Abstract
Common variable immunodeficiency (CVID), the most common symptomatic primary antibody deficiency, is accompanied in some patients by a duodenal inflammation and malabsorption syndrome known as CVID enteropathy (E-CVID). The goal of this study was to investigate the immunological abnormalities in CVID patients that lead to enteropathy as well as the contribution of intestinal microbiota to this process. We found that, in contrast to noE-CVID patients (without enteropathy), E-CVID patients have exceedingly low levels of IgA in duodenal tissues. In addition, using transkingdom network analysis of the duodenal microbiome, we identified Acinetobacter baumannii as a candidate pathobiont in E-CVID. Finally, we found that E-CVID patients exhibit a pronounced activation of immune genes and down-regulation of epithelial lipid metabolism genes. We conclude that in the virtual absence of mucosal IgA, pathobionts such as A. baumannii, may induce inflammation that re-directs intestinal molecular pathways from lipid metabolism to immune processes responsible for enteropathy.
Introduction
Common variable immunodeficiency (CVID), the most frequently occurring form of symptomatic immunodeficiency, is a heterogeneous disease that is most likely due to many different genetic defects [1–4]. With the advent of early gamma globulin replacement therapy, CVID patients are relatively free from life-threatening infections and have a more prolonged survival [1, 3, 5]. The latter, however, has revealed that CVID can be associated with other problems not due to infectious agents. One of these, CVID enteropathy, is characterized by villous atrophy, malabsorption and diarrhea and occurs in up to 15% of CVID patients in symptomatic form and perhaps in a larger percentage in enteropathy since this manifestation of the disease has not been coupled to a particular asymptomatic form [6],[7]. No genetic differences accounting for the hypogammaglobulinemia in the subset of patients that develop enteropathy and those that do not have been identified in prior studies or in the present study (see below); thus, a genetic basis for the occurrence of enteropathy is not yet known.
The villous atrophy accompanying CVID enteropathy is histologically similar to that in celiac disease and, as such, is characterized by a lamina propria mononuclear cell infiltration and increased numbers of intra-epithelial lymphocytes; however, the prominent plasma cell infiltration seen in celiac disease is not present [7]. Studies conducted by Malamut et al. [7], showed that intraepithelial lymphocytes (IELs) in celiac disease differed from those in CVID enteropathy with regard to some natural killer cell markers and the TCRγ/δ phenotype characteristic of that in celiac disease was generally not present in CVID. These differences between CVID and celiac disease were corroborated by recent work showing that the two diseases are also quite distinct in gene profiling studies [8]. Finally, studies of the immunologic features of CVID enteropathy conducted by Mannon et al.[9] revealed that lamina propria T cells extracted from patients with this disease produce increased amounts of IL-12 and IFN-γ but little if any IL-17. These findings correlate with recent studies showing that increased numbers of innate (CD3 -) ILC3 cells producing IFN-γ are present in mucosal tissues of patients with CVID but not patients with IBD; thus, these cells may also contribute to the Th1 cytokine bias associated with enteropathy [10].
In previous work we have shown using systems biology methodology that total immunoglobulin and IgA deficiency in mice are associated with a remarkable up-regulation of gut epithelial interferon-inducible genes accompanied by a down-regulation of GATA4-related metabolic genes [11]. Since these changes were not seen in germ-free mice it was clear that they were dependent on one or more organisms in the gut microbiota. These studies therefore suggested that CVID enteropathy may be characterized by the presence of an immune defect that leads to colonization with an organism that is responsible for these multiple gene expression changes. To address this possibility we conducted immunologic studies of CVID patients with and without enteropathy as well as control individuals and found that patients with enteropathy but not those without enteropathy exhibited the interferon gene/metabolic gene imbalance mentioned above. In addition, patients with CVID enteropathy could be distinguished from those without enteropathy by the presence of vanishingly low mucosal IgA levels, irrespective of accompanying serum IgA levels. We then used these immunologic data and accompanying microbiome data to conduct a systems biology and transkingdom network analysis [12] for the identification of a possible candidate bacterium capable of causing villous atrophy and malabsorption in the absence of mucosal IgA secretion.
Results
General clinical immunological and genetic characteristics of the patients under study
The patient/control groups studied here consisted of biopsies of 7 patients with E-CVID, 8 patients with noE-CVID and 7 control individuals; the relevant patient clinical history, histopathologic information accompanying these patient/control groups are provided in the Methods section. All patients underwent peripheral blood B and T cell phenotyping. In evaluation of their B cells, E-CVID patients exhibited a decrease in total B cells as signified by expression of CD20: mean absolute number CD20+ B cells of 47 cells/uL in E-CVID patients as compared with 158 cells/uL in noE-CVID patients with normal control patients 111 cells/uL. In addition, E-CVID patients exhibited a decreased number of memory B cells as signified by the number of CD20/CD27+ cells: mean absolute number of CD20/CD27+ cells of 11.8 cells/uL in E-CVID patients as compared with 17.2 cells/uL in noE-CVID patients with normal control patients 28 cells/uL. This apparent difference in memory B cells in E-CVID patient group as compared to noECVID patient group, however, did not reach statistical significance. In evaluation of their T cells, no significant differences was observed among E-CVID patients, noE-CVID patients and normal controls with respect to absolute number of CD3+ T cells: mean absolute cell number for E-CVID patients: 1150 cells/uL, noE-CVID patients: 1332 cells/uL and normal control patients: 1268 cells/uL. However, in analysis of T cell subsets, E-CVID patients exhibited a decreased CD4+/CD8+ ratio as compared to no-ECVID patients. This was reflected in the mean absolute cell number of CD4+ T cells of 463 cells/uL in E-CVID patients as compared to 762 cells/uL in noE-CVID patients and CD8+ T cells of 899 cells/uL in E-CVID patients as compared to 529 cells/uL in noE-CVID patients. This apparent increase of CD8+ T cells in the E-CVID group as compared to the noE-CVID group, however, did not reach statistical significance.
As mentioned in the Introduction, no differences in the presence of genes previously associated with CVID with and without enteropathy has yet been reported. To examine this possibility in the present group of patients the latter were screened at the NIH with targeted Ion Torrent AmpliSeq methodology (see Methods section) which included exome sequencing of genes known to be associated with CVID. These included LRBA, PIK3, CTLA-4, NFKB1, NFKB2, PIK3R1, IKZF1 (IKAROS), ICOS, TNFSF12 (TWEAK), CD19, TNFRSF7 (CD27), IL-21R, TNFRSF13B (TACI), TNFRSF13C (BAFF-R), and TNFSF13 (APRIL). Neither CVID patients with enteropathy (E-CVID patients) nor CVID patients without enteropathy (noE-CVID patients) exhibited deleterious mutations in these specific genes.
Gene expression signature of CVID enteropathy and its accompanying inflammatory and lipid metabolism defects
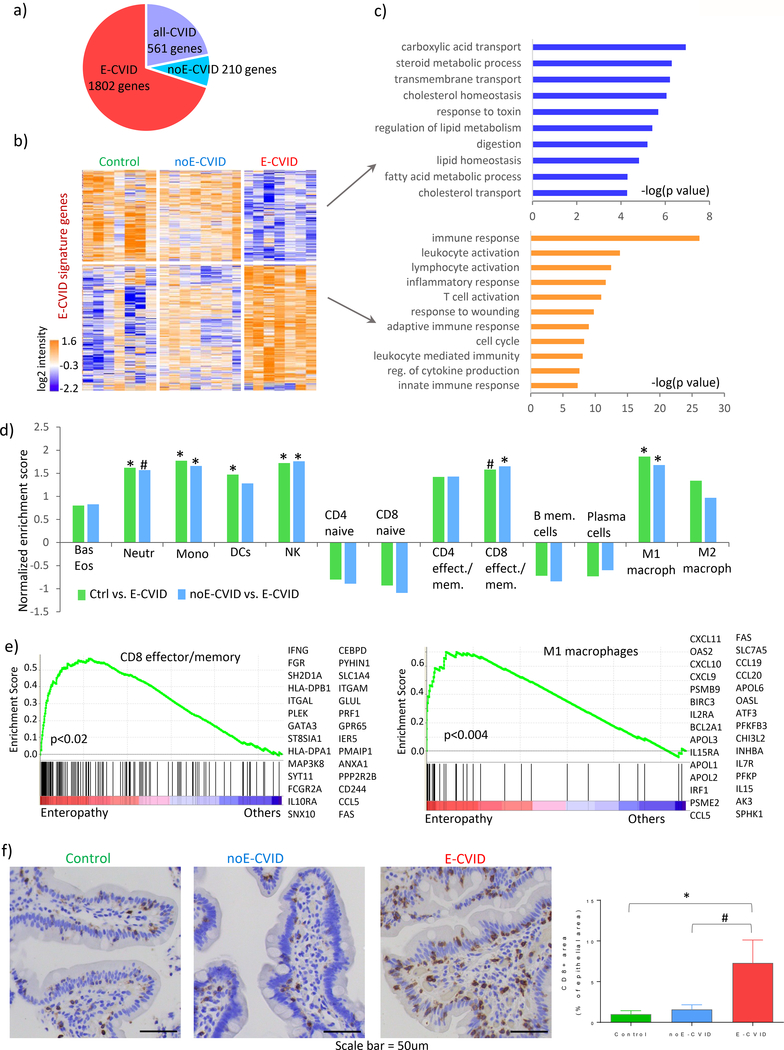
We started by defining the global gene expression profiles of duodenal biopsy tissues obtained from CVID patients with and without enteropathy as well as in control individuals using RNA sequencing (see Methods). This analysis revealed 2573 genes that were differentially expressed among the three groups (Figure 1a and Supplementary Table S1). Analyzing these gene expression profiles using pairwise comparisons between genes, we then defined gene signatures characteristic of CVID with enteropathy (E-CVID; 1802 genes), CVID without enteropathy (noE-CVID; 210 genes) and a general CVID signature common to both patient groups (all-CVID, 561 genes) (Figures 1a-b, Supplementary Figure S1). Next, focusing on the gene expression signature in the various groups (Figure 1b), we identified the down-regulated and up-regulated genes corresponding to various Gene Ontology categories as defined in Figure 1c. We found that in the E-CVID group but not in the noE-CVID group, down-regulated genes consisted of those relating to epithelial cell metabolic functions such as lipid metabolism and absorption whereas up-regulated genes related to immune responses and lymphocyte activation (Figure 1c, upper and lower panels respectively and Supplementary Table S2). Thus, these data expanded our previously reported results about differential regulation of immunity and lipid metabolism in humans with immunodeficiency in that they established for the first time that this phenomenon is a specific feature of CVID enteropathy and is not present in other CVID patients or controls. In addition, the difference in the gene signature in the E-CVID and noE-CVID patients was not influenced by the presence of norovirus infection as no statistically significant differences in gene expression were found in norovirus-positive patient samples relative to the expression in other E-CVID patients.
Figure 1.
