Abstract
miRNAs have shown promise as potential biomarkers for acute myocardial infarction (AMI). However, the current used quantitative real-time PCR (qRT-PCR) allows solely for relative expression of nucleic acids and it is susceptible to day-to-day variability, which has limited the validity of using the miRNAs as biomarkers. In this study we explored the technical qualities and diagnostic potential of a new technique, chip-based digital PCR, in quantifying the miRNAs in patients with AMI and ischaemia-reperfusion injury (I/R). In a dilution series of synthetic C.elegans-miR-39, chip-based digital PCR displayed a lower coefficient of variation (8.9% vs 46.3%) and a lower limit of detection (0.2 copies/μL vs 1.1 copies/μL) compared with qRT-PCR. In the serum collected from 24 patients with ST-elevation myocardial infarction (STEMI) and 20 patients with stable coronary artery disease (CAD) patients after percutaneous coronary intervention (PCI), we used qRT-PCR and multiplexed chip-based digital PCR to quantify the serum levels of miRNA-21 and miRNA-499 as they have been validated in AMI in prior studies. In STEMI, I/R injury was assessed via measurement of ST-segment resolution (ST-R). Chip-based digital PCR revealed a statistical significance in the difference of miR-21 levels between stable CAD and STEMI groups (118.8 copies/μL vs 59 copies/μL; P=0.0300), whereas qRT-PCR was unable to reach significance (136.4 copies/μL vs 122.8 copies/μL; P=0.2273). For miR-499 levels, both chip-based digital PCR and qRT-PCR revealed statistically significant differences between stable CAD and STEMI groups (2 copies/μL vs 8.5 copies/μL, P=0.0011; 0 copies/μL vs 19.4 copies/μL; P<0.0001). There was no association between miR-21/499 levels and ST-R post-PCI. Our results show that the chip-based digital PCR exhibits superior technical qualities and promises to be a superior method for quantifying miRNA levels in the circulation, which may become a more accurate and reproducible method for directly quantifying miRNAs, particularly for use in large multi-centre clinical trials.
Keywords: ST-segment elevation myocardial infarction, ichaemia-reperfusion injury, micro-RNAs, chip-based digital PCR, qRT-PCR
Introduction
micro-RNAs (miRNAs) are small non-coding RNAs (∼22 nucleotide-long) involved in the post-transcriptional regulation of genetic expression via alteration of messenger RNA translation1. miRNAs are known to be released into the circulation in both physiological and pathological states1. Given their proposed physiological roles, as well as their ability to be detected in a wide range of bodily fluids and their remarkable stability in the circulation, miRNAs have become attractive candidates as novel biomarkers2. In recent years, a variety of miRNAs have demonstrated potential as possible biomarkers for myocardial damage2,3,4. Of particular interest for this study are miRNA-21 and miRNA-499. miRNA-21 is known to have a plethora of functions in health and disease, specifically involved in both enhancing the function of stem and progenitor cells in AMI and contributing to post-AMI fibrogenesis and cellular hypertrophy5,6,7. miRNA-499 is found in myocardial and skeletal muscle, and has been physiologically linked to cardiac differentiation and I/R injury8,9,10. Both miRNAs have been validated as upregulated in AMI, and miRNA-499 has even shown potential as a biomarker in I/R injury11,12,13,14,15,16. Despite showing promise, a number of issues have arisen which have limited the validity of using these and other miRNAs as biomarkers17. Foremost amongst these limitations is the current method for quantifying miRNAs, quantitative real-time PCR (qRT-PCR)18. qRT-PCR allows solely for relative expression of nucleic acids and it is susceptible to day-to-day variability, limiting its ability to be used as a diagnostic medium or in validating miRNAs in large multi-centre clinical trials18. Digital PCR is an end-point analysis, which is able to absolutely quantify nucleic acids directly within a sample by partitioning a sample into mutliple smaller reactions, whereby each division may or may not contain a nucleic acid19. Current methods to allow for this include multi-well plates, capillaries, oil emulsions and arrays of miniaturised chambers19. Digital PCR has thus far exhibited a number of unique advantages over qRT-PCR, including decreased variability, increased day-to-day reproducibility and superior sensitivity20,21,22. It can be proposed that with these advantages, digital PCR would offer greater comparability and hence greater utility as a diagnostic machine and in large multi-centre clinical trials.
At current, there are a number of comercially available digital PCR systems, each utilising a different method to achieve absolute quantification. QuantStudio® 3D digital PCR is a relatively novel method of digital PCR, which loads a PCR sample onto a microchip, partitioning the sample across 20 000 reaction wells, essentially allowing 20 000 separate PCR reactions to take place23. The QuantStudio® system uses a Poisson statistical analysis of fluorescent signals from positive and negative wells to allow for absolute quantification, without need for referencing to a standard control23. A small number of studies have demonstrated its promising use as a robust tool for quantitative analysis of nucleic acids for a wide range of biomedical applications22,23,24,25. At current there is little evidence with regard to the technical precision of this system, and to the best of our knowledge, none examining its use in diagnosing AMI with validated miRNAs. This project aims to address the current limitations of using miRNAs as valid biomarkers for AMI and reperfusion injury, and to explore the role of chip-based digital PCR in quantifying these miRNAs.
Materials and methods
Synthetic oligonucleotide dilution series of C.elegans-miR-39
A dried 5′-phosphorylated synthetic oligonucleotide for Caenorrhabditis elegans-miR-39 (C.elegans-miR-39) (Integrated DNA Technologies) (5′-phos--3′), with a known starting concentration of 7.6 nmol was centrifuged dry before being diluted in molecular grade Tris-EDTA buffer (Thermo Fisher Scientific) to a final concentration of 10 pmol/μL. A 12-step dilution series beginning with 2500 copies/μL to 0 copies/μL was performed for C.elegans-miR-39. Each diluted sample then underwent reverse transcription using 4.16 μL/well nuclease-free water (Applichem Panreac), 1.50 μL/well RT buffer, 0.15 μL/well 100 nmol/L dNTP, 0.19 μL/well RNAse inhibitor, 1.00 μL/well multi-scribe reverse transcriptase and 3 μL/well specific RT primer (Applied Biosystems, Inc, ID: 000200). For each diluent, 10 μL master-mix and 5 μL sample were combined for each diluent. Samples underwent a 15 μL thermal cycling protocol using the C1000 Touch™ Thermal Cycler (BioRad) at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and were then held at 4°C.
