Abstract
Personalized medicine is partly based on biomarker-guided diagnostics, therapy and prognosis, which is becoming an unavoidable concept in modern cardiology. However, the clinical significance of single biomarker studies is rather limited. A promising novel approach involves combining multiple markers into a multiplex panel, which could refine the management of a particular patient with cardiovascular pathology. Two principally different assay formats have been developed to facilitate simultaneous quantification of multiple antigens: planar array assays and microbead assays. These approaches may help to better evaluate the complexity and dynamic nature of pathologic processes and offer substantial cost and sample savings compared with traditional enzyme-linked immunosorbent assay (ELISA) measurements. However, a multiplex multimarker approach cannot become a generally disseminated method until analytical problems are solved and further studies confirming improved clinical outcomes are accomplished. These drawbacks underlie the fact that a limited number of systematic studies are available regarding the use of a multiplex biomarker approach in cardiovascular medicine to date. Our perspective underscores the significant potential of the use of the multiplex approach in a wider conceptual framework under the close cooperation of clinical and experimental cardiologists, pathophysiologists and biochemists so that the personalized approach based on standardized multimarker testing may improve the management of various cardiovascular pathologies and become a ubiquitous partner of population-derived evidence-based medicine.
Keywords: cardiac biomarkers, multimarker approach, Luminex, planar array assays, microbead assays, cardiac troponins, personalized medicine
Introduction
We are living in a time of large clinical trials, and evidence-based medicine seems to be the dominant therapeutic approach to patients with cardiovascular disease. On the other hand, we are becoming aware that any population is different from the subset of patients in clinical trials given that the average patient is older, suffers from a wide variety of non-cardiovascular disorders and takes a number of different therapies that interfere with cardiovascular treatment. Thus, personalized medicine is a broad and rapidly spreading concept of medicine in the 21st century and can be defined as follows: Personalized medicine is considered to be the tailoring of medical treatment to the special characteristics of an individual patient. The principle aim is not to create drugs or treatment strategies that are unique to a particular patient but rather to refine the approach to the individual patient (or group of patients) based on the precise characterization of his pathologic condition or his response to a specific treatment. The management of a precisely defined cohort of patients can then be focused on those who will benefit, sparing expenses and side effects for those who will not1. To date, the use of personalized medicine in cardiology has been limited to only several fields, eg, genetic diseases, pharmacogenomics and biomarker research2:
- Pharmacogenomics in cardiovascular medicine has revealed polymorphisms affecting responses to various drugs, such as warfarin, β-blockers, ACE inhibitors, angiotensin II receptor blockers, diuretics or statins. However, only the influence of CYP2C9 and VKORC1 polymorphisms on warfarin susceptibility has been established in clinical practice.
- Plasma natriuretic peptides (BNP and NT-proBNP) not only serve as principle biomarkers in the diagnosis of heart failure but also reflect the efficiency of heart failure treatment. Moreover, the vasodilator and diuretic natures of natriuretic peptides are being considered in clinical trials.
- Findings of increased levels of tumor necrosis factor (TNFα) or other cytokines in patients with heart failure induced sophisticated approaches to reduce inflammation (now considered an important goal in the pathophysiology of a failing heart), such as transcriptional/translational approaches, targeted anticytokine treatment or immunomodulation therapy. Improving the knowledge on the pathophysiological background of the particular cardiovascular disorder may refine the therapeutic intervention.
Numerous studies searching for various biochemical markers have emerged in the last two decades. In a narrow sense, biomarkers are circulating molecules that should provide pathophysiologic insights and aid to establish a diagnosis, refine the prognosis and modify the treatment. The ideal biomarker exhibits high sensitivity and/or specificity that reflects the severity of the disease. The ideal biomarker helps in clinical decision making, and its levels are reduced with effective therapy. However, the clinical significance of biomarker studies is limited in general given that they focus on individual biomarkers representing only one of several features within a particular pathologic condition3,4. It is important to gain comprehensive information on the pathologic mechanisms underlying the various alterations of the diseased cardiovascular system, such as inflammation, oxidative stress, apoptosis, or neurohumoral activation, resulting in hypertension, coronary heart disease, atrial fibrillation or heart remodeling and failure. One promising approach for maximizing the utility of cardiovascular biomarkers involves combining multiple markers into a multimarker panel to increase their diagnostic and prognostic value in the setting of primary or secondary prevention5.