Gene expression and cellular characterization of CVID enteropathy. (a) Number of genes assigned to each of the three signatures (E-CVID, noE-CVID and all-CVID) defined as described in Methods and Supplementary Figure 1. (b) heatmap of genes in E-CVID profile; columns, samples (Control n=7, noE-CVID n=8, E-CVID n=7), rows, genes; log2 normalized intensities of gene expression across all samples are represented. (c) Enriched Gene Ontology terms (Biological Process) in E-CVID profile (p-values obtained using DAVID bioinformatics resources). (d) Gene set enrichment analysis (GSEA) for hematopoietic cell types in E-CVID vs. other groups; *p<0.05; # p<0.1 by permutation test implemented in GSEA. (e) GSEA leading edge scores and genes for CD8 effector/memory cells and M1 macrophages. Leading edge plots for other enriched populations are in Supplementary Figure S2. (f) Representative images and quantitative analysis of CD8 staining in biopsies from Control (n=4), noE-CVID (n=6), E-CVID (n=7), Mann-Whitney one-sided test *p<0.05; #p<0.1, mean, SEM shown. Abbreviations: E-CVID, enteropathy associated with common variable immunodeficiency; noE-CVID, CVID without enteropathy; Ctrl, Control; Bas, basophils, Eos, eosinophils; Neutr, neutrophils; Mono, monocytes; DCs, dendritic cells; NK, natural killer cells.
We next used a gene expression approach to estimate which cells were present in the small intestinal infiltrate in patients with E-CVID as compared to patients with noE-CVID or controls. This consisted of a gene set enrichment analysis (GSEA) in which we determined the array of hematopoietic cell signatures as defined previously in the D-MAP study (Supplementary Table S3 [13, 14]) that were enriched in the intestinal gene pool of E-CVID patients compared to the pools of noE-CVID patients or controls. We found that the gene signatures of variety of hematopoietic cells were enriched in the intestinal gene pool of the E-CVID patients (Figure 1d), most prominently natural killer cells, M1 macrophages, neutrophils and CD8+ T effector/memory lymphocytes (Figure 1d and Supplementary Figure S2a, S2b). Furthermore, we observed a trend toward a reduced presence for memory B cells, plasma cells, naïve CD8+ and CD4+ cells in biopsies from E-CVID patients compared to biopsies from noE-CVID patients or controls. Finally, the results with respect to CD8+ cells were not influenced by the presence of norovirus infection.
To validate one of the most important inferences about the infiltrating cells derived from the above gene expression analysis, we evaluated levels of CD8+ T cell staining in biopsies of tissues obtained from representative patients. The results confirmed the gene expression analysis in showing increased infiltration of CD8+ T cells in biopsies of E-CVID patients compared to biopsies of noE-CVID patients (Figure 1f). Interestingly, this result is compatible with peripheral (circulating) patient cell lymphophenotyping in that E-CVID patients exhibited an increase in the absolute numbers of CD8+ T cells compared to noE-CVID patients. However, this association is tentative because the increase in peripheral cells was not statistically significant. Taken together, these results demonstrate that CVID enteropathy is characterized by a complex Th1/cytotoxic T cell-related infiltration.
Finally, we used GSEA to demonstrate that among the top “leading edge” genes expressed in CD8+ T cells were those encoding IFNG and HLA class II molecules and for genes expressed in M1 macrophages there were those encoding several chemokines such as CXCL9, CXCL10, CCL19, CCL20, CCL5 as well as transcription factor IRF1 (Figure 1e). Since expression of these genes is known to be associated with features of an inflammatory process, their expression by CD8+ T cells and macrophages in E-CVID tissue suggested that these cells are participating or mediating intestinal inflammation (Figure 1f; Supplementary Table S4).
Intestinal IgA levels discriminate CVID patients with and without enteropathy
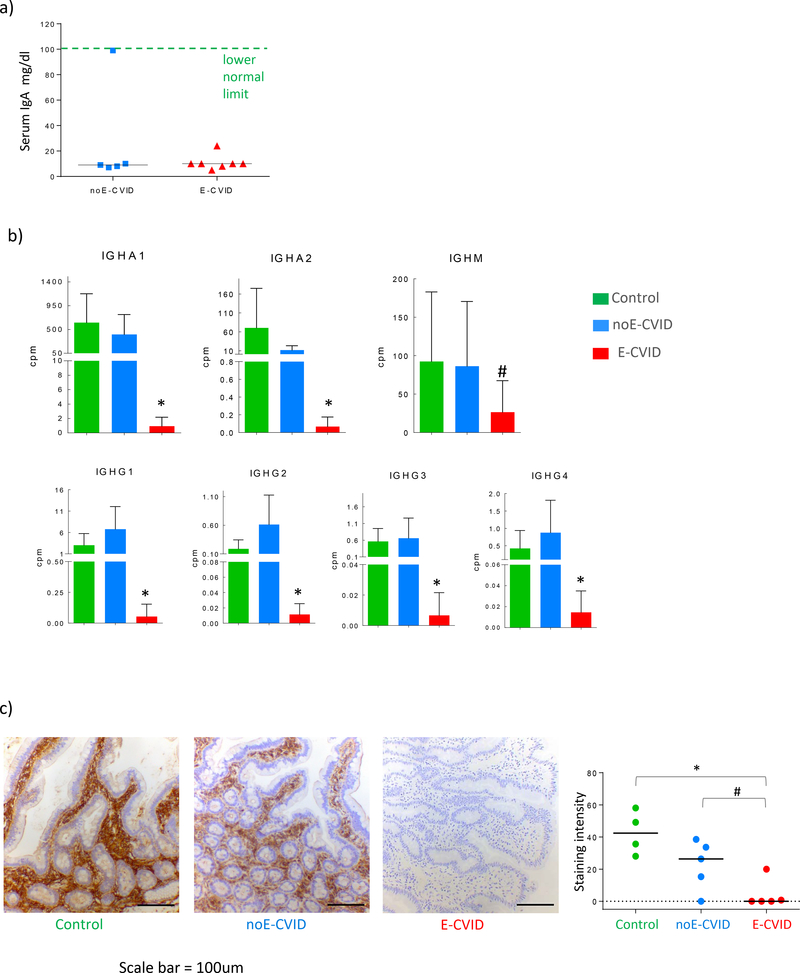
The trend noted above indicating that E-CVID patients’ gene expression exhibited decreased enrichment for memory B cells and plasma cell signatures compared with expression in noE-CVID patients suggested to us that these cells might be more depleted in the mucosa of the E-CVID group than in the noE-CVID group despite the fact that serum immunoglobulins and IgA levels are similarly low in the two groups (Figure 2a). To examine this possibility, we extracted the immunoglobulin transcript levels from the total duodenal RNAseq data and found that mRNA levels of both IgA subclasses (encoded by the IGHA1 and IGHA2 genes) in the mucosa from E-CVID patients were strikingly decreased compared to levels in the mucosa from noE-CVID patients and the levels in the latter group were much closer to those in control individuals (Figure 2b). Similarly, levels of gene expression of each of the four subclasses of IgG were also decreased in the E-CVID mucosa compared to noE-CVID mucosa and again levels in noE-CVID were similar to those in controls (Figure 2b). Finally, transcript levels of IgM were also low but in this case the error bars were quite large and the result was therefore ambiguous. It should be noted that overall copy numbers of transcripts coding for IGHA1 was ~10 times higher than for IgAH2 and ~100 times higher than for IgG genes across all three groups indicating that IgA overall (and IgA1 in particular) is the dominant gut immunoglobulin not only in the control small intestine but also in noE-CVID patient small intestine.
Figure 2.
Intestinal IgA levels discriminate patients with and without CVID enteropathy. (a) Levels of total IgA in serum of CVID patients. (b) Expression of genes coding for IgA1, IgA2, IgM, IgG1, IgG2, IgG3, IgG4 in duodenal biopsies of CVID patients with enteropathy (n=6), noE-CVID (n=8) and Control subjects (n=7); *p<0.05; # p<0.1, one-sided t-test, comparing E-CVID to other groups, mean/SD shown. (c) Representative images of IgA immunohistochemistry in duodenal biopsies and quantitative analysis, *p<0.05; # p<0.1 Mann-Whitney one-sided test comparing E-CVID to other groups; cpm stands for counts per million.
To corroborate these mRNA data at the protein level we performed immunohistochemistry staining of IgA in tissues from E-CVID patients, noE-CVID patients and controls; subsequently, one of us (MQ, a gastrointestinal pathologist) evaluated the stained tissues in a blinded fashion. We found that cells expressing IgA were absent in duodenal tissues from 6/7 E-CVID patients and infrequently present (at best) in the remaining E-CVID patient. In contrast, cells expressing IgA were present in the tissues of all but one noE-CVID patients, albeit with reduced levels of IgA staining intensity compared to controls (Figure 2c). Interstitial IgA was not observed in tissues from patients in any group indicating that IgA+ plasma cells were indeed the source of the staining. These results thus confirmed the mRNA data showing that E-CVID can be differentiated from noE-CVID on the basis of mucosal IgA expression despite similarity in circulating IgA levels. Taken together with the mRNA data they open the door to the possibility that the absence of IgA in the duodenum of E-CVID patients subject the latter to possible colonization/infection with organisms that induce inflammatory T cell and macrophage responses that result in villous atrophy.