For qRT-PCR, 7.67 μL/well nuclease-free water, 10 μL/well universal PCR master mix and 1 μL/well of a specific 20× hydrolysis primer/probe (Applied Biosystems, Inc. ID: 000200) were combined and partitioned into 18.67 μL/well with 1.33 μL/well RT product. Each sample was prepared in duplicate. Samples then underwent a 20 μL qRT-PCR protocol using the C1000 Touch™ Thermal Cycler (BioRad) at 95°C for 10 min, 95°C for 15 s and 60°C for 60 s, before being repeated for 39 more cycles and held at 4°C. All qRT-PCR data was analysed using CFX Manager™ (BioRad).
For chip-based digital PCR, 1.525 μL/well nuclease-free water, 7.25 μL/well Quantstudio® 3D supermix (ThermoFisher) and 0.725 μL/well 20× specific hydrolysis primer/probe were combined before being partitioned into 9.5 μL/well with 5 μL/well RT product. Thereafter, 14.5 μL of each sample were loaded onto a chip and transferred to the ProFlex™ 2× Flat PCR System (ThermoFisher) and underwent thermal cycling set for 14.5 μL at 96°C for 10 min, 60°C for 2 min, 98°C for 30 s, with the cycling being repeated for 39 more cycles, with a final extension at 60°C for 2 more minutes and then held at 10 °C. Chips were then read by the QuantStudio™ 3D Digital PCR Instrument (ThermoFisher).
The technical qualities of chip-based digital PCR versus qRT-PCR were compared statistically by a number of methods.
The coefficient of variation was calculated by the following equation:
The CV% was calculated for each dilution of the oligonucleotide for chip-based digital PCR. Values with no-call were set at 0 copies/μL.
Before calculating the CV% for each diluent in qRT-PCR, the Ct values were converted to absolute copies using the Pfaffl analysis26:
Where E target represents the efficiency of each individual qRT-PCR run according to the slope of the line of best fit:
The control for the Pfaffl equation was calculated by calculating the mean of the highest concentration (i.e. 2500 copies/μL), and comparing that to each sample. Each sample could then be converted to absolute copies by the following equation:
The qRT-PCR data could then be analysed for CV% and compared with the absolute quantification determined by chip-based digital PCR. Reduction in CV% for each diluent across each day was calculated by the equation:
Lower limit of linear range (LLLR) was determined by runs-testing27, removing successive diluton points until the P-value was >0.05 (Prism Version 5.0c software). The limit of detection (LOD) was defined as the lowest concentration that could be quantified, and was calculated by the following equation28:
 |
The limit of quantification (LOQ) was defined as the lowest concentration tested that remained above or equal to both the LLLR and the LOD.
Linearity for chip-based digital PCR and qRT-PCR for the dilution series was assessed using the r-value for the line of best fit (GraphPad Prism v5.0).
Values of quantification are presented as log transformed values. Calculations for CV%, LOD, LOQ, LLLR and linearity were calculated according to the absolute quantification reported.
Patient population
In this single-centre prospective study, peripheral venous blood was obtained from 24 STEMI patients and 20 patients with stable CAD enrolled between October 2013 and August 2015. Blood was obtained 4.75 (IQR: 3-7) h after PCI. Patients with STEMI as evidenced by ST-elevation >0.1 mV in at least two contiguous leads were included in this study. Major exclusion criteria included previous heart failure (known LVEF <30%), and cardiogenic shock. The final study population consisted of 44 patients. The protocol of this study conforms to the ethical guidelines of the 1975 Declaration of Helsinki and was henceforth approved by the institutional ethical commitee of the Universitätsklinikum Freiburg (CF16/1546-2016000808). Written informed consent was obtained from all patients.
Percutaneous coronary intervention
PCI was performed with standard catheters. All STEMI patients received aspirin (minimum of 250 mg) and an ADP receptor blocker (prasugrel 60 mg, ticagrelor 180 mg or clopidogrel 600 mg). Unfractionated heparin (5000 U) was administered prior to angiography. Eptifibatide was given at the discretion of the cardiologist.
miRNA extraction
Before undergoing miRNA extraction, all serum samples were spiked with 5 μL of 9.635×105 copies/μL C.elegans-miR-39 to act as an endogenous control. Serum samples underwent miRNA extraction according to the manufacturer's instructions (Qiagen).
Reverse transcription, qRT-PCR and Quantstudio® 3D Digital PCR
Samples underwent reverse transcription, qRT-PCR and chip-based digital PCR according to the above-mentioned protocols. Every sample also underwent separate RT and qRT-PCR and chip-based digital PCR for C.elegans-miR-39 to act as an endogenous control. Chip-based digital PCR was performed multiplexed by using a mastermix composed of 0.8 μL/well nuclease-free water, 7.25 μL/well Quantstudio® 3D supermix (ThermoFisher), 0.725 μL/well specific FAM labelled hydrolysis probe and primer set for either hsa-miR-21-5p or hsa-miR-499-5p (ThermoFisher) and 0.725 μL/well specific VIC labelled hydrolysis probe and primer set for C.elegans-miR-39 VIC labelled (ThermoFisher; ID: 000200). We then normalised the data across samples using a median normalisation procedure (normalisation factor = median value of all C.elegans-miR-39 measurements/C.elegans-miR-39 value for the given sample)26.