Two methodological multimarker panel approaches
Two basic assay formats have been developed to facilitate simultaneous quantification of multiple antigens: planar array assays and microbead assays. In the first approach, different capture antibodies are spotted at defined positions on a two-dimensional array (Biochip Array Technology – Randox, Crumlin, United Kingdom). In the second approach, the capture antibodies are conjugated to different populations of microbeads that can be distinguished by their fluorescence intensity in a flow cytometer (Luminex - Austin, Texas, USA)6.
Biochip Array Technology
This method works by combining a panel of related tests on a single biochip with a single set of reagents, controls and calibrators (Figure 1). Only one single undivided sample is used. Each biochip includes 25 discrete test regions, and each region holds an individual test. Thus, 23 tests can be performed, with two reserved for internal quality control, representing another unique Biochip Array Technology feature. The results are read by a CCD camera and custom image-processing software6.
Figure 1.
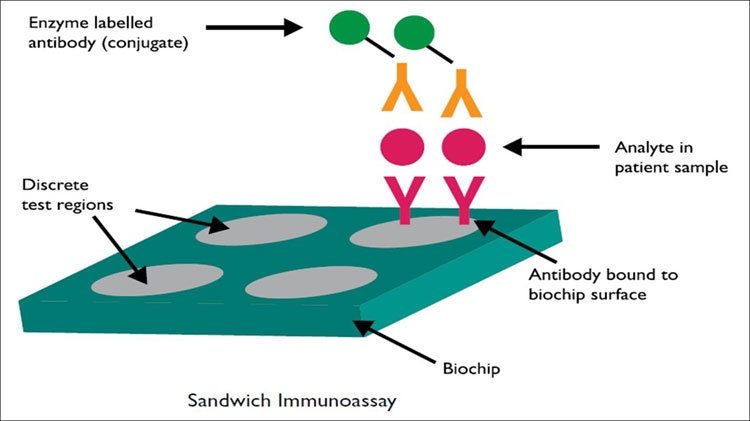
Principle of the sandwich biochip assay. Evidence Investigator Operation Manual, Picture is published with the agreement of Randox company. In a sandwich immunoassay, the solid phase (biochip) of the assay is spotted with antibodies. When the antigen is added in the first step, the antigen binds to the antibody. Then, a second antibody or conjugate is added, and the conjugate is labeled with an enzyme soluble substrate to produce a chemiluminescent signal.
Luminex® xMAP® technology
This is a unique combination of flow cytometry and sandwich immunoassays. This technique involves 100 distinctly colored bead sets created using two fluorescent dyes at distinct ratios. Each bead set can be coated with a reagent specific to a particular bioassay, allowing the capture and detection of specific analytes from a sample (Figure 2). The use of different colored beads enables the simultaneous detection of many analytes (up to 100) in the same sample. Imaging or laser excitation is used to determine the different assays by bead colors and analyte concentration by measuring the reporter dye fluorescence. The main advantages of multiplex assay compared with ELISA are the low sample volume (20 μL), the identical conditions of analysis, cost and time savings, and the comprehensive information obtained about the pathologic process6.
Figure 2.