The Bacterial Microbiome in the CVID Duodenum
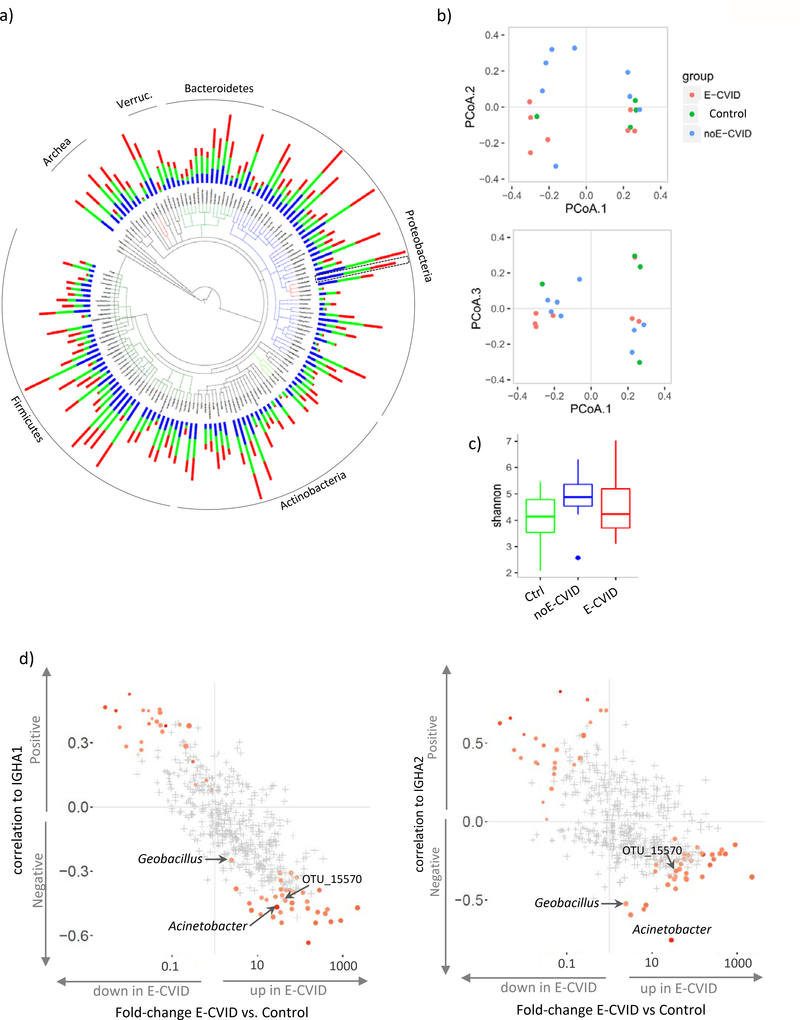
We next turned our attention to studies characterizing the bacterial microbiome associated with duodenal tissue in CVID. Recognizing that duodenal tissues contain an overwhelming number of human cells compared to microbial cells, we first used a targeted gene approach in which bacterial genes were identified by sequencing 16S ribosomal RNA genes present in bacterial but not in mammalian genomes. Despite previous reports of bacterial overgrowth in fecal material of CVID patients [15], we did not observe significant differences in bacterial abundance in duodenal biopsies from the E-CVID, noE-CVID and control groups (Supplementary Figure S3). Tissues from each of these groups contained bacterial taxa dominated by members of the Proteobacteria, Firmicutes, and Actinobacteria phyla and expressing abundant bacteria in the Gammaproteobacteria and Bacilli classes (Figure 3a; Supplementary Table S5). Importantly, there were not significant differences between the groups in respect to overall composition as illustrated by PCoA (Figure 3b) or global diversity evaluated by the Shannon index (Figure 3c). Thus, while comparison of abundance of Operational Taxonomic Units (OTUs) between the groups indicated differential representation of some OTUs at the nominal p value of <0.05, none of differences achieved significance after multiple comparison correction (i.e. low FDR).
Figure 3.
Microbiome analysis of duodenal biopsies. (a) Tree-of-life plot at the genus level; bars represent relative frequencies; red, E-CVID, blue, noE-CVID, green, Control; Acinetobacter genus is indicated by dashed box as it will be focus of the analysis in Fig 4. (b) Principal coordinate analysis of microbiota composition in biopsies. (c) Shannon index estimating microbiome diversity in groups of samples; blue dot is an outlier sample, no significant differences detected between groups by one-sided t test. (d) Correlations between OTU abundances and levels of IGHA1 (left) or IGHA2 (right) mRNA plotted against change in OTU Frequency in ECVID vs. all other samples. OTU, operational taxonomic unit. Red color indicates OTUs that pass FDR 0.2 with stronger intensity corresponding to lower FDR. OTUs marked in grey present FDR above 0.2. E-CVID, enteropathy associated with common variable immunodeficiency; noE-CVID, CVID without enteropathy; Ctrl, Control subjects.
In view of the above results, we sought to narrow the universe of bacteria with the potential of causing enteropathy in CVID. First, we took into consideration our results showing that low mucosal IgA levels discriminated between patients with and without enteropathy (Figure 2). Second, we reasoned that since IgA is known to be protective against pathogens and can regulate commensal microbiota [16–18], bacterial species potentially contributing to disease in E-CVID patients would have increased abundance in E-CVID tissue compared to control tissue and would be negatively correlated with IgA levels. To select such bacterial candidates, we combined the results of correlations between bacterial abundances and IgA gene expression with the results of enrichment of bacteria in E-CVID using a recently developed statistical approach [19, 20]. Thus, out of 582 OTUs detected in duodenal biopsies, we found 45 OTUs (FDR<20%) that were enriched with bacteria potentially involved in enteropathy (red dots in right lower corners in both panels of Figure 3d, Supplementary Table S6).
Transkingdom Network Analysis Identifies Candidate Microbes Capable of Driving CVID Enteropathy
To further identify the microbe or microbes driving CVID enteropathy, we employed transkingdom network analysis, an analytic method that we recently developed [20, 21] and successfully used to identify microbiota causing changes in host phenotypes [12, 22]. Using this method of analysis, we integrated abundances of microbial OTUs with mRNA levels of host genes present in the E-CVID signature and thus formed a network in which microbes with strong connections to altered host genes were revealed. The resulting network (Figure 4A) consisted of 1474 human genes and 72 OTUs (45 potential causal and 27 potential protective bacteria, Supplementary Table S7) from the microbial candidates previously identified by their association to mucosal IgA levels and abundancies in E-CVID (Figure 3d).
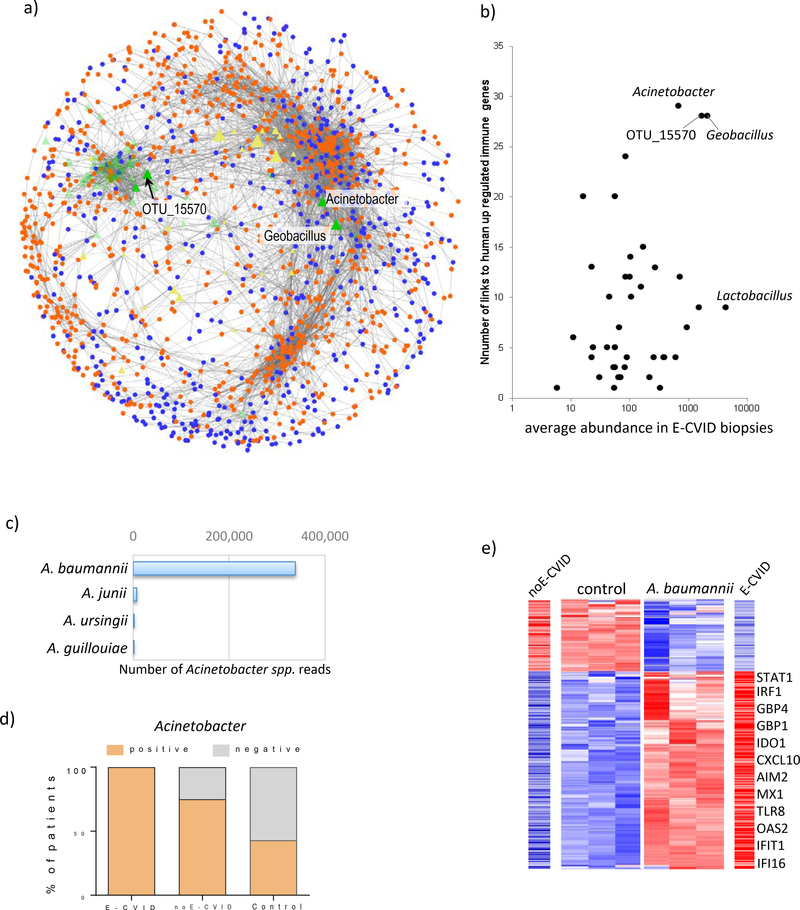
Figure 4.
Transkingdom network analysis identifies Acinetobacter baumannii as a candidate microbe driving CVID enteropathy. (a) Transkingdom network reconstructed from levels of expression of human genes from E-CVID signature and candidate microbes selected based on analysis in Fig. 3d; orange and blue indicate up-regulated and downregulated genes in E-CVID, respectively; green and yellow are bacterial OTUs increased or decreased in E-CVID, respectively. (b) Number of connections to genes up-regulated in E-CVID and average abundance in E-CVID biopsies are plotted for each OTU identified by analysis in Fig. 3d. (c) Number of reads corresponding to species of Acinetobacter genus in biopsies based on shotgun metagenomics. (d) Number of biopsy samples positive or negative for Acinetobacter in each patient group (p<0.04, Chi-square test). (e) Gene expression from E-CVID signature as assessed by RNAseq in THP-1 cells after 6 h of incubation with A. baumannii. Representative genes are indicated and levels of expression of the same genes in biopsies with E-CVID or noE-CVID are shown; full list of genes is in Supplementary Table S13.
Among the OTUs that were both most abundant and highly connected to human up-regulated genes (mostly pro-inflammatory) in the majority of patients with E-CVID were OTUs from the Acinetobacter genus, Geobacillus genus and otu_15570 with an ambiguous taxonomy (Figure 4b, Supplementary Table S8). The number of edges (i.e. links) in the network connecting Acinetobacter, Geobacillus and otu_15570 with human genes was 29, 28, 28 and fold-change of enrichment in E-CVID over other groups was 28, 3, 44 respectively (Figure 3d). Thus, these three OTUs emerged as possible microbes causing E-CVID (Figure 4b).
Of these three OTUs, Acinetobacter was the most promising since this genus harbors several known pathobionts (commensal microorganisms that can become pathogens under favorable circumstances [23, 24]) whereas Geobacillus and the otu_15570 bacterium have not been previously associated with human disease. In further studies to identify organisms in these groups at the species level we employed shotgun RNAseq rather than 16S rRNA methodology since the latter was not sufficient to support species resolution. Using this approach, we verified all sequences belonging to the three genera and assigned them to species when possible. We found that whereas shotgun RNAseq analysis did not support species resolution for Geobacillus and otu_15570 (Supplementary Figure S4, and Supplementary Table S9) it did reveal that Acinetobacter baumannii is the predominant species in the Acinetobacter organism genus identified in duodenal biopsies (Figure 4c; Supplementary Table S10). This bacterium has been found in the human intestine and can be a cause of hospital-acquired nosocomial infection not involving the GI tract, presumably as a result of its resistance to antibiotic treatment [25, 26], [27]. Furthermore, analysis of the duodenal microbiome of patients/controls in all 3 study groups by 16S rRNA sequencing of duodenal biopsies showed that Acinetobacter was present in the duodenal microbiome in all 7 E-CVID patients, 6/8 noE-CVID patients and 3/7 controls (Figure 4d). Furthermore, direct association with IgA expression levels showed that Acinetobacter had the most significant negative correlation with IGHA1 and IGHA2 (FDR=0.016, with r=−0.76, r=−0.47, respectively) among all detectable taxa. Taking these results together with the outcome of the transkingdom network analysis, it became reasonable to postulate that A. baumannii, acting as a pathobiont, was a significant factor in the development of enteropathy in CVID patients.
