as antipyretic and analgesic agents and have been recorded in the Chinese pharmacopeia (CPC, 2010).2,3 In USA and Europe, black cohosh (Caulerpa racemosa [L.] Nutt.) is widely used as a herbal dietary supplement for the relief of menopause-related symptoms, with a clinical history over the past 40 years.4,5 The cycloartane triterpene glycosides are considered to be the main bioactive components of Cimicifuga yunnanensis.6 Although more than 40 triterpenes and triterpene glycosides have been isolated from Cimicifuga, the antitumor effect of this herb and its compounds have not been explored yet.6–9
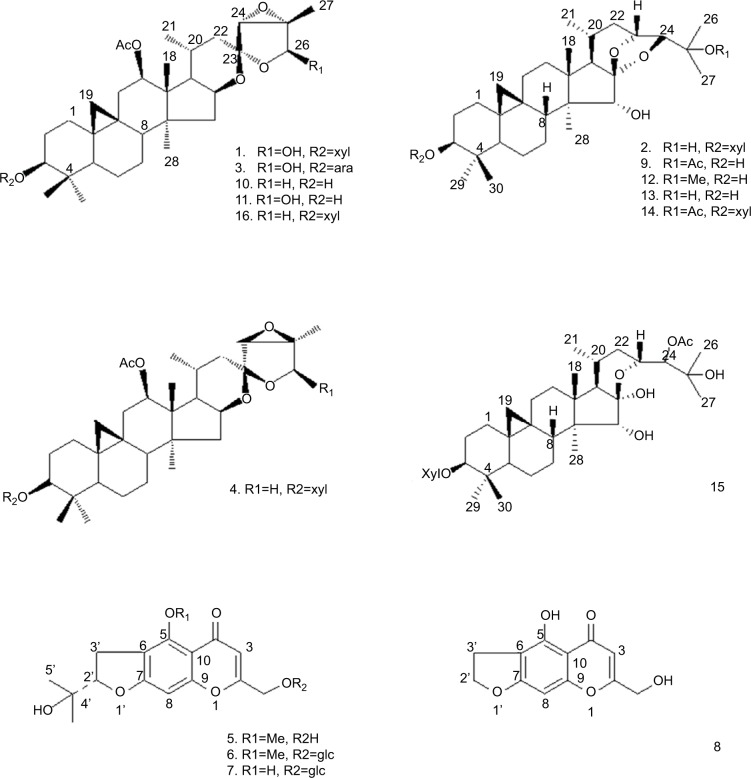
In searching for novel antitumor agents from natural products, we found that the ethanol extracts from the rhizomes of C. yunnanensis exhibited antitumor effect on cancer cells. To evaluate the anticancer activities of C. yunnanensis’ rhizome, we performed bioassay-guided fractionation on the extracts and isolated 16 purified components (1–16; Figure 1) elucidated as cycloartane triterpenes and chromones. Structural elucidation was achieved by using spectroscopic data (nuclear magnetic resonance [NMR] spectroscopy and mass spectrometry) and related references. For cellular-level antitumor effect study, we performed MTT assay and found that, among these compounds, 23-epi-26-deoxyactein (4) and cimigenol (13) have dramatic antitumor effect on breast cancer cells and especially on triple-negative breast cancer (TNBC) cell line, MDA-MB-231, compared with other compounds. We further identified the in vivo antitumor effect of these two compounds on MDA-MB-231 cells and found that 23-epi-26-deoxyactein and cimigenol inhibited the subcutaneous growth and intra-hepatic growth of MDA-MB-231 cells.
Figure 1.
Structures of compounds 1–16.
Note: The structure of all compounds was tested using NMR spectral data analysis. Abbreviation: NMR, nuclear magnetic resonance.
Materials and methods
Plant material
The plant material of C. yunnanensis Hsiao, in the Ranunculaceae family and Cimicifuga genus, which consisted of the underground part of the plant, was collected by Yang Lixin, Prof Pei Sheng Ji, Prof Li Bin Jun, and Chen Guiqing of the Applied Ethnobotany Research Group in the Laboratory of Ethnobotany, on August 29, 2006, in Gezi Village, Shigu Township of Yulong Country, Yunnan Province, People’s Republic of China, along the Yangtze river at an altitude of 1,647 m. The precise location was N26°58′13″, E99°57′19″. The specimen number is 6829 and is deposited in the evidential herbarium at the Laboratory of Ethnobotany. The green part of the plant was air-dried on a paper box at 23°C for 15 days and was subsequently stored and crushed at room temperature.
General Methods
The NMR spectra were obtained using Bruker AM 400 NMR at 400 MHz (1H) and 100 MHz (13C) with tetramethylsilane (TMS) as the internal standard. Mass data were obtained on VG Auto Spec-3000 spectrometers. Silica gel (200–300 mesh) for column chromatography (CC) and GF254 (Qingdao Marine Chemical Factory, Qingdao City, Shandong Province, People’s Republic of China), D101 macroporous resin (Nankai Chemical Factory, Osaka, Japan), Lichroprep RP-18 (40–63 µm; EMD Millipore, Billerica, MA, USA), and Sephadex LH-20 (Pharmacia Fine Chemical Co., Ltd.) were used for CC. Fractions were monitored by thin layer chromatography (TLC), and spots were visualized by heating TLC sprayed with 10% H2SO4.
Plant extraction and purification
The dried, milled rhizome of C. yunnanensis (2.3 kg) was extracted with 95% EtOH (×4). The extract was evaporated under reduced pressure to yield a syrup-like residue (about 380 g). The syrup was suspended in H2O and extracted successively with EtOAc (4 mL × 2,000 mL). After removing the solvent under reduced pressure at 60°C, the residue (180 g) was subjected to D101 macroporous resin CC (Ø 300 mm × 2,000 mm) using MeOH/H2O mixtures of increasing polarity (50:50, 90:10, 100:0, each 3,000 mL) to give three fractions. Fraction 2 (90 g) was repeatedly subjected to silica gel CC eluted with CHCl3/MeOH (9:1, 5:1, 3:1, 3:2, each 1,000 mL) to give four subfractions (A–D). Subfraction B was rechromatographied on silica gel CC eluted with CHCl3/MeOH (9:1) to give two compounds: 1 (500 mg), 2 (250 mg). Subfractions C and D were rechromatographied on silica gel CC eluted with CHCl3/MeOH (5:1), then RP-18 using MeOH–H2O (6:4) to afford two compounds: 3 (24 mg), 4 (49 mg). Fraction F3 (70 g) was repeatedly subjected to silica gel CC eluted with CHCl3/MeOH (9:1, 5:1, 3:1, each 1,000 mL) to give three subfractions (E–G). Subfraction E was purified on Sephadex LH-20 (MeOH) and silica gel CC to give four compounds: 5 (60 mg), 6 (43 mg), 7 (60 mg), 8 (160 mg). Subfraction F was rechromatographied on RP-18 using MeOH–H2O (1:1, 2:1, 3:1, 4:1, each 1,000 mL) as an elution to give four subfractions F1–F4. Subfraction F2, after purified on silica gel CC (Ø 15 mm × L 400 mm) eluted with CHCl3/MeOH (7:1) yielded compounds 9 (17 mg) and 10 (15 mg). Subfraction F3 after purified on silica gel CC (Ø 15 mm × L 400 mm) eluted with CHCl3/MeOH (9:1) yielded compounds 11 (14 mg), 12 (13 mg). Subfraction G was rechromatographied on RP-18 using MeOH–H2O (1:1, 2:1, 3:1, 4:1, each 1,000 mL) and purified on silica gel CC eluted with CHCl3/MeOH yielded compounds 13 (54 mg), 14 (47 mg), 15 (15 mg), and 16 (35 mg). All compounds were tested by HPLC before using NMR spectral data analysis.
HPLC analysis
To determine the purity of all compounds, compounds 1–16 were subjected to XTerra Rp18 (5 µm, 3.9 internal diameter (i.d.) × 150 mm; Waters Corporation, Milford, Massachusetts, USA) column in HPLC (LC-20AT; Shimadzu, Kyoto, Japan). The samples were dissolved in MeOH (0.2 mg/mL) and injected into column (10 µL) in a mobile phase with the flow rate of 1 mL/min. The samples were eluted with a linear gradient from the initial 30% acetonitrile in 0.1% formic acid to 80% acetonitrile in 0.1% formic acid for 40 minutes and additional elution with 100% acetonitrile for 10 minutes.
Chemical structure of compounds 1–16
The bioactive-guided purification of the crude extract on silica gel yielded 16 pure bioactive compounds 1–16, whose chemical structures (Figure 1) were identified by means of 1H and 13C NMR and fast Atom Bombardment mass (FAB-MS) data. These data were in accordance with previously reported data on literatures.
Cell lines and cell culture
Human breast cancer cells, MCF-7 (an endocrine-dependent breast cancer cell line), MDA-MB-231 (TNBC), and SK-BR3 (a HER2-positive breast cancer cell line), were all purchased and originated from the Type Culture Collection of the Chinese, Academy of Sciences (Shanghai, People’s Republic of China). Among these cells, MCF-7 and MDA-MB-231 were the gift from Dr Fan Feng from research center for clinical and translational medicine, the 302nd Hospital of Chinese PLA, Beijing, People’s Republic of China.10,11 Cells were cultured in a humidified atmosphere of 5% CO2 in air and fed with the culture medium of DMEM (Thermo Fisher Scientific, Waltham, MA, USA), 10% FBS (Thermo Fisher Scientific), and 1% penicillin–streptomycin solution (Thermo Fisher Scientific). The usage of cell lines or experiment protocols was all approved by the ethics committee of the Kunming Institute of Botany, Chinese Academy of Sciences.
MTT assay on breast cancer cells
The MTT assay was used to measure the cytotoxicity of compounds 1–16 on breast cancer cells following the methods described in the studies by Zhang et al11 and Ma et al.12 Adriamycin was chosen as a positive control, and the concentrations of compounds used in MTT experiments were 0.08, 0.4, 2, 10, and 50 µg/mL. Inhibition rates were calculated as follows: (OD 490 nm of control group – OD 490 nm of administration group)/(OD 490 nm of control group – OD 490 nm of blank group)×100%. IC50 value of compounds was calculated based on Inhibitory rates (IR) of indicated concentration of compounds on cells. To evaluate the bioactive components of four compounds, the in vivo antitumor effect was identified.
Subcutaneous tumor formation
Animal experiments were approved by the Animal Care Committee of Kunming Institute of Botany, People’s Republic of China. Four- to five-week-old female mice with severe combined immunodeficiency (SCID) were inoculated with 1×107 MDA-MB-231 cells. Animal studies were carried out in accordance with the UK Animals (Scientific Procedures) Act, 1986, and associated guidelines. When the volume of tumors reached about 300 mm3, mice were randomly allocated into four groups: solvent control, 23-epi-26-deoxyactein, cimigenol, or adriamycin. For each treatment, sorafenib (1.5 mg/kg) or rifampicin (2 mg/kg) was given via oral gavage administration twice per week for 3 weeks. Tumor size was monitored every week by measuring the length and width using a caliper after compound injection.13,14 The volume of tumors was calculated as follows: width2× length/213,14. Finally, tumors were dissected out and weighed.
Transwell experiments
MDA-MB-231 cells were cultured and treated with solvent control, compound 4, compound 13, or adriamycin for 24 hours. Then, the cells were harvested for transwell analysis. Transwell analysis was performed following the methods described in the studies by Li et al15 and Zhang et al.16 The relative invasion or migration cell number inhibition was calculated by (OD 546 nm of administration group)/(OD 546 nm of control group). The inhibition rate of compounds on MDA-MB-231 cells’ in vitro invasion or migration was calculated as follows: (relative invasion cell number of control group – relative invasion cell number of administration group)/(relative invasion cell number of control group) or (relative migration cell number of control group – relative migration cell number of administration group)/(relative migration cell number of control group).
Breast cancer liver metastasis formation
To prepare a breast cancer liver metastasis model, MDA-MB-231 cells were cultured and injected into live nude mice via hepatic portal vein (1×104 per animal). After 1-week growth, mice were divided into four groups and treated with 1) control; 2) 2 mg/kg compound 4; 3) 2 mg/kg compound 13; or 4) 2 mg/kg compound 4; or Adriamycin for 2 weeks (once per day). Finally, mice were analyzed by positron emission tomography/computed tomography (PET/CT) screening.17–20 The 18F-fluorodeoxyglucose (18F-FDG) image of nodules formed by MDA-MB-231 cells was quantificationally analyzed by Image J software following the methods described by Wu et al,21 Shao et al,22 and Xie et al.23
Statistical analyses
All statistical significance analyses were performed by Bonferroni correction with or without two-way ANOVA using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA). The IC50 values of compounds were calculated by Origin 6.1 software.