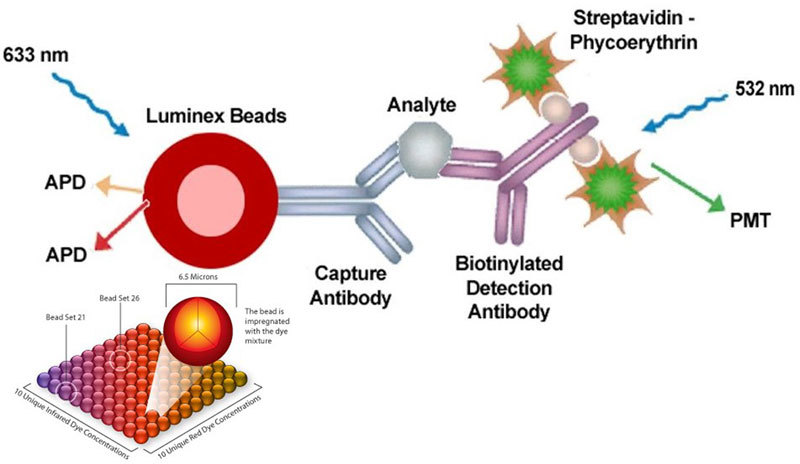
Principle of Luminex. Picture is published with the agreement of Merck company. The Luminex® xMAP® technology is based on polystyrene or paramagnetic microspheres or beads that are internally dyed with red and infrared fluorophores of differing intensities. Individual bead sets are then coated with a capture antibody qualified for one specific analyte. Imaging or laser excitation is used to determine the different assays by bead colors and analyte concentration by measuring the reporter dye fluorescence.
Experimental studies using a multiplex biomarker approach
The data from experiments exploiting multiplex assay are rather sparse. However, Millipore released the Rat CVD 1 Panel, providing an important tool for studies on the pathogenesis of cardiovascular disease that can simultaneously detect 10 biomarkers: B-type natriuretic peptide (BNP), interleukin-6 (IL-6), monocyte chemoattractant protein-1 (MCP-1), plasminogen activator inhibitor 1 (PAI-1), tissue inhibitor of matrix metalloproteinases type I (TIMP-1), tumor necrosis factor alpha (TNFα), cardiac troponin I (cTnI), cardiac troponin T (cTnT), vascular endothelial growth factor (VEGF), and von Willebrand Factor (vWF). To the best of our knowledge, the Rat CVD panel was used for the first time in experimental cardiology studies in our laboratory. We aimed to evaluate the relationship of selected biomarkers describing the remodeling process of the hypertensive heart in L-NAME-induced hypertension in two subsequent periods of hypertension development (4 and 7 weeks of L-NAME-treatment). The BNP, TNFα and VEGF plasma levels did not differ significantly among groups. However, as measured by a highly sensitive ELISA, cTnT was markedly increased in L-NAME groups compared with the control7. The cardiac biomarkers cTnI, cTnT, and FABP3 were also measured in serum using rat kits on the Meso Scale Discovery (MSD) platform to detect myocardial alterations in the model of imatinib-induced cardiotoxicity in rats8.
Others studies using Luminex technology in experimental cardiology have predominantly focused on cytokine spectrum mapping. Four murine animal models of various cardiovascular pathologies (banding of the ascending aorta or the pulmonary artery, myocardial infarction and a cardiomyopathy model with inducible cardiomyocyte-specific knockout of the sarcoplasmic reticulum Ca2+-ATPase (SERCA2KO)) were compared to establish whether the expression of 25 cytokines is dependent on the etiology of myocardial hypertrophy and heart failure (HF). Circulating cytokine levels were not increased in mice 1 week after AB; however, substantial myocardial hypertrophy was present. After 1 week of AMI, only interleukin (IL)-18 was increased. In SERCA2KO mice with HF, the circulating levels of IL-1α, IL-2, IL-3, IL-6, IL-9, IL-10, IL-12p40, eotaxin, granulocyte-colony stimulating factor (G-CSF), interferon-γ, monocyte chemoattractant protein-1 and macrophage inflammatory protein-1β were increased. In mice with PB, the IL-1α, IL-6, G-CSF and monokine induced by gamma-interferon levels were increased. The serum levels of cytokines in mice with HF varied depending on the etiology. Increased serum levels of several cytokines were observed in models with increased right ventricular afterload, suggesting that the cytokine responses result primarily from systemic congestion9.
These studies indicate the possible use of the multimarker approach in experimental cardiology, thus facilitating translational medicine. In the past, the diagnosis of myocardial injury in the experiments was based mainly on histological examination at the end of experiments. However, myocardial biopsies are rarely performed in practice. Currently, we have the opportunity to monitor the same biomarkers using the same methods in both experimental and clinical medicine.
Clinical studies using a multimarker panel approach
Single biomarker studies have tended to focus only on single aspects of the pathologic process but have gradually started to include serial measurement of biomarkers to provide a more dynamic view of the pathologic process. Combining different biomarkers from distinct pathophysiological pathways can overcome some of the limitations of single marker measurements. Unfortunately, the first clinical studies using combinations of biomarkers for the prediction of both primary and secondary cardiovascular disease presented statistically significant results but usually only modest clinical improvements in prediction compared with a model with traditional risk factors10.
Blankenberg et al 11 evaluated thirty biomarkers from different pathophysiological pathways (lipid metabolism, inflammation, hemodynamic physiology, vascular function, oxidative stress, coagulation, renal function, angiogenesis, and myocardial necrosis) in 7915 people using single assays. No single biomarker consistently improved the risk estimation. However, the addition of a biomarker score, such as NT-proBNP, CRP and hs cTnI, to a conventional risk model improved the 10-year risk estimation for cardiovascular events.
Recent projects using multiplex panels have facilitated the “big data” concept. A hallmark of this approach is the combination of distinct sources and types of data. Historically, prediction models have relied on a limited number of specified parameters. Such models generally lack precision. These models may function in populations but not in individual patients. Big data analytics evaluate patterns associated with an outcome directly from a full range of associations and interactions among the data. The computer algorithm creates a unique phenotype by processing all of the data sources, compares the phenotype with numerous additional patients, suggests the patient's diagnosis, and indicates individualized risk contributing to decision making for therapeutic options12.
Halim et al 13 used a nested case-control design to examine the association of circulating proteins with the risk of death or AMI in a subset of patients enrolled in the MURDOCK Horizon 1 Cardiovascular Disease Study. More than 500 selected people underwent invasive coronary angiography at baseline. The investigators used techniques such as penalized logistic regression to select 6 of 53 proteins (matrix metalloproteinase-3, NT-proBNP, IL-6, intercellular adhesion molecule-1, sCD40L and IGFBP2) associated with death or AMI. In the model conditioned to include all clinical risk factors, only sCD40L was retained in ≥85% of the samples. In the model with simultaneous evaluation of biomarkers and clinical risk factors, all 6 biomarkers were retained14.
Furthermore, serum samples from 8401 participants in the Outcome Reduction with Initial Glargine Intervention trial were assayed for 237 biomarkers linked to CVD based on other research. The primary objectives of the ORIGIN study were to determine whether insulin glargine-mediated normoglycemia can reduce cardiovascular morbidity and/or mortality in people at high risk for vascular disease with either impaired fasting glucose (IFG), impaired glucose tolerance (IGT) or early type 2 diabetes. Ten novel biomarkers were identified in patients with dysglycemia that were each independent determinants of CV composite outcomes of nonfatal AMI, nonfatal stroke or CV death15. Moreover, 9 biomarkers were identified as independent determinants of the expanded composite outcomes, including heart failure hospitalization and revascularization. Although NT-proBNP, a well-known risk marker, was consistently identified in all analyses and perhaps serves as a good positive control, angiopoeitin-2 and glutathione S transferase-α were also consistently selected across multiple different modeling conditions, suggesting that these proteins may represent biological pathways contributing to CVD risk16.
Vistnes et al 17 demonstrated that the lack of pro-inflammatory cytokine mobilization (low levels of MCP1 and IL-1β and low IL-1β/IL-1ra ratios) predicts poor prognosis in patients with acute heart failure. Although the multiple biomarker approach is becoming popular, it must be emphasized that systematic data on clinical studies with multiplex panels are lacking, and such studies are eagerly awaited.
A number of factors could distort the results of these studies, eg, the type of population investigated, the biomarkers tested and their timing, the duration of follow-up, the choice of primary and secondary endpoints in a particular clinical study, and the statistical methods used10. Additionally, there are several challenges to overcome prior to the widespread integration of multiplex immunoassays in clinical practice: biomarker validation and standardization of immunoassay design; availability, stability, specificity and cross-reactivity of reagents; assay automation; and the use of new biostatistical methods for transformation of raw data into diagnostic results18.
Future perspectives
Biomarkers have become a widely accepted tool for specification of diagnosis, treatment and prognosis in patients with cardiovascular diseases. At present, the goal is not to search for novel molecules that could offer more diagnostic benefits. Our aim should be to identify an optimal combination of already well-established biomarkers in such a way that could yield the most comprehensive and precise information on cardiovascular pathology in a given patient. Such an approach may induce the potential for better evaluation of the complexity and dynamic nature of pathologic processes and offer substantial cost and sample savings compared with traditional ELISA measurements. The multimarker strategy involves employing a set of pathophysiologically different biomarkers and their interplay such that each marker may contribute independently by providing complementary or additional information, which represents a sophisticated approach to individualizing pharmacological intervention and outcome prediction. However, the multimarker approach is not expected to become a generally disseminated method until analytical problems are solved and further studies confirming improved clinical outcomes are accomplished.
For a particular cardiovascular pathology, the multiplex set should implement biomarkers respecting the following attributes:
1) The principle biomarker for the determination of diagnosis have to be addressed.
2) The biomarker (or a combination of them) characterizing the severity of the disease and the prognostic implications ought to be defined.
3) A cluster of biomarkers that reflects the impact of therapeutic interventions should be suggested.
4) A set of biomarkers reflecting peripheral organ damage, which is valuable in the advanced period of the particular disease, could be delineated.
Conclusions
How the measurement of multiple prognostic or diagnostic biomarkers should be implemented into modern clinical practice remains an open problem and may be expected one of the principle questions of future studies. This perspective has no ambition to serve as an overview of the current topic. The goal of this perspective is to consider the multiplex approach in a wider conceptual framework under the bilateral interactions between clinical and experimental cardiologists. The close cooperation of pathophysiologists, clinicians and biochemists is necessary for the implementation of a personalized approach based on multiplex biomarker panels in the management of various cardiovascular pathologies in everyday clinical practice, and this approach could serve as a logical partner of population-based evidence in large clinical trials.
Acknowledgements
Supported by the program PROGRES Q 40/5 and grant VEGA 1/0071/15.
References
- 1.US President's Council of Advisors on Science and Technology (PCAST) Priorities for Personalized Medicine. Washington DC: Executive Office of the President of United States; 2008.