We next conducted in vitro experiments to determine if A. baumannii can be a contributing pathobionts in E-CVID. Based on the supposition that macrophages, cells that we found were highly enriched in E-CVID mucosa (Figure 1d and 1e), would be the primary cells responding to A. baumannii and triggering a pro-inflammatory reaction in the CVID intestine, we determined whether A. baumannii can induce macrophages to display inflammatory alterations similar to those observed in CVID enteropathy tissues. In this experiment, we co-cultured THP-1 macrophages with A. baumannii or with a control bacterium for 6 h and then determined gene alterations occurring in the macrophages. The control bacterium used in this experiment was Lactobacillus plantarum, a bacterium that was the most abundant in duodenal biopsies from E-CVID patients among those potentially controlled by IgA, but did not fulfill other criteria for candidate pathobionts (see Figure 4b and more details in M&M). We found that A. baumannii strongly induced expression of interferon type I (IFNB1) and CXCL9, the latter a chemokine involved in M1/Th1 activation (Figure S6). In addition, using global gene expression analysis, we found that A. baumannii induced alterations in the expression of 252 genes (185 Up- and 67 Down-regulated) that were highly concordant with the CVID enteropathy expression profile whereas Lactobacillus plantarum did not (Figure 4e). Up-regulated genes included those coding for proteins involved in immune responses such as intracellular microbial nucleic acid sensors (AIM2, IFI16, TLR8), numerous IFN-dependent genes (GBP1, IRF1, STAT1, MX1, IDO1, TAP1, IFI44, OAS2, IFIT1, RSAD2), chemokines (CCL2, CXCL1, CXCL10), and many other innate immune response genes (Figure 4e). These in vitro data therefore show that A. baumannii can induce an immune response in macrophages consistent with the induction of the Th1-driven inflammation that has been found in E-CVID and thus support the possibility that this bacterium can be a pathobiont contributing to CVID enteropathy.
The Interferon Response in E-CVID Drives Inhibition of Lipid Metabolism in ECVID Epithelial Cells
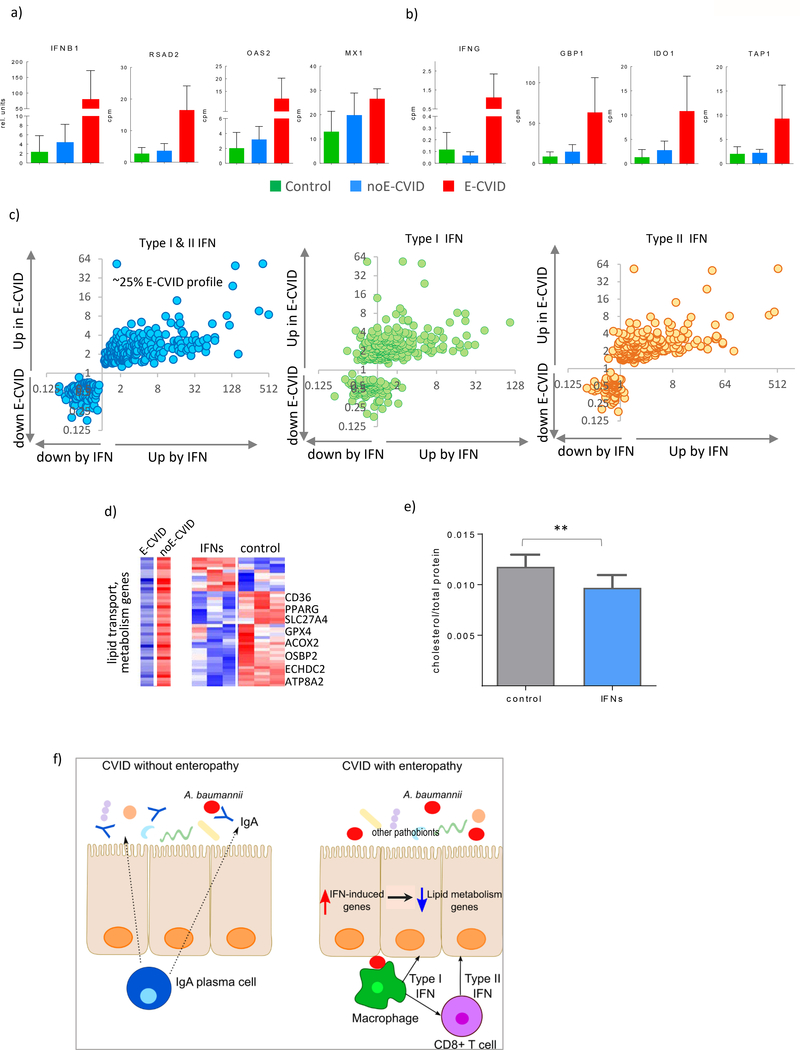
In further studies we examined the relation between the nature of the abnormal immune response dominating enteropathy in E-CVID and lipid malabsorption. As mentioned above, in our previous studies in humans [9] and in mouse model [11], metabolic alterations in epithelial functions were associated with elevations in both type I and type II interferons. Therefore, we further investigated this phenomenon and observed elevated expression of several genes known to be induced by IFN type I (OAS2, MX1, RSAD2) or type II (GBP1, IDO1, TAP1) as well as increased expression of both types of IFNs themselves in E-CVID tissue (Figure 5a,b). Next, to evaluate the effects of IFNs on epithelium we cultured a human duodenal epithelial cell line (FHs 74 int) in the presence of the two types of IFNs together or separately and then assessed gene expression changes for the genes from the E-CVID profile we have established in this work (Fig. 1b). To take a full advantage of this in vitro model and the same time to reduce its potential limitations we focused our analysis on gene expression signature discovered evaluating patients’ biopsies. We found that alterations in the expression of approximately 25% of the genes in the E-CVID gene signature (p<0.05, FDR=15%) were consistent with the direct effect of IFNs on the epithelial cells (Figure 5c; left panel). Interestingly, treatment with individual interferon types regulated significantly fewer genes than both interferons together with 70 genes regulated by type I and 232 by type II interferon whereas these interferons together regulated 343 genes (Figure 5c).
Figure 5.
Interferons Drive Inhibition of Lipid Metabolism in Epithelial Cells. (a) Expression of IFNB1 and IFN type I dependent genes (RSAD2, OAS2, MX1) in duodenal biopsies from of CVID patients with (red), without (blue) enteropathy and control subjects (green), all genes passed false discovery rate <0.03 comparing E-CVID to other two groups. (b) expression of IFNG and IFN type II dependent genes (GBP1, IDO, TAP1); (c) Gene expression ratios between E-CVID and control individuals (y axis) and between interferon treated versus untreated human epithelial cells (x axis; n=3 per group), left panel both types of interferons; middle panel interferon type I; right panel interferon type II. (d) Expression of genes defined as lipid metabolism by Gene Ontology and downregulated in E-CVID, representative genes are indicated; left panel: median values in E-CVID and noE-CVID tissues; right panel: individual values in cells treated and untreated with both interferons, red is high and blue is low expression; (e) intracellular cholesterol levels in epithelial cells treated or untreated with both types of interferons data presented as mean±s.e.m.; each column number of biological replicates equals 14 (**p<0.0001 by one-tailed Wilcoxon test); (f) model of CVID enteropathy.
While most of the interferon-regulated genes (234 genes) were among those up-regulated in the gene signature, there were 109 downregulated genes enriched for several metabolic processes (Supplementary Table S11). Furthermore, as in the case of the up-regulated genes, the magnitude of downregulation was much less pronounced with individual IFNs than with both of IFNs (Supplementary Figure S8). In an additional analysis focusing on lipid metabolism we found that among genes belonging to the group down-regulated by IFNs in epithelial cell culture, the majority (32 out of 43, 74%) had decreased expression after interferon stimulation similar to that observed in biopsies from CVID patients with enteropathy (Figure 5d; Supplementary Table S11). These genes are known to be involved in various aspects of enterocyte lipid metabolism such as fatty acid uptake, transport and biosynthesis (CD36, SLC27, ECHDC2, ACSF2), fatty acid oxidation (ACOX1, ACOX2), cholesterol binding (OSBP2, OSBPL7, HSD17B2), and phospholipid transport (ATP8A2, ATP8B2). To test if these effects of IFNs on lipid metabolic gene expression translates into functional alterations we measured intracellular cholesterol levels. In fact, as predicted by the transcriptomic alterations, we observed a consistent reduction in intracellular cholesterol levels after IFN treatment (Figure 5e). Taken together, these results suggested that the upregulation of interferon-related genes in CVID drives changes in epithelial cell metabolism in this disease.
Discussion
Gastrointestinal (GI) symptoms are diverse and relatively frequent in CVID patients despite IgG-replacement therapy. It has been reported that up to 50% of these patients exhibit various degrees of GI symptomatology and up to 15% of the patients exhibit CVID-associated enteropathy characterized by villous atrophy, malabsorption and severe diarrhea that is potentially life threatening and usually treatable only with further immunosuppression [7, 9].
A central question that we aimed to address in the present study is why some CVID patients develop enteropathy and others do not. Although it was widely recognized that plasma cell levels and serum IgA levels were decreased in CVID patients, no clear distinction with respect to these features of immunodeficiency was attached to the presence or absence of enteropathy [1, 8]. In the current study we made the unexpected finding that while CVID with and without enteropathy did indeed have diminished serum IgA levels, most if not all of the CVID patients with enteropathy exhibited dramatically decreased IgA expression in the duodenal intestinal tissues compared to their counterparts without enteropathy. This finding strongly suggests that patients with enteropathy exhibit more mucosal immunodeficiency than those without enteropathy and are therefore more at risk for a form of gut infection that could cause enteropathy as extensively discussed below. Prior studies partially support this conclusion in that they showed that CVID patients with relatively low IgA levels were more prone to develop various bacterial and viral infections than other CVID patients [28],[29]. However, these studies did not show that low mucosal IgA levels was associated with enteropathy; this discrepancy may be related to the sensitivity of the staining procedures used in the prior studies as well as the fact that they did not evaluate patients by assessing mucosal Ig mRNA levels (which in this study corroborated the tissue staining findings).
Finally, it should be noted that while defective mucosal IgA responses may define the pre-condition central to development of enteropathy in CVID at the level of immune function it does not define the underlying immunologic abnormality necessary for the development of this pre-condition. A possibility that requires future exploration in this regard is that those CVID patients with enteropathy have in common a genetically determined defect in B cell trafficking that impairs the migration of IgA as well as IgG-producing B cells into the mucosal tissues.
One possible caveat to the above conclusion is that enteropathy only rarely occurs in patients with selective IgA deficiency. However, this can be explained by the fact that patients with this immunodeficiency almost invariable have B cells that produce IgA, albeit at a level lower than that in control individuals. In addition, it has been reported that these patients produce IgG and IgM in the mucosal tissues that partially compensate for the lack of IgA [30],[31]. A second possible caveat is that enteropathy has not been reported in X-linked immunodeficiency patients who have as profound a deficiency in mucosal IgA levels as CVID patients with enteropathy. A possible explanation for this discrepancy is that CVID patients have an accompanying immunologic abnormality that pre-disposes them to the intense Th1-mediated immune response in the upper intestine that is the actual cause of the villous atrophy and resulting malabsorption. Evidence supporting this view comes from previous studies showing that cells from all CVID cells exhibit an IFN-γ signature as well as the present study which shows that duodenal tissue from CVID patients with enteropathy exhibit overexpression of IFN genes (see further discussion below). This T cell abnormality could be a primary defect accompanying the disease or, as suggested recently, an abnormality secondary to lack of B cell regulation of Th1 T cell development [32]. In any case, one is led to the conclusion that, while a B cell abnormality manifesting as mucosal IgA deficiency sets the stage for enteropathy-causing infection in CVID and serves as a risk factor for the latter sustained inflammation due to a robust T cell response is also necessary for the development of the main manifestation of enteropathy, villous atrophy and malabsorption.
The above explanation of the immunologic conditions allowing the development of enteropathy introduces the question as to which organism could be causing CVID enteropathy. Early on, infection with Giardia lamblia was noted to occur with increased frequency in CVID and was suggested to be a cause of CVID enteropathy; however, it became apparent that while eradication of this organism in CVID patients does resolve symptoms of the infection it does not reverse co-existing enteropathy; in addition, in the past decade most patients with enteropathy are free of Giardia infection [9]. In a more recent study of infection in CVID patients it was found that sporadic infection with a variety of bacterial and viral pathogens does occur in CVID, but no evidence that these were causative of enteropathy and in most cases the infection did not cause symptomatic disease [33]. A possible exception to this trend is the possibility that CVID is caused by norovirus infection since this organism has been found in stool and biopsies of some (but not all) patients with this condition [29, 33, 34]. Our molecular analysis of organisms associated with the duodenal specimens showed that CVID patients with and without enteropathy exhibited a relatively low norovirus positivity in biopsies (Supplementary Figure S9). This suggests that norovirus alone cannot explain the presence of enteropathy, at least in our patient cohort. It remains possible, however, that this agent is playing a role as an adjuvant to the causative organism as it can induce intestinal inflammation in humans and mice (reviewed in [35]). Finally, it should be noted that duodenal biopsy specimens were also evaluated by PCR and whole genome RNA sequencing for the presence of enterovirus, adenovirus, astrovirus and rotovirus; only enterovirus was detected in one tissue specimen that happened to be positive for norovirus.
In another study [8], it was found that the fecal microbiome of CVID patients exhibited a “dysbiosis” characterized by multiple alterations in the composition of the microbial community including both increases and decreases of several taxa compared to those in control individuals. In general, however, these changes in fecal microbiome did not differentiate patients with enteropathy from those without enteropathy and did not identify a specific organism as a cause of the enteropathy. In studies of the gut microbiome carried out here and focusing on the microbial community in the duodenum of CVID patients with and without enteropathy we did not observe a changed level of diversity compared to controls. However, a more complex systems biology analysis of these data yielded more positive results. As detailed in Results, this involved first narrowing the list of possible organisms to those that both negatively correlated with intestinal IgA levels and that are enriched in CVID enteropathy. Second, this involved selecting among the organisms meeting these criteria to a transkingdom network analysis in which possible causative organisms are identified on the basis of their closeness (i.e., linkage) to the up-regulated host gene expression profile associated with enteropathy. Out of three different taxa that have higher chance to be pathobionts potentially contributing to enteropathy in patients with CVID one organism (Acinetobacter baumannii) could be identified to the species level.
A. baumannii fulfilled several of the criteria one might expect of an enteropathy-inducing organism. In fact, in vitro culture of this organism, induced Th1/M1-like gene expression profile in a human macrophage line that was in fact very similar to that observed in tissue from CVID with enteropathy (Figure 5f). A. baumannii also has clinical features that would characterize a possible organism causative of enteropathy. First, it is a notably antibiotic-resistant bacterium, which would fit with the fact that CVID enteropathy does not usually respond to antibiotic therapy. Second, although A. baumannii has been found in the intestine of healthy individuals [36, 37] and we detected this bacterium in both control individuals and patients with CVID without enteropathy, this microbe has been associated with wound, lung and blood infections in soldiers and patients in intensive care units [38–42]. Interestingly, in a very recent study it was found that an Acinetobacter genus was associated with inflammatory cellular and gene expression profiles in IBD patients [43]; this provides further support for the potential role of these bacteria in gut inflammation
The above studies supporting the idea that A. baumannii is acting as a pathobiont in CVID enteropathy notwithstanding, the possibility exists that other bacteria could also have this role. This follows from the fact that the transkingdom network analysis identified other possible pathobionts in CVID, including, Geobacillus, a bacterium that shares some genetic properties with segmented filamentous bacteria (SFB) and that is well known as an inducer of pro-inflammatory cytokines in the mouse small intestine [44]. Thus, while our study, together with previous mouse data [11], supports the concept that CVID enteropathy is possibly caused by a pathobiont in the upper GI tract and identified A. baumannii as one such pathobiont, it is likely that other pathobionts will be identified which also have this role and are the cause of enteropathy in CVID patients in which this organism is not found.
In the present studies we performed extensive global gene expression profiling that confirmed our previous finding that a Th1-like inflammatory process underlies CVID enteropathy [9]. In addition, it led to the further insight that both type I and II interferons are involved (Fig. 5). Interestingly, these findings coincide with a previous study showing up-regulation of IFN-driven gene expression in whole blood cell populations is a feature of CVID patients with a variety of inflammatory complications including those with and without gastrointestinal inflammation [45]. Confirming our previous mouse studies we identify in humans that the shift from metabolic to immune/pro-inflammatory in the small intestine is associated with the development of enteropathy. In addition, we now show that the type I and type II interferons above are responsible for this shift and that, for instance, lipid transport gene expression and intracellular cholesterol levels are directly and adversely affected by interferons. It should be noted, however, that while these metabolic alterations may contribute to the malabsorption characterizing CVID enteropathy the latter is mainly due to cytotoxic effects on epithelial cells arising from the inflammation.
Overall, in this work, we present evidence that CVID enteropathy is uniquely characterized by a severe deficit in mucosal IgA secretion, an abnormality that sets the stage for possible colonization and infection by one or more commensal organisms acting as pathobionts in the IgA-deficient mucosal environment. We have identified A. baumannii as one such possible pathobionts. CVID enteropathy is also uniquely characterized by a gene profile in which IFN gene expression is greatly increased whereas metabolic gene expression is greatly decreased. This abnormality may contribute to the development of enteropathy by facilitating the robust interferonmediated response necessary to induce villous atrophy and malabsorption.
Material and Methods
Patients
Eligible patients older than 18 years of age had a diagnosis of common variable immunodeficiency according to established criteria of a medical history of mucosal infections, decreased immunoglobulin levels 2 standard deviations below the norm and decreased humoral vaccine challenge responses. CVID patients with a concomitant history of chronic diarrhea or unexplained weight loss greater than 5% of total body weight over the previous 6 months with changes in more than one parameter consisting of body mass index, serum albumin, serum protein and number of liquid stools were classified in the CVID enteropathy cohort of patients (N=7, 4 males, 3 females). CVID patients without enteropathy constituted the no enteropathy CVID group (N=8, 2 male, 6 females). Additional control patients consisted of patient population without acute disease, CVID or GI pathology disease but, could include patients with irritable bowel syndrome (N=7, 3 males, 4 females). Patients were additionally classified based upon duodenal histopathology (see Supplementary Table S14 and Supplementary Table S15). All eligible patients had negative stool testing for pathogens (Salmonella, Shigella, Yersinia, Campylobacter, Vibrio, Escherichia coli O157/H7, Cryptosporidia, Cyclospora, Microsporidia, and Giardia) and Clostridia difficile toxin, negative hydrogen breath test, were on no immunomodulatory agents for the preceding 4 months, and were human immunodeficiency virus (HIV) negative. CVID patients with enteropathy or CVID patients without enteropathy had received antibiotic therapy in the year prior to this study for a variety of clinical conditions which included small bowel intestinal overgrowth, C. difficile intestinal infection, sinusitis, and pulmonary prophylaxis treatment. Patients in these groups did not receive any antibiotic therapy in the three months prior to analysis (except for one patient in each of the CVID with enteropathy and without enteropathy groups which received antibiotics on a continued basis for lung prophylaxis treatment). The enteropathy in the CVID patient who receiving continued antibiotic therapy did not have a change in his enteropathy. All protocol and informed consent procedures were approved by the National Institute of Allergy and Infectious Disease and Oregon State University Institutional Review Boards.
Patient Peripheral Cell Lymphophenotyping
Peripheral blood cells (3 mL EDTA anticoagulated samples) were stained for flow cytometry using a whole- blood lysis method and analyzed on a CANTO2 (Becton Dickinson Biosciences, San Jose, CA). CD3+T cells and T-cell subsets were identified by directly conjugated monoclonal antibodies anti-CD3, anti-CD4, anti- CD8. B cells and B cell subsets were identified by directly conjugated monoclonal antibodies anti-CD20; and memory B cells with anti-CD27; directly conjugated, murine IgG1 was used to ascertain background staining. All monoclonal antibodies were obtained from Becton Dickinson Biosciences (San Diego, CA). Lymphocyte gate was established using forward and side scatter with list mode parameters collected for 106 lymphocytes. To calculate absolute numbers of each lymphocyte subset, the proportion of cells staining positive was multiplied by the absolute peripheral blood lymphocyte count. Peripheral blood obtained from study control individuals was stained and analyzed to establish comparison normal control values.
Exome Sequencing
Targeted exome sequencing was performed at the National Institutes of Health facility for genes known to be correlated with CVID using the Ion Torrent AmpliSeq RDY Exome Kit (Life Technologies) and the Ion Chef and Proton instruments (Life Technologies). Briefly, 100 ng of gDNA was used as the starting material for the AmpliSeq RDY Exome amplification step following the manufacturer’s protocol. Library templates were clonally amplified and enriched using the Ion Chef and the Ion PI Hi-Q Chef Kit (Chef package version IC.4.4.2, Life Technologies), following the manufacturer’s protocol. Enriched, templated Ion Sphere Particles were sequenced on the Ion Proton sequencer using the Ion PI chip v3 (Life Technologies). Read mapping and variant calling were performed using the Ion Torrent Suite software v4.4.2. In short, sequencing reads were mapped against the UCSC hg19 reference genome using the Torrent Mapping Alignment Program (TMAP) map4 algorithm. SNPs and INDELS were called by the Torrent Variant Caller plugin (v.4.414–1) using the ‘Generic-Proton-Germ Line – Low Stringency’ configuration. Only reads that were unambiguously mapped were used for variant calling. Variants were annotated using ANNOVAR (http://annovar.openbioinformatics.org/). Data mining, biological interpretation, and candidate gene discovery were performed using various online tools, including The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov), and GeneCards (http://www.genecards.org). Target coverage was evaluated using the Torrent Coverage Analysis plugin (v.4.414–1), and the output was further evaluated using in-house, custom Perl scripts.
Endoscopic Biopsy
Endoscopic biopsy specimens were fixed in 10% formalin prior to paraffin embedding and routine staining with H&E. Biopsy specimens were reviewed by a gastrointestinal pathologist who was masked to the clinical status of the patients. Biopsy specimens were labeled as nodular lymphoid hyperplasia (NLH) if germinal centers were present, intraepithelial lymphocytosis if there were greater than 5 lymphocytes per 20 epithelial cells, and noted for apoptosis if there were greater than 5 apoptotic epithelial cells per 10 glands. CVID patients with enteropathy were noted to exhibit concomitant histologic changes in the duodenum which included villus blunting, flattening/atrophy, an increased number of intraepithelial lymphocytes, the appearance of increased polymorphonuclear cells and lymphocytes in the lamina propria.
Immunohistochemistry and image analysis
Paraffin embedded biopsy tissues were and cut into 5 μm sections. Antigen retrieval was performed at 90°C for 20 min and sections were blocked with bovine serum albumin following by staining with anti-IgA (Abcam ab124716) or anti-CD8 (Dako M710301–2) both at a dilution of 1:100. Sections were developed with 3,3’-diaminobenzidine (DAB), and counterstained with hematoxylin by HistoServ (Germantown, MD). Slides were imaged using a Leica DM E microscope. For IgA quantification, DAB+ area and total tissue area were quantified from 5–7 randomly selected fields representative of total tissue area. For CD8+ area, the epithelial layer was manually selected from 5–7 randomly fields and DAB+ area and total tissue area were quantified from this selected epithelial area. All image selection, color deconvolution and area quantifications were performed using Fiji [46].
Shotgun RNA sequencing and 16S rRNA gene sequencing
Total RNA from biopsies was isolated using TotalPrep RNA/DNA kit (Qiagen) after mechanical homogenization with Omni rotor followed by Qiashredder (Qiagen). For analysis of bacterial 16S rRNA gene expression, 1 microgram of total RNA was reverse transcribed using VILO cDNA synthesis kit with random primers (Invitrogen) and this cDNA was used for qPCR with universal bacterial primers and for 16S rRNA library prep and then sequenced on MiSeq instrument according to a previous protocol [22]. For human gene expression and shotgun metagenomics, libraries were prepared using ScriptSeq™ Complete Gold Kit (Epicentre/Illumina) according to manufacturer’s instructions including removal of rRNA. These libraries were sequenced on HiSeq2000 instrument (Illumina) with four samples per lane resulting in ~50*10^6 reads per sample.
RNA-seq gene expression analysis
Forward read sequences with at least one ambiguous nucleotide were filtered out by prinseq. Trimmometic was used to trim Illumina adaptor sequences (parameters: seed mismatches:1, palindrome clip threshold: 10 and simple clip threshold: 10), to remove leading and trailing low-quality bases (below quality 3), to scan the read with a 4-base wide sliding window, and to cut when the average quality per base drops below 20 and to drop reads that below 60 bases long. Reads were aligned to human genome and transcriptome (ENSEMBLE release-70) using Tophat with default parameters. Number of reads per million for human genes were counted using HTSeq. Genes that were detected in at least three samples were included in downstream analysis using BRB ArrayTools and normalized by quantile normalization.
For signature finding, differentially expressed genes were defined by comparing three groups using univariate test requiring p<0.05 and FDR <0.2, followed by pairwise comparisons using t-test, FDR<0.1 and Venn Diagram analysis. See details in Supplementary Figure 1.
Gene Ontology enrichment analysis of Biological Processes (BP_FAT) was performed using DAVID Bioinformatics Resources 6.7 with threshold gene count 2 and enrichment score p-value <0.01[47].
For immunoglobulin expression analysis we retrieved quantile normalized gene expression levels for IGHA1, IGHA2, IGHM, IGHG1, IGHG2, IGHG3, IGHG4.
Cell type enrichment analysis
For M1 and M2 microphages, we used the gene signatures from work of Martinez and co-workers [14]. For all other cell types, we inferred gene signatures using data from D-map [13]. The signature of a given cell type was defined as a group of genes with at least two-fold higher expression (FDR<0.1) than in other cell types (e.g. T lymphocytes vs other hematopoietic cells), and followed by subdivision of this parent signature by comparing a given cell type with closely related cell types (e.g. naïve vs memory T lymphocytes). Enrichment of gene signatures was performed using Gene Set Enrichment Analysis (GSEA) 2–2.2.3 with default parameters [48].
Microbiome 16S gene analysis
The raw forward-end fastq reads were quality-filtered, demultiplexed and analyzed using ‘quantitative insights into microbial ecology’ (QIIME). For quality filtering, the default parameters of QIIME were maintained in which reads containing ambiguous base calls and containing fewer than 187 nt (75% of 250 nt) of consecutive high-quality base calls, were discarded. Additionally, reads with three consecutive low-quality bases were truncated. The samples sequenced were demultiplexed using 12 bp barcodes, allowing 1.5 errors in the barcode. UCLUST was used to choose OTUs with a threshold of 97% sequence similarity against Green gene database (version gg_12_10) A representative set of sequences from each OTU were selected for taxonomic identification of each OTU by selecting the cluster seeds. The Greengenes OTUs (version gg_12_10) reference sequences (97% similarity) were used for taxonomic assignment using BLAST with E_value 0.001. Raw read counts of OTUs were normalized against total number of reads that passed quality filtration to generate relative abundance of OTUs. DNA amount of an OTU in a sample was calculated by multiplying relative abundance of the OTU by total amount of bacterial DNA in the corresponding sample. Differentially abundant OTUs were identified using univariate-test in BRB array tools’ ‘class comparison between groups of arrays’ modules. Beta diversity was calculated using QIIME script beta_diversity.py using option -m binary_pearson followed by principal_coordinates.py to generate principle coordinates. Alpha diversity was calculated using QIIME script alpha_diversity.py for calculating shannon index on randomly subsampling 15003 sequences for each sample. Tree of life graph was generated using the online tool iTOL (http://itol.embl.de/).
Network analysis and causal inference
Overall procedure is described in [49]. In brief, human gene–gene network was reconstructed using following steps. First, for each pair of genes/OTUs, three Pearson correlation coefficients and p-values are calculated using abundance levels of each of the three groups of E-CVID, NoE-CVID, Controls. Second, Fisher combined p-value is calculated from three p-values using Fisher’s combined probability test. FDR value is calculated from the combined p-value. The sign of correlation coefficients in three groups should be consistent (all positive correlation or all negative correlation across three groups). Also, we selected correlations that had sign of correlation consistent with possibility of causal relations between genes[19]. Third, human gene-gene network was generated by requiring all p-values of correlation coefficient from three groups <0.05 and combined FDR<0.1. Three human gene expression networks corresponding to three groups of samples were generated by selection and inclusion of gene pairs as following: Individual p-value of correlation within each group was smaller than 0.2; combined FDR smaller than 0.15.
The bi-partite betweenness centrality calculation is similar to conventional betweenness centrality except that shortest paths are calculated only between nodes belonging to two different groups of nodes and not between any nodes within the same subnetwork [12, 20]:
Where s and t are two nodes belonging to two different subnetworks, σst is the total number of shortest paths from node s to node t and is σst(v) the number of those paths that pass through v.
We used the scaled betweenness centrality for each network (by requiring individual p-value <0.05, 0.1 or 0.2) using the formula:
Reconstruction and interrogation of transkingdom network
To select OTUs potentially IgA dependent and differentially represented in E-CVID, Pearson correlation was calculated between OTU levels and expressions of IgA1 and IgA2, respectively. P values from two correlation analysis were integrated with t-test result from comparisons between E-CVID and HV using Fisher’s test followed by FDR calculations. OTUs that had FDR <=20% were selected for further integration into transkingdom network.
For human gene-microbe transkingdom network reconstruction, we applied the same procedure as for the gene-gene network requiring FDR <0.1. In order to identify potential candidate microbes that promote inflammation we interrogated a transkingdom network that represent a statistical model of interaction between microbiome and host transcriptome [20]. We thought that commonly used network parameters such degree of a given node would not be appropriate to ask our biological question. Therefore, in order to rank OTUs by strength of their potential influence on the host we focused our analysis and counted number of direct links (edges) between a given OTU and up-regulated genes in transkingdom network.
Identification of species for OTUs that are potential stimulators of human up-regulated immune genes
Transkingdom network analysis revealed several OTU candidates that are abundant in E-CVID and had many connections to human up-regulated immune genes. They are also potential regulators of human immune gene expression. However, 16S-based OTU taxa assignment of these candidates didn’t reach species level, thus limited our ability to experimentally test their functions. To better characterize the candidate OTUs, we utilized shotgun metatranscriptomic data derived from the same samples to profile abundance of species from the same most detailed taxa level (genus/family/order) of the candidate OTUs. We assign reads from shotgun metatranscriptomic to species based on our previous procedure [12]. Briefly, we aligned reads to reference protein database then used MEGAN5 to transfer the taxa of proteins to the reads that aligned to them. For the OTU denovo96904 (assigned to genus Geobacillus), we found that there are four Geobacillus species with similar abundance detected in the samples (Supplementary Figure S4). In this case there was not enough information to determine which species denovo96904 corresponds to. For the OTU denovo15570 (assigned to order Stramenopiles), we didn’t detect species from this order using shotgun metatranscriptomic data. We then took the representative 16s rRNA amplicon sequence of this OTU to search against NCBI nt database to figure out which species it came from. The result (Supplementary Table S9) indicated that there were many species sharing the exact same 16s rRNA region, including Cyclotella and uncharacterized bacterium. Thus, we were unable to obtain species information for the two candidates except the Acinetobacter OTU denovo56853 (Acinetobacter baumannii).
Technical parameters utilized for shotgun analysis of RNA-seq data to establish individual microbes corresponding to candidate OTUs
Sequences with at least one ambiguous nucleotide were filtered out by prinseq. Trimmomatic was used to filter out Illumina adaptor sequences (under parameters: seed mismatches: 1, palindrome clip threshold: 10 and simple clip threshold: 10), to remove leading and trailing low quality bases (below quality 3), to scan the read with a 4-base wide sliding window, and to cut when the average quality per base drops below 20 and to drop reads that are below 60 bases long. Human sequences were filtered out by aligning reads against human genome (hg19) using Bowtie2 under default parameters.
Around 82% of original reads passed filter steps and subject to downstream analysis. Taxonomic assignment of reads was carried out using DIAMOND alignment against the integrated nonredundant protein database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/nr.tar.gz). DIAMOND alignment hits with e-values larger than 0.001 were filtered out, and for each read, top 20 hits were retained to distinguish taxonomic groups. The taxonomical level of each read was determined by the lowest common ancestor (LCA)-based algorithm that was implemented in MEGAN4. In this algorithm, if a read had significant alignment hit in many species, it was assigned to the LCA instead of a species. Megan4 parameters were set to: min support=1, min score=50, min complexity=0.44, top percent=10, win score=0.
Selection of control bacterium
To select bacterial candidate that would serve as a control for A. baumannii as common commensal bacteria in duodenum that does not contribute to E-CVID according to our analysis. We wanted this bacterium being similar A. baumannii to for all features except inflammation promotion properties. Thus, top OTU (operational taxonomic unit) was dramatically enriched on biopsies from E-CVID patients (fold change 2225.3), negatively correlates with IgA genes(r=−0.351, r=−0.47; FDR<0.087), but did not connect much with proinflammatory genes in the network (Figure 4b; and Supplementary Figure S5). This candidate was OTU from Lactobacillus genus. Shotgun RNA-SEQ taxa assignment indicates multiple species with similar number of reads assigned. Thus, for each species detected by shotgun RNA-SEQ, we correlated their abundance across samples with the abundance of the candidate lactobacillus OTU and found the species Lactobacillus plantarum has highest correlation to the candidate OTU. Thus, we selected this species as negative control in regulating human up-regulated in E-CVID genes.