Results
The identification of compounds from C. yunnanensis
By bioactive-guided purification, from the crude extracts of C. yunnanensis, 16 pure bioactive compounds 1–16 (Figure 1 and Table S1) were identified as follows: actein (1),24 cimigenol-3-O-β-d-xylopyranoside (23R,24S) (2),25 26-deoxyacetylacteol-3-O-α-l-arabinopyranoside (3),26 23-epi-26-deoxyactein (4),27 cimifugin (5),28 cimifugin glucoside (6),28 norcimifugin glucoside (7),29 norkhellol (8),30 25-O-acetylcimigenol (9),31 26-deoxyacetylacteol (10),25 acetylacteol (11),25 25-O-methylcimigenol (12),26 cimigenol (13),31 25-O-acetylcimigenol-3-O-β-d-xylopyranoside (14),29 24-O-acetylhydroshengmanol-3-O-β-d-xylopyranoside (15),32 26-deoxyactein (16)33 by using 1H and 13C NMR FAB-MS spectroscopic data and referring previously reported data. The purity of compounds 1–16 was determined by HPLC. They showed single peak in XTerra Rp18 column with retaining times. Among them, compounds 1, 3, 5–12, and 16 are new isolated chemicals in this genus. The HPLC results of compounds 1–16 are shown in Figure S1.
The in vitro antitumor effect of selective compounds on different breast cancer cells
All isolated compounds were screened for in vitro antitumor activity. The IC50 values of these compounds were evaluated based on MTT assay results as summarized in Table 1. Compounds 4 and 13 have most dramatic cytotoxic effect on breast cancer cells. The IC50 values of compound 13 against human breast cancer cell lines, MCF-7, MDA-MB-231, and SK-BR3, were 0.1 µg/mL, 0.32 µg/mL, and 0.21 µg/mL, respectively, whereas IC50 values of compound 4 on these cell lines were 3.1 µg/mL, 2.5 µg/mL, and 5.5 µg/mL, respectively. Other compounds, eg, 10, 12, and 16, showed moderate antitumor effect against breast cancer cells. Since TNBC represents the most therapeutically intractable type compared with endocrine-related breast cancer or HER-positive breast cancer, we asked if those novel compounds have antitumor effect of compounds on TNBC cell line. Notably, we found that compounds 4 and 13 have promising antitumor effect on MDA-MB-231, a highly aggressive TNBC cell line.
Table 1.
IC50 values of compounds 1–16 on breast cancer cells, MCF-7, MDA-MB-231, or Sk-Br3
| Compounds | IC50 (μg/mL) | ||
|---|---|---|---|
|
| |||
| MCF-7 | MDA-MB-231 | SK-BR3 | |
|
| |||
| 1. Actein | – | 5.4 | – |
| 2. Cimigenol-3-O-β-d-xylopyranoside (23R,24S) | – | 4.1 | 7.42 |
| 3. 26-Deoxyacetylacteol-3-O-α-l-arabinopyranoside | 8.57 | – | – |
| 4. 23-Epi-26-deoxyactein | 3.1 | 2.5 | 5.5 |
| 5. Cimifugin | – | – | – |
| 6. Cimifugin glucoside | 4.78 | 8.46 | – |
| 7. Norcimifugin glucoside | – | – | – |
| 8. Norkhellol | – | – | – |
| 9. 25-O-acetyl-cimigenol | – | – | – |
| 10. 26-Deoxyacetylacteol | 1.32 | 3.52 | 4.33 |
| 11. Acetylacteol | – | – | – |
| 12. 25-O-Methyl-cimigenol | 1.56 | 3.79 | 6.75 |
| 13. Cimigenol | 0.1 | 0.32 | 0.21 |
| 14. 25-O-Acetylcimigenol-3-O-β-d-xylopyranoside | – | – | – |
| 15. 24-O-Acetylhydroshengmanol-3-O-β-d-xylopyranoside | – | – | – |
| 16. 26-Deoxyactein | 2.77 | 5.46 | – |
| Adriamycin | 1.7 | 2.4 | 1.90 |
The in vivo antitumor effect of selective compounds on TNBC subcutaneous tumor models
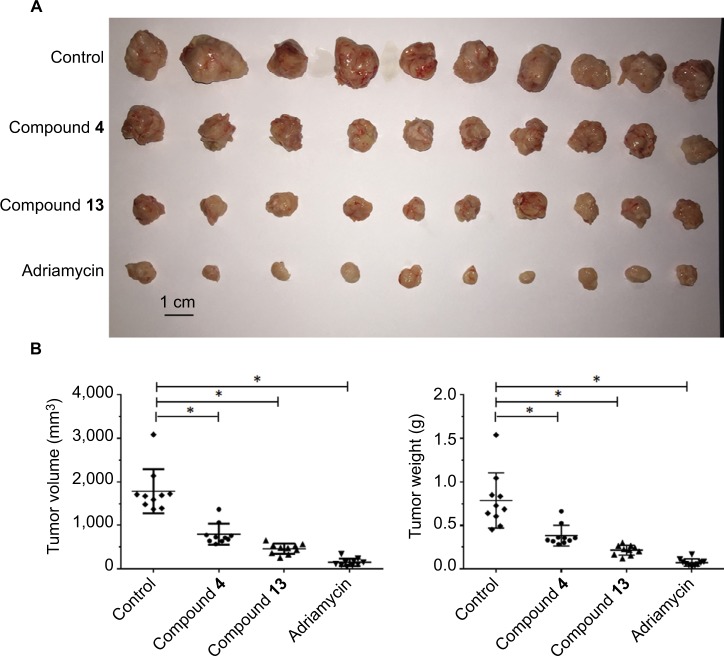
Next, we evaluated the in vivo antitumor effect of the selective compounds on tumors established by TNBC MDA-NB-231 cells. For MDA-231 subcutaneous tumor model, tumor-bearing nude mice received compound 4 or 13, and adriamycin was used as positive control, for 3 weeks. At the end point, tumor volume and weight were measured. As shown in Figure 2, compound 4 or 13 could inhibit the subcutaneous growth of MDA-MB-231 cells in nude mice. However, the antitumor effects of the selective compounds were lower than the positive control, adriamycin. Overall, these results revealed the in vivo antitumor activation of selective compounds on MDA-MB-231 cells.
Figure 2.
Compound 4 or 13 inhibited the subcutaneous growth of MDA-MB-231 cells.
Notes: MDA-MB-231-H cells were injected into nude mice to form subcutaneous tumors. Next, mice were divided into four groups: 1) solvent control group; 2) compound 4 group; 3) compound 13 group; (4) adriamycin group. Results are shown as follows: (A) photographs, (B) tumor volumes, and (C) tumor weights. *P<0.05.
The inhibitory activation of selective compounds on TNBC cells’ in vitro invasion or migration
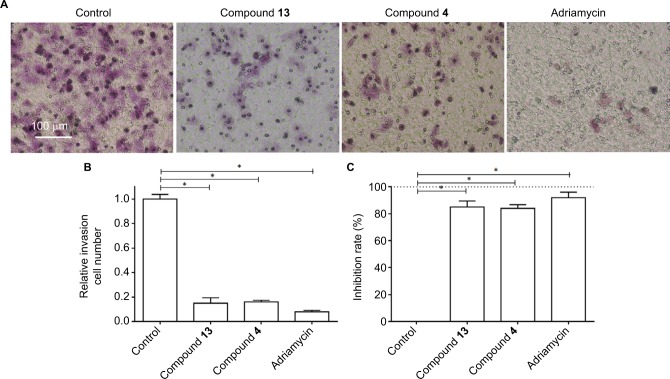
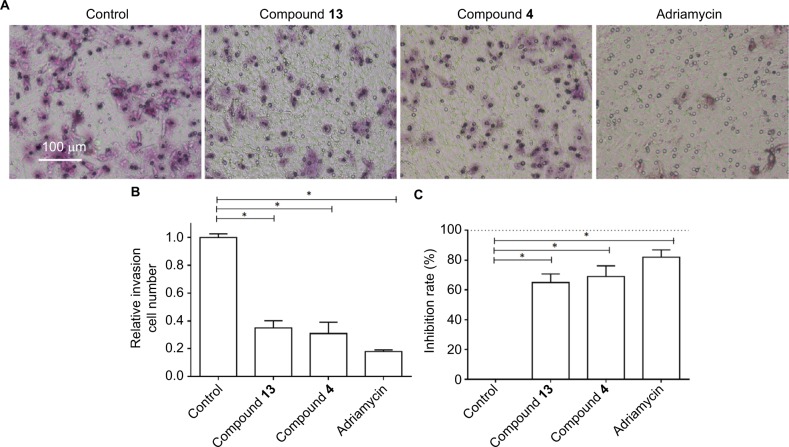
Based on the fact that highly aggressive ability is one of the foremost features of TNBC cells, the transwell experiments were performed to examine the in vitro invasion or migration of MDA-MB-231 cells. Compound 4 or 13 could inhibit the in vitro invasion (Figure 3) or migration (Figure 4) of MDA-MB-231 cells. However, the antitumor effects of the selective compounds were lower than the positive control, adriamycin (Figures 3 and 4). Therefore, these results further revealed the antitumor effect of compounds and showed that compounds could inhibit the in vitro migration or invasion of MDA-MB-231 cells.
Figure 3.
Compound 4 or 13 inhibited the in vitro invasion of MDA-MB-231 cells.
Notes: MDA-MB-231 cells were treated with IC50 of compound 4, compound 13, or adriamycin for 24 hours. Then, the cells were harvested for transwell experiments to reveal the in vitro invasion of MDA-MB-231 cells. Results are shown as follows: (A) photographs, (B) relative invasion number, or (C) inhibition rate of compound 4, compound 13, or adriamycin on MDA-MB-231’s in vitro invasion. *P<0.05.
Figure 4.
Compound 4 or 13 inhibited the in vitro migration growth of MDA-MB-231 cells.
Notes: MDA-MB-231 cells were treated with IC50 of compound 4, compound 13, or adriamycin for 24 hours. Then, the cells were harvested for transwell experiments to reveal the in vitro migration of MDA-MB-231 cells. Results are shown as follows: (A) photographs, (B) relative migration number, or (C) inhibition rate of compound 4, compound 13, or adriamycin on MDA-MB-231’s in vitro migration. *P<0.05.
The inhibitory activation of selective compounds on TNBC cells’ liver metastasis
Next, based on the highly aggressive feature of MDA-MB-231 cells, the breast cancer liver metastasis assay was performed. MDA-MB-231 cells were injected into liver organs via hepatic portal vein and formed multi-nodules of tumors. The treatment of compound 4, compound 13, or adriamycin was performed after the tumors were established and lasted for 2 weeks. Intrahepatic growth of MDA-MB-231 cells in nude mice was detected by PET/CT screening at the end point, and tumor volume and weight were measured. As shown in Figures 5 and 6, compound 4 and compound 13 have a good antitumor effect on MDA-MB-231’ intrahepatic growth in nude mice. However, the antitumor effects of the selective compounds were lower than the positive control, adriamycin (Figures 5 and 6). Therefore, selected compounds could effectively inhibit the liver metastasis of MDA-MB-231 cells.
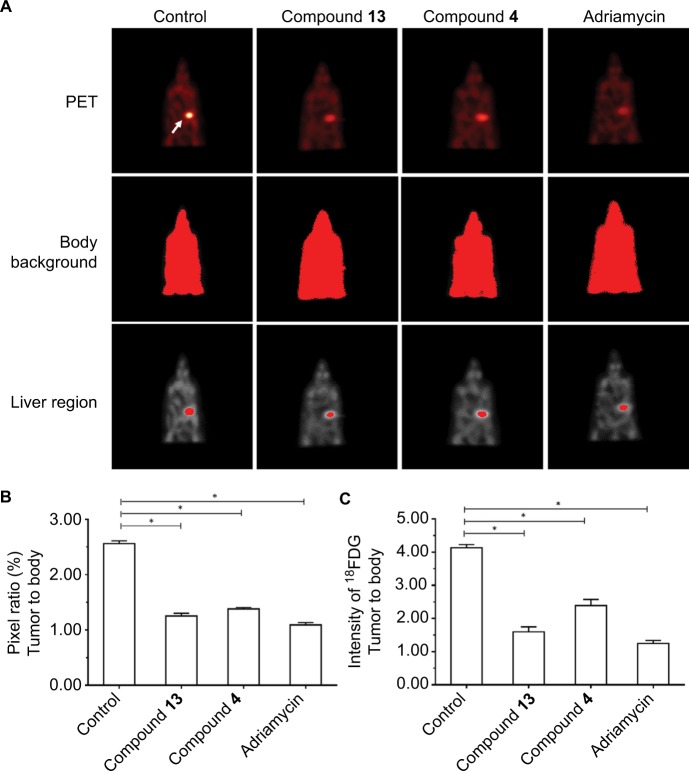
Figure 5.