- 2.Johnson JA, Cavallari LH. Pharmacogenetics and cardiovascular disease – implication for personalized medicine. Pharmacol Rev. 2013;65:987–1009. doi: 10.1124/pr.112.007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamcova M, Lencova-Popelova O, Jirkovsky E, Mazurova Y, Palicka V, Simko F, et al. Experimental determination of diagnostic window of cardiac troponins in the development of chronic anthracycline cardiotoxicity and estimation of its predictive value. Int J Cardiol. 2015;201:358–67. doi: 10.1016/j.ijcard.2015.07.103. [DOI] [PubMed] [Google Scholar]
- 4.Adamcova M, Popelova-Lencova O, Jirkovsky E, Simko F, Gersl V, Sterba M. Cardiac troponins: Translational biomarkers in cardiology: Theory and practice of cardiac troponin high-sensitivity assays. Biofactors. 2016;42:133–48. doi: 10.1002/biof.1261. [DOI] [PubMed] [Google Scholar]
- 5.Taqui S, Daniels LB. Putting it into perspective: multimarker panels for cardiovascular disease risk assessment. Biomark Med. 2013;7:317–27. doi: 10.2217/bmm.13.15. [DOI] [PubMed] [Google Scholar]
- 6.Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56:314–8. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamcova M, Ruzickova S, Simko F. Multiplexed immunoassays for simultaneous quantification of cardiovascular biomarkers in the model of HG-nitro-L-arginine methylester (L-NAME) hypertensive rat. J Physiol Pharmacol. 2013;64:211–7. [PubMed] [Google Scholar]
- 8.Herman E, Knapton A, Zhang J, Estis J, Todd J, Lipshultz S. The utility of serum biomarkers to detect myocardial alterations induced by Imatinib in rats. Pharmacol Res Perspect. 2014;2:e00015. doi: 10.1002/prp2.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vistnes M, Wæhre A, Nygard S, Sjaastad I, Andersson KB, Husberg C, et al. Circulating cytokine levels in mice with heart failure are etiology dependent. J Appl Physiol. 2010;108:1357–64. doi: 10.1152/japplphysiol.01084.2009. [DOI] [PubMed] [Google Scholar]
- 10.Lori LD, Maisel AS. Multiple marker approach to risk stratification in patients with stable coronary artery disease: to have or have not. Eur Heart J. 2010;31:2980–3. doi: 10.1093/eurheartj/ehq336. [DOI] [PubMed] [Google Scholar]
- 11.Blankenberg S, Zeller T, Saarela O, Havulinna AS, Kee F, Tunstall-Pedoe H, et al. Contribution of 30 biomarkers to 10-year cardiovascular risk estimation in 2 population cohorts: the Monica, Risk, Genetics, Archiving, and Monograph (MORGAM) biomarker project. Circulation. 2010;121:2388–97. doi: 10.1161/CIRCULATIONAHA.109.901413. [DOI] [PubMed] [Google Scholar]
- 12.Shah RU, Rumsfeld JS. Big data in cardiology. Eur Heart J. 2017;38:1865–7. doi: 10.1093/eurheartj/ehx284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halim SA, Neely ML, Pieper KS, Shah SH, Kraus WE, Hauser ER, et al. Simultaneous consideration of multiple candidate protein biomarkers for long-term risk for cardiovascular events. Circ Cardiovasc Genet. 2015;8:168–77. doi: 10.1161/CIRCGENETICS.113.000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta DK, Wang TJ. Biomarker discovery: searching for quality in quantity. Circ Cardiovasc Genet. 2015;8:8–10. doi: 10.1161/CIRCGENETICS.115.000973. [DOI] [PubMed] [Google Scholar]
- 15.Gerstein HC, Paré G, McQueen MJ, Haenel H, Lee SF, Pogue J, et al. Outcome reduction with initial glargine intervention trial investigators: Identifying novel biomarkers for cardiovascular events or death in people with dysglycemia. Circulation. 2015;132:2297–304. doi: 10.1161/CIRCULATIONAHA.115.015744. [DOI] [PubMed] [Google Scholar]
- 16.de Lemos JA, Rohatgi A, Ayers CR. Applying a big data approach to biomarker discovery: running before we walk? Circulation. 2015;132:2289–92. doi: 10.1161/CIRCULATIONAHA.115.019648. [DOI] [PubMed] [Google Scholar]
- 17.Vistnes M, Hoiseth AD, Rosjo H, Nygard S, Pettersen E, Soyseth V, et al. Lack of pro-inflammatory cytokine mobilization predicts poor prognosis in patients with acute heart failure. Cytokine. 2013;61:962–9. doi: 10.1016/j.cyto.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era : Potentials and pitfalls. Proteomics Clin Appl. 2015;9:406–22. doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]