Bacterial cultures
Acinetobacter baumannii (ATCC 17978 or AB5075 from A. baumannii Transposon Mutant Library, UW Genome Sciences) was streaked out from −80°C on Luria-Bertani (LB) agar (BD) plate and incubated overnight at 37°C. To determine the optical density (OD600) and its corresponding colony forming units (CFU), one colony from this plate was inoculated in 10 ml of LB broth (Fisher BioReagents), and incubated at 37°C with shaking at 200 rpm. Every 2 hours, OD600 of the broth was verified and bacteria were plated in LB agar in serial dilutions. After overnight incubation, the CFU was calculated and used to establish a growth curve for the relationship between OD600 and CFU. For co-incubation with human cells, one colony from the LB agar plate was inoculated in 10 ml LB broth, incubated at 37°C with shaking at 200 rpm for 8–9 hours and OD600 was checked. This OD600 was used to interpolate the CFU in the broth using the established growth curve.
Lactobacillus plantarum (DSM 12028) was streaked out from −80°C on de Man, Rogosa and Sharpe (MRS) agar (Hardy Diagnostics) plate and incubated overnight in anaerobic conditions using GasPak EZ Gas Generating Container Systems (BD, Catalog# 260001) at 37°C. The correspondence between CFU and OD600 was established using the same method as for A. baumannii except using MRS medium and anaerobic conditions without shaking. To co-incubate the cells, one colony from the agar plate was inoculated in 10 ml MRS broth (Hardy Diagnostics) and incubated anaerobically as above for 8–9 hours. OD600 was checked and used to determine CFU.
Co-incubation of bacteria with THP-1 Cells
Human myelomonocytic leukemia cells line THP-1 (ATCC TIB-202) was grown in RPMI 1640 medium (Gibco/Thermo Fisher Scientific, Catalog # 11875093) with 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin (P/S). For differentiation, 2.5×10^5 cells per well (500 ul per well) were seeded in 24 well plate for 48 hours with phorbol 12-myristate 13-acetate (PMA-Sigma) at 50ng/ml, after which medium was changed and followed by 24-hour resting period. After medium was removed, 2.5×10^5 CFU of either bacterium (1:1 MOI, multiplicity of infection) suspended in 500 ul of RPMI (without P/S) were added to cells. As a negative control, RPMI (without P/S) was put in the control wells. After 6 hours of co-incubation, medium was removed from all wells, cells were lysed using RLT buffer (Qiagen) and stored immediately in −80C for RNA isolation. RNA was extracted using RNeasy, QIAshredder and DNase in QIAcube (all from Qiagen) automated system as per the recommended protocol. cDNA synthesis was performed using qScript cDNA synthesis kit (Quantabio) as per the recommended protocol. Quantitative PCR (qPCR) was done using Perfecta SYBR Green FastMix (Quantabio) and Applied Biosystems RT-PCR platform. TMEM59 was used as the control gene selected from RNAseq data on the basis of low variability across stimulated and control cells. RNA library prep for sequencing was performed as described below for intestinal epithelial cells.
Intestinal epithelial cell cultures
Human epithelial cell line (FHs 74 Int) was acquired from ATCC and maintained according to ATCC protocol using Hybri-Care Medium (cat# ATCC® 46-X™) with addition of 10% FBS (ATCC, cat# 30–2020, lot: 62144240), 5% Penicillin Streptomycin (Corning, REF 30–002-CI) and 30 ng/ml epidermal growth factor (EGF; Tonbo Biosciences, cat# 21–8356-M001). Passages used in experiments were P5–7.
FHs 74 Int maintained in Hybri-Care Medium (10% FBS, 5% penicillin Streptomycin, and 30 ng/ml EGF) was split according to ATCC protocol and seeded into 96 wells plates with 20 000 cells/well density (200 ul of media per well). 24 h after seeding into plate cells were stimulated with one of the following: interferon alpha (Universal Type I Interferon, Pbl assay science, cat# PHC4044), IFN gamma (Recombinant Human Interferon – gamma, Gibco, cat# PHC4031), IFN alpha and IFN gamma, or media (negative control). Stimulation concentrations of interferons used were: 500 units/mL for interferon alpha, 100 units/mL for IFN gamma. Cells were collected by lysis with RLT buffer (Qiagen, cat# 79216) 48 h after stimulation. RNA was extracted with RNeasy Mini Kit (Qiagen, cat# 74106) according to the manufacturer’s protocol, DNase treatment was not included as additional step.
For RNA sequencing library preparation, 500 to 1000 ng of total RNA was used with Wafergen Biosystems PrepX RNA-Seq Sample and Library Preparation Kits for the Apollo 324 NGS Library Prep System and sequenced on Illumina HiSeq 3000 (150 bp, single end) in the pool of total 28 samples per HiSeq lane. RNAseq data processing was performed as described above for biopsies. For comparison to E-CVID, expression of genes from E-CVID signature was compared between unstimulated cells and cells stimulated with both interferons as specified above. Genes that passed p value of 0.05 (FDR<0.15) and presented same direction of change as in E-CVID were selected as concordant.
Cells were lysed 48 h after IFNs stimulation using RLT Buffer (Qiagen). Concentrations of RNA measured with Qubit RNA BR Assay Kit (Life Technologies, cat#Q10211). cDNA prepared using iScript (Rio-Rad, cat# 1708891). Quantitative Real-Time PCR was done for the samples using QuantiFast SYBR Green PCR Kit (Qiagen, cat# 204056). qRT PCR set up used: samples were heated to 95°C, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. House-keeping gene used for normalization (SRSF11) was selected according to the procedure described in previous publication [50]. For list of primers see Supplementary Table S12.
Measurements of total cholesterol concentrations in FHs 74 Int cells
Before been collected, cells in each well were washed with PBS and then lysed with Lysis Buffer from Amplex® Red Cholesterol Assay Kit (Invitrogen, cat#A12216), sonicated on ice 2 times 10 s each. Total cholesterol concentrations were measured using Amplex® Red Cholesterol Assay Kit (Invitrogen, cat# A12216) and MyQubit Amplex® Red Cholesterol Assay Protocol (MAN0007532). Protein concentrations were measured in the same lysates using Qubit® Protein Assay Kit (Life Technologies, cat# Q33211) following the manufacturer’s protocol (MAN0002349). Normalization of cholesterol concentration levels was done by dividing acquired total cholesterol concentrations by protein concentrations for the same sample.
Norovirus detection in duodenal biopsies
Norovirus strains were genotyped by partial nucleotide sequencing of the polymerase gene. Briefly, total RNA isolated from duodenal biopsies was reverse transcribed and fragments of the norovirus polymerase gene were amplified using primers designed to detect both sapo- and noroviruses [51]. In case of PCR products of expected size, they were purified using PCR MinElute kit (Qiagen) and sequenced using Sanger sequencing in ABI 3730 machine to confirm the norovirus. Derived nucleotide sequences were submitted to the online Norovirus Genotyping Tool (http://www.rivm.nl/mpf/norovirus/typingtool). In addition, to identify closely related strains, the nucleotide sequences were compared with those available in GenBank using BLAST software (http://blast.ncbi.nlm.nih.gov).
Data Sharing
All sequencing data were deposited to European Nucleotide Archive under accession number PRJEB18593.
Supplementary Material
Acknowledgements
This research was supported by the grant NIH U01 AI109695 and by the Intramural Research Program of the NIH, NIAID (MY, IJF, WS). NS was also supported by NIH R01 DK103761. We thank Oregon State University Center for Genome Research and Biocomputing (CGRB) for sequencing services and technical support.
This project also has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bonilla FA, Barlan I, Chapel H, Costa-Carvalho BT, Cunningham-Rundles C, de la Morena MT, Espinosa-Rosales FJ, Hammarstrom L, Nonoyama S, Quinti I, Routes JM, Tang ML, Warnatz K, International Consensus Document (ICON): Common Variable Immunodeficiency Disorders, The journal of allergy and clinical immunology. In practice, 4 (2016) 38–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ameratunga R, Brewerton M, Slade C, Jordan A, Gillis D, Steele R, Koopmans W, Woon ST, Comparison of Diagnostic Criteria for Common Variable Immunodeficiency Disorder, Frontiers in Immunology, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jolles S, The variable in common variable immunodeficiency: a disease of complex phenotypes, The journal of allergy and clinical immunology. In practice, 1 (2013) 545–556; quiz 557. [DOI] [PubMed] [Google Scholar]
- [4].Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, Kim CE, Hou C, Wang F, Chiavacci R, Kugathasan S, Sleasman JW, Baldassano R, Perez EE, Chapel H, Cunningham-Rundles C, Hakonarson H, Genome-wide association identifies diverse causes of common variable immunodeficiency, The Journal of allergy and clinical immunology, 127 (2011) 1360–1367.e1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, Fieschi C, Thon V, Abedi MR, Hammarstrom L, Common variable immunodeficiency disorders: division into distinct clinical phenotypes, Blood, 112 (2008) 277–286. [DOI] [PubMed] [Google Scholar]
- [6].Mouillot G, Carmagnat M, Gerard L, Garnier JL, Fieschi C, Vince N, Karlin L, Viallard JF, Jaussaud R, Boileau J, Donadieu J, Gardembas M, Schleinitz N, Suarez F, Hachulla E, Delavigne K, Morisset M, Jacquot S, Just N, Galicier L, Charron D, Debre P, Oksenhendler E, Rabian C, Group DS, B-cell and T-cell phenotypes in CVID patients correlate with the clinical phenotype of the disease, Journal of clinical immunology, 30 (2010) 746–755. [DOI] [PubMed] [Google Scholar]
- [7].Malamut G, Verkarre V, Suarez F, Viallard JF, Lascaux AS, Cosnes J, Bouhnik Y, Lambotte O, Bechade D, Ziol M, Lavergne A, Hermine O, Cerf-Bensussan N, Cellier C, The enteropathy associated with common variable immunodeficiency: the delineated frontiers with celiac disease, The American journal of gastroenterology, 105 (2010) 2262–2275. [DOI] [PubMed] [Google Scholar]
- [8].Jorgensen SF, Troseid M, Kummen M, Anmarkrud JA, Michelsen AE, Osnes LT, Holm K, Hoivik ML, Rashidi A, Dahl CP, Vesterhus M, Halvorsen B, Mollnes TE, Berge RK, Moum B, Lundin KE, Fevang B, Ueland T, Karlsen TH, Aukrust P, Hov JR, Altered gut microbiota profile in common variable immunodeficiency associates with levels of lipopolysaccharide and markers of systemic immune activation, Mucosal immunology, 9 (2016) 1455–1465. [DOI] [PubMed] [Google Scholar]
- [9].Mannon PJ, Fuss IJ, Dill S, Friend J, Groden C, Hornung R, Yang Z, Yi C, Quezado M, Brown M, Strober W, Excess IL-12 but not IL-23 accompanies the inflammatory bowel disease associated with common variable immunodeficiency, Gastroenterology, 131 (2006) 748–756. [DOI] [PubMed] [Google Scholar]
- [10].Cols M, Rahman A, Maglione PJ, Garcia-Carmona Y, Simchoni N, Ko HB, Radigan L, Cerutti A, Blankenship D, Pascual V, Cunningham-Rundles C, Expansion of inflammatory innate lymphoid cells in patients with common variable immune deficiency, The Journal of allergy and clinical immunology, 137 (2016) 1206–1215 e1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, Fraser-Liggett C, Matzinger P, Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut, Nature medicine, 17 (2011) 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, Schuster M, Hsiao W, Matzinger P, Shulzhenko N, Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks, Gut, 64 (2015) 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL, Densely interconnected transcriptional circuits control cell states in human hematopoiesis, Cell, 144 (2011) 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Martinez FO, Gordon S, Locati M, Mantovani A, Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression, Journal of immunology (Baltimore, Md. : 1950), 177 (2006) 7303–7311. [DOI] [PubMed] [Google Scholar]
- [15].Washington K, Stenzel TT, Buckley RH, Gottfried MR, Gastrointestinal pathology in patients with common variable immunodeficiency and X-linked agammaglobulinemia, The American journal of surgical pathology, 20 (1996) 1240–1252. [DOI] [PubMed] [Google Scholar]
- [16].Macpherson AJ, Slack E, The functional interactions of commensal bacteria with intestinal secretory IgA, Current opinion in gastroenterology, 23 (2007) 673–678. [DOI] [PubMed] [Google Scholar]
- [17].Pabst O, New concepts in the generation and functions of IgA, Nature reviews. Immunology, 12 (2012) 821–832. [DOI] [PubMed] [Google Scholar]
- [18].Mantis NJ, Forbes SJ, Secretory IgA: arresting microbial pathogens at epithelial borders, Immunological investigations, 39 (2010) 383–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yambartsev A, Perlin MA, Kovchegov Y, Shulzhenko N, Mine KL, Dong X, Morgun A, Unexpected links reflect the noise in networks, Biology direct, 11 (2016) 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dong X, Yambartsev A, Ramsey SA, Thomas LD, Shulzhenko N, Morgun A, Reverse enGENEering of Regulatory Networks from Big Data: A Roadmap for Biologists, Bioinformatics and biology insights, 9 (2015) 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Greer R, Dong X, Morgun A, Shulzhenko N, Investigating a holobiont: Microbiota perturbations and transkingdom networks, Gut microbes, 7 (2016) 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Greer RL, Dong X, Moraes AC, Zielke RA, Fernandes GR, Peremyslova E, Vasquez-Perez S, Schoenborn AA, Gomes EP, Pereira AC, Ferreira SR, Yao M, Fuss IJ, Strober W, Sikora AE, Taylor GA, Gulati AS, Morgun A, Shulzhenko N, Akkermansia muciniphila mediates negative effects of IFNgamma on glucose metabolism, Nature communications, 7 (2016) 13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Peleg AY, Seifert H, Paterson DL, Acinetobacter baumannii: emergence of a successful pathogen, Clinical microbiology reviews, 21 (2008) 538–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Antunes LC, Visca P, Towner KJ, Acinetobacter baumannii: evolution of a global pathogen, Pathogens and disease, 71 (2014) 292–301. [DOI] [PubMed] [Google Scholar]
- [25].Pistiki A, Galani I, Pyleris E, Barbatzas C, Pimentel M, Giamarellos-Bourboulis EJ, In vitro activity of rifaximin against isolates from patients with small intestinal bacterial overgrowth, International journal of antimicrobial agents, 43 (2014) 236–241. [DOI] [PubMed] [Google Scholar]
- [26].Corbella X, Pujol M, Ayats J, Sendra M, Ardanuy C, Dominguez MA, Linares J, Ariza J, Gudiol F, Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii, Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 23 (1996) 329–334. [DOI] [PubMed] [Google Scholar]
- [27].Winkelstein JA, Marino MC, Lederman HM, Jones SM, Sullivan K, Burks AW, Conley ME, Cunningham-Rundles C, Ochs HD, X-linked agammaglobulinemia: report on a United States registry of 201 patients, Medicine, 85 (2006) 193–202. [DOI] [PubMed] [Google Scholar]
- [28].Herbst EW, Armbruster M, Rump JA, Buscher HP, Peter HH, Intestinal B cell defects in common variable immunodeficiency, Clinical and experimental immunology, 95 (1994) 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van de Ven AA, Janssen WJ, Schulz LS, van Loon AM, Voorkamp K, Sanders EA, Kusters JG, Nierkens S, Boes M, Wensing AM, van Montfrans JM, Increased prevalence of gastrointestinal viruses and diminished secretory immunoglobulin a levels in antibody deficiencies, Journal of clinical immunology, 34 (2014) 962–970. [DOI] [PubMed] [Google Scholar]
- [30].Klemola T, Immunohistochemical findings in the intestine of IgA-deficient persons: number of intraepithelial T lymphocytes is increased, Journal of pediatric gastroenterology and nutrition, 7 (1988) 537–543. [DOI] [PubMed] [Google Scholar]
- [31].Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, Pons N, Le Chatelier E, Levenez F, Kennedy S, Galleron N, de Barros JP, Malphettes M, Galicier L, Boutboul D, Mathian A, Miyara M, Oksenhendler E, Amoura Z, Dore J, Fieschi C, Ehrlich SD, Larsen M, Gorochov G, Microbial ecology perturbation in human IgA deficiency, Science translational medicine, 10 (2018). [DOI] [PubMed] [Google Scholar]
- [32].Vlkova M, Ticha O, Nechvatalova J, Kalina T, Litzman J, Mauri C, Blair PA, Regulatory B cells in CVID patients fail to suppress multifunctional IFN-gamma+ TNFalpha+ CD4+ T cells differentiation, Clinical immunology, 160 (2015) 292–300. [DOI] [PubMed] [Google Scholar]
- [33].Jorgensen SF, Reims HM, Frydenlund D, Holm K, Paulsen V, Michelsen AE, Jorgensen KK, Osnes LT, Bratlie J, Eide TJ, Dahl CP, Holter E, Tronstad RR, Hanevik K, Brattbakk HR, Kaveh F, Fiskerstrand T, Kran AB, Ueland T, Karlsen TH, Aukrust P, Lundin KE, Fevang B, A Cross-Sectional Study of the Prevalence of Gastrointestinal Symptoms and Pathology in Patients With Common Variable Immunodeficiency, The American journal of gastroenterology, 111 (2016) 1467–1475. [DOI] [PubMed] [Google Scholar]
- [34].Woodward JM, Gkrania-Klotsas E, Cordero-Ng AY, Aravinthan A, Bandoh BN, Liu H, Davies S, Zhang H, Stevenson P, Curran MD, Kumararatne D, The role of chronic norovirus infection in the enteropathy associated with common variable immunodeficiency, The American journal of gastroenterology, 110 (2015) 320–327. [DOI] [PubMed] [Google Scholar]
- [35].Karst SM, Wobus CE, A working model of how noroviruses infect the intestine, PLoS pathogens, 11 (2015) e1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hakansson A, Molin G, Gut microbiota and inflammation, Nutrients, 3 (2011) 637–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chiu CM, Huang WC, Weng SL, Tseng HC, Liang C, Wang WC, Yang T, Yang TL, Weng CT, Chang TH, Huang HD, Systematic analysis of the association between gut flora and obesity through high-throughput sequencing and bioinformatics approaches, BioMed research international, 2014 (2014) 906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sebeny PJ, Riddle MS, Petersen K, Acinetobacter baumannii skin and soft-tissue infection associated with war trauma, Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 47 (2008) 444–449. [DOI] [PubMed] [Google Scholar]
- [39].Davis KA, Moran KA, McAllister CK, Gray PJ, Multidrug-resistant Acinetobacter extremity infections in soldiers, Emerging infectious diseases, 11 (2005) 1218–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maragakis LL, Perl TM, Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options, Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 46 (2008) 1254–1263. [DOI] [PubMed] [Google Scholar]
- [41].Garnacho-Montero J, Ortiz-Leyba C, Fernandez-Hinojosa E, Aldabo-Pallas T, Cayuela A, Marquez-Vacaro JA, Garcia-Curiel A, Jimenez-Jimenez FJ, Acinetobacter baumannii ventilator-associated pneumonia: epidemiological and clinical findings, Intensive care medicine, 31 (2005) 649–655. [DOI] [PubMed] [Google Scholar]
- [42].Young LS, Sabel AL, Price CS, Epidemiologic, clinical, and economic evaluation of an outbreak of clonal multidrug-resistant Acinetobacter baumannii infection in a surgical intensive care unit, Infection control and hospital epidemiology, 28 (2007) 1247–1254. [DOI] [PubMed] [Google Scholar]
- [43].Tang MS, Bowcutt R, Leung JM, Wolff MJ, Gundra UM, Hudesman D, Malter LB, Poles MA, Chen LA, Pei Z, Neto AG, Abidi WM, Ullman T, Mayer L, Bonneau RA, Cho I, Loke P, Integrated Analysis of Biopsies from Inflammatory Bowel Disease Patients Identifies SAA1 as a Link Between Mucosal Microbes with TH17 and TH22 Cells, Inflamm Bowel Dis, 23 (2017) 1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Petersen JA CR, Specificity of the Adaptive Immune Response to the Gut Microbiota, in: Fagarasan A CA (Ed.) In Advances in Immunology, Mucosal Immunity., vol. 107, Academic Press; 2010. [DOI] [PubMed] [Google Scholar]
- [45].Park J, Munagala I, Xu H, Blankenship D, Maffucci P, Chaussabel D, Banchereau J, Pascual V, Cunningham-Rundles C, Interferon signature in the blood in inflammatory common variable immune deficiency, PloS one, 8 (2013) e74893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nature methods, 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nature protocols, 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [48].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mine KL, Shulzhenko N, Yambartsev A, Rochman M, Sanson GF, Lando M, Varma S, Skinner J, Volfovsky N, Deng T, Brenna SM, Carvalho CR, Ribalta JC, Bustin M, Matzinger P, Silva ID, Lyng H, Gerbase-DeLima M, Morgun A, Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer, Nature communications, 4 (2013) 1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shulzhenko N, Yambartsev A, Goncalves-Primo A, Gerbase-DeLima M, Morgun A, Selection of control genes for quantitative RT-PCR based on microarray data, Biochemical and biophysical research communications, 337 (2005) 306–312. [DOI] [PubMed] [Google Scholar]
- [51].Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X, Genetic diversity among sapoviruses, Archives of virology, 149 (2004) 1309–1323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.