Compound 4 or 13 inhibited the MicroPET signaling mice with the intrahepatic growth of MDA-MB-231 cells in the liver region.
Notes: MDA-MD-231 cells were injected into nude mice’s liver via hepatic portal vein. Next, the mice were divided into four groups: 1) solvent control group; 2) compound 4 treatment group; 3) compound 13 treatment group; 4) adriamycin treatment group. The tumor nodules formed by MHCC97-H cells in mice’s liver were examined by MicroPET scanning. Results are shown as follows: (A) MicroPET results from animals, (B) pixel ratio of tumor to body, or (C) intensity of tumor to body. Arrow indicates the tumor tissues in the liver. *P<0.05.
Abbreviation: PET, positron emission tomography.
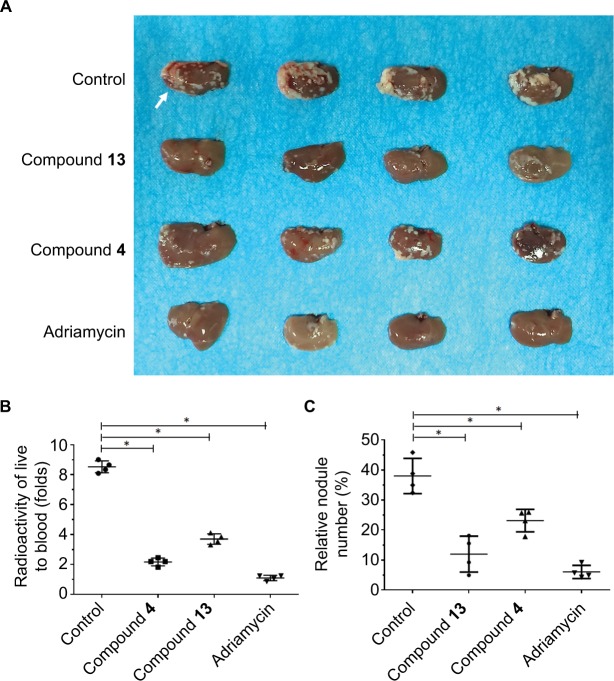
Figure 6.
Compound 4 or 13 inhibited the intrahepatic growth of MDA-MB-231 cells.
Notes: MDA-MD-231 cells were injected into nude mice’s liver via hepatic portal vein. Next, mice were divided into four groups: 1) solvent control group; 2) compound 4 treatment group; 3) compound 13 treatment group; 4) adriamycin treatment group. The tumor nodules formed by MHCC97-H cells in the mice’s liver were examined by MicroPET scanning. After MicroPET scanning, the liver organs in mice were harvested. Results are shown as follows: (A) tumor organs or (B) radioactivation of tumor tissue to blood or (C) relative nodule formed by MDA-MB-231 cells in the liver organ. White arrow in Figure 6 indicated the nodules formed by MDA-MB-231 cells in nude mice’s liver organ.*P<0.05.
Abbreviation: PET, positron emission tomography.
Discussion
As the secondary metabolic agents preserved through natural selection in the process of the biological evolution during millions of years, natural products have chemical and biological diverse features.34,35 With the characteristics of biological activity or chemical–structural diversity, natural product not only provides us with novel antitumor drugs but also inspires us in terms of chemical synthesis and structural modification, which can be identified as the precursor of semisynthesis.36,37 Several important and effective chemotherapeutic drugs are natural products or with structurally natural modifications, such as doxorubicin,37 paclitaxel,38 and etoposide.39,40 In this study, we show the potential use of isolated compounds 4 and 13 from C. yunnanensis Hsiao as treatment options for breast cancer, especially in TNBC. Structurally, cycloartane triterpenoids seem to have higher anticancer activities than chromones. These data suggest that triterpenoid saponin from Cimicifuga might be helpful for its anticancer activity. Next, we will try to find the structure–activity relation of these compounds.
Breast cancer is one of the foremost threatens of female health nowadays. There are three major pathological types of human breast cancer: 1) endocrine-related breast cancer;41–43 2) HER2-positive breast cancer;44–46 and 3) TNBC.47–49 Among these types, estrogen receptor antagonists have good antitumor effects in endocrine-related breast cancer treatment.50–52 Molecular targets agents, eg, trastuzumab, significantly improved the prognosis and quality of life of HER2-positive breast cancer patients.53–57 However, the treatment option for TNBC is still limited.58 Certain molecular targeted agents are effective in TNBC treatment; however, the cost of the molecular targeted therapy became a heavy medical burden.58–60 In this study, we show that compounds 4 and 13 could inhibit the in vitro and in vivo growth of TNBC cell line, MDA-NB-231. Although the in vitro antitumor effect of compound 13 or 4 was better than that of adriamycin, the in vivo effect of these compounds was lower than adriamycin. Based on the fact that the solubility and absorbency of adriamycin are very good, the solubility and absorbency of selective compounds would be the limit of their in vivo application. Therefore, the solubility and bioavailability may limit the in vivo application of the isolated compounds treating TNBC. Further improvement in chemical properties by chemical modifications and novel formulations of these compounds are urgently needed for the next step to improve the solubility and bioavailability of compounds.
Conclusion
In this study, our results suggested that the selective compounds isolated from C. yunnanensis Hsiao could be promising new agents for TNBC treatment.
Supplementary materials
HPLC results of compounds 1–16.
Note: All compounds were examined by HPLC to achieve quality control of the chemicals.
Table S1.
Chemical structure of compounds 1–16
| Compound 1 (actein): white powder, C37H56O11, FAB-MS m/z: 676 [M]+; 13C-NMR (100 MHz, C5D5N) δc: 32.0 (t, C-1), 29.9 (t, C-2), 88.2 (d, C-3), 41.3 (s, C-4), 47.1 (d, C-5), 20.5 (t, C-6), 25.8 (t, C-7), 45.8 (d, C-8), 20.2 (s, C-9), 26.8 (s, C-10), 36.8 (t, C-11), 77.2 (t, C-12), 47.9(s, C-13), 48.8 (s, C-14), 43.7 (t, C-15), 73.1 (d, C-16), 56.5 (d, C-17), 13.7 (q, C-18), 29.5 (t, C-19), 26.1 (d, C-20), 21.1 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 63.5 (d, C-24), 65.7 (s, C-25), 98.2 (d, C-26), 13.2 (q, C-27), 19.7 (q, C-28), 26.8 (q, C-29), 15.4 (q, C-30), 170.7 (s, COCH3), 21.8 (q, COCH3), 107.6 (d, C-1’), 75.6 (d, C-2’), 78.6 (d, C-3’), 71.3 (d, C-4’), 67.1 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.23 (1H, d, J=4.0 Hz, H-19α), 0.56 (1H, d, J=4.0 Hz, H-19β), 0.78 (3H, s, H-28), 0.84 (3H, s, H-30), 0.97 (3H, d, J=9.3 Hz, H-21), 1.28 (3H, s, H-29), 1.34 (3H, s, H-18), 1.61 (1H, s, H-27), 1.76 (1H, s, H-26), 2.14 (3H, s, COCH3), 3.70 (1H, dd, J=4.2, 8.9 Hz, H-3), 3.90 (1H, s, H-24), 4.11 (1H, t, J=6.7 Hz, H-2’), 4.33 (1H, dd, J=2.7, 7.6 Hz, H-3’), 4.78 (1H, d, J=7.1 Hz, H-1’) |
| Compound 2 (cimigenol-3-O-β-d-xylopyranoside(23R,24S)): white powder, C35H56O9, FAB-MS m/z: 620 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 32.6 (t, C-1), 30.3 (t, C-2), 88.7 (d, C-3), 41.5 (s, C-4), 47.7 (d, C-5), 21.2 (t, C-6), 26.5 (t, C-7), 48.8 (d, C-8), 20.2 (s, C-9), 26.8 (s, C-10), 26.6 (t, C-11), 34.2 (t, C-12), 42.0 (s, C-13), 47.4 (s, C-14), 80.4 (d, C-15), 112.1 (s, C-16), 59.7 (d, C-17), 19.7 (q, C-18), 31.0 (t, C-19), 24.2 (d, C-20), 19.7 (q, C-21), 38.3 (t, C-22), 72.0 (s, C-23), 90.3 (d, C-24), 71.4 (s, C-25), 25.5 (q, C-26), 25.9 (q, C-27), 12.0 (q, C-28), 27.3 (q, C-29), 15.6 (q, C-30), 107.8 (d, C-1’), 75.7 (d, C-2’), 78.8 (d, C-3’), 71.1 (d, C-4’), 67.3 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.30 (1H, d, J=3.3 Hz, H-19α), 0.54 (1H, d, J=3.3 Hz, H-19β), 0.87 (3H, d, J=6.3 Hz, H-21), 1.08 (3H, s, H-30), 1.31 (3H, s, H-29), 1.46 (3H, s, H-27), 1.48 (3H, s, H-28), 1.49 (3H, s, H-26), 1.49 (3H, s, H-18), 3.76 (1H, dd, J=4.0, 12.0 Hz, H-3), 4.07 (1H, t, J=7.8 Hz, H-2’), 4.19 (1H, t, J=8.6 Hz, H-3’), 4.37 (1H, dd, J=5.0, 11.2 Hz, H-4’), 4.74 (1H, d, J=9.0 Hz, H-23), 4.89 (1H, d, J=7.4 Hz, H-1’) |
| Compound 3 (26-deoxyacetylacteol-3-O-α-l-arabinopyranoside): white powder, C37H56O10, FAB-MS m/z: 660 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 31.8 (t, C-1), 28.8 (t, C-2), 89.1 (d, C-3), 40.7 (s, C-4), 46.9 (d, C-5), 20.3 (t, C-6), 25.6 (t, C-7), 45.8 (d, C-8), 19.9 (s, C-9), 27.5 (s, C-10), 36.5 (t, C-11), 74.6 (d, C-12), 47.5 (s, C-13), 48.5 (s, C-14), 43.7 (t, C-15), 72.3 (d, C-16), 55.9 (d, C-17), 14.3 (q, C-18), 30.0 (t, C-19), 22.9 (d, C-20), 21.0 (q, C-21), 37.5 (t, C-22), 105.4 (s, C-23), 61.8 (d, C-24), 62.5 (s, C-25), 68.0 (t, C-26),13.0 (q, C-27), 19.8 (q, C-28), 25.5 (q, C-29), 15.0 (q, C-30), 170.9 (s, COCH3), 21.7 (q, COCH3), 104.5 (d, C-1’), 73.2 (d, C-2’), 77.2 (d, C-3’), 69.7 (d, C-4’), 63.0 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.36 (1H, d, J=4.0 Hz, H-19α), 0.70 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-30), 0.88 (3H, s, H-28), 0.94 (3H, d, J=6.5 Hz, H-21), 0.97 (3H, s, H-29), 1.37 (1H, s, H-5), 1.51 (3H, s, H-18), 1.24 (3H, s, H-27), 2.02 (3H, s, COCH3), 3.20 (1H, dd, J=4.3, 11.3 Hz, H-3), 3.33 (1H, s, H-24), 3.52 (1H, d, J=6.8 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26), 4.48 (1H, d, J=5.5 Hz, H-1’), 4.86(1H, t, J=8.6 Hz, H-3’) |
| Compound 4 (23-epi-26-deoxyactein): white powder, C37H56O10, FAB-MS m/z: 659 [M-1]+, 13C-NMR (100 MHz, C5D5N) δc: 31.9 (t, C-1), 29.4 (t, C-2), 88.1 (d, C-3), 41.2 (s, C-4), 47.0 (d, C-5), 20.3 (t, C-6), 25.7 (t, C-7), 45.6 (d, C-8), 20.2 (s, C-9), 26.7 (s, C-10), 36.6 (t, C-11), 77.1 (d, C-12), 47.5 (s, C-13), 48.8 (s, C-14), 44.2 (t, C-15), 74.5 (d, C-16), 56.2 (d, C-17), 14.3 (q, C-18), 29.9 (t, C-19), 23.3 (d, C-20), 21.3 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 62.3 (d, C-24), 62.5 (s, C-25), 68.1 (t, C-26),13.5 (q, C-27), 19.7 (q, C-28), 25.6 (q, C-29), 15.3 (q, C-30), 170.6 (s, COCH3), 21.6 (q, COCH3), 107.5 (d, C-1’), 75.6 (d, C-2’), 78.6 (d, C-3’), 71.3 (d, C-4’), 67.1 (t, C-5’);1H-NMR (400 MHz, C5D5N) δH: 0.16 (1H, d, J=4.0 Hz, H-19α), 0.51 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-28), 0.98 (3H, s, H-30), 1.01 (3H, d, J=6.5 Hz, H-21), 1.23 (1H, s, H-5), 1.29 (3H, s, H-29), 1.41 (3H, s, H-18), 1.46 (3H, s, H-27), 2.13 (3H, s,COCH3), 3.44 (1H, dd, J=4.3, 11.6 Hz, H-3), 3.66 (1H, s, H-24), 3.74 (1H, d, J=9.0 Hz, H-26), 4.04 (1H, d, J=10.4 Hz, H-26), 4.06 (1H, t, J=10.4 Hz, H-2’), 4.17 (1H, t, J=8.6 Hz, H-3’), 4.82 (1H, d, J=7.5 Hz, H-1’) |
| Compound 5 (cimifugin): colorless needless, C16H18O6, FAB-MS m/z: 305 [M-1]+; 13C-NMR (100 MHz, CD3OD) δc: 168.5 (s, C-2), 109.2 (d, C-3), 179.5 (s, C-4), 166.9 (s, C-5), 118.1 (s, C-6), 160.9 (s, C-7), 94.4 (d, C-8), 156.9 (s, C-9), 112.1 (s, C-10), 92.5 (d, C-2′), 28.7 (t, C-3′), 72.1 (s, C-4′), 25.3 (q, C-5′), 25.4 (q, C-6′), 60.9 (t, CH2OH), 61.1 (q, OCH3); 1H-NMR (400 MHz, CD3OD) δH: 1.22, 1.28 (each 3H, s, H-5′, 6′), 3.30 (2H, m, H-3′), 3.90 (3H, s, OCH3), 4.41 (2H, s, CH2OH), 4.71 (1H, t, J=8.5, 8.7Hz, H-2′), 6.17 (1H, s, H-3), 6.48 (1H, s, H-8) |
| Compound 6 (cimifugin glucoside): white powder, C22H28O11, FAB-MS m/z: 467 [M-1]+; 13C-NMR (125 MHz, CD3OD) δc: 167.1 (s, C-2), 110.9 (d, C-3), 179.6 (s, C-4), 165.1 (s, C-5), 118.5 (s, C-6), 161.1 (s, C-7), 94.6 (d, C-8), 157.0 (s, C-9), 112.4 (s, C-10), 92.7 (d, C-2′), 28.8 (t, C-3′), 72.3 (s, C-4′), 25.5 (q, C-5′), 25.4 (q, C-6′), 62.8 (t, CH2OR), 61.1 (q, OCH3), Glc: 104.1 (d, C-1), 74.9 (d, C-2), 78.2 (d, C-3), 71.6 (d, C-4), 78.0 (d, C-5), 67.3 (t, C-6); 1H-NMR (500 MHz, CD3OD) δH: 1.30, 1.40 (each 3H, s, H-5′,6′), 3.27 (2H, m, H-3′), 3.91 (3H, s, OCH3), 4.41 (1H, d, J=7.7 Hz, H-1′), 4.74 (1H, t, J=9.0 Hz, H-2′), 6.34 (1H, s, H-3), 6.55 (1H, s, H-8) |
| Compound 7 (norcimifugin glucoside): yellow powder, C21H26O10, FAB-MS m/z: 454 [M]+; 13C-NMR (125 MHz, C5D5N) δc: 168.2 (s, C-2), 106.6 (d, C-3), 184.2 (s, C-4), 167.8 (s, C-5), 110.7 (s, C-6), 159.7 (s, C-7), 93.1 (d, C-8), 157.4 (s, C-9), 108.4 (s, C-10), 90.1 (d, C-2′), 27.3 (t, C-3′), 72.3 (s, C-4′), 25.3 (q, C-5′), 25.2 (q, C-6′), 67.4 (t, CH2OR), Glc:104.1 (d, C-1), 75.0 (d, C-2), 78.1 (d, C-3), 71.5 (d, C-4), 77.9 (d, C-5), 62.7 (t, C-6); 1H-NMR (500 MHz, C5D5N) δH: 1.25, 1.30 (each 3H, s, H-5′,6′), 3.27 (2H, m, H-3′), 4.44 (1H, d, J=7.7 Hz, H-1′), 4.64 (1H, t, J=9.0 Hz, H-2′), 6.34 (1H, s, H-3), 6.55 (1H, s, H-8) |
| Compound 8 (norkhellol): colorless needless, C12H8O5, FAB-MS m/z: 232 [M]+;13C-NMR (100 MHz, C5D5N) δc: 172.6 (s, C-2), 105.5 (d, C-3), 184.8 (s, C-4), 159.2 (s, C-5), 113.3 (s, C-6), 155.8 (s, C-7), 91.1 (d, C-8), 154.6 (s, C-9), 106.4 (s, C-10), 145.9 (d, C-2′), 104.6 (d, C-3′), 60.8 (t, CH2OH); 1H-NMR (400 MHz, C5D5N) δH: 4.75 (2H, d, J=4.8 Hz, H-3), 6.82 (1H, s, H-8), 7.16 (1H, d, J=3.0 Hz, H-3′), 7.57 (1H, s, H-3), 7.85 (1H,d, J=2.4 Hz, H-2′) |
| Compound 9 (25-O-acetyl-cimigenol): white powder, C32H50O6, FAB-MS m/z: 530 [M]+, 13C-NMR (125 MHz, C5D5N) δc: 32.7 (t, C-1), 31.2 (t, C-2), 78.1 (d, C-3), 41.2 (s, C-4), 47.3 (d, C-5), 21.4 (t, C-6), 26.5 (t, C-7), 48.8 (d, C-8), 19.7 (s, C-9), 27.0 (s, C-10), 26.6 (t, C-11), 34.1 (t, C-12), 41.9 (s, C-13), 47.5 (s, C-14), 80.2 (d, C-15), 112.5 (s, C-16), 59.5 (d, C-17), 19.6 (q, C-18), 31.3 (t, C-19), 24.0 (d, C-20), 19.6 (q, C-21), 38.0 (t, C-22), 71.8 (s, C-23), 86.8 (d, C-24), 83.2 (s, C-25), 24.1 (q, C-26), 23.4 (q, C-27), 11.9 (q, C-28), 26.2 (q, C-29), 14.9 (q, C-30), 170.3 (s, COCH3), 21.6 (q, COCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.33 (1H, d, J=3.8 Hz, H-19α), 0.59 (1H, d, J=3.8 Hz, H-19β), 0.85 (3H, d, J=6.4 Hz, H-21), 1.09 (3H, s, H-29), 1.09 (3H, s, H-18), 1.15 (3H, s, H-30), 1.16 (3H, s, H-28), 1.68 (3H, s, H-27), 1.69 (3H, s, H-26), 1.96 (3H, s, COCH3), 3.53 (1H, dd, J=4.5, 11.4 Hz, H-3), 4.59 (1H, d, J=9.0 Hz, H-23) |
| Compound 10 (26-deoxyacetylacteol): white powder, C32H48O, FAB-MS /z + 6 m : 528 [M], 13C-NMR (125 MHz, CD3Cl) δc: 31.9 (t, C-1), 30.2 (t, C-2),77.1 (d, C-3), 40.4 (s, C-4), 46.6 (d, C-5), 20.5 (t, C-6), 26.8 (t, C-7), 45.9 (d, C-8), 19.8 (s, C-9), 28.9 (s, C-10), 36.5 (t, C-11), 78.5 (d, C-12), 48.5 (s, C-13), 47.5 (s, C-14), 43.7 (t, C-15), 74.6 (d, C-16), 56.0 (d, C-17), 13.9 (q, C-18), 30.1 (t, C-19), 28.9 (d, C-20), 21.0 (q, C-21), 37.5 (t, C-22), 105.4 (s, C-23), 62.5 (d, C-24), 61.8 (s, C-25), 68.1 (t, C-26), 13.1 (q, C-27), 19.8 (q, C-28), 25.4 (q, C-29), 14.3 (q, C-30), 170.8 (s, COCH3), 21.7 (q, COCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.34 (1H, d, J=4.4 Hz, H-19α), 0.68 (1H, d, J=4.4 Hz, H-19β), 0.79 (3H, s, H-28), 0.87 (3H, s, H-30), 0.93 (3H, d, J=6.5 Hz, H-21), 0.96 (1H, s, H-5), 1.23 (3H, s, H-29),1.26 (3H, s, H-18), 1.50 (3H, s, H-27), 2.01 (3H, s, COCH3), 3.27 (1H, dd, J=3.4, 10.8 Hz, H-3), 3.52 (1H, d, J=10.4 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26) |
| Compound 11 (acetylacteol): white powder, C32H48O7, FAB-MS m/z: 544 [M]+; 13C-NMR (100 MHz, C5D5N) δc: 32.2 (t, C-1), 29.9 (t, C-2), 77.7 (d, C-3), 41.0 (s, C-4), 46.9 (d, C-5), 20.7 (t, C-6), 25.7 (t, C-7), 46.0 (d, C-8), 20.0 (s, C-9), 26.2 (s, C-10), 36.9 (t, C-11), 77.3 (t, C-12), 48.8(s, C-13), 47.8 (s, C-14), 43.6 (t, C-15), 73.1 (d, C-16), 56.5 (d, C-17), 13.6(q, C-18), 29.5 (t, C-19), 26.0 (d, C-20), 21.0 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 63.5 (d, C-24), 65.6 (s, C-25), 98.4 (d, C-26), 13.1 (q, C-27), 19.6 (q, C-28), 25.9 (q, C-29), 14.8 (q, C-30), 170.7 (s, COCH3), 21.7 (q, COCH3). 1H-NMR (400 MHz, C5D5N) δH: 0.28 (1H, d, J=4.0 Hz, H-19α), 0.67 (1H, d, J=4.0 Hz, H-19β), 0.84 (3H, s, H-28), 0.96 (3H, d, J=7.1 Hz, H-21), 1.00 (3H, s, H-30), 1.19 (1H, s, H-5), 1.39 (3H, s, H-18), 1.78 (3H, s, H-27), 1.81 (3H, s, H-29), 2.15(3H, s, OAc), 3.48 (1H, dd, J=4.0,11.0 Hz, H-3), 3.93(1H, s, H-24), 5.73 (1H, s, H-26) |
| Compound 12 (25-O-methyl-cimigenol): white powder, C31H50O6, FAB-MS m/z: 501 [M]+, 13C-NMR (125 MHz, C5D5N) δc: 32.7 (t, C-1), 31.3 (t, C-2), 78.1 (d, C-3), 41.1 (s, C-4), 47.6 (d, C-5), 21.4 (t, C-6), 26.6 (t, C-7), 48.7 (d, C-8), 19.6 (s, C-9), 27.1 (s, C-10), 26.6 (t, C-11), 34.2 (t, C-12), 42.0 (s, C-13), 47.4 (s, C-14), 80.1 (d, C-15), 112.0 (s, C-16), 59.5 (d, C-17), 19.6 (q, C-18), 31.1 (t, C-19), 24.1 (d, C-20), 19.5 (q, C-21), 38.3 (t, C-22), 71.7 (s, C-23), 88.3 (d, C-24), 76.2 (s, C-25), 26.5 (q, C-26), 26.5 (q, C-27), 11.8 (q, C-28), 26.2 (q, C-29), 14.8 (q, C-30), 22.2 (q, OCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.37 (1H, d, J=3.9 Hz, H-19α), 0.65 (1H, d, J=3.8 Hz, H-19β), 0.89 (3H, d, J=6.4 Hz, H-21), 1.10 (3H, s, H-30), 1.20 (3H, s, H-18), 1.21 (3H, s, H-29), 1.21 (3H, s, H-28), 1.26(3H, s, OCH3), 1.27 (3H, s, H-26), 1.28 (3H, s, H-27), 3.53 (1H, dd, J=4.5, 11.3 Hz, H-3), 4.59 (1H, d, J=9.0 Hz, H-23) |
| Compound 13 (cimigenol): white powder, C30H48O5, FAB-MS m/z: 511 [M + Na]+, 13C-NMR (125 MHz, CD3OD) δc: 31.9 (t, C-1), 30.7 (t, C-2), 78.2 (d, C-3), 40.2 (s, C-4), 46.8 (d, C-5), 20.7 (t, C-6), 25.8 (t, C-7), 48.0 (d, C-8), 19.5 (s, C-9), 26.5 (s, C-10), 25.0 (t, C-11), 33.4 (t, C-12), 41.0 (s, C-13), 46.7 (s, C-14), 79.6 (d, C-15), 111.2 (s, C-16), 58.8 (d, C-17), 19.0 (q, C-18), 29.6 (t, C-19), 23.3 (d, C-20), 18.8 (q, C-21), 37.5 (t, C-22), 71.5 (s, C-23), 89.1 (d, C-24), 71.0 (s, C-25), 30.4 (q, C-26), 25.0 (q, C-27), 10.7 (q, C-28), 24.1 (q, C-29), 13.7 (q, C-30); 1H-NMR (500 MHz, CD3OD) δH: 0.34 (1H, d, J=4.1 Hz, H-19α), 0.60 (1H, d, J=4.1 Hz, H-19β), 0.84 (3H, d, J=6.5 Hz, H-21), 1.09 (3H, s, H-30), 1.19 (3H, s, H-29), 1.19 (3H, s, H-18), 1.28(3H, s, H-28), 1.56 (1H, dd, J=3.4, 12.9 Hz, H-5), 2.11 (3H, s, H-26), 2.12 (3H, s, H-27), 3.20 (1H, dd, J=4.4, 10.7 Hz, H-3), 4.25 (1H, d, J=9.1 Hz, H-23) |
| Compound 14 (25-O-acetylcimigenol-3-O-β- -xylopyranoside): white powder, C37H58O, FAB-MS /z + 10 m : 622 [M], 13C-NMR (100 MHz, CD3OD) δc: 32.6 (t, C-1), 30.2 (t, C-2), 89.4 (d, C-3), 41.3 (s, C-4), 47.9 (d, C-5), 21.2 (t, C-6), 26.6 (t, C-7), 48.7 (d, C-8), 20.1 (s, C-9), 26.8 (s, C-10), 26.6 (t, C-11), 34.1 (t, C-12), 42.0 (s, C-13), 47.5 (s, C-14), 80.3 (d, C-15), 112.4 (s, C-16), 59.7 (d, C-17), 19.2 (q, C-18), 29.7 (t, C-19), 24.1 (d, C-20), 19.0 (q, C-21), 38.0 (t, C-22), 70.6 (s, C-23), 87.4 (d, C-24), 83.6 (s, C-25), 24.1 (q, C-26), 21.3 (q, C-27), 11.0 (q, C-28), 25.3 (q, C-29), 14.8 (q, C-30), 171.5 (s, COCH3), 21.6 (q, COCH3), 106.5 (d, C-1’), 74.7 (d, C-2’), 77.2 (d, C-3’), 72.2 (d, C-4’), 65.9 (t, C-5’); 1H-NMR (400 MHz, CD3OD) δH: 0.39 (1H, d, J=3.5 Hz, H-19α), 0.65 (1H, d, J=3.5 Hz, H-19β), 0.88 (3H, d, J=6.5 Hz, H-21), 0.90 (3H, s, H-30), 1.00 (3H, s, H-28), 1.03 (3H, s, H-18), 1.11 (3H, s, H-29), 1.40 (3H, s, H-26), 1.47 (3H, s, H-27), 1.95 (3H, s, COCH3), 3.21 (1H, t, J=6.5 Hz, H-2’), 3.30 (1H, t, J=6.9 Hz, H-3’), 3.82 (1H, dd, J=5.0, 9.7 Hz, H-3), 4.30 (1H, d, J=7.1 Hz, H-23), 4.39 (1H, d, J=5.9 Hz, H-1’), 4.47 (1H, dd, J=8.9, 18.0 Hz, H-4’) D |
| Compound 15 (24-O-acetylhydroshengmanol-3-O-β-d-xylopyranoside): white powder, C37H60O11, FAB-MS m/z: 679 [M-1]+, 13C-NMR (100 MHz, CD3OD) δc: 32.5 (t, C-1), 30.1 (t, C-2), 89.1 (d, C-3), 41.2 (s, C-4), 47.5 (d, C-5), 20.8 (t, C-6), 27.0 (t, C-7), 52.7 (d, C-8), 19.6 (s, C-9), 27.0 (s, C-10), 27.0 (t, C-11), 37.1 (t, C-12), 48.9 (s, C-13), 49.5 (s, C-14), 86.6 (d, C-15), 105.0 (s, C-16), 70.6 (d, C-17), 19.6 (q, C-18), 31.3 (t, C-19), 34.1 (d, C-20), 21.0 (q, C-21), 43.0 (t, C-22), 74.8 (s, C-23), 82.7 (d, C-24), 84.0 (s, C-25), 25.2 (q, C-26), 26.9 (q, C-27), 13.3 (q, C-28), 23.7 (q, C-29), 14.7 (q, C-30), 171.9 (s, COCH3), 20.2 (q, COCH3), 106.7 (d, C-1’), 77.3 (d, C-2’), 77.8 (d, C-3’), 72.4 (d, C-4’), 66.0 (t, C-5’); 1H-NMR (400 MHz, CD3OD) δH: 0.17 (1H, d, J=4.0 Hz,H-19α), 0.50 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-30), 0.98 (3H, s, H-28), 1.30 (3H, s, H-18), 1.34 (3H, s, H-29), 1.33 (3H, d, J=7.6 Hz, H-21), 1.68 (3H, s, H-26), 1.74 (3H, s, H-27), 2.06 (3H, s, COCH3), 4.20 (1H, s, H-15), 4.84 (1H, d, J=7.5 Hz, H-1’) |
| Compound 16 (26-deoxyactein): colorless needles, C37H56O10, FAB-MS m/z: 660 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 31.9 (t, C-1), 29.9 (t, C-2), 88.1 (d, C-3), 41.2 (s, C-4), 46.9 (d, C-5), 20.1 (t, C-6), 25.9 (t, C-7), 45.8 (d, C-8), 20.4 (s, C-9), 26.7 (s, C-10), 36.7 (t, C-11), 77.0 (d, C-12), 48.2 (s, C-13), 47.9 (s, C-14), 43.7 (t, C-15), 73.0 (d, C-16), 56.4 (d, C-17), 13.8 (q, C-18),29.5 (t, C-19), 25.7 (d, C-20), 21.0 (q, C-21), 36.7(t, C-22), 105.9 (s, C-23), 63.3 (d, C-24), 63.3 (s, C-25), 68.7 (t, C-26),13.5 (q, C-27), 19.5 (q, C-28), 25.5 (q, C-29), 15.3 (q, C-30), 170.6 (s, COCH3), 21.7 (q, COCH3), 107.6 (d, C-1’), 76.0 (d, C-2’), 78.7 (d, C-3’), 71.3 (d, C-4’), 67.2 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.17 (1H, d, J=3.8 Hz, H-19α), 0.52 (1H, d, J=3.8 Hz, H-19β), 0.97 (3H, s, H-30), 0.83 (3H, s, H-28), 1.00 (3H, d, J=6.5 Hz, H-21), 1.28 (3H, s, H-29), 1.23 (1H, s, H-5), 1.40 (3H, s, H-18), 1.24 (3H, s, H-27), 2.02 (3H, s, COCH3), 3.44 (1H, dd, J=3.4, 10.8 Hz, H-3), 3.33 (1H, s, H-24), 3.62 (1H, d, J=9.0 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26), 4.82 (1H, d, J=7.5 Hz, H-1’), 4.10 (1H, t, J=8.6 Hz, H-3’) |
Abbreviation: NMR, nuclear magnetic resonance.
Acknowledgments
This work was financially supported by the Natural Science Foundation (20702055 and 20872148) and Henan Academy of Sciences Research and Development Project (10KF13003). We thank Dr Feng at the Research Center for Clinical and Translational Medicine, 302 Hospital of Chinese PLA, Beijing, 100039, PR China for his help and advice.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Disch L, Forsch K, Siewert B, Drewe J, Fricker G. In Vitro and In Situ Characterization of Triterpene Glycosides From Cimicifuga racemosa Extract. J Pharm Sci. 2017;106(12):3642–3650. doi: 10.1016/j.xphs.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 2.Yue GG, Xie S, Lee JK, et al. New potential beneficial effects of actein, a triterpene glycoside isolated from Cimicifuga species, in breast cancer treatment. Sci Rep. 2016;6:35263. doi: 10.1038/srep35263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nian Y, Wang HY, Zhou L, Su J, Li Y, Qiu MH. Cytotoxic cycloartane triterpenes of the traditional Chinese medicine “shengma” (Cimicifuga dahurica) Planta Med. 2013;79(1):60–69. doi: 10.1055/s-0032-1328019. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman S. A review of the effectiveness of Cimicifuga racemosa (black cohosh) for the symptoms of menopause. J Womens Health. 1998;7(5):525–529. doi: 10.1089/jwh.1998.7.525. [DOI] [PubMed] [Google Scholar]
- 5.McKenna DJ, Jones K, Humphrey S, Hughes K. Black cohosh: efficacy, safety, and use in clinical and preclinical applications. Altern Ther Health Med. 2001;7(3):93–100. [PubMed] [Google Scholar]
- 6.Nian Y, Zhang YL, Chen JC, Lu L, Qiu MH, Qing C. Cytotoxic chemical constituents from the roots of Cimicifuga foetida. [corrected] J Nat Prod. 2010;73(2):93–98. doi: 10.1021/np9003855. [DOI] [PubMed] [Google Scholar]
- 7.Zhu N, Jiang Y, Wang M, Ho CT. Cycloartane triterpene saponins from the roots of Cimicifuga foetida. J Nat Prod. 2001;64(5):627–629. doi: 10.1021/np000483k. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Yan J, Nian Y, Zhou L, Zhang H, Qiu M. New triterpene diglycosides from the rhizome of Cimifuga foetida. Molecules. 2008;13(8):1712–1721. doi: 10.3390/molecules13081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Chen JC, Song HJ, Li Y, Nian Y, Qiu MH. Five new triterpene bisglycosides with acyclic side chains from the rhizomes of Cimicifuga foetida L. Chem Pharm Bull (Tokyo) 2010;58(5):729–733. doi: 10.1248/cpb.58.729. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Feng F, Zhang F, et al. LINE-1 ORF-1p functions as a novel HGF/ETS-1 signaling pathway co-activator and promotes the growth of MDA-MB-231 cell. Cell Signal. 2013;25(12):2652–2660. doi: 10.1016/j.cellsig.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F, Feng F, Yang P, et al. Four-and-a-half-LIM protein 1 down-regulates estrogen receptor α activity through repression of AKT phosphorylation in human breast cancer cell. Int J Biochem Cell Biol. 2012;44(2):320–326. doi: 10.1016/j.biocel.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Ma H, Yao Y, Wang C, et al. Transcription factor activity of estrogen receptor α activation upon nonylphenol or bisphenol A treatment enhances the in vitro proliferation, invasion, and migration of neuroblastoma cells. Onco Targets Ther. 2016;9:3451–3463. doi: 10.2147/OTT.S105745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia H, Yang Q, Wang T, et al. Rhamnetin induces sensitization of hepatocellular carcinoma cells to a small molecular kinase inhibitor or chemotherapeutic agents. Biochim Biophys Acta. 2016;1860(7):1417–1430. doi: 10.1016/j.bbagen.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 14.An L, Li DD, Chu HX, et al. Terfenadine combined with epirubicin impedes the chemo-resistant human non-small cell lung cancer both in vitro and in vivo through EMT and Notch reversal. Pharmacol Res. 2017;124:105–115. doi: 10.1016/j.phrs.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Kang L, Zhao W, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Li D, Jiang Q, et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of Sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9(7):743. doi: 10.1038/s41419-018-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Bai Z, Feng F, et al. Cross-talk between EPAS-1/HIF-2α and PXR signaling pathway regulates multi-drug resistance of stomach cancer cell. Int J Biochem Cell Biol. 2016;72:73–88. doi: 10.1016/j.biocel.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8(7):e2928. doi: 10.1038/cddis.2017.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Fan Z, Liang C, et al. A signature motif in LIM proteins mediates binding to checkpoint proteins and increases tumour radiosensitivity. Nat Commun. 2017;8:14059. doi: 10.1038/ncomms14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Fan Z, Kang L, et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123(2):630–645. doi: 10.1172/JCI64265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M, Zhao G, Zhuang X, et al. Triclosan treatment decreased the antitumor effect of sorafenib on hepatocellular carcinoma cells. Onco Targets Ther. 2018;11:2945–2954. doi: 10.2147/OTT.S165436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao Z, Li Y, Dai W, et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188–200. doi: 10.1016/j.phrs.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Xie H, Tian S, Yu H, et al. A new apatinib microcrystal formulation enhances the effect of radiofrequency ablation treatment on hepatocellular carcinoma. Onco Targets Ther. 2018;11:3257–3265. doi: 10.2147/OTT.S165000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang DQ, Yang CR. Studies on the chemical constituents from Souliea Vaginata. Nat Prod Res Dev. 1995;7(1):42–44. [Google Scholar]
- 25.Li CJ, Chen DH, Xiao PG. Chemical constituents of traditional Chinese drug “sheng-ma” (Cimicifuga dahurica) Yao Xue Xue Bao. 1993;28(10):777–781. Chinese. [PubMed] [Google Scholar]
- 26.Kadota S, Li JX, Tanaka K, Namba T. Constituents of cimicifugae rhizoma II. Isolation and structures of new cycloartenol triterpenoids and related compounds from Cimicifuga foetida L. Tetrahedron. 1995;51(4):1143–1166. [Google Scholar]
- 27.Cj L. Chemical constituents of Cimicifuga foetida L. Chinese Traditional and Herbal Drugs. 1995;26:318, 288–289. Chinese. [Google Scholar]
- 28.Sun LR. A New cycloartane Triterpenoid from the rhizome of Cimicifuga foetida Collected in Dali. Acta Botanica Yunnanica. 2005;27:331–336. [Google Scholar]
- 29.Bansi L. An antilinflammatory active Furochromme, Norcimifugin from Cimicifuga foetida: Isolation, Characterization, total Synthesis and Anti-inflammatory. Indian J Chem. 1998;37(9):881–893. [Google Scholar]
- 30.Baba K, Hata K, Kimura Y, Matsuyama Y, Kozawa M. Chemical studies of Angelica japonica A. Gray. I. On the constituents of the ethyl acetate extract of the root. Chem Pharm Bull. 1981;29(9):2565–2570. [Google Scholar]
- 31.Sakurai N, Inoue T, Nagai M. Studies on the Chinese crude drug “Shôma.” II. Triterpenes of Cimicifuga dahurica Maxim. Yakugaku Zasshi. 1972;92(6):724–728. doi: 10.1248/yakushi1947.92.6_724. Japanese. [DOI] [PubMed] [Google Scholar]
- 32.Sakurai N, Kimura O, Inoue T, Nagai M. Studies on the Chinese crude drug “Shoma.” V. Structures of 24-O-acetylhydroshengmanol xyloside and 22-hydroxycimigenol xyloside. Chem Pharm Bull. 1981;29(4):955–960. [Google Scholar]
- 33.Chen SN, Fabricant DS, Lu ZZ, Fong HH, Farnsworth NR. Cimiracemosides I-P, new 9,19-cyclolanostane triterpene glycosides from Cimicifuga racemosa. J Nat Prod. 2002;65(10):1391–1397. doi: 10.1021/np0200818. [DOI] [PubMed] [Google Scholar]
- 34.Tang JJ, He QR, Dong S, et al. Diversity Modification and Structure-Activity Relationships of Two Natural Products 1β-hydroxy Alantolactone and Ivangustin as Potent Cytotoxic Agents. Sci Rep. 2018;8(1):1722. doi: 10.1038/s41598-018-20192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao F, Yang K, Wu C, et al. New natural inhibitors of hexokinase 2 (HK2): Steroids from Ganoderma sinense. Fitoterapia. 2018;125:123–129. doi: 10.1016/j.fitote.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Fujie A. The path to producing pharmaceuticals from natural products uncovered by academia-from the perspective of a science coordinator. Biosci Biotechnol Biochem. 2017;81(1):38–42. doi: 10.1080/09168451.2016.1248367. [DOI] [PubMed] [Google Scholar]
- 37.Maffioli SI, Zhang Y, Degen D, et al. Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase. Cell. 2017;169(7):1240–1248.e23. doi: 10.1016/j.cell.2017.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargiu AV, Ramaswamy VK, Malvacio I, Malloci G, Kleinekathöfer U, Ruggerone P. Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochimica Biophys Acta Gen Subj. 2018;1862(4):836–845. doi: 10.1016/j.bbagen.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Kim SW, Hasanuzzaman Md, Cho M, et al. Role of 14-3-3 sigma in over-expression of P-gp by rifampin and paclitaxel stimulation through interaction with PXR. Cell Signal. 2017;31:124–134. doi: 10.1016/j.cellsig.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 40.McGuire J, Utset-Ward TJ, Reed DR, Lynch CC. Re-calculating! Navigating through the osteosarcoma treatment roadblock. Pharmacol Res. 2017;117:54–64. doi: 10.1016/j.phrs.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Xia E, Zhou X, Bhandari A, Zhang X, Wang O. Synaptopodin-2 plays an important role in the metastasis of breast cancer via PI3K/Akt/mTOR pathway. Cancer Manag Res. 2018;10:1575–1583. doi: 10.2147/CMAR.S162670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Zhang WW, Peng F, Sun JY, He ZY, Wu SG. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. doi: 10.2147/CMAR.S155923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh E, Wang Q, Zhang R, et al. Vertebral fractures among breast cancer survivors in China: a cross-sectional study of prevalence and health services gaps. BMC Cancer. 2018;18(1):104. doi: 10.1186/s12885-018-4014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aghazadeh S, Yazdanparast R. Activation of STAT3/HIF-1α/Hes-1 axis promotes trastuzumab resistance in HER2-overexpressing breast cancer cells via down-regulation of PTEN. Biochim Biophys Acta Gen Subj. 2017;1861(8):1970–1980. doi: 10.1016/j.bbagen.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Vega JF, Ramos J, Cruz VL, et al. Molecular and hydrodynamic properties of human epidermal growth factor receptor HER2 extracellular domain and its homodimer: Experiments and multi-scale simulations. Biochim Biophys Acta Gen Subj. 2017;1861(9):2406–2416. doi: 10.1016/j.bbagen.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Mishra R, Hanker AB, Garrett JT. Genomic alterations of ERBB receptors in cancer: clinical implications. Oncotarget. 2017;8(69):114371–114392. doi: 10.18632/oncotarget.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bimonte S, Barbieri A, Cascella M, et al. The effects of naloxone on human breast cancer progression: in vitro and in vivo studies on MDA. MB231 cells. Onco Targets Ther. 2018;11:185–191. doi: 10.2147/OTT.S145780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cevatemre B, Erkısa M, Aztopal N, et al. A promising natural product, pristimerin, results in cytotoxicity against breast cancer stem cells in vitro and xenografts in vivo through apoptosis and an incomplete autophagy in breast cancer. Pharmacol Res. 2018;129:500–514. doi: 10.1016/j.phrs.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 49.Hasanpourghadi M, Pandurangan AK, Mustafa MR. Modulation of oncogenic transcription factors by bioactive natural products in breast cancer. Pharmacol Res. 2018;128:376–388. doi: 10.1016/j.phrs.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Marsh LA, Carrera S, Shandilya J, et al. BASP1 interacts with oestrogen receptor α and modifies the tamoxifen response. Cell Death Dis. 2017;8(5):e2771. doi: 10.1038/cddis.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Zhou C, Jiang H, et al. ZEB1 induces ER-α promoter hyper-methylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8(4):e2732. doi: 10.1038/cddis.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah N, Mohammad AS, Saralkar P, et al. Investigational chemotherapy and novel pharmacokinetic mechanisms for the treatment of breast cancer brain metastases. Pharmacol Res. 2018;132:47–68. doi: 10.1016/j.phrs.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinert T, Barrios CH. Overall survival and progression-free survival with endocrine therapy for hormone receptor-positive, HER2-negative advanced breast cancer: review. Ther Adv Med Oncol. 2017;9(11):693–709. doi: 10.1177/1758834017728928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pivot X, Bondarenko I, Nowecki Z, et al. Phase III, Randomized, Double-Blind Study Comparing the Efficacy, Safety, and Immunogenicity of SB3 (Trastuzumab Biosimilar) and Reference Trastuzumab in Patients Treated With Neoadjuvant Therapy for Human Epidermal Growth Factor Receptor 2-Positive Early Breast Cancer. J Clin Oncol. 2018;36(10):968–974. doi: 10.1200/JCO.2017.74.0126. [DOI] [PubMed] [Google Scholar]
- 55.Aghazadeh S, Yazdanparast R. Activation of STAT3/HIF-1α/Hes-1 axis promotes trastuzumab resistance in HER2-overexpressing breast cancer cells via down-regulation of PTEN. Biochim Biophys Acta Gen Subj. 2017;1861(8):1970–1980. doi: 10.1016/j.bbagen.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Evans MK, Sauer SJ, Nath S, Robinson TJ, Morse MA, Devi GR. X-linked inhibitor of apoptosis protein mediates tumor cell resistance to antibody-dependent cellular cytotoxicity. Cell Death Dis. 2016;7:e2073. doi: 10.1038/cddis.2015.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Centelles MN, Wright M, Gedroyc W, Thanou M. Focused ultrasound induced hyperthermia accelerates and increases the uptake of anti-HER-2 antibodies in a xenograft model. Pharmacol Res. 2016;114:144–151. doi: 10.1016/j.phrs.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 58.Madrid-Paredes A, Cañadas-Garre M, Sánchez-Pozo A, et al. ABCB1 C3435T gene polymorphism as a potential biomarker of clinical outcomes in HER2-positive breast cancer patients. Pharmacol Res. 2016;108:111–118. doi: 10.1016/j.phrs.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Gollamudi J, Parvani JG, Schiemann WP, Vinayak S. Neoadjuvant therapy for early-stage breast cancer: the clinical utility of pertuzumab. Cancer Manag Res. 2016;8:21–31. doi: 10.2147/CMAR.S55279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong H, Yan T, Zhang W, et al. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC results of compounds 1–16.
Note: All compounds were examined by HPLC to achieve quality control of the chemicals.
Table S1.
Chemical structure of compounds 1–16
| Compound 1 (actein): white powder, C37H56O11, FAB-MS m/z: 676 [M]+; 13C-NMR (100 MHz, C5D5N) δc: 32.0 (t, C-1), 29.9 (t, C-2), 88.2 (d, C-3), 41.3 (s, C-4), 47.1 (d, C-5), 20.5 (t, C-6), 25.8 (t, C-7), 45.8 (d, C-8), 20.2 (s, C-9), 26.8 (s, C-10), 36.8 (t, C-11), 77.2 (t, C-12), 47.9(s, C-13), 48.8 (s, C-14), 43.7 (t, C-15), 73.1 (d, C-16), 56.5 (d, C-17), 13.7 (q, C-18), 29.5 (t, C-19), 26.1 (d, C-20), 21.1 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 63.5 (d, C-24), 65.7 (s, C-25), 98.2 (d, C-26), 13.2 (q, C-27), 19.7 (q, C-28), 26.8 (q, C-29), 15.4 (q, C-30), 170.7 (s, COCH3), 21.8 (q, COCH3), 107.6 (d, C-1’), 75.6 (d, C-2’), 78.6 (d, C-3’), 71.3 (d, C-4’), 67.1 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.23 (1H, d, J=4.0 Hz, H-19α), 0.56 (1H, d, J=4.0 Hz, H-19β), 0.78 (3H, s, H-28), 0.84 (3H, s, H-30), 0.97 (3H, d, J=9.3 Hz, H-21), 1.28 (3H, s, H-29), 1.34 (3H, s, H-18), 1.61 (1H, s, H-27), 1.76 (1H, s, H-26), 2.14 (3H, s, COCH3), 3.70 (1H, dd, J=4.2, 8.9 Hz, H-3), 3.90 (1H, s, H-24), 4.11 (1H, t, J=6.7 Hz, H-2’), 4.33 (1H, dd, J=2.7, 7.6 Hz, H-3’), 4.78 (1H, d, J=7.1 Hz, H-1’) |
| Compound 2 (cimigenol-3-O-β-d-xylopyranoside(23R,24S)): white powder, C35H56O9, FAB-MS m/z: 620 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 32.6 (t, C-1), 30.3 (t, C-2), 88.7 (d, C-3), 41.5 (s, C-4), 47.7 (d, C-5), 21.2 (t, C-6), 26.5 (t, C-7), 48.8 (d, C-8), 20.2 (s, C-9), 26.8 (s, C-10), 26.6 (t, C-11), 34.2 (t, C-12), 42.0 (s, C-13), 47.4 (s, C-14), 80.4 (d, C-15), 112.1 (s, C-16), 59.7 (d, C-17), 19.7 (q, C-18), 31.0 (t, C-19), 24.2 (d, C-20), 19.7 (q, C-21), 38.3 (t, C-22), 72.0 (s, C-23), 90.3 (d, C-24), 71.4 (s, C-25), 25.5 (q, C-26), 25.9 (q, C-27), 12.0 (q, C-28), 27.3 (q, C-29), 15.6 (q, C-30), 107.8 (d, C-1’), 75.7 (d, C-2’), 78.8 (d, C-3’), 71.1 (d, C-4’), 67.3 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.30 (1H, d, J=3.3 Hz, H-19α), 0.54 (1H, d, J=3.3 Hz, H-19β), 0.87 (3H, d, J=6.3 Hz, H-21), 1.08 (3H, s, H-30), 1.31 (3H, s, H-29), 1.46 (3H, s, H-27), 1.48 (3H, s, H-28), 1.49 (3H, s, H-26), 1.49 (3H, s, H-18), 3.76 (1H, dd, J=4.0, 12.0 Hz, H-3), 4.07 (1H, t, J=7.8 Hz, H-2’), 4.19 (1H, t, J=8.6 Hz, H-3’), 4.37 (1H, dd, J=5.0, 11.2 Hz, H-4’), 4.74 (1H, d, J=9.0 Hz, H-23), 4.89 (1H, d, J=7.4 Hz, H-1’) |
| Compound 3 (26-deoxyacetylacteol-3-O-α-l-arabinopyranoside): white powder, C37H56O10, FAB-MS m/z: 660 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 31.8 (t, C-1), 28.8 (t, C-2), 89.1 (d, C-3), 40.7 (s, C-4), 46.9 (d, C-5), 20.3 (t, C-6), 25.6 (t, C-7), 45.8 (d, C-8), 19.9 (s, C-9), 27.5 (s, C-10), 36.5 (t, C-11), 74.6 (d, C-12), 47.5 (s, C-13), 48.5 (s, C-14), 43.7 (t, C-15), 72.3 (d, C-16), 55.9 (d, C-17), 14.3 (q, C-18), 30.0 (t, C-19), 22.9 (d, C-20), 21.0 (q, C-21), 37.5 (t, C-22), 105.4 (s, C-23), 61.8 (d, C-24), 62.5 (s, C-25), 68.0 (t, C-26),13.0 (q, C-27), 19.8 (q, C-28), 25.5 (q, C-29), 15.0 (q, C-30), 170.9 (s, COCH3), 21.7 (q, COCH3), 104.5 (d, C-1’), 73.2 (d, C-2’), 77.2 (d, C-3’), 69.7 (d, C-4’), 63.0 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.36 (1H, d, J=4.0 Hz, H-19α), 0.70 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-30), 0.88 (3H, s, H-28), 0.94 (3H, d, J=6.5 Hz, H-21), 0.97 (3H, s, H-29), 1.37 (1H, s, H-5), 1.51 (3H, s, H-18), 1.24 (3H, s, H-27), 2.02 (3H, s, COCH3), 3.20 (1H, dd, J=4.3, 11.3 Hz, H-3), 3.33 (1H, s, H-24), 3.52 (1H, d, J=6.8 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26), 4.48 (1H, d, J=5.5 Hz, H-1’), 4.86(1H, t, J=8.6 Hz, H-3’) |
| Compound 4 (23-epi-26-deoxyactein): white powder, C37H56O10, FAB-MS m/z: 659 [M-1]+, 13C-NMR (100 MHz, C5D5N) δc: 31.9 (t, C-1), 29.4 (t, C-2), 88.1 (d, C-3), 41.2 (s, C-4), 47.0 (d, C-5), 20.3 (t, C-6), 25.7 (t, C-7), 45.6 (d, C-8), 20.2 (s, C-9), 26.7 (s, C-10), 36.6 (t, C-11), 77.1 (d, C-12), 47.5 (s, C-13), 48.8 (s, C-14), 44.2 (t, C-15), 74.5 (d, C-16), 56.2 (d, C-17), 14.3 (q, C-18), 29.9 (t, C-19), 23.3 (d, C-20), 21.3 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 62.3 (d, C-24), 62.5 (s, C-25), 68.1 (t, C-26),13.5 (q, C-27), 19.7 (q, C-28), 25.6 (q, C-29), 15.3 (q, C-30), 170.6 (s, COCH3), 21.6 (q, COCH3), 107.5 (d, C-1’), 75.6 (d, C-2’), 78.6 (d, C-3’), 71.3 (d, C-4’), 67.1 (t, C-5’);1H-NMR (400 MHz, C5D5N) δH: 0.16 (1H, d, J=4.0 Hz, H-19α), 0.51 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-28), 0.98 (3H, s, H-30), 1.01 (3H, d, J=6.5 Hz, H-21), 1.23 (1H, s, H-5), 1.29 (3H, s, H-29), 1.41 (3H, s, H-18), 1.46 (3H, s, H-27), 2.13 (3H, s,COCH3), 3.44 (1H, dd, J=4.3, 11.6 Hz, H-3), 3.66 (1H, s, H-24), 3.74 (1H, d, J=9.0 Hz, H-26), 4.04 (1H, d, J=10.4 Hz, H-26), 4.06 (1H, t, J=10.4 Hz, H-2’), 4.17 (1H, t, J=8.6 Hz, H-3’), 4.82 (1H, d, J=7.5 Hz, H-1’) |
| Compound 5 (cimifugin): colorless needless, C16H18O6, FAB-MS m/z: 305 [M-1]+; 13C-NMR (100 MHz, CD3OD) δc: 168.5 (s, C-2), 109.2 (d, C-3), 179.5 (s, C-4), 166.9 (s, C-5), 118.1 (s, C-6), 160.9 (s, C-7), 94.4 (d, C-8), 156.9 (s, C-9), 112.1 (s, C-10), 92.5 (d, C-2′), 28.7 (t, C-3′), 72.1 (s, C-4′), 25.3 (q, C-5′), 25.4 (q, C-6′), 60.9 (t, CH2OH), 61.1 (q, OCH3); 1H-NMR (400 MHz, CD3OD) δH: 1.22, 1.28 (each 3H, s, H-5′, 6′), 3.30 (2H, m, H-3′), 3.90 (3H, s, OCH3), 4.41 (2H, s, CH2OH), 4.71 (1H, t, J=8.5, 8.7Hz, H-2′), 6.17 (1H, s, H-3), 6.48 (1H, s, H-8) |
| Compound 6 (cimifugin glucoside): white powder, C22H28O11, FAB-MS m/z: 467 [M-1]+; 13C-NMR (125 MHz, CD3OD) δc: 167.1 (s, C-2), 110.9 (d, C-3), 179.6 (s, C-4), 165.1 (s, C-5), 118.5 (s, C-6), 161.1 (s, C-7), 94.6 (d, C-8), 157.0 (s, C-9), 112.4 (s, C-10), 92.7 (d, C-2′), 28.8 (t, C-3′), 72.3 (s, C-4′), 25.5 (q, C-5′), 25.4 (q, C-6′), 62.8 (t, CH2OR), 61.1 (q, OCH3), Glc: 104.1 (d, C-1), 74.9 (d, C-2), 78.2 (d, C-3), 71.6 (d, C-4), 78.0 (d, C-5), 67.3 (t, C-6); 1H-NMR (500 MHz, CD3OD) δH: 1.30, 1.40 (each 3H, s, H-5′,6′), 3.27 (2H, m, H-3′), 3.91 (3H, s, OCH3), 4.41 (1H, d, J=7.7 Hz, H-1′), 4.74 (1H, t, J=9.0 Hz, H-2′), 6.34 (1H, s, H-3), 6.55 (1H, s, H-8) |
| Compound 7 (norcimifugin glucoside): yellow powder, C21H26O10, FAB-MS m/z: 454 [M]+; 13C-NMR (125 MHz, C5D5N) δc: 168.2 (s, C-2), 106.6 (d, C-3), 184.2 (s, C-4), 167.8 (s, C-5), 110.7 (s, C-6), 159.7 (s, C-7), 93.1 (d, C-8), 157.4 (s, C-9), 108.4 (s, C-10), 90.1 (d, C-2′), 27.3 (t, C-3′), 72.3 (s, C-4′), 25.3 (q, C-5′), 25.2 (q, C-6′), 67.4 (t, CH2OR), Glc:104.1 (d, C-1), 75.0 (d, C-2), 78.1 (d, C-3), 71.5 (d, C-4), 77.9 (d, C-5), 62.7 (t, C-6); 1H-NMR (500 MHz, C5D5N) δH: 1.25, 1.30 (each 3H, s, H-5′,6′), 3.27 (2H, m, H-3′), 4.44 (1H, d, J=7.7 Hz, H-1′), 4.64 (1H, t, J=9.0 Hz, H-2′), 6.34 (1H, s, H-3), 6.55 (1H, s, H-8) |
| Compound 8 (norkhellol): colorless needless, C12H8O5, FAB-MS m/z: 232 [M]+;13C-NMR (100 MHz, C5D5N) δc: 172.6 (s, C-2), 105.5 (d, C-3), 184.8 (s, C-4), 159.2 (s, C-5), 113.3 (s, C-6), 155.8 (s, C-7), 91.1 (d, C-8), 154.6 (s, C-9), 106.4 (s, C-10), 145.9 (d, C-2′), 104.6 (d, C-3′), 60.8 (t, CH2OH); 1H-NMR (400 MHz, C5D5N) δH: 4.75 (2H, d, J=4.8 Hz, H-3), 6.82 (1H, s, H-8), 7.16 (1H, d, J=3.0 Hz, H-3′), 7.57 (1H, s, H-3), 7.85 (1H,d, J=2.4 Hz, H-2′) |
| Compound 9 (25-O-acetyl-cimigenol): white powder, C32H50O6, FAB-MS m/z: 530 [M]+, 13C-NMR (125 MHz, C5D5N) δc: 32.7 (t, C-1), 31.2 (t, C-2), 78.1 (d, C-3), 41.2 (s, C-4), 47.3 (d, C-5), 21.4 (t, C-6), 26.5 (t, C-7), 48.8 (d, C-8), 19.7 (s, C-9), 27.0 (s, C-10), 26.6 (t, C-11), 34.1 (t, C-12), 41.9 (s, C-13), 47.5 (s, C-14), 80.2 (d, C-15), 112.5 (s, C-16), 59.5 (d, C-17), 19.6 (q, C-18), 31.3 (t, C-19), 24.0 (d, C-20), 19.6 (q, C-21), 38.0 (t, C-22), 71.8 (s, C-23), 86.8 (d, C-24), 83.2 (s, C-25), 24.1 (q, C-26), 23.4 (q, C-27), 11.9 (q, C-28), 26.2 (q, C-29), 14.9 (q, C-30), 170.3 (s, COCH3), 21.6 (q, COCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.33 (1H, d, J=3.8 Hz, H-19α), 0.59 (1H, d, J=3.8 Hz, H-19β), 0.85 (3H, d, J=6.4 Hz, H-21), 1.09 (3H, s, H-29), 1.09 (3H, s, H-18), 1.15 (3H, s, H-30), 1.16 (3H, s, H-28), 1.68 (3H, s, H-27), 1.69 (3H, s, H-26), 1.96 (3H, s, COCH3), 3.53 (1H, dd, J=4.5, 11.4 Hz, H-3), 4.59 (1H, d, J=9.0 Hz, H-23) |
| Compound 10 (26-deoxyacetylacteol): white powder, C32H48O, FAB-MS /z + 6 m : 528 [M], 13C-NMR (125 MHz, CD3Cl) δc: 31.9 (t, C-1), 30.2 (t, C-2),77.1 (d, C-3), 40.4 (s, C-4), 46.6 (d, C-5), 20.5 (t, C-6), 26.8 (t, C-7), 45.9 (d, C-8), 19.8 (s, C-9), 28.9 (s, C-10), 36.5 (t, C-11), 78.5 (d, C-12), 48.5 (s, C-13), 47.5 (s, C-14), 43.7 (t, C-15), 74.6 (d, C-16), 56.0 (d, C-17), 13.9 (q, C-18), 30.1 (t, C-19), 28.9 (d, C-20), 21.0 (q, C-21), 37.5 (t, C-22), 105.4 (s, C-23), 62.5 (d, C-24), 61.8 (s, C-25), 68.1 (t, C-26), 13.1 (q, C-27), 19.8 (q, C-28), 25.4 (q, C-29), 14.3 (q, C-30), 170.8 (s, COCH3), 21.7 (q, COCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.34 (1H, d, J=4.4 Hz, H-19α), 0.68 (1H, d, J=4.4 Hz, H-19β), 0.79 (3H, s, H-28), 0.87 (3H, s, H-30), 0.93 (3H, d, J=6.5 Hz, H-21), 0.96 (1H, s, H-5), 1.23 (3H, s, H-29),1.26 (3H, s, H-18), 1.50 (3H, s, H-27), 2.01 (3H, s, COCH3), 3.27 (1H, dd, J=3.4, 10.8 Hz, H-3), 3.52 (1H, d, J=10.4 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26) |
| Compound 11 (acetylacteol): white powder, C32H48O7, FAB-MS m/z: 544 [M]+; 13C-NMR (100 MHz, C5D5N) δc: 32.2 (t, C-1), 29.9 (t, C-2), 77.7 (d, C-3), 41.0 (s, C-4), 46.9 (d, C-5), 20.7 (t, C-6), 25.7 (t, C-7), 46.0 (d, C-8), 20.0 (s, C-9), 26.2 (s, C-10), 36.9 (t, C-11), 77.3 (t, C-12), 48.8(s, C-13), 47.8 (s, C-14), 43.6 (t, C-15), 73.1 (d, C-16), 56.5 (d, C-17), 13.6(q, C-18), 29.5 (t, C-19), 26.0 (d, C-20), 21.0 (q, C-21), 37.6 (t, C-22), 105.9 (s, C-23), 63.5 (d, C-24), 65.6 (s, C-25), 98.4 (d, C-26), 13.1 (q, C-27), 19.6 (q, C-28), 25.9 (q, C-29), 14.8 (q, C-30), 170.7 (s, COCH3), 21.7 (q, COCH3). 1H-NMR (400 MHz, C5D5N) δH: 0.28 (1H, d, J=4.0 Hz, H-19α), 0.67 (1H, d, J=4.0 Hz, H-19β), 0.84 (3H, s, H-28), 0.96 (3H, d, J=7.1 Hz, H-21), 1.00 (3H, s, H-30), 1.19 (1H, s, H-5), 1.39 (3H, s, H-18), 1.78 (3H, s, H-27), 1.81 (3H, s, H-29), 2.15(3H, s, OAc), 3.48 (1H, dd, J=4.0,11.0 Hz, H-3), 3.93(1H, s, H-24), 5.73 (1H, s, H-26) |
| Compound 12 (25-O-methyl-cimigenol): white powder, C31H50O6, FAB-MS m/z: 501 [M]+, 13C-NMR (125 MHz, C5D5N) δc: 32.7 (t, C-1), 31.3 (t, C-2), 78.1 (d, C-3), 41.1 (s, C-4), 47.6 (d, C-5), 21.4 (t, C-6), 26.6 (t, C-7), 48.7 (d, C-8), 19.6 (s, C-9), 27.1 (s, C-10), 26.6 (t, C-11), 34.2 (t, C-12), 42.0 (s, C-13), 47.4 (s, C-14), 80.1 (d, C-15), 112.0 (s, C-16), 59.5 (d, C-17), 19.6 (q, C-18), 31.1 (t, C-19), 24.1 (d, C-20), 19.5 (q, C-21), 38.3 (t, C-22), 71.7 (s, C-23), 88.3 (d, C-24), 76.2 (s, C-25), 26.5 (q, C-26), 26.5 (q, C-27), 11.8 (q, C-28), 26.2 (q, C-29), 14.8 (q, C-30), 22.2 (q, OCH3); 1H-NMR (500 MHz, C5D5N) δH: 0.37 (1H, d, J=3.9 Hz, H-19α), 0.65 (1H, d, J=3.8 Hz, H-19β), 0.89 (3H, d, J=6.4 Hz, H-21), 1.10 (3H, s, H-30), 1.20 (3H, s, H-18), 1.21 (3H, s, H-29), 1.21 (3H, s, H-28), 1.26(3H, s, OCH3), 1.27 (3H, s, H-26), 1.28 (3H, s, H-27), 3.53 (1H, dd, J=4.5, 11.3 Hz, H-3), 4.59 (1H, d, J=9.0 Hz, H-23) |
| Compound 13 (cimigenol): white powder, C30H48O5, FAB-MS m/z: 511 [M + Na]+, 13C-NMR (125 MHz, CD3OD) δc: 31.9 (t, C-1), 30.7 (t, C-2), 78.2 (d, C-3), 40.2 (s, C-4), 46.8 (d, C-5), 20.7 (t, C-6), 25.8 (t, C-7), 48.0 (d, C-8), 19.5 (s, C-9), 26.5 (s, C-10), 25.0 (t, C-11), 33.4 (t, C-12), 41.0 (s, C-13), 46.7 (s, C-14), 79.6 (d, C-15), 111.2 (s, C-16), 58.8 (d, C-17), 19.0 (q, C-18), 29.6 (t, C-19), 23.3 (d, C-20), 18.8 (q, C-21), 37.5 (t, C-22), 71.5 (s, C-23), 89.1 (d, C-24), 71.0 (s, C-25), 30.4 (q, C-26), 25.0 (q, C-27), 10.7 (q, C-28), 24.1 (q, C-29), 13.7 (q, C-30); 1H-NMR (500 MHz, CD3OD) δH: 0.34 (1H, d, J=4.1 Hz, H-19α), 0.60 (1H, d, J=4.1 Hz, H-19β), 0.84 (3H, d, J=6.5 Hz, H-21), 1.09 (3H, s, H-30), 1.19 (3H, s, H-29), 1.19 (3H, s, H-18), 1.28(3H, s, H-28), 1.56 (1H, dd, J=3.4, 12.9 Hz, H-5), 2.11 (3H, s, H-26), 2.12 (3H, s, H-27), 3.20 (1H, dd, J=4.4, 10.7 Hz, H-3), 4.25 (1H, d, J=9.1 Hz, H-23) |
| Compound 14 (25-O-acetylcimigenol-3-O-β- -xylopyranoside): white powder, C37H58O, FAB-MS /z + 10 m : 622 [M], 13C-NMR (100 MHz, CD3OD) δc: 32.6 (t, C-1), 30.2 (t, C-2), 89.4 (d, C-3), 41.3 (s, C-4), 47.9 (d, C-5), 21.2 (t, C-6), 26.6 (t, C-7), 48.7 (d, C-8), 20.1 (s, C-9), 26.8 (s, C-10), 26.6 (t, C-11), 34.1 (t, C-12), 42.0 (s, C-13), 47.5 (s, C-14), 80.3 (d, C-15), 112.4 (s, C-16), 59.7 (d, C-17), 19.2 (q, C-18), 29.7 (t, C-19), 24.1 (d, C-20), 19.0 (q, C-21), 38.0 (t, C-22), 70.6 (s, C-23), 87.4 (d, C-24), 83.6 (s, C-25), 24.1 (q, C-26), 21.3 (q, C-27), 11.0 (q, C-28), 25.3 (q, C-29), 14.8 (q, C-30), 171.5 (s, COCH3), 21.6 (q, COCH3), 106.5 (d, C-1’), 74.7 (d, C-2’), 77.2 (d, C-3’), 72.2 (d, C-4’), 65.9 (t, C-5’); 1H-NMR (400 MHz, CD3OD) δH: 0.39 (1H, d, J=3.5 Hz, H-19α), 0.65 (1H, d, J=3.5 Hz, H-19β), 0.88 (3H, d, J=6.5 Hz, H-21), 0.90 (3H, s, H-30), 1.00 (3H, s, H-28), 1.03 (3H, s, H-18), 1.11 (3H, s, H-29), 1.40 (3H, s, H-26), 1.47 (3H, s, H-27), 1.95 (3H, s, COCH3), 3.21 (1H, t, J=6.5 Hz, H-2’), 3.30 (1H, t, J=6.9 Hz, H-3’), 3.82 (1H, dd, J=5.0, 9.7 Hz, H-3), 4.30 (1H, d, J=7.1 Hz, H-23), 4.39 (1H, d, J=5.9 Hz, H-1’), 4.47 (1H, dd, J=8.9, 18.0 Hz, H-4’) D |
| Compound 15 (24-O-acetylhydroshengmanol-3-O-β-d-xylopyranoside): white powder, C37H60O11, FAB-MS m/z: 679 [M-1]+, 13C-NMR (100 MHz, CD3OD) δc: 32.5 (t, C-1), 30.1 (t, C-2), 89.1 (d, C-3), 41.2 (s, C-4), 47.5 (d, C-5), 20.8 (t, C-6), 27.0 (t, C-7), 52.7 (d, C-8), 19.6 (s, C-9), 27.0 (s, C-10), 27.0 (t, C-11), 37.1 (t, C-12), 48.9 (s, C-13), 49.5 (s, C-14), 86.6 (d, C-15), 105.0 (s, C-16), 70.6 (d, C-17), 19.6 (q, C-18), 31.3 (t, C-19), 34.1 (d, C-20), 21.0 (q, C-21), 43.0 (t, C-22), 74.8 (s, C-23), 82.7 (d, C-24), 84.0 (s, C-25), 25.2 (q, C-26), 26.9 (q, C-27), 13.3 (q, C-28), 23.7 (q, C-29), 14.7 (q, C-30), 171.9 (s, COCH3), 20.2 (q, COCH3), 106.7 (d, C-1’), 77.3 (d, C-2’), 77.8 (d, C-3’), 72.4 (d, C-4’), 66.0 (t, C-5’); 1H-NMR (400 MHz, CD3OD) δH: 0.17 (1H, d, J=4.0 Hz,H-19α), 0.50 (1H, d, J=4.0 Hz, H-19β), 0.83 (3H, s, H-30), 0.98 (3H, s, H-28), 1.30 (3H, s, H-18), 1.34 (3H, s, H-29), 1.33 (3H, d, J=7.6 Hz, H-21), 1.68 (3H, s, H-26), 1.74 (3H, s, H-27), 2.06 (3H, s, COCH3), 4.20 (1H, s, H-15), 4.84 (1H, d, J=7.5 Hz, H-1’) |
| Compound 16 (26-deoxyactein): colorless needles, C37H56O10, FAB-MS m/z: 660 [M]+, 13C-NMR (100 MHz, C5D5N) δc: 31.9 (t, C-1), 29.9 (t, C-2), 88.1 (d, C-3), 41.2 (s, C-4), 46.9 (d, C-5), 20.1 (t, C-6), 25.9 (t, C-7), 45.8 (d, C-8), 20.4 (s, C-9), 26.7 (s, C-10), 36.7 (t, C-11), 77.0 (d, C-12), 48.2 (s, C-13), 47.9 (s, C-14), 43.7 (t, C-15), 73.0 (d, C-16), 56.4 (d, C-17), 13.8 (q, C-18),29.5 (t, C-19), 25.7 (d, C-20), 21.0 (q, C-21), 36.7(t, C-22), 105.9 (s, C-23), 63.3 (d, C-24), 63.3 (s, C-25), 68.7 (t, C-26),13.5 (q, C-27), 19.5 (q, C-28), 25.5 (q, C-29), 15.3 (q, C-30), 170.6 (s, COCH3), 21.7 (q, COCH3), 107.6 (d, C-1’), 76.0 (d, C-2’), 78.7 (d, C-3’), 71.3 (d, C-4’), 67.2 (t, C-5’); 1H-NMR (400 MHz, C5D5N) δH: 0.17 (1H, d, J=3.8 Hz, H-19α), 0.52 (1H, d, J=3.8 Hz, H-19β), 0.97 (3H, s, H-30), 0.83 (3H, s, H-28), 1.00 (3H, d, J=6.5 Hz, H-21), 1.28 (3H, s, H-29), 1.23 (1H, s, H-5), 1.40 (3H, s, H-18), 1.24 (3H, s, H-27), 2.02 (3H, s, COCH3), 3.44 (1H, dd, J=3.4, 10.8 Hz, H-3), 3.33 (1H, s, H-24), 3.62 (1H, d, J=9.0 Hz, H-26), 4.02 (1H, d, J=10.4 Hz, H-26), 4.82 (1H, d, J=7.5 Hz, H-1’), 4.10 (1H, t, J=8.6 Hz, H-3’) |
Abbreviation: NMR, nuclear magnetic resonance.