Ischaemia-reperfusion injury
ECGs were analysed within the STEMI group both before and 2.25 h (1.5–3.5) after PCI. In keeping with prior studies, we used Schröder's criteria and defined ST-segment resolution ≥70% as complete resolution29,30,31. Patients who fulfilled this criterion were defined as adequately reperfused, whereas those who did not fulfill this criterion were defined as having suffered significant I/R injury. Four of the STEMI patients had to be excluded from analysis, due to a lack of required ECGs in the hospital's database.
Statistical analysis
Continuous variables are presented as mean±standard deviation (SD) if found to follow a Gaussian distribution according to a D'Agostino-Pearson omnibus normality test, or as median and interquartile range if found to follow a non-Gaussian distritbution. Normally distributed variables were compared using the Student's t-test and non-normally distributed variables using the Mann-Whitney U test. Categorical variables are presented as percentages. A Chi-square analysis was performed to compare categorical variables.
A receiver operating characteristic (ROC) analyis was performed for sensitivity and specificity, with the endpoint analysis being the area under the curve (AUC).
miR-21/499 levels between STEMI patients deemed to have suffered I/R injury compared to those who reached adequate reperfusion were compared using a Mann-Whitney U test, and ROC analyses were performed for each miRNA.
Analyses for correlations between clinical characteristics and miR-21 and miR-499 levels were performed by Pearson analysis for parametric data or a Spearman test for non-parametric data.
A P-value of ≤0.05 was considered statistically significant for all analyses except for the analysis of correlation. This particular analysis involved multiple comparisons, thus requiring correction via Bonferroni's method. This set the new statistically significant P-value at 0.0029 for the correlation analysis.
All analyses were performed using Graph Pad Prism v5.0 (GraphPad Software, Inc, La Jolla, CA, USA).
Results
Dilution series of synthetic C.elegans-miR-39
Chip-based digital PCR demonstrated a lower coefficient of variation between most dilution points (Figure 1A). Chip-based digital PCR had an average CV% of 8.9%, compared to an average CV% of 46.3% for qRT-PCR, culminating in an average decrease in CV% of 70.7%. Chip-based digital PCR also demonstrated greater linearity, with an r 2 of 0.9969 compared to an r 2 of 0.9787 for qRT-PCR (Figure 1B).
Figure 1.
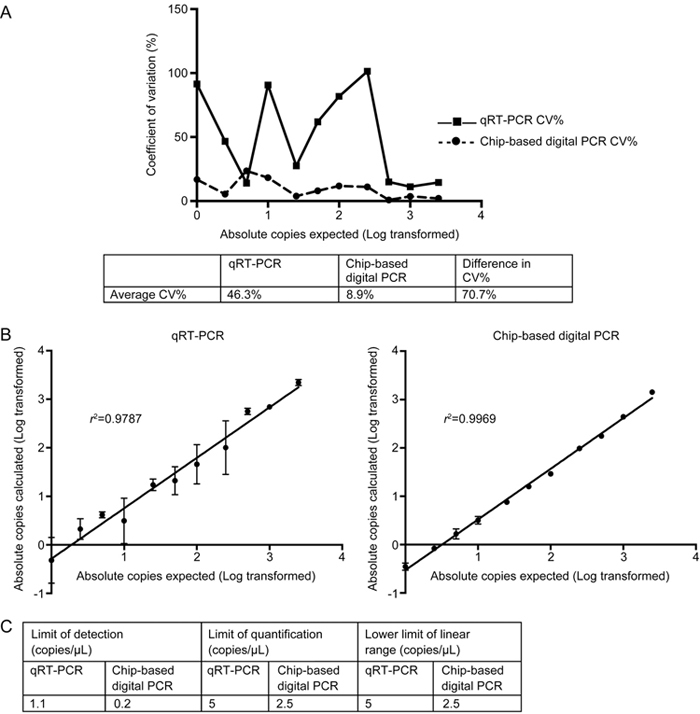
Technical qualities of chip-based digital PCR compared to qRT-PCR, including coefficient of variation (CV%), linearity, limit of detection (LOD), limit of quantification (LOQ) and lower limit of linear range (LLLR) for a dilution series of C.elegans-miR-39 performed in duplicate. (A) Trendline comparing coefficient of variation (CV%) for qRT-PCR and chip-based digital PCR at different concentration points with corresponding table demonstrating average CV% and difference in CV% (B) Duplicate measurements are presented log transformed as absolute copies for each dilution point reported for qRT-PCR and chip-based digital PCR with corresponding linearity and goodness-of-fit (r 2-value). Data are presented as mean±SD (C) LOD, LOQ and LLLR for qRT-PCR and chip-based digital PCR.
Finally, chip-based digital PCR demonstrated a superior LOD, LOQ and LLLR compared to qRT-PCR (Figure 1C).
Through absolute quantification, we noted measurements by chip-based digital PCR corresponding to 57.6% of the expected copy number.
Patient characteristics
The patient population consisted of 73% men and the average age across all patients was 65.7±SD 13.4 years. Table 1 presents the baseline characteristics of STEMI and stable CAD groups. A higher level of the cardiac biomarkers CK and cTnT at peak were demonstrated in STEMI patients. STEMI patients were more likely to receive prasugrel, ticagrelor and eptifibatide. Stable CAD patients were more likely to be overweight. There were no other significant differences in baseline clinical characteristics.
Table 1.
Main patient characteristics.
| Characteristic | All Patients (n=44) | STEMI Patients (n=24) | Stable CAD Patients (n=22) | P-value |
|---|---|---|---|---|
| Age | 65.7±13.4 | 64.9±13.8 | 66.7±13.1 | 0.6651 |
| Men | 73% | 79% | 65% | 0.3837 |
| DM | 32% | 25% | 40% | 0.1171 |
| HTN | 57% | 54% | 60% | 0.7249 |
| Dyslipidaemia | 50% | 38% | 60% | 0.0833 |
| Smoker | 48% | 38% | 55% | 0.1671 |
| Family history | 34% | 33% | 35% | 0.8887 |
| Overweight | 36% | 21% | 55% | 0.0010** |
| Aspirin | 84% | 96% | 70% | 0.1418 |
| Clopidogrel | 34% | 25% | 40% | 0.1171 |
| Prasugrel | 32% | 54% | 5% | 0.0001*** |
| Ticagrelor | 25% | 42% | 5% | 0.0001*** |
| Eptifibatide | 9% | 17% | 0% | 0.0001*** |
| Rivaroxaban | 7% | 4% | 10% | 0.1675 |
| Statin | 61% | 54% | 70% | 0.3031 |
| Number of coronaries affected | ||||
| 1 | - | 42% | - | - |
| 2 | - | 16% | - | - |
| 3 | - | 42% | - | - |
| Diseased coronary | ||||
| LAD | - | 71% | - | - |
| LCA | - | 4% | - | - |
| RCA | - | 63% | - | - |
| Other | - | 58% | - | - |
| Serum creatinine (mg/dL) | 0.98 (0.875–1.165) | 1.02 (0.905–1.24) | 0.93 (0.85–1.01) | 0.1376 |
| CK Peak (ng/dL) | 1214.3 +/− 1515.8 | 1198 (339.75–2536.25) | 126 (88.5–159) | 0.0001*** |
| cTnT Peak (ng/dL) | 0.3105 (0.0775–4.535) | 1.695 (0.153–5.4475) | 0.018 (0.011–0.02375) | 0.0001*** |
| CK-MB Peak (ng/dL) | - | 196 (58–542) | - | - |
| LVEF(%) | 48% (40%–50%) | 48.75% (43.1%–50%) | 45% (32.75%–55%) | 0.6780 |
| CRP (mg/L) | 7 (3–42.75) | 10.5 (4-50.5) | 3 (3–6) | 0.0853 |
| Time blood taken after PCI (h) | 5 +/− 2.7 | 5.4 +/− 3 | 4.5 +/− 2.2 | 0.3279 |
| Anterior Infarction | - | 29% | - | - |
DM: Diabetes mellitus; HTN: Hypertension; LAD: Left anterior descending; LCA: Left circumflex artery; RCA: Right coronary artery; CK: Creatine kinase cTnT: Cardiac troponin T;CK-MB: Creatine kinase myocardial-band; LVEF: Left ventricular ejection fraction; CRP: C-reactive protein; PCI: Percutaneous coronary intervention.
** P<0.01.
*** P<0.001.
Chip-based digital PCR for the diagnosis of AMI
Chip-based digital PCR demonstrated superior sensitivity and specificity for miR-21 but not for miR-499 (Figures 2 and 3).
Figure 2.

Quantification of circulating miRNA-21-5p (A). Duplicate measurements of miR-21 for each patient sample represented as median and IQR between stable CAD and STEMI groups with corresponding P-value for qRT-PCR (blue) and chip-based digital PCR (red) as calculated by a Mann-Whitney U test. (B) ROC analysis from data in A, representing the senstivity and specificity for cTnT, qRT-PCR and chip-based digital PCR with corresponding area under the curve values. (C) Median and IQR of all measurements by qRT-PCR and chip-based digital PCR for miRNA-21 for STEMI patients and patients with stable CAD given in copies/μL.
Figure 3.
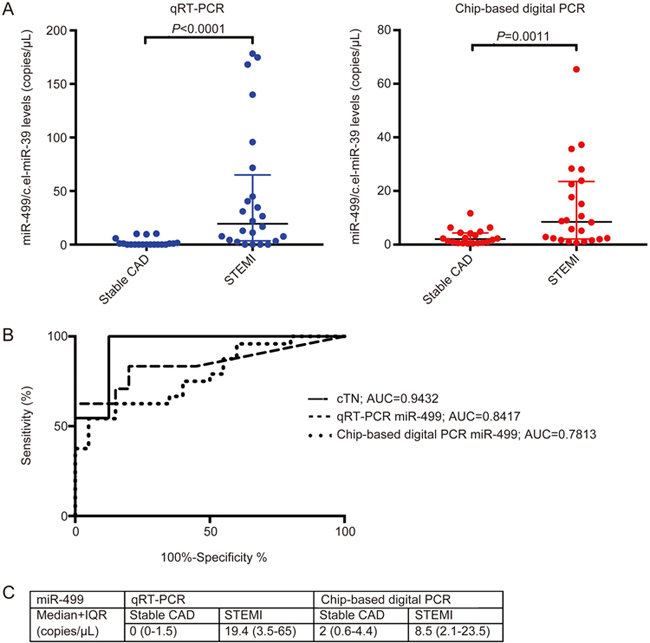
Quantification of circulating miRNA-499-5p (A). Duplicate measurements of miR-499 for each patient sample represented as median and IQR between stable CAD and STEMI groups with corresponding P-value for qRT-PCR (blue) and chip-based digital PCR (red) as calculated by a Mann-Whitney U test. (B) ROC analysis from data in A, representing the senstivity and specificity for cTnT, qRT-PCR and chip-based digital PCR with corresponding area under the curve values. (C) Median and IQR of all measurements by qRT-PCR and chip-based digital PCR for miR-499 for STEMI patients and patients with stable CAD given in copies/μL.
For miR-21, chip-based digital PCR demonstrated significant differences in median levels between stable CAD and STEMI groups (118.8 copies/μL vs 59 copies/μL; P=0.0300), whereas qRT-PCR was unable to reach significance (136.4 copies/μL vs 122.8 copies/μL; P=0.2273).
For miR-499, qRT-PCR demonstrated superior significance (0 copies/μL vs 19.4 copies/μL; P<0.0001 for qRT-PCR; 2 copies/μL vs 8.5 copies/μL; P=0.0011 for chip-based digital PCR).
A ROC analysis showed that chip-based digital PCR better classified STEMI using miR-21 (AUC of 0.6917 vs AUC 0.6083 for qRT-PCR), but not for miR-499 (AUC of 0.7813 vs AUC of 0.8417 for qRT-PCR). No single miRNA analysed was able to outcompete peak cTnT in diagnostic utility (AUC of 0.9432 for cTnT).
Negative and non-template controls for each miRNA were run on chip and did not show any positive results.
Chip-based digital PCR for the diagnosis of ischaemia-reperfusion injury
Neither chip-based digital PCR nor qRT-PCR were able to demonstrate significant differences in miR-21 or miR-499 levels between patients deemed to be successfully reperfused and those who suffered I/R injury (Figure 4). A ROC analysis demonstrated that miR-499 was most accurately able to differentiate successful reperfusion from having relevant reperfusion injury (AUC of 0.7467, supplementary Figure S1).
Figure 4.
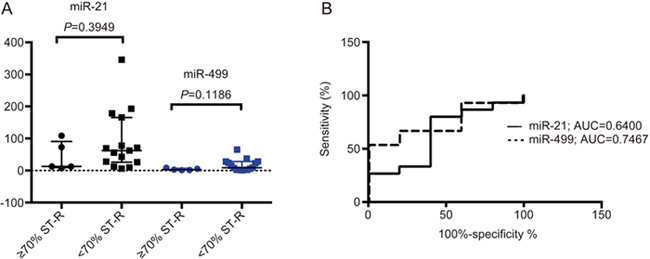
Chip-based digital PCR results of miRNA-21 and 499 levels in diagnosing ischaemia-reperfusion injury as defined by ST-segment resolution (ST-R) <70%. (A) Median and IQR of miR-21 (black) and miR-499 (blue) levels with corresponding significance values calculated via a Mann-Whitney U test. (B) ROC analysis of miR-21 and miR-499 levels in diagnosing I/R injury.
Correlation analysis
Table 2 displays the correlation analyses for main clinical data for miR-21 and miR-499. After Bonferroni's correction, the only clinical correlations to reach statistical significance included a positive correlation with miR-499 with CK as quantified by both qRT-PCR and chip-based digital PCR and miR-499 with CK-MB and eptifibatide as quantified by chip-based digital PCR alone. In a subgroup analysis in STEMI patients comparing miR-499 levels to eptifibatide use, those who received eptifibatide had significantly higher miR-499 levels (29.12 copies/μL vs 5.5 copies/μL; P=0.0177), and eptifibatide use demonstrated a significantly positive correlation with miR-499 levels (r=0.1668; P=0.0476). These results are shown in supplementary Figure S2.
Table 2.
Correlation analysis for miRNA-21 and miRNA-499.
| Characteristic | miRNA-21 | miRNA-499 | ||
|---|---|---|---|---|
| qRT-PCR | Chip-based digital PCR | qRT-PCR | Chip-based digital PCR | |
| Age | P=0.5259 | P=0.2925 | P=0.4583 | P=0.1686 |
| DM | P=0.4186 | P=0.2331 | P=0.6590 | P=0.9482 |
| Smoking | P=0.3888 | P=0.4722 | P=0.4981 | P=0.5758 |
| Aspirin | P=0.6604 | P=0.3315 | P=0.2368 | P=0.2205 |
| Clopidogrel | P=0.5949 | P=0.9036 | P=0.0566 | P=0.0966 |
| Prasugrel | P=0.0947 | P=0.4382 | P=0.0045 | P=0.0073 |
| Ticagrelor | P=0.4132 | P=0.4364 | P=0.1547 | P=0.5186 |
| Eptifibatide | P=0.5161 | P=0.5768 | P=0.0933 | P=0.0012 |
| Rivaroxaban | P=0.1663 | P=0.3014 | P=0.6577 | P=0.6067 |
| Statin | P=0.4476 | P=0.5391 | P=0.5789 | P=0.8316 |
| Creatinine | P=0.5248 | P=0.0429 | P=0.6878 | P=0.6690 |
| CK | P=0.0045 | P=0.0367 | P<0.0001 | P=0.0027 |
| cTnT | P=0.0471 | P=0.1801 | P=0.0803 | P=0.2136 |
| CK-MB | P=0.0071 | P=0.0800 | P=0.2151 | P=0.0004 |
| CRP | P=0.0580 | P=0.7150 | P=0.8451 | P=0.3048 |
| LVEF% | P=0.4348 | P=0.5248 | P=0.1492 | P=0.9088 |
| Time blood taken for miRNA analysis after PCI (h) | P=0.5272 | P=0.6715 | P=0.0142 | P=0.8285 |
DM: Diabetes mellitus; CK: Creatine kinase; cTnT: Cardiac troponin T; CK-MB: Creatine kinase myocardial-band; CRP: C-reactive protein; LVEF%: Left ventricular ejection fraction (%); PCI: Percutaneous coronary intervention.
Discussion
Digital PCR is the first PCR method able to directly and absolutely quantify nucleic acids within a biological sample19. Given its reputed advantages over traditional qRT-PCR, digital PCR could be a superior method for the detection of miRNAs for both diagnostic purposes and in validating miRNA expressions in large multi-centre studies. The recently developed Quantstudio® 3D digital PCR is one of a number of digital PCR systems, which specifically utilises a chip-based method in order to partition a sample into 20 000 reaction wells, thereby permitting absolute quantification23. Other currently available systems favour an oil-emulsion droplet generation to allow for absolute quantification. At present there are very few studies investigating the use of chip-based digital PCR system in quantifying miRNAs, and, to the best of our knowledge, none that have investigated its utility for diagnosing AMI or reperfusion injury with miRNAs. With this report, we aim to validate chip-based digital PCR for the quantification of miRNAs using a combination of both synthetic oligonucleotides and validated human cardiovascular miRNAs extracted from patient serum for AMI and reperfusion injury. For patient samples we used a multiplexed PCR protocol combining the normalistion miRNA (C.elegans-miR-39) and the target miRNA.
Chip-based digital PCR demonstrated efficient technical qualities in a dilution series of synthetic C.elegans-miR-39
In a dilution series of synthetic C.elegans-miR-39, chip-based digital PCR demonstrated lower variability, higher linearity and superior sensitivity compared to qRT-PCR. Through absolute quantification, we observed that the copy numbers actually detected were lower than the numbers that were expected. This has been demonstrated in a number of other papers examining absolute quantification as a method for PCR, and is most likely due to suboptimal reverse transcription21,23. Ultimately, chip-based digital PCR demonstrated superior technical characteristics indicating that it could become a method of choice for more accurate quantification of miRNAs.
miRNA-21/499 levels were significantly dysregulated in STEMI patients compared to patients with stable CAD
Both miR-21 and miR-499 were found to be significantly dysregulated in STEMI patients compared to patients with stable CAD as quantified by chip-based digital PCR. miR-21 levels were found to decrease in STEMI patients by both PCR systems. This is in contrast to other studies, which have shown that miR-21 levels increase in the circulation as a result of infarction11,12. This might be explained by the difference between the times blood was taken in past studies compared to this study. Past studies have analysed blood at the time of admission, whereas this study analysed blood 5±SD 2.7 h after PCI11,12. It is therefore possible that miR-21 levels could decrease below normal levels according to its kinetics, or decrease as a result of therapeutic success. According to a murine model, both Dong et al and Gu et al demonstrated that elevated miR-21 is beneficial for cardiomyocyte survival, significantly decreasing the size of the infarct32,33. However, Liu et al revealed that long-term elevation in miR-21 levels is deleterious to post-infarct repair, contributing to left-ventricular remodelling via its role in cardiac fibrosis34. Given the above information and the knowledge that miRNAs have endocrine-like roles, it is feasible that miR-21 levels are released during the acute phase of AMI, possibly via the rupture of necrotic/apoptotic cardiomyocytes, while after successful reperfusion, and the offset of cardiomyocyte death, miR-21 might translocate to cardiac fibroblasts to contribute to post-infarct repair, hence leading to a decrease in serum levels. However, this needs to be verified in both larger time-course studies in patients and potentially animal experimentation. miR-499 levels, on the other hand, were found to increase in STEMI patients, which is consistent with the current literature12,14,16. In past studies, miR-499 has shown as much as 80 to 3×105-fold differences between AMI and control groups12,16. In this study, qRT-PCR and chip-based digital PCR only demonstrated approximately 21 and 4-fold difference between mean values respectively. The reason for this may once again be due to the time blood was drawn, this being post-PCI, whereas the aforementioned studies analysed blood at admission. In Chen's study of miR-499 kinetics, there was a 58% decrease in miR-499 levels 24-h post-PCI and 48% lower levels in those who received PCI compared to those who did not14. Therefore, the lower levels found in our study may be due to therapeutic reperfusion and might also be a marker of therapeutic success. This evidence also gives credence to the hypothesis that miR-21 levels decrease after PCI indicating therapeutic intervention.
Chip-based digital PCR more accurately classified STEMI cases from stable CAD patients for miRNA-21, but not for miRNA-499
For miRNA-21, chip-based digital PCR, unlike qRT-PCR, demonstrated significanct differences between patient groups and exhibited superior sensitivity and specificity as demonstrated by ROC analysis, giving clinically relevant evidence to its diagnostic potential. Conversely, for miR-499, qRT-PCR reached superior significance. This discrepency might be explained by the fact that miR-499 is normally expressed at levels below the limit of detection of qRT-PCR13,14. This was verified in our experiments, whereby qRT-PCR only detected miR-499 in 45% of patients with stable CAD and 83% of STEMI patients, whereas chip-based digital PCR was able to quantify levels in 100% of the patients analysed, even at low concentrations. Our experiments on synthetic C.elegans-miR-39 also agreed with past studies that demonstrated superior sensitivity for detecting miRNAs using digital PCR21,35. Therefore, this system might be a better reflection of actual miRNA-499 levels, and might be better able to quantify both this miRNA and others that are normally expressed at minimal levels within the circulation. However, this needs to be validated in larger studies. Ultimately, chip-based digital PCR exhibited promising potential for quantifying miRNA-21 and miRNA-499 levels for analysis in cardiovascular biology. We demonstrated that Quantstudio® 3D digital PCR is a robust system with clinical diagnostic potential and technical qualities for use in large multi-centre miRNA studies.
Chip-based digital PCR did not demonstrate increases of miRNA-21 or miRNA-499 in ischaemia-reperfusion injury
In our study, neither miRNA-21 nor miRNA-499 demonstrated a significant association with significant I/R injury after PCI (as defined by lack of ≥70% ST-segment resolution). miRNA-499 as measured by chip-based digital PCR showed the greatest trend, which is in keeping with past studies that suggested a role of miRNA-499 in cardiac I/R injury10. A study with a larger patient cohort utilising other markers of reperfusion injury is required.
miRNA-499 demonstrated significant correlation with myocardial necrosis markers
In a correlation analysis miR-499 demonstrated a significant positive correlation with both peak CK and CK-MB levels as measured by chip-based digital PCR. Both CK and CK-MB have been associated with size of infarct and prognosis after AMI36,37,38,39. This indicates that miR-499 possibly has similar utility. miR-499 presents itself as a particularly interesting biomarker for AMI. miR-499 is normally expressed at low levels, and has high myocardial specificity13,14. Furthermore, past studies have demonstrated significant increases in miR-499 levels post-AMI, which significantly decrease after therapeutic reperfusion12,13,14,16. We therefore propose that miR-499 might serve as both a sensitive and specific prognostic and therapeutic marker. However, further studies are needed looking at both short-term and long-term outcomes, as well as the size of the infarct in order to validate this hypothesis. It must also be mentioned that there was a positive correlation between miR-499 and eptifibatide use. This is most likely explainable by the fact that only patients with STEMI received eptifibatide, and miR-499 levels were significantly increased in the STEMI population. Furthermore, in a subgroup analysis, those who received eptifibatide had significantly higher miR-499 levels, and eptifibatide use was positively correlated with miR-499. In our study, eptifibatide was given at the discretion of the treating cardiologist. As per the 2017 European Society of Cardiology guidelines for STEMI, GP IIb/IIIa inhibitors such as eptifibatide should be reserved for bailout if there is evidence of large thrombus on angiography, no-reflow or other thrombotic complications40. Consequently, it can be argued that miR-499 levels were higher in more complicated STEMI cases, giving further evidence to miR-499's validity as a clinically interesting biomarker for AMI.
Limitations
Despite demonstrating promising technical qualities and potential for more accurate miRNA quantification, our study has a number of limitations. In the synthetic dilution series we detected absolute copy numbers lower than the copy numbers expected, possibly suggesting suboptimal assay design and/or reverse transcription. Furthermore, the patient cohort was relatively small and there was no healthy control group. The limited size in the STEMI subgroup might explain why miRNA-499 failed to reach significance in association with reperfusion injury. Furthermore, only one surrogate marker of reperfusion injury was assessed, this being ECG post PCI. Other markers such as cardiac MRI must be integrated into future studies. Moreover, a true association between miRNA-21/499 and the size of infarct and prognosis could not be directly assessed, as these parameters were not analysed. Finally, we only compared qRT-PCR to one method of digital PCR, this being the Quantstudio® 3D system. This was sufficient to support the use of chip-based digital PCR; however, further studies comparing a variety of digital PCR methods would be more clinically advantageous. Ultimately, our findings need to be verified in studies with larger patient cohorts and further investigation into the physiology of miRNA-21 and miRNA-499 must be undertaken.
In conclusion, we have demonstrated that chip-based digital PCR has a number of promising characteristics allowing its potential use as a more accurate method for validating miRNA levels, particularly in large multi-centre clinical trials. Chip-based digital PCR exhibited low variability, high linearity and remarkable sensitivity and showed promise as a diagnostic instrument. These results warrant further investigation and validation in larger more comprehensive studies. Nevertheless, this relatively novel instrument has shown potential as a more accurate and reproducible method for quantifying miRNAs in the circulation.
Author contribution
Marcus HORTMANN designed research; Samuel ROBINSON, Marie FOLLO, David HAENEL, Maximilian MAULER, Daniela STALLMANN, Lukas Andreas HEGER, and Marcus HORTMANN performed research; Marie FOLLO, Marcus HORTMANN, Samuel ROBINSON contributed new analytical tools and reagents; Samuel ROBINSON, Marcus HORTMANN, Ingo AHRENS, Marie FOLLO, Daniel DUERSCHMIED, Thomas HELBING, Karlheinz PETER, Christoph BODE analyzed data; Samuel ROBINSON and Marcus HORTMANN wrote the paper.
Abbreviations
AMI: Acute myocardial infarction; AUC: Area under the curve; CAD: Coronary artery disease; C.elegans-miR-39: Caenorrhabditis elegans miRNA-39; CK: Creatine kinase; CK-MB:Creatine kinase myocardial-band; cTnT: Cardiac troponin T; CV%: Coefficient of variation; ECG: Electrocardiogram; IQR: Interquartile range; I/R: Ischaemia-reperfusion; LLLR: Lower limit of linear range; LOD: Limit of detection; LOQ: Limit of quantification; LVEF: Left ventricular ejection fraction; miRNAs:micro-RNAs; miR-21: miRNA-21; miR-499: miRNA-499; PCI: Percutaneous coronary intervention; qRT-PCR: Quantitative real-time polymerase chain reaction; ROC: Receiver operating characteristic; SD: Standard deviation; STEMI: ST-elevation myocardial infarction; ST-R: ST-resolution.
Acknowledgements
The authors declare that discounted consumables related to the use of the QuantStudio® 3D Digital PCR system were offered by ThermoFisher.
References
- 1.Mohr AM, Justin LM. Overview of microRNA biology. Seminars in Liver Disease. 2014;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimmeler S, Zeiher AM. Circulating microRNAs: novel biomarkers for cardiovascular diseases? Eur Heart J. 2010;31:2705–7. doi: 10.1093/eurheartj/ehq221. [DOI] [PubMed] [Google Scholar]
- 3.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;11:2383–90. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 5.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 6.Hu S, Huang M, Nguyen PK, Gong Y, Li Z, Jia F, et al. Novel MicroRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation. 2011;124:27–34. doi: 10.1161/CIRCULATIONAHA.111.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 8.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinrich EM, Dimmeler S. MicroRNAs and stem cells control of pluripotency reprogramming and lineage commitment. Circ Res. 2012;110:1014–22. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 10.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, et al. miR-499 regulated mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–8. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, et al. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One. 2014;9:9. doi: 10.1371/journal.pone.0105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivieri F, Antonicelli R, Lorenzi M, D'Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531–6. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 13.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 14.Xi C, Zhang L, Su T, Li H, Huang Q, Wu D, et al. Kinetics of plasma microRNA-499 expression in acute myocardial infarction. J Thorac Dis. 2015;7:890–6. doi: 10.3978/j.issn.2072-1439.2014.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Y, Shao J, Bai X, Zhang G. Expression of plasma microRNA-1/21/ 208a/499 in myocardial ischemic reperfusion injury. Cardiology. 2015;130:237–41. doi: 10.1159/000371792. [DOI] [PubMed] [Google Scholar]
- 16.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–67. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 17.Schulte C, Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease - Summing up the facts. Cardiovasc Diagnost Ther. 2015;5:17–36. doi: 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koshiol J, Wang E, Zhao Y, Marincola F, Landi MT. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol Biomark Prevent. 2010;19:907–11. doi: 10.1158/1055-9965.EPI-10-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanagal-Shamanna R. Digital PCR: principles and applications. Methods Mol Biol. 2016;1392:43–50. doi: 10.1007/978-1-4939-3360-0_5. [DOI] [PubMed] [Google Scholar]
- 20.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput Droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindson CM, Chevillet JR, Briggs HA, Gallichotte E, Ruf IK, Hindson BJ, et al. Absolute quantification by Droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–5. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosman KJ, Nijhuis M, van Ham PM, Wensing AMJ, Vervisch K, Vandekerckhove L, et al. Comparison of digital PCR platfroms and semi nested qPCR as a tool to detemine the size of the HIV reservoir. Sci Rep. 2015;5:13811. doi: 10.1038/srep13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conte D, Verri C, Borzi C, Suatoni P, Pastorino U, Sozzi G, Fortunato O. Novel method to detect microRNAs using chip-based QuantStudio 3D digital PCR. BMC Genomics. 2015;16:849. doi: 10.1186/s12864-015-2097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Park Y, We Y, Han DJ, Seo JW, Moon H, et al. Evaluation of digital PCR as a technique for monitoring acute rejection in kidney transplantation. Genomics Inform. 2017;15:2–10. doi: 10.5808/GI.2017.15.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinz E, Leiherer A, Lang AH, Drexel H, Muendlein A. Accurate quantitation of JAK2 V617F allele burden by array-based digital PCR. Int Jnl Lab Hem. 2015;37:217–24. doi: 10.1111/ijlh.12269. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW. Quantification strategies in real-time PCR. A-Z of quantitative PCR [serial on the Internet]. 2004 Jun [cited 2016 Aug 4]; Chapter 3. Available from: http://gene-quantification.org/chapter-3-pfaffl.pdf. (4 August 2016).
- 27.Massart DL. Data Handling in Science and Technology.In: Massart DL, Vandeginste BGM, Buydens LMC, De Jong S, Lewi PJ, Smereys-Verbeke J, editors. Handbook of Chemometrics and Qualimetrics. Amsterdam:Elsevier Science B.V.; 1998. Part A.
- 28.Shrivastava A, Gupta VB. Methods for the determination of limit of detection and limit of quantification of the analytical methods. Chron Young Sci. 2011;2:21–5. doi: 10.4103/2229-5186.79345. [DOI] [Google Scholar]
- 29.Schröder R, Wegscheider K, Schroder K, Dissmann R, Meyer-Sabellek W, for the INJECT Trial Group. SExtent of early ST segment elevation resolution: a strong predictor of outcome in patients with acute myocardial infarction and a sensitive measure to compare thrombolytic regimens. J Am Coll Cardiol 1995; 26: 1657–64 [DOI] [PubMed]
- 30.de Lemos JA, Braunwald E. ST segment resolution as a tool for assessing the efficacy of reperfusion therapy. J Am Coll Cardiol. 2001;38:1283–94. doi: 10.1016/S0735-1097(01)01550-9. [DOI] [PubMed] [Google Scholar]
- 31.de Roeck L, Vandamme S, Everaert BR, Hoymans V, Haine S, Vandendriessche T, et al. Adiponectin and ischemia-reperfusion injury in ST segment elevation myocardial infarction. Eur Heart J Acute Cardiovasc Care. 2016;5:71–6. doi: 10.1177/2048872615570770. [DOI] [PubMed] [Google Scholar]
- 32.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–25. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu GL, Xu XL, Sun XT, Zhang J, Guo CG, Wang CS, et al. Cardioprotective effect of microRNA-21 in murine myocardial infarction. Cardiovasc Ther. 2015;33:109–17. doi: 10.1111/1755-5922.12118. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Dong Y, Chen S, Zhang G, Zhang M, Gong Y, Li X. Circulating micro-RNA-146a and microRNA-21 predict left ventricular remodeling after ST-elevation myocardial infarction. Cardiology. 2015;132:233–41. doi: 10.1159/000437090. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Li N, Jiang F. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomarker Insights. 2013;8:127–36. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di CA, Plewka M, Werren M, Badano LP, Fresco C, Fioretti PM. Estimation of infarct size by single measurements of creatine kinase levels in patients with a first myocardial infarction. J Cardiovasc Med. 2006;7:340–6. doi: 10.2459/01.JCM.0000223256.01439.1b. [DOI] [PubMed] [Google Scholar]
- 37.Savonitto S, Granger CB, Ardissino D, Gardner L, Cavallini C, Galvani M, et al. The prognostic value of creatine kinase elevations extends across the whole spectrum of acute coronary syndromes. J Am Coll Cardiol. 2002;39:22–9. doi: 10.1016/S0735-1097(01)01706-5. [DOI] [PubMed] [Google Scholar]
- 38.Dohi T, Maehara A, Brener SJ, Genereux P, Gershlick AH, Mehran R, et al. Utility of peak creatine kinase-MB measurements in predicting myocardial infarct size, left ventricular dysfunction, and outcome after first anterior wall acute myocardial infarction (from the INFUSE-AMI trial) Am J Cardiol. 2015;115:563–70. doi: 10.1016/j.amjcard.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Nienhuis MB, Ottervanger JP, de Boer MJ, Dambrink JE, Hoorntje JCA, Gosselink M, et al. Prognostic importance of creatine kinase and creatine Kinase-MB after primary percutaneous coronary intervention for ST-elevation myocardial infarction. Am Heart J. 2008;155:673–9. doi: 10.1016/j.ahj.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation. Eur Heart J. 2017;00:1–66. doi: 10.1016/j.rec.2017.11.010. [DOI] [PubMed] [Google Scholar]


