Abstract
Pervasive transcription of the human genome is responsible for the production of a myriad of non-coding RNA molecules (ncRNAs) some of them with regulatory functions. The pivotal role of ncRNAs in cardiovascular biology has been unveiled in the last decade, starting from the characterization of the involvement of micro-RNAs in cardiovascular development and function, and followed by the use of circulating ncRNAs as biomarkers of cardiovascular diseases. The human non-coding secretome is composed by several RNA species that circulate in body fluids and could be used as biomarkers for diagnosis and outcome prediction. In cardiovascular diseases, secreted ncRNAs have been described as biomarkers of several conditions including myocardial infarction, cardiac failure, and atrial fibrillation. Among circulating ncRNAs, micro-RNAs (miRNAs), long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) have been proposed as biomarkers in different cardiovascular diseases. In comparison with standard biomarkers, the biochemical nature of ncRNAs offers better stability and flexible storage conditions of the samples, and increased sensitivity and specificity. In this review we describe the current trends and future prospects of the use of the ncRNA secretome components as biomarkers of cardiovascular diseases, including the opening questions related with their secretion mechanisms and regulatory actions.
Keywords: circulating ncRNA, miRNA, lncRNA, circRNA, biomarker, cardiovascular diseases, myocardial infarction, cardiac failure, atrial fibrillation
Non-coding transcripts in cardiovascular physiopathology
The dynamic transcriptomic output of the human genome composed by coding and non-coding RNA (ncRNA) transcripts constitutes a molecular fingerprint of the physiological state and characteristics of a particular cell or tissue1. Coding transcripts or messenger RNAs (mRNAs) are devoted to the production of proteins by ribosome translation, whereas ncRNAs are untranslated RNAs that usually perform regulatory functions controlling the flow of information from the genome2. Nc-RNAs have been described as “architects” of eukaryotic genomic complexity since they are responsible for the establishment of an additional layer of regulation of the genomic output that enables the integration of complex suites of genomic expression at the cellular level1. Nc-RNAs can be classified into small (<200 nt) or large (>200 nt) families. With some exceptions, small RNAs are tipically generated from genomic loci by transcription and further processing by specific nucleases, whereas large ncRNAs are produced by the same transcription and splicing machinery employed to produce coding mRNAs3.
The essential role of ncRNAs in cardiovascular pathophysiology started to be recognized early at the beginning of the discovery of ncRNAs with the characterization of ncRNAs necessary for proper heart function and physiology4. In the last decade, the entangled regulatory network controlled by ncRNAs in cardiovascular diseases (CVDs) is starting to be unveiled5,6,7. The main groups of ncRNAs described to be involved in cardiovascular physiology and disease are: microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs).
Micro-RNAs (miRNAs)
Among small ncRNAs, micro-RNAs (miRNAs) are the most abundant family in the human genome, which contains more than 2000 different loci for the production of miRNAs8. MiRNAs are short ncRNAs (18-22 mer) generated by a complex metabolic pathway that involves the transcription of specific genomic loci and the processing of the generated transcripts by specialized RNA endonucleases9. They act as negative regulators of gene expression by binding to the 3′-UTR region of coding transcripts and recruiting specific silencing proteins that form the RISC (RNA Induced Silencing Complex). In healthy cells, miRNAs act as modulators to maintain protein levels within the physiological homeostatic intervals. The regulatory activity of a miRNA depends on the presence of its cognate mRNA target, and for this reason the same miRNA could have distinct regulatory effects in different cellular types10. In a pathological context, an ectopic expression of a specific miRNA out of its original tissue can lead to regulation of non-natural target transcripts and to a functional disequilibrium11,12.
Cardiac physiology requires a delicate balance between chemical and electrical stimuli that are responsible for the complicated mechanism of contraction. In consequence, a tight regulation of these synchronous processes is required, and miRNAs seemed to be ideal regulators to exert the control over the levels of sarcomeric proteins, ion transporters and contractility regulators13. In fact, cardiovascular biology was a pioneer area in the study of the function of miRNAs and their roles in heart development and maintenance of contractility. The pathophysiological role of miRNAs in the myocardium was initially inferred by the study of aberrant expression patterns of these small ncRNAs in specific cardiac pathologies such as hyperthropy and heart failure5, myocardial infarction14 or cardiac myopathies6. Further studies based on the selective manipulation of miRNA levels in animal models showed their intrinsic relevance in cardiovascular biology and their importance not only as regulatory molecules but also as potential therapeutic targets7.
There are three cardiac specific human miRNAs highly expressed in myocardium and with less extend in striated muscle, which are located within introns of genes encoding sarcomeric proteins: hsa-miR-499a-5p, located in the intron 19 of MYH7B gene encoding for the cardiac Myosin heavy chain 7B (Genomic coordinates, GRCh38, chr20: 34990376-34990497); hsa-miR-208a-3p, located in the intron 29 of MYH6 gene encoding for the cardiac Myosin heavy chain 6 or “fast myosin” and predominantly expressed in fetal myocardium (Genomic coordinates, GRCh38, chr14: 23388596-23388666); and hsa-miR-208b-3p, generated by the intron 31 of the MYH7 gene encoding for the cardiac Myosin heavy chain 7 or “slow myosin”, which is mainly expressed in myocardium in the adulthood (Genomic coordinates, GRCh38, chr14: 23417987-23418063). The roles of miR-208 family and miR-499a-5p in humans appeared to be redundant, being involved in the regulation of heart contractility and function15. Aberrant expression of cardiac-specific miRNAs in animal models resulted in abnormal myocardium function, leading to cardiac hypertrophy, arrhythmias and fibrosis16. Additional muscle-specific and cardiac-enriched miRNAs such as miR-1 were characterized to be essential for proper heart development in mouse models4 but also were involved in the onset and progression of cardiac conditions such as hypertrophy17.
In the last decade, the availability of transcriptomic data supported by the development of the high-throughput sequencing techniques, allowed the characterization of the functional role of hundreds of miRNAs in the onset and progression of CVDs. The last version of HMDD2.0 database, an experimentally-supported database of miRNA-disease associations18, contains references for 165 different miRNAs related with CVDs, which genomic loci are widespread across the human genome (Figure 1A). It is very interesting to observe that the miRNAs more often referred as related with CVDs are not cardiac-specific or even muscle-enriched species (Figure 1B). In fact, many miRNAs previously characterized as onco-miRNAs such as hsa-miR-21-5p or hsa-miR-126-3p are key players in CVD, regulating myocardial fibrosis11, response to ischaemia19, ventricular remodeling20, heart failure21, arrhythmia12 and even controlling the brain-heart interaction axis22.
Figure 1.
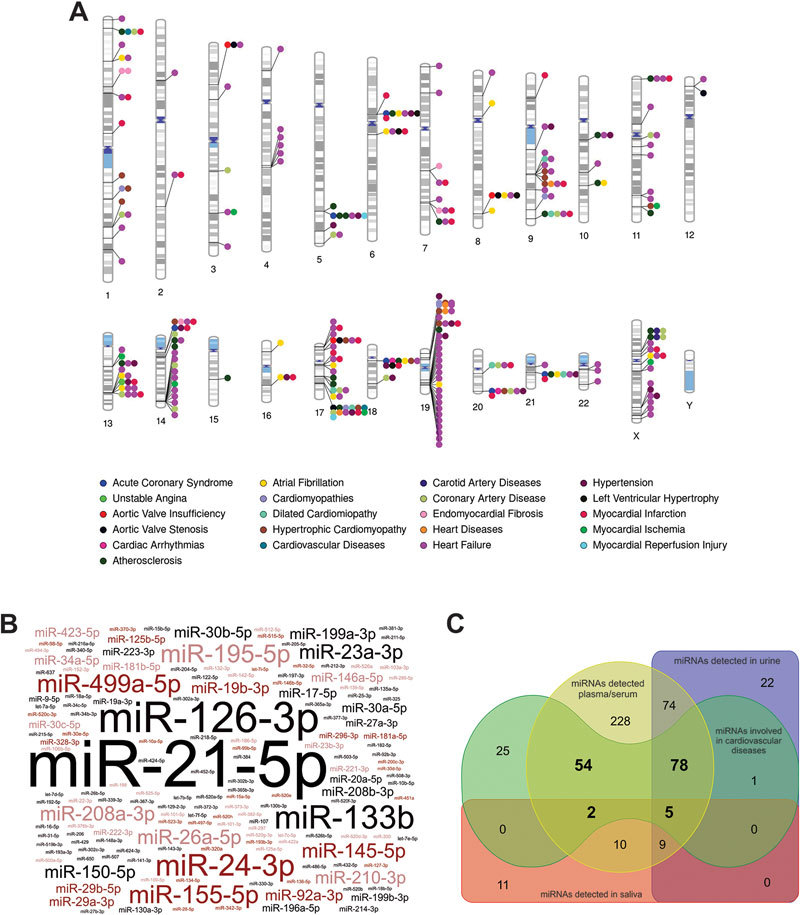
Human miRNAs involved in cardiovascular diseases (CVDs) and their presence in biological fluids. Panel A, human chromosomal ideogram depicting the genomic location of key miRNAs loci and their involvement in CVDs obtained from HMDD 2.0 database18 (Supplementary Table S1 for more details) and generated by the Phenogram web application (http://visualization.ritchielab.psu.edu/phenograms/plot). Panel B, word-cloud representation of the different human miRNAs showing experimental evidence of involvement in CVDs and generated by the Wordart web application (http://wordart.com). The size of miRNA names is proportional to the number of PUBMED references demonstrating their functional involvement at the cellular or organ level in CVDs. Panel C, Venn diagram showing the presence of miRNAs involved in CVDs that can be also detected in plasma/serum, urine and saliva as compiled in miRandola 2017 database for circulating miRNAs116 and generated by the DrawVenn application (http://bioinformatics.psb.ugent.be/webtools/Venn). For clarity reasons, miRNA nomenclature from miRBase 21.0 has been abbreviated by removing the specific human (hsa) prefix.
In complex eukaryotic organisms as humans and due to the redundancy of the regulatory cellular circuits, it is very difficult to find a “master” miRNA regulator that may be individually responsible for the control of a particular biological pathway. Instead, a complex network of entangled regulatory interactions is expected, involving many miRNAs that would work together supporting the regulatory effect over the metabolic pathway. This is also true for CVDs, where complex association of the regulatory effects of several miRNAs can control cardiovascular function4, disease progression23 and clinical secondary complications24. Moreover CVDs are complex clinical scenarios frequently driven by chronic conditions such as atherosclerosis, and miRNAs could be important players in the progression of this chronic condition by regulating plaque initiation, remodeling and rupture, being considered as a risk modulating factors25.
Interestingly, a considerable fraction of the miRNAs involved in CVDs can be detected as a circulating species in several human biofluids (blood, urine and saliva), which could suggest their possible application as biomarkers (Figure 1C).
Long non-coding RNAs (lncRNAs)
Large ncRNAs, also known as long non-coding RNAs or lncRNAs are non-coding RNA transcripts >200 nt and generated from transcriptional units that resemble protein coding genes but lacking their coding potential2. Following the last version of Gencode database (v.27.0, January 2017, GRch38)26, the estimated number of these ncRNA species in the human genome is ∼15 000. These transcripts have initially been suggested to represent only the bystander's transcription within protein-coding regions. However, they are known to be very relevant players in the regulation of cell functions since they can interact with genomic DNA as well as RNA functioning as flexible molecular scaffolds for the recruitment of chromatin modifying enzymes and transcription factors, driving their location to the correct functional localization27. LncRNAs have also been shown regulate the activity of other ncRNAs, specifically miRNAs, by acting as 'sponges' that titrate miRNAs away from natural mRNA targets (thereby acting as competing endogenous RNAs; ceRNAs)28. Loss-of-function experiments have provided evidence of lncRNAs functional importance in the regulation of gene expression patterns that control many cell functions ranging from cell pluripotency, differentiation and survival29. LncRNAs have tendency to be very specific in their expression pattern and can exert they regulatory function in cis, modulating nearby genes on the same allele, or in trans by affecting genes at long genomic distances. LncRNAs are in general actively regulated and may function predominantly in embryonic development. However most lncRNAs evolve rapidly in terms of sequence and expression levels, tissue specificities and probably tridimensional structure of lncRNAs would be conserved among close related species.
The evidence of the important role of lncRNAs in cardiac function was firstly described in the case of Braveheart lncRNA (lncRNA-Bvht), a mouse lncRNA with an essential role in the establishment of cardiovascular lineage during heart development by regulating the specific recruitment of CM cellular precursors30. LncRNA-Bvht interacts with SUZ12, a component of polycomb-repressive complex 2 (PRC2), during CM (CM) differentiation, suggesting that lncRNA-Bvht mediates epigenetic regulation of cardiac commitment, and also has a role in maintaining cardiac fate in neonatal CMs. LncRNA-Bvht is also involved in the differentiation of mesenchimal mouse stem cells to CMs31. The heart-related lncRNA-fendrr was also characterized in mouse models as modulator of proper heart differentiation by controlling pluripotency, lineage commitment, and cell differentiation at the chromatin level32. Non-species conservation of lncRNAs has prevented the easy discovery of the corresponding human counterparts of these cardiac-related lncRNAs. Recently, lncRNA-HBL1 (Heart Brake LncRNA 1) has been described as regulator of CM development from human induced pluripotent stem cells (hiPSCs). Overexpression of lncRNA-HBL1 repressed CM differentiation from hiPSCs, by a mechanism involving the sequestration of hsa-miR-133.
Dysregulation of lncRNA global expression pattern has been described in some cardiovascular conditions as ischemic heart failure either in mouse models or humans34. Moreover, some lncRNAs related with cardiac pathophysiology are specific of the cell type involved in a particular condition. In mouse models, Viereck and coworkers have recently identified lncRNA-chast (cardiac hypertrophy-associated transcript) as a regulator of cardiac hypertropy at the CM level, and validated the results in human embryonic stem cell-derived CMs upon hypertrophic stimuli35. At the fibroblast level, lncRNA-wisper (Wisp2 super-enhancer-associated RNA) is a cardiac fibroblast-enriched lncRNA that regulates cardiac fibrosis after injury, and its expression was correlated with cardiac fibrosis both in a mouse model of MI and in hearts from human aortic stenosis patients36.
Circular RNAs (circRNAs)
CircRNAs are a family of ncRNAs generated from RNA transcripts by non-canonical back-splicing events that inversely connect exon boundaries37. Despite of other putative regulatory functions, circRNAs act as scavengers to capture other RNA molecules, behaving as molecular sponges that control the levels of other regulatory proteins or RNAs including miRNAs. The expression pattern of circRNAs is highly dependent on the organism and cell type, showing a low degree of conservation between species. In human heart tissue, abundance of circRNAs is generally related with their cognate mRNAs, being the most abundant cardiac-expressed circRNAs generated from key cardiac transcripts including TTN, RYR2 and DMD genes38. The role of circRNAs in cardiac disease conditions is starting to be unveiled by recent reports that showed their important functions as modulators of miRNA levels. In animal models some circRNAs have been recently characterized as promoters of cardiac fibrosis by sponging miR-141 or miR-26b-5p39,40, as protectors for cardiac hypertrophy by capturing miR-22341, as mediators of CM death by decreasing the levels of miR-652-3p42 and as promoters of myocardial infarction by sequestering miR-7a43. In all the described situations, circRNAs are essential players in controlling the levels of endogenous miRNAs, which enhance their importance in cardiac physiology. Function of circRNAs as molecular buffers modulating miRNA levels and their role in the onset and progression of cardiac conditions are essentially uncharacterized in humans, since all the already published studies have been performed in animal models.
The human non-coding RNA secretome
Introduction
NcRNAs can be detected in extracellular medium and circulating biofluids in the human body. The presence of circulating RNA species was initially characterized in specific families of tumors, being described as products of the tumor-induced cellular apoptosis and subsequent lysis44. However human and other eukaryotic cells are able to actively secrete RNAs using specific secretory cellular mechanisms. The first evidence for this phenomenon was described almost ten years ago by Valadi and coworkers45. The authors were able to show that exosomes derived from a mouse and a human mast cell lines (MC/9 and HMC-1 cell lines), as well as primary bone marrow-derived mouse mast cells, contain RNA. Among the exosome-contained RNA species, the authors identified several coding mRNA transcripts as well as miRNAs. RNA-containing exosomes were quickly recognized as a putative mechanism for the interchange of genetic functional information between cells45. Extracellular and functionally active RNAs can be considered as “genetic hormones” that could be produced in a particular cell and reach a distant target by using body fluids. This idea has been elegantly described and demonstrated in a mouse model specifically knocked-out at the Dicer enzyme locus in the adipose tissue. Adipose tissue Dicer knock-out mice suffered a severe lipodystrophy that could be reverted by the injection of serum exosomes from normal mice, showing the intrinsic relevance of circulating miRNA in the overall organism homeostasis46.
The searching and characterization of extracellular RNAs has grown exponentially in the last decade because their intrinsic regulatory functions, but also because they may can be used as biomarkers of disease and response to treatment. Among secreted regulatory RNAs, miRNAs are the most widely studied group as biomarkers in CVDs, however there is a recent increasing interest in the study of other circulating RNAs such as lncRNAs and circRNAs.
Extracellular vesicles as ncRNA conveyors
Extracellular vesicles (EVs) are small cell-derived structures (diameter up to 4000 nm) enclosed by lipid bilayer. They are released by cells within the cardiovascular system both in physiological and pathological conditions including MI, stroke and coronary artery diseases47. Their presence has been demonstrated in all biological fluids, including blood, urine, saliva and cerebrospinal fluids, while their number and molecular composition can vary depending of the origin and type of cell from which they were released48.
Plasma EVs were identified in 1967 as a subcellular fraction that originates from platelets and promotes coagulation, termed “platelet dust”. Nowadays, it is recognised that platelet-derived EVs contribute in about 25% of all circulating EVs in blood, whereas the remaining part of circulating EVs is released from other blood cells (leukocytes, vascular endothelium) and tissues, including myocardium. Two types of EVs which differ in size, mechanism of generation and specific molecule signature are described as involved in ncRNA transfer between cells: exosomes and microvesicles (Figure 2)49. Smaller exosomes are the most homogenous vesicles, 30–100 nm in diameter, which are released by exocytosis of multivesicular bodies (MVBs). On the contrary, bigger vesicles called microvesicles or ectosomes are more heterogeneous in size and composition. They are 100–1000 nm in diameter and they are released by budding from plasma membrane upon cell activation and expose cell-specific CD50.
Figure 2.
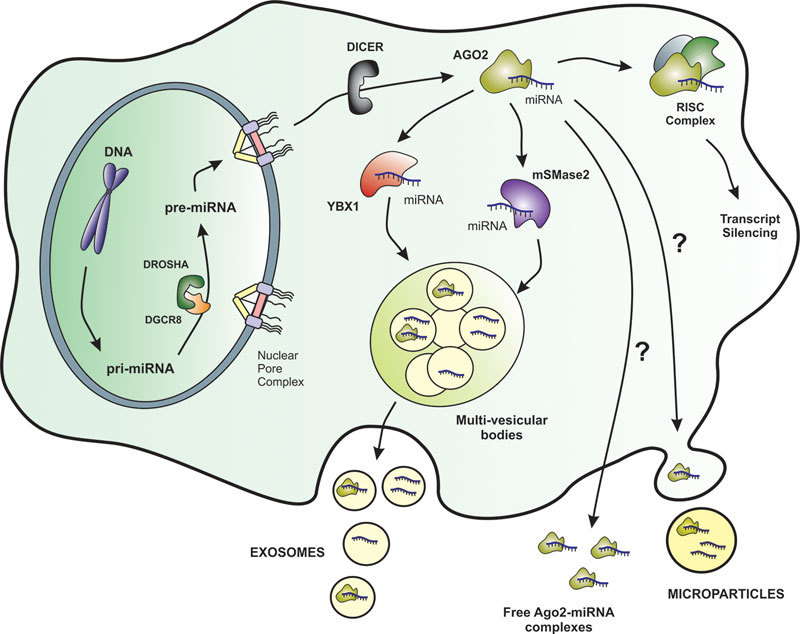
Biosynthesis and proposed secretion pathways of miRNAs in human cells. The miRNA precursors are synthesized in the nucleus a processed by specific nucleases to engage the RISC complex and exert their negative regulatory activity over mRNA transcripts. MiRNA secretion routes are still not totally clear, but at least two RNA-binding proteins, YBX1 and mSMase2, have been described to be involved in the specific recruitment of miRNAs into multi-vesicular bodies and further into exosomes. However, miRNAs can be also found in biofluids in the form of free Ago2-complexes, within microparticles, and complexed with plasma proteins, and in these cases the secretion routes are not known. The secretion pathways for other ncRNAs such as lncRNAs and circRNAs remain uncharacterized.
Exosome formation and RNA sorting
In the process of exosome formation, four protein complexes called endosomal sorting complexes required for transport (ESCRT) play an important role in the every step of vesicle secretion: cargo sorting, intraluminal vesicle (ILV) formation and release to the MVB. ILVs are formed by ESCRT machinery which consist of four escortin complexes: ESCRT-0 responsible for cargo clustering, ESCRT-1/ ESCRT-2 responsible for induction and bud formation and ESCRT-3 controlling vesicle scission and accessory proteins (VPS4 ATPases) which allow dissociation and recycling of the ESCRT machinery51. Despite the ESCRT activity is confirmed in protein sorting, the mechanisms of membrane sorting are still not revealed in this complicated machinery. The lipid cargo depends on sphingolipid ceramide presence and sphingomyelinase activity. Moreover, the neural sphingomyelinase 2 (nSMase2)-dependent pathway was reported to be related to miRNA secretion into exosomes and overexpression of nSMase2 increased the number of exosomal miRNAs, on the other hand its inhibition reduced the number of exosomal miRNAs (Figure 2)52. Another important issue is exosome release to the extracellular space, which is regulated by a family of small G-proteins–Rab GTP-ases (Rab27, Rab11 and Rab35) and effector proteins Sl4 and Slac 2b. MVB fusion with plasma membrane is controlled by soluble NFS-Attachement Protein Receptor (SNARE) complexes, however their role in this process is not fully understood51.
The RNA content of exosomes mainly consists of small ncRNAs (miRNAs), some miRNAs are enriched in exosomes while others are depleted, showing the presence of a specific machinery involved in the active sorting of specific types of miRNAs into exosomes (Figure 2). Among them miR-222 and miR-143 are relatively most abundant in ischaemic cardiac exosomes53. Still unraveled is the process of miRNA sorting to MVB compartments and the critical role of miRNA effector complexes is proposed. Among them, the ubiqiunated proteins Ago2, which belongs to Argonaute family, and GW1822 (P-body protein) are abundantly present in secreted exosomes and modulate miRNA activity54. Another possible mechanism is the contribution of hnRNPA2B1 ribonucleoprotein by the recognition of specific EXO-motif (GGAG) present in RTS sequences that could controls the loading of these miRNA into exosomes. Moreover, hnRNPA2B1 in exosomes is sumoylated, and this post-traslational modification controls the binding of hnRNPA2B1 to miRNA being essential for the proper miRNA loading into exosomes55. Recently, YBX1 (RNA-binding Y-box protein I) has been also described as essential player for the correct sorting and selective enclosure within exosomes of specific miRNAs in HEK293T cells and in vitro models (Figure 2)56.
Ectosome release and miRNA transfer
Ectosomes are generated by outward budding and the fusion of the plasma membrane. Two negatively charged phospholipids regulate ectosome release: phosphatidylserine (PS) and fosphatidylethyoamine (PE) which are vertically transferred by two ATP-dependent aminophospholipid transferases: flipases and flopases. Cytoskeletal reorganization upon ectosome release is induced by calpain and gelsolin activity, by destabilizing actin cytoskeleton. These specific pathways of cytoskeleton reorganization and contraction driving to ectosome release can be different in platelets and endothelial cells (ECs), but the final vesicle budding is completed via a non-muscle myosin II motor protein-dependent pathway57.
In CVDs, ectosomes play an important role in miRNA transfer. TNF-α-stimulated endothelium release ectosomes with down-regulated miR-126-3p and miR-21-5p and up-regulated miR-155-5p58 (Figure 1B). It has been reported that miR-126-3p is transferred between EC via ectosomes, contributing to endothelium integrity repair and down-regulating SPRED-1 transcripts in hyperglycemic conditions59. In contrast to healing endothelial-derived ectosomes, platelet-derived ones transfer miR-233 which down-regulates IGF-1 receptor expression on the surface of ECs and promotes apoptosis induced by advanced glycation end products. Interestingly, it has been reported that this miR-223 is delivered to EC in platelet ectosomes in complexes with Ago2 protein60. Plausibly, Ago2 contributes in miRNA sorting not only to exosomes but also to ectosomes, which was observed also in endothelial ectosomes carrying miR-126-3p to smooth muscle cells9.
Regardless of several next generation sequencing studies showing distinct miRNA-enrichment signatures in different subtypes of EVs, the mechanisms of ectosome miRNA sorting and their roles in cardiovascular diseases are still unraveled. One mechanistic hypothesis assumes the plausible role of Ago2 in miRNA sorting, which was reported in endothelial ectosomes carrying miR-126-3p to smooth muscle cells61. However, the number of ectosome-enriched miRNAs is smaller when compared with the exosome-enriched ones.
Low and high density lipoproteins in miRNA transfer
Although the majority of miRNAs is transferred by exosome- and or ectosome dependent systems, several studies suggest that circulating high-density lipoprotein (HDL) and low-density lipoproteins (LDL) can bind and transport endogenous miRNAs. Namely, miR-223, and delivers those to hepatocytes and endothelial cell changing their inflammatory status62. Notably, miR-486 and miR-92a have the highest levels in differs HDL subfractions from patients with coronary artery disease63. Most of the work on lipoprotein miRNAs has been addressed to HDL transport; however, each of these three studies also analyzed LDL-miRNAs associations. Generally, LDL levels were found to be considerably lower than HDL levels63. LDL/Lp(a) apheresis has an impact on differences in plasma levels in miR-451a, miR-16, miR-19a/b, miR-223 and miR-185 and is associated with lipid homeostasis and vascular status64. Differently from the above reported observations, lipoprotein-associated miRNAs are not efficiently delivered to vascular cells (endothelial and smooth muscle cells) suggesting that the lipoprotein-associated pool of miRNAs does not have the regulating function of that cells65.
Biomarkers from the human ncRNA secretome in cardiovascular diseases
Myocardial infarction
Myocardial infarction (MI) is a leading cause of mortality in western countries, characterized by localized heart tissue damage with cell death due to a lack of blood flow to the myocardium. Despite of all the new methods and techniques of interventional cardiology that are able to stabilize the patients after cardiac crisis, there is a current unmet medical need of selective biomarkers for the risk stratification of patients and their therapeutic response after the infarction. Besides of the electrophysiological parameters, classical MI biomarkers included the serum levels of cardiac troponins (cardiac troponin T and I) and the creatine kinase MB (CK-MB), an isoenzyme of creatine kinase specifically expressed in cardiac muscle66.
Circulating cell-free muscle and myocardial enriched miRNAs as miR-1 were early described as biomarkers for acute MI67. Cardiac specific miRNAs expressed from cardiac myosin genes (hsa-miR-208a/b and hsa-miR-499) were characterized as biomarkers of myocardial damage and severity of an infarction68. Additional data obtained from patients and animal models showed a positive correlation of muscle and myocardial circulating miRNAs with the course of acute MI with segment-T elevation (STEMI). In fact, the circulating levels of miR-1, miR-133a, miR-133b and miR-499-5p followed the same increasing pattern as observed for Troponin-T concentration in STEMI patients and left-ventricular ejection fraction (LVEF), being considered to be correlated with the degree of myocardial damage and necrosis after infarction69. Interestingly, those pioneer studies in the early 2010 showed that the levels of circulating non-muscle miRNAs such as the liver hsa-miR-122-5p or the pancreatic-specific hsa-miR-375 in STEMI patients followed the opposite pattern to the muscle and cardiac-specific miRNAs, being downregulated in this cohort of patients69. These results were in contradiction with those found in animal models of cardiogenic shock, where the plasma levels of hepatic scr-miR-122-5p showed a dramatic increase after the external cardiac intervention, and could be explained due to the time of sample collection after the infarction70. In fact, recent studies showed that plasma levels of hsa-miR-122-5p measured at less than 8 h after infarction showed the same increased patterns that in the animal model, and could be used in combination with other cardiac-specific miRNAs such as hsa-miR-133b to successfully stratify the risk STEMI patients71. Levels of hsa-miR-133b in MI are determined by the infarct related coronary artery (IRA) occlusion regardless ST-elevation. Patients with the occluded IRA had higher levels of hsa-miR-133a, hsa-miR-133b then patients with patent IRA, but no difference in troponin T levels. These data suggest that circulating miRNA elevations in MI may indicate patients requiring urgent coronary revascularization72.
Circulating miRNAs were also used to predict the individual risk for future fatal acute MI in healthy individuals73. The HUNT study, performed over 112 health participants, described 10 plasmatic miRNAs differentially expressed between fatal cases and controls. The best model for predicting future fatal MI include the levels of a miRNA panel composed by hsa-miR-106a-5p, hsa-miR-424-5p, hsa-let-7g-5p, hsa-miR-144-3p and hsa-miR-660-5p, providing 77.6% correct risk assessment for both genders, and 74.1% and 81.8% for men and women, respectively73. Other circulating miRNAs such as hsa-miR-192-5p, hsa-miR-194-5p and hsa-miR-34a-5p were also demonstrated to be good risk assessment predictors for heart failure after MI74. Interestingly, these miRNAs are expressed in a p53-dependent manner, which connect them with other miRNAs already characterized as drivers of CVDs that also appeared misregulated in cancer75. Additional studies of circulating miRNAs and different forms of MI completed the universe of more than 60 different miRNAs from diverse origins that can be used for the diagnosis and risk stratification (Figure 3 and Supplementary Table S2).
Figure 3.
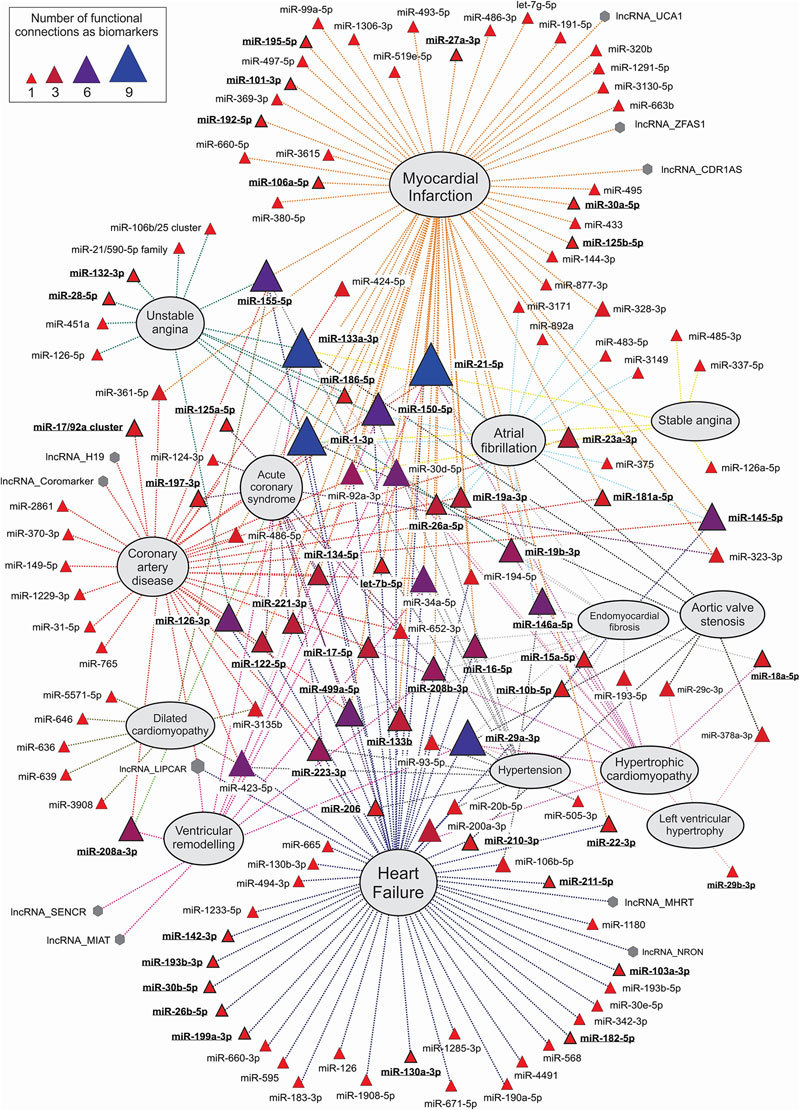
Overall landscape of plasma/serum ncRNAs biomarkers of cardiovascular diseases (CVDs). The functional network was constructed by manual text mining on bibliography, and connects every ncRNA with a particular CVD where it has been described as functional biomarker in any of the following items: onset, progression, prognostic, diagnostic or therapeutic response to treatments (see Supplementary Table S2 for more details). Network was firstly produced by Navigator software117 and further manually refined by graphical editing. MiRNAs are represented as triangles and lncRNAs as hexagons. The size of each ncRNA symbol is proportional to the number of functional relationships established as biomarker with cardiovascular conditions. The names of miRNAs also experimentally characterized as main functional players in cardiovascular conditions are highlighted in bold and underlined text. For clarity reasons, miRNA nomenclature from miRBase 21.0 has been abbreviated by removing the specific human (hsa) prefix.
Recently, some circulating lncRNAs were also described as potential biomarkers for acute MI. The circulating Zinc finger antisense 1 (ZFAS1) and CDR1 antisense (CDR1AS), showed significant differential expression between acute MI patients and control subjects, being inversely correlated between them. Similar changes of circulating ZFAS1 and CDR1AS were consistently observed also in mouse models. Reciprocal changes of circulating ZFAS1 and CDR1AS independently predict acute MI76. Another lncRNA, designed as urothelial carcinoma-associated 1 (UCA1) is known to be expressed in bladder and lung cancers, being a predictive biomarker for such kind of tumors. In healthy individuals, UCA1 is also specifically expressed in heart of adult. The level of plasma UCA1 is decreased at the early state of acute MI patients and increased at day 3 after the crisis. UCA1 circulating levels are inversely associated with the expression of hsa-miR-177.
Heart failure
Heart failure is a complex disease often caused by other conditions, characterized by the existence of a decreased heart pumping efficiency, insufficient to support all the needs of the body and lungs. This condition has a very diverse pathophysiology which can involve a dysfunction either at the systolic or diastolic levels78. The existence of contributing factors to the heart failure due to previous CVDs such as atrial fibrillation and MI, and pharmacological treatments, makes the biomarker discovery in this area extremely challenging79. For diagnosis, non-invasive cardiac imaging techniques such as ultrasound scanning, cardio computer tomography (cardioCT) or positron emission tomography (PET) and the determination of cardiac electrophysiological parameters, have to be supported with standard biochemical biomarkers as circulating levels of the beta-natriuretic peptide B (BNP). Elevated BNP levels are useful to distinguish different form of acute heart failure from other dyspnea-causing conditions80. Interestingly, the first evidence that links a circulating ncRNA to heart failure syndrome was obtained from a study to find circulating miRNA biomarkers to stratify different types of dyspnea. Tijsen and coworkers discovered a panel of 6 upregulated circulating miRNAs linked to the dyspneic phenotype, where only hsa-miR-423-5p was strongly correlated with heart failure patients81. This circulating miRNA was previously characterized as upregulated within the myocardium in chronic heart failure patients82.
There are at least 50 different circulating miRNAs and 3 circulating lncRNAs described as biomarkers of different manifestations of heart failure (Figure 3, Supplementary Table S2). Comparative screening studies between myocardial and circulating miRNAs in heart failure allowed demonstrating that miRNAs are more sensitive than classical biomarkers of heart injury and failure as BNP or cardiac troponins. In humans, some heart and muscle-specific circulating miRNAs levels are increased in heart failure, and their levels were reverted after left ventricular assist device (LVAD) support83. In a rat model of heart failure induced by hypertension, Dickinson and coworkers demonstrate that circulating miRNAs can be used as biomarkers of therapeutic response and disease progression. Screening for circulating miRNA in this model showed that circulating levels of rno-miR-16, rno-miR-20b, rno-miR-93, rno-miR-106b, rno-miR-223, and rno-miR-423-5p were significantly increased in response to hypertension-induced heart failure, while this effect was reverted after treatment with ACE inhibitors84.
Recently, a more detailed comparative study was performed in a reduced cohort of patients with chronic heart failure, showing that 50% of the miRNAs with altered levels in the myocardium of patients remained unaltered in circulation, whereas circulating miRNAs from non-cardiac origin showed more variability in heart failure patients. Interestingly, four fibroblast-enriched miRNAs (hsa-miR-660-3p, -665, -1285-3p and -4491) were significantly upregulated simultaneously in heart tissue and plasma samples during heart failure, being correlated with the compromise of cardiac function quantified as LVEF85. In patients with systolic dysfunction during heart failure, the left ventricular mass (LVM) is a classical prognosis parameter, that has been also positively correlated with the circulating levels of hsa-miR-155-5p and hsa-miR-59586. Moreover, circulating miRNAs have been also used for patient stratification in different degrees of ventricular malfunction. In plasma, the levels of four circulating microRNAs (hsa-miR-125a-5p, -190a-5p, -550a-5p, and -638) distinguished heart failure with reduced vs preserved left ventricular ejection fraction, showing stronger discriminative power than N-terminal pro-brain natriuretic peptide (NT-proBNP)87.
Other circulating ncRNAs such as lncRNAs were studied as possible biomarkers of heart failure. In patients with heart failure induced by a previous MI, the levels of circulating mitochondrial lncRNA LIPCAR (uc022bqs.1) were shown to be correlated with the patients developing cardiac remodeling, being an independent prediction risk factor for survival rate88. Recently, the non-coding repressor of NFAT (NRON) and myosin heavy-chain-associated RNA transcripts (MHRT) were demonstrated to be upregulated in plasma of heart failure patients and proposed as new predictive biomarkers for the disease89.
Cardiomyopathies
Cardiomyopathies are a group of cardiac diseases that are characterized by morphological and functional abnormalities in the myocardium. They can be classified as primary or intrinsic cardiomyopathies when they have their origin in a myocardial dysfunction or physical change, and secondary or extrinsic cardiomyopathies, when their causing factors are extrinsic to the heart90. Intrinsic cardiomyopathies can have a genetic inherited basis or being acquired in response to a stress over the myocardium. Inherited cardiomyopathies are the most frequent form of the disease and in consequence, genetic tests are the most prevalent form of diagnosis, together with cardiac imaging techniques. However, patients with cardiomyopathies usually undergo series of biochemical tests for the detection of biomarkers that can aid to the diagnosis of conditions that can increase myocardial stress and assess secondary organ dysfunction (for a comprehensive review about these biomarkers see91).
Deregulation of the myocardial expression levels of some ncRNAs as miRNAs was early demonstrated in cardiomyopathies6, and the possible value of circulating miRNAs as biomarkers also investigated by several research groups. Initial studies for the use of circulating miRNAs as biomarkers and performed in dilated cardiomyopathy patients were not conclusive, since they showed increased levels of some plasmatic miRNAs as hsa-miR-423-5p that were not correlated with the severity of the cardiomyopathy itself but with the cases of heart failure associated with the primary condition92. In subsequent studies, it was possible to demonstrate that cardiomyopathies are characterized by increased circulating levels of both cardiac- and non-cardiac-specific miRNAs93. Cardiac remodeling associated with hypertrophic cardiomyopathy appeared to be responsible for the release of miRNAs into the bloodstream. However, correlation with left ventricular hypertrophy parameters is significant for only a few miRNAs (hsa-miR-199a-5p, -27a-3p, and -29a-5p), whereas only hsa-miR-29a-5p appeared to be associated with both cardiac hypertrophy and fibrosis which could suggest its use as a biomarker for myocardial remodeling assessment93.
In the context, circulating miRNAs have been employed in the risk stratification of specific groups such as pediatric patients suffering genetically inherited dilated cardiomyopathy and the need for a potential heart transplantation. In fact, a molecular signature of four circulating miRNAs was able to differentiate those children with recovery potential from those that would need a heart transplant. Two miRNAs were significantly up-regulated (hsa-miR-155-5p and hsa-miR-636) and two miRNAs were down-regulated (hsa-miR-646 and hsa-miR-639) in children with genetic dilated cardiomyopathy who were transplanted or died compared with patients who recovered their ventricular function94.
In dilated cardiomyopathy the other most common presentation of the disease, characterized by an enlargement of the ventricular cavity, circulating miRNAs have been shown to be useful for the diagnosis of cardiac fibrosis. Fang and coworkers, demonstrated that a panel of eight circulating miRNAs (hsa-miR-15a-5p, -17-5p, -18a-5p, -193-5p, -19b-3p, -200a-3p, -21-5p and -29a-3p) offered increased sensitivity and specificity to diagnose fibrosis originated as a complication of dilated cardiomyopathy95.
Coronary artery disease and atherosclerosis
In general, coronary artery disease and atherosclerosis is associated with endothelial dysfunction and impaired lipid metabolism, in both miRNAs have their significant impact96. The role of miRNAs in atherosclerosis and its attendant clinical complications has been described thoroughly by Feinberg and Moore96. MiRNAs can regulate development and progression of atherosclerotic plaque on different pathways: regulation of lipid homeostasis, cytokine responsiveness, leukocyte recruitment and vascular smooth muscle cell function97. In situ, the miRNA expression profile is changing in early coronary atherosclerotic plaques in carotid endarterectomy specimens from patients with asymptomatic and symptomatic MI97. Another candidate miRNA, hsa-miR-370 is associated with lipid metabolism and its plasma levels are increased in CVD patients both with normo- and hyperlipidemia98. Interestingly, hyperlipidemic conditions favor the increase of hsa-miR-122-5p levels, designate this miRNA as a prognostic biomarker of AMI71,98. Circulating hsa-miR-208a-3p levels, which is specifically expressed in the heart muscle, are significantly higher in plasma of CVD patients and correlates with the severity of coronary atherosclerosis99.
Type 2 diabetes mellitus induced atherosclerosis, increased levels of circulation hsa-miR-21-5p, hsa-miR-218-5p and hsa-miR-211 was confirmed, designate them as useful biomarkers for detecting the progression of atherosclerosis100 (Figure 1B). As a negative biomarker, miR-126-3p should be mentioned. Decreased levels of circulating hsa-miR-126-3p were observed in stable CVD and increased expression of hsa-miR-126 in circulating microvesicles was significantly associated with a lower incidence of major adverse cardiovascular events rate101. Two miRNAs associated with CVD progression are highly abundant in HDL fraction: hsa-miR-223 and hsa-miR-126-3p, but the increase in HDL-miRNA in CAD progression associated with hsa-miR-126-3p only97.
Atherosclerotic plaque destabilization can be predicted by several miRNAs related with matrix metalloproteinase expression (hsa-miR-21-5p) or apoptosis (hsa-miR-135a/ hsa-miR-147), however the last ones were overexpressed in peripheral blood mononuclear cells (PBMCs) only102. In patients with non-calcified coronary artery lesions, higher miR-21-5p in macrophages and lower hsa-miR-21-5p serum levels appears to be a unique signature associated with coronary plaque instability103 (Figure 1B). Interestingly, hsa-miR-21-5p (together with hsa-miR-126-5p and five others) is upregulated in microvesicles obtained from unstable CVD, showing their cell origin104. Thus the approach to use miRNA signature in circulating EVs to predict disease (CVD), seems to be the more appropriate approach then analysis of total miRNAs isolated from PBMCs.
In terms of atherosclerosis biomarkers, some issues must be raised. Smoking, which is a major risk factor for the development of CVD, leads to an increase of circulating leukocyte-derived microvesicles (ectosomes) and an increase in pro-atherogenic hsa-miR-29b content. Meanwhile, smoking reduces platelet-derived microvesicles and hsa-miR-223 related to them105.
Very recently, the combined use of miRNAs and circRNAs as biomarkers for carotide plaque rupture was also described. In a validation study composed by a cohort of 112 patients, Bazán and coworkers demonstrated that the ratio of serum circR-284/hsa-miR-221 was significantly elevated in the acutely symptomatic patients with high-grade carotid disease, and exhibited favorable characteristics to be consider as a biomarker of carotid plaque rupture and stroke106.
Other cardiovascular conditions
Cardiac ischemia can lead to severe functional complications and finally to failure of the myocardium. Among the different classes of complications derived from cardiac ischemias, anginas are the most prevalent in western countries. Angina pectoris is characterized by a chest pain, and can be stable or unstable depending on its progression dynamics; whereas stable angina symptoms are confined in a short time lapse, unstable angina is characterized by a reduction in coronary flow that could lead to an ischemia, myocardium necrosis and subsequent heart attack107. There is a current medical need for diagnosis biomarkers allowing the differentiation between stable and unstable forms of angina. Plasma circulating miRNA signatures have been recently proposed as a diagnosis biomarker for stable vs unstable angina. In patients with stable coronary artery disease, 3 circulating miRNAs were consistently upregulated hsa-miR-337-5p, hsa-miR-433, and hsa-miR-485-3p when compared with healthy individuals. From those miRNAs, the plasma levels of hsa-miR-337-5p were positively correlated with stable angina events108. Using a profiling-validation-replication model, Zeller and coworkers found an additional group of circulating miRNAs (hsa-miR-132-3p, -150-5p and -186-5p) that showed high discriminatory power between patients with stable vs unstable angina109. Niculescu and coworkers described the use of the HDL-associate miRNAs hsa-miR-92a-3p and hsa-miR-486 as discriminating biomarkers useful to distinguish between stable and vulnerable coronary artery disease patients63. Screening studies performed by low-density qPCR arrays also described a circulating miRNA signature, consisting of the hsa-miR-106b/25 cluster, hsa-miR-17/92a cluster, hsa-miR-21/590-5p family, hsa-miR-126a-5p and hsa-miR-451a, that predicted a negative outcome in patients with coronary artery disease104.
Cardiac arrhythmias are also sources of cardiovascular complications, especially when they appear associated to other heart functional defects. In general, the most prevalent form of cardiac arrhythmia in western countries is atrial fibrillation (AF). Pioneer studies in the field of circulating ncRNAs and AF showed that miRNAs contained in platelets could be used as biomarkers of the disease110. When AF appeared as a complication of a heart surgery, the levels of two circulating miRNAs (hsa-miR-23a-3p and -26a-5p) were significantly decreased in those patients showing post-operative AF after coronary bypass intervention, suggesting that those circulating miRNA could be involved in the underlying biology of the development of this complication111. Moreover, and as a part of the inflammatory response associated with AF, patients with the established chronic fibrillation showed decreased levels of circulating hsa-miR-150-5p, that were negatively correlated with the C-reactive protein levels, a classical biomarker of the disease112. Genome-wide studies such as the miRythm, demonstrated a correlation between the levels of myocardial and plasma miRNAs in patients with AF after catheter ablation. This study was composed by a cohort of 112 participants with AF and 99 without AF after the surgery, where 31 patients were considered for the screening of miRNA levels in atrial tissue and in plasma. The main findings suggested the correlation between circulating and heart tissue levels of hsa-miR-150-5p and hsa-miR-21-5p in patients with AF113. These miRNAs have been already described as regulators of atrial remodeling and fibrosis11. Interestingly, the circulating miRNAs showed a specific pattern of differential regulation in AF patients when the coronary circulation was considered. A study by Xu and coworkers supported by the idea that coronary circulation would better reflect the state and metabolic level of myocardial miRNA in those patients, demonstrated that the levels of circulating miRNAs in coronary blood are different to those observed in peripheral samples. In AF patients, coronary circulating hsa-miR-892a and hsa-miR-3149 were up-regulated and hsa-miR-3171 was down-regulated when compared with healthy individuals, suggesting their possible use as a biomarker of the disease114.
Limitations and future directions
From the data presented in this review, it is clear that the potential of circulating ncRNAs as non-invasive biomarkers of CVDs is enormous, either alone or combined with classic biochemical or electrophysiological biomarkers (Figure 3). They offer the sensitivity and specificity required to any biomarker however their use in the clinical practice for diagnosis, prognosis or therapeutic response analysis is still not implemented. The reasons for this lack of implementation and inclusion within the clinical guidelines are diverse, but all of them have a technical background. Firstly, there is not a standard protocol for the processing of the biofluids in order to extract circulating nucleic-acids. Some protocols are based on the use of phenol-containing reagents, whereas others are phenol-free protocols which rely on the use of ion-exchange chromatographic separation. The yield and purity of each method are different and also the potential contaminants found. Secondly, the small amounts of recovered nucleic acids from biofluids make their quantification a challenging task. Classical spectrophotometric approach for the quantification of nucleic acids is often useless when dealing with the small amounts of sample obtained, and the alternative methods (spectrofluorimetry and capillary electrophoresis) are expensive and in some cases time consuming. Directly related with the quantification of the sample, another technical thread is the technique employed for the profiling of the circulating nucleic acids. For screening, next generation sequencing (NGS) or low-density qPCR arrays are preferred, whereas for direct sampling the only available technique is qPCR. The development of modified oligonucleotides, namely Taqman or LNA probes, has made qPCR a routine technique but it remains a challenge for the quantification of low-abundance species. Quantitative PCR needs also proper normalization controls and protocols, which in the case of biofluids are not consensual among the scientific community and need to be standardized. In the particular case of next-generation sequencing, there is also not a standard protocol for library preparation and pre-amplification of the initial sample which hardly condition the possibility of comparing results obtained by the different approaches and sequencing platforms. In both quantification strategies, NGS and qPCR the reverse-transcription reaction required for the cDNA synthesis is often challenging due to the low-molecular weight contaminants present in the biofluids that could inhibit the reaction.
There are, of course, new trends and fields to explore in this area. Nowadays the preferred fluid for ncRNA quantification is blood, but other fluids remained unexplored. Saliva, for instance, could be a very interesting field to investigate since it is the body fluid that contains a higher number of circulating ncRNA species and it is easy to collect. However, the natural composition of salivar fluid, enriched in polysaccharides, makes it very difficult to process for RNA extraction. Another noninvasive source of circulating ncRNA species is urine. It is easy to collect and reach in extracellular vesicles having both protein and miRNA cargo. A cardio-enriched hsa-miR-1 is present in urine from patients after open-heart surgeries and its levels are correlated with cardiac troponin I115.
Many interesting topics related with this field have arisen in the last decade and remained also almost unexplored in the biomarker field. One of the most interesting fields of future studies is the compartmentalization of ncRNAs as circulating species within fluids, and the determination of the different populations of ncRNAs in different compartments. This idea has been started to be explored in the blood with the determination of ncRNA subpopulations associated with circulating bodies, namely microparticles, exosomes, ectosomes, platelets and lipoproteins, but it will require further investigation. We expect that a deeper knowledge in this field will allow the clinicians to study determined subpopulations of ncRNAs associated to a compartment that would be more relevant for a particular CVD.
The progress in the ncRNA field brought out the generation of huge data which need big repositories enable ncRNA sequences classification and categorization. Some projects have been established as for example the miRandola 2017 database, to feel the gap between circulating miRNA presence and pathology116. miRandola is a comprehensive manually curated classification of different extracellular circulating non-coding RNA types. This database classifies extracellular RNAs based on their extracellular form: Argonaute 2 protein, exosome, microvesicle, microparticle, membrane vesicle, high density lipoprotein and other circulating forms. This kind of knowledge repository can serve as information source not only for laboratory testing, but may be evaluated during data mining or machine learning.
Finally, and understanding that the presence of ncRNAs in the fluids is a natural process not only related with cell death but also with active secretion mechanisms, there is a clear mechanistic need that dissect the cellular pathway(s) employed to export different ncRNAs to the extracellular medium, and which are the reasons and specificity of this process. The information available in the literature about this topic is disperse and only covers a few possible partners that will collaborate in the secretion of ncRNA species. Only if we understand the mechanism of ncRNA sorting and introduction into the cell secretion machinery, we will be able to correlate them with a possible function. Recent literature point out the real action of ncRNAs as “genetic hormones” with a potential to regulate distant target cells by being transported within circulating biofluids in a real “organ cross-talk”. There is still a long road to pave, but a fundamented physiological insight to this circulating ncRNA species is needed in order to understand their abilities, roles and potential uses as diagnosis tools and therapeutic targets for CVDs.
Conclusion
The use of circulating ncRNAs as biomarkers for CVDs offers increased advantages over the classical biomarkers; however there is still a current medical need for a translation from the bench to the bedside and their inclusion in medical guidelines. Among circulating ncRNAs, plasma/serum miRNAs have been described as potential biomarkers in many cardiac pathologies, with applications in diagnosis, prognosis and therapeutic response. Other circulating ncRNAs species as lncRNAs and circRNAs are also promising biomarkers in the CVD field, however their physiological roles in the context of CVDs remain largely unknown. Current technical limitations in the extraction and quantification procedures for circulating ncRNAs need to be circumvented in order to implement solid protocols for their consideration as clinically useful biomarkers. Moreover the future detailed knowledge of the presence of these molecular species in several circulating compartments (platelets, microparticles, microvesicles) and their relationships with CVDs are also needed to establish reliable biomarkers that could be useful in the clinical practice.
Electronic supplementary material
miRNAs in cardiovascular_pathologies.
Circulating ncRNAs as biomarkers in cardiac pathologies.
Electronic supplementary material
Supplementary Information is available at the website of Acta Pharmacologica Sinica. Molecular Psychiatry website 10.1038/aps.2018.35
References
- 1.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–91. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 3.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–71. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barwari T, Joshi A, Mayr M. MicroRNAs in cardiovascular disease. J Am Coll Cardiol. 2016;68:2577–84. doi: 10.1016/j.jacc.2016.09.945. [DOI] [PubMed] [Google Scholar]
- 8.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuo Y, Gao G, Shi JA, Zhou X, Wang X. miRNAs: biogenesis, origin and evolution, functions on virus-host interaction. Cell Physiol Biochem. 2013;32:499–510. doi: 10.1159/000354455. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22:1243–54. doi: 10.1101/gr.132514.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam O, Lohfelm B, Thum T, Gupta SK, Puhl SL, Schafers HJ, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 12.Wei XJ, Han M, Yang FY, Wei GC, Liang ZG, Yao H, et al. Biological significance of miR-126 expression in atrial fibrillation and heart failure. Braz J Med Biol Res. 2015;48:983–9. doi: 10.1590/1414-431X20154590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci (Lond) 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 14.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–73. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–4. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, et al. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–9. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramanujam D, Sassi Y, Laggerbauer B, Engelhardt S. Viral Vector-Based targeting of miR-21 in cardiac nonmyocyte cells reduces pathologic remodeling of the heart. Mol Ther. 2016;24:1939–48. doi: 10.1038/mt.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiang L, Hong L, Ningfu W, Huaihong C, Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. Int J Cardiol. 2013;168:2082–8. doi: 10.1016/j.ijcard.2013.01.160. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Cui C, Yang X, Xu J, Venkat P, Zacharek A, et al. MiR-126 affects brain-heart interaction after cerebral ischemic stroke. Transl Stroke Res. 2017;8:374–85. doi: 10.1007/s12975-017-0520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortez-Dias N, Costa MC, de Sousa J, Fiuza M, Gallego J, Nobre A, et al. Expanding the functional role of miRNAs in the establishment of permanent atrial fibrillation. Int J Cardiol. 2016;222:340–1. doi: 10.1016/j.ijcard.2016.07.296. [DOI] [PubMed] [Google Scholar]
- 24.Moraes LN, Fernandez GJ, Vechetti-Junior IJ, Freire PP, Souza RWA, Villacis RAR, et al. Integration of miRNA and mRNA expression profiles reveals microRNA-regulated networks during muscle wasting in cardiac cachexia. Sci Rep. 2017;7:6998. doi: 10.1038/s41598-017-07236-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowara M, Cudnoch-Jedrzejewska A, Opolski G, Wlodarski P. MicroRNA regulation of extracellular matrix components in the process of atherosclerotic plaque destabilization. Clin Exp Pharmacol Physiol. 2017;44:711–8. doi: 10.1111/1440-1681.12772. [DOI] [PubMed] [Google Scholar]
- 26.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32:211–24. doi: 10.1016/j.tig.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–24. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataoka M, Huang ZP, Wang DZ. Build a braveheart: the missing linc (RNA) Circ Res. 2013;112:1532–4. doi: 10.1161/CIRCRESAHA.113.301519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou J, Long H, Zhou C, Zheng S, Wu H, Guo T, et al. Long noncoding RNA brave heart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res Ther. 2017;8:4. doi: 10.1186/s13287-016-0454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Li Y, Lin B, Sheng Y, Yang L. HBL1 is a human long noncoding rna that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev Cell. 2017;42:333–48. doi: 10.1016/j.devcel.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14:183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 36.Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med 2017; 9. pii: eaai9118. doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed]
- 37.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 39.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487:769–75. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–20. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappl G, Hasselmann DO, Rossler M, Ugurel S, Tilgen W, Reinhold U. Detection of tumor-associated circulating mRNA in patients with disseminated malignant melanoma. Ann N Y Acad Sci. 2001;945:189–91. [PubMed] [Google Scholar]
- 45.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 46.Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–5. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepien E, Stankiewicz E, Zalewski J, Godlewski J, Zmudka K, Wybranska I. Number of microparticles generated during acute myocardial infarction and stable angina correlates with platelet activation. Arch Med Res. 2012;43:31–5. doi: 10.1016/j.arcmed.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 48.van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 49.Waldenstrom A, Ronquist G. Role of exosomes in myocardial remodeling. Circ Res. 2014;114:315–24. doi: 10.1161/CIRCRESAHA.114.300584. [DOI] [PubMed] [Google Scholar]
- 50.Kim DK, Lee J, Simpson RJ, Lotvall J, Gho YS. EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Semin Cell Dev Biol. 2015;40:4–7. doi: 10.1016/j.semcdb.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–59. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ribeiro-Rodrigues TM, Laundos TL, Pereira-Carvalho R, Batista-Almeida D, Pereira R, Coelho-Santos V, et al. Exosomes secreted by cardiomyocytes subjected to ischaemia promote cardiac angiogenesis. Cardiovasc Res. 2017;113:1338–50. doi: 10.1093/cvr/cvx118. [DOI] [PubMed] [Google Scholar]
- 54.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 55.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 2016; 5. pii: e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed]
- 57.Betapudi V, Lominadze G, Hsi L, Willard B, Wu M, McCrae KR. Anti-beta2GPI antibodies stimulate endothelial cell microparticle release via a nonmuscle myosin II motor protein-dependent pathway. Blood. 2013;122:3808–17. doi: 10.1182/blood-2013-03-490318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexy T, Rooney K, Weber M, Gray WD, Searles CD. TNF-alpha alters the release and transfer of microparticle-encapsulated miRNAs from endothelial cells. Physiol Genomics. 2014;46:833–40. doi: 10.1152/physiolgenomics.00079.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen F, Yang X, Hoelscher M, Cattelan A, Schmitz T, Proebsting S, et al. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128:2026–38. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
- 60.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–61. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 61.Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, et al. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circ Res. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niculescu LS, Simionescu N, Sanda GM, Carnuta MG, Stancu CS, Popescu AC, et al. MiR-486 and miR-92a identified in circulating HDL discriminate between stable and vulnerable coronary artery disease patients. PLoS One. 2015;10:e0140958. doi: 10.1371/journal.pone.0140958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dlouha D, Blaha M, Blaha V, Fatorova I, Hubacek JA, Stavek P, et al. Analysis of circulating miRNAs in patients with familial hypercholesterolaemia treated by LDL/Lp(a) apheresis. Atheroscler Suppl. 2017;30:128–34. doi: 10.1016/j.atherosclerosissup.2017.05.037. [DOI] [PubMed] [Google Scholar]
- 65.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, et al. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:1392–400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 66.Hachey BJ, Kontos MC, Newby LK, Christenson RH, Peacock WF, Brewer KC, et al. Trends in use of biomarker protocols for the evaluation of possible myocardial infarction. J Am Heart Assoc 2017; 6. [DOI] [PMC free article] [PubMed]
- 67.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 69.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersson P, Gidlof O, Braun OO, Gotberg M, van der Pals J, Olde B, et al. Plasma levels of liver-specific miR-122 is massively increased in a porcine cardiogenic shock model and attenuated by hypothermia. Shock. 2012;37:234–8. doi: 10.1097/SHK.0b013e31823f1811. [DOI] [PubMed] [Google Scholar]
- 71.Cortez-Dias N, Costa MC, Carrilho-Ferreira P, Silva D, Jorge C, Calisto C, et al. Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ J. 2016;80:2183–91. doi: 10.1253/circj.CJ-16-0568. [DOI] [PubMed] [Google Scholar]
- 72.Gacon J, Kablak-Ziembicka A, Stepien E, Enguita FJ, Karch I, Derlaga B, et al. Decision-making microRNAs (miR-124, -133a/b, -34a and -134) in patients with occluded target vessel in acute coronary syndrome. Kardiol Pol. 2016;74:280–8. doi: 10.5603/KP.a2015.0174. [DOI] [PubMed] [Google Scholar]
- 73.Bye A, Rosjo H, Nauman J, Silva GJ, Follestad T, Omland T, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals - The HUNT study. J Mol Cell Cardiol. 2016;97:162–8. doi: 10.1016/j.yjmcc.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res. 2013;113:322–6. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 75.Evans S, Mann DL. Circulating p53-responsive microRNAs as predictive biomarkers in heart failure after acute myocardial infarction: the long and arduous road from scientific discovery to clinical utility. Circ Res. 2013;113:242–4. doi: 10.1161/CIRCRESAHA.113.301951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X, et al. Circulating long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. Biomