Abstract
In the recent issue of Nature Biotechnology, Frock et al. (2015) developed an elegant technique to capture translocation partners that can be utilized to determine off-target regions of genome-editing endonucleases as well as endogenous mutators at nucleotide resolution.
The two most prevalent genome-editing technologies—TALEN (transcription activator-like effector nucleases) and CRISPR (custered, regularly interspaced, short palindromic repeats)—garnered attention in recent years due to their seemingly endless potential ranging from excising HIV out of the human genome, to curing leukemia and modifying the DNA of human embryos to eradicate genetic diseases before a baby is born. Both TALEN and CRISPR use endonucleases that can target almost any region in the genome, generating a double-strand break (DSB) that will be subsequently processed by DNA repair systems. Unless the DNA ends are “sutured” in an error-prone manner so as to evade recognition by sequence-specific endonucleases, error-free products will undergo reiterative cleavage-repair cycles. However, despite their therapeutic potential, one caveat to genome editing is the presence of off-target cleavage sites. Up until this point, the precise frequency and location of such collateral damage remained unclear. The report by Frock et al. provides a comprehensive map of unintended engineered endonuclease cleavage sites with unprecedented sensitivity, as well as a method to uncover genome instability originating from endogenous DSBs (Frock et al., 2015).
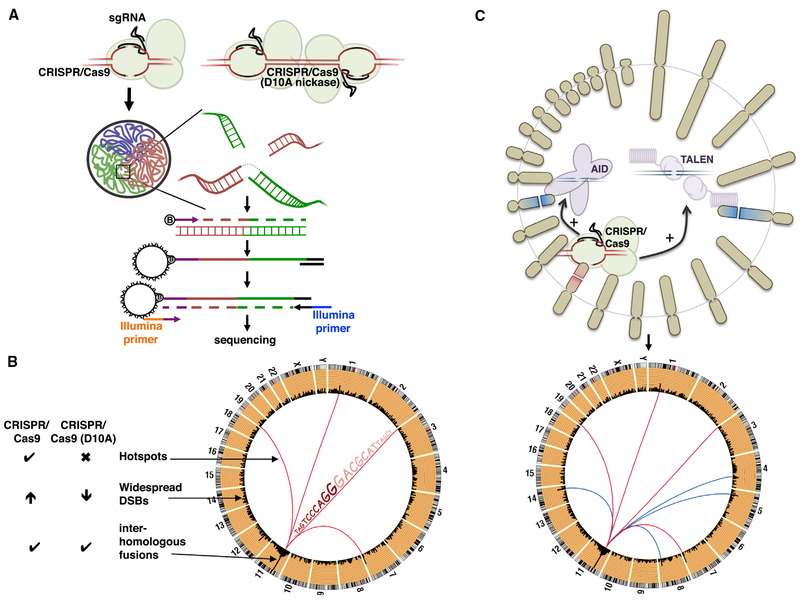
Alt and colleagues had already developed a technique called high-throughput, genome-wide, translocation sequencing (HTGTS), which relies on the generation of bait DSBs at a defined location, and the subsequent capture of “prey” DSBs that together form a hybrid DNA fragment (translocation) (Chiarle et al., 2011). Frock et al. (2015) improved their original published approach by incorporating linear-amplification-PCR (LAM-PCR) prior to the enrichment step and the subsequent next-generation sequencing of the hybrid fragment (Figure 1A). Such mapping of prey sequences in the fusion products allows for determination of uncharacterized DSBs at base-pair resolution and calculation of the frequency of these breaks (Figure 1B) (Frock et al., 2015).
Figure 1. Translocations Discovered by LAM-PCR HTGTS.
(A) Top: CRISPR/Cas9 wild-type (left) and D10A nickase (right) generates DSBs and DNA nicks, respectively, leading to on-target and potentially off-target DSBs; aberrant fusions of the DNA ends can cause chromosomal translocations. Bottom: scheme of LAM-PCR HTGTS technology to detect novel translocation partners. Cartoon illustrates the arrangement of chromosomes into three distinct territories within the nucleus. The highlighted square indicates the enlarged view of the on-target (red) and off-target (green) DSBs that are in proximity to each other, thus promoting translocations. Following DNA extraction and shearing, a biotinylated primer specific to the bait DSB is extended, amplifying the hybrid fragment on-target-off-target (bait-prey). The product is subsequently purified with streptavidin and ligated to an adaptor, allowing PCR amplification followed by the identification of the off-targets by next-generation sequencing.
(B) Circos plot illustrating the different type of translocations discovered by the HTGTS method, including off-targets DSB “hotspots,” widespread DSBs reflecting non-specific and endogenous DSBs, and inter-homologous fusions involving DSBs generated on both homologous chromosomes. CRISPR/Cas9 (D10A nickase) suppresses translocation hotspots and reduces widespread DSBs in contrast to wild-type CAS9, although inter-homologous fusions at the bait chromosome still persist.
(C) Combination of a known DNA break instigator (CRISPR/Cas9; leading to translocation pattern depicted by red lines, bottom) together with an uncharacterized genomic mutator (e.g., TALEN or AID, top) reveal novel translocation partner induced by the latter (blue lines, bottom).
As a proof of principle, Frock et al. (2015) utilized the CRISPR/Cas9 system targeting a region of the human RAG1 locus and two previously described TALENs targeting ATM and C-MYC loci. The detection of off-targets by LAMPCR HTGTS relies on the presence of highly enriched clusters of genomic bait (on-target)-prey (off-target) junctions, which in turn are designated as translocation “hotspots.” Consistent with the idea that the fusion of two DSBs requires their physical juxtaposition (Zhang et al., 2012), HTGTS not only detected off-target events, but also reveals a dramatic enrichment of translocation signals within the vicinity of the bait region, indicative of preferential DNA end joining within spatially proximal regions. As a striking example, the authors demonstrated that ionizing radiation treatment leads to extensive translocations specifically in cis along the chromosome where the bait is located—presumably due to random DNA breaks generated within proximity of the bait. Even though homologous chromosomes do not necessarily occupy identical nuclear territories, the authors uncovered a characteristic signature of chromosomal breaks generated by genome-editing nucleases: inversional translocations generating dicentric chromosomes instigated by the generations of two DSBs at identical sites in each homologous chromosome.
In addition to high-frequency trans-location hotspots, LAM-PCR HTGTS also detected rare DSBs that tended to be spread throughout the genome(Figure 1B). The origin of this damage remains unclear, reflecting either non-specific damage associated with the nuclease or spontaneous DSBs arising from damage-labile regions such as fragile sites (Barlow et al., 2013). In an effort to reduce the off-target effect of the CRISPR/Cas9 system, Frock et al. (2015) tested the paired nickase system based on the Cas9 D10A mutant that generates staggered DSBs (Figures 1A and B). Such an approach suppressed off-target translocation hot-spots, but nevertheless the inter-homologous fusions—involving DSBs generated on both homologous chromosomes surrounding the bait site—still persisted.
As this technique efficiently captures DNA ends, pairing the CRISPR/Cas9 system with uncharacterized DSB instigators could thereby reveal novel off-targets mediated by the latter. Indeed, the group validated this approach and revealed regions that are commonly mis-targeted by TALEN and I-SceI meganuclease (Frock et al., 2015). HTGTS and other related techniques have also been used to map off-target sites of physiological mutators such as RAG and AID recombinases that primarily target antigen receptor loci but occasionally cause translocations with oncogenes (Figure 1C) (Chiarle et al., 2011; Klein et al., 2011; Zhang et al., 2012).
HTGTS technology utilizes a bait region to instigate a DSB by well-characterized exogenous DNA cutter leading to a clean and efficient DNA break at a single, predefined position. It remains to be seen if the bait end could be derived from DNA ends instigated by endogenous mutators whereby the cut sites may be relatively imprecise and cover broader regions. It would also be interesting to apply the method to map regions targeted by chemotherapeutic agents. For example, topoisomerase poisons are potent and widely used anticancer drugs that stabilize the normally transient topoisomerase-induced DSBs. However, these drugs are also associated with therapy-related secondary acute leukemia often bearing 11q23 translocations involving the MLL gene. Since chromo-some translocations represent a critical early event in the development of these leukemias, LAM-PCR HTGTS could be used to map hotspots for topoisomerase-associated breaks, which eventually might suggest ways to protect the MLL gene against fragility, and reduce the occurrence of this secondary cancer. On a more basic level, nucleotide level detection of breakpoint junctions can allow for in depth analysis of mechanisms of end-joining repair in human cells (Ghezraoui et al., 2014).
Translocation formation is dependent on the frequency of DSBs at each partner chromosome and the frequency at which the loci are in contact. Since the spectrum of translocations is influenced by the pre-existing spatial juxtaposition of translocation partners, the number of the off-target DSBs could be under-represented by LAM-PCR HTGTS. For example, the assay does not detect uncaptured DSBs (e.g., low-frequency non-proximal free ends) or other DNA lesions such as nicks that could eventually lead to DSBs. Because of the influences of 3D conformation on translocations, sensitive methods to directly determine unresolved DNA damage would be useful. Indeed, two groups have developed methods to map integration of foreign DNA-bait sequences into off-target DSBs (Tsai et al., 2015; Wang et al., 2015), and others have directly detected the DNA ends in situ (Crosetto et al., 2013; Kato et al., 2012).
In summary, sophisticated genome-editing nucleases do not always work with laser-sharp precision. One important question that remains is how to minimize the deleterious effect of the technologies. While it is clear that the CRISPR-Cas9 nickase is an improvement, inter-homologous fusions still pose a threat. Developing an approach to preferentially target one homologous chromosome at a time—mimicking allelic exclusion during programmed DNA damage and repair in lymphocyte development—might provide an answer.
REFERENCES
- Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. (2013). Cell 152, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. (2011). Cell 147, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. (2013). Nat. Methods 10, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, and Alt FW (2015). Nat. Biotechnol 33, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. (2014). Mol. Cell 55, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato L, Begum NA, Burroughs AM, Doi T, Kawai J, Daub CO, Kawaguchi T, Matsuda F, Hayashizaki Y, and Honjo T (2012). Proc. Natl. Acad. Sci. USA 109, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. (2011). Cell 147, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). Nat. Biotechnol 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin RJ, and Yee JK (2015). Nat. Biotechnol 33, 175–178. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, and Dekker J (2012). Cell 148, 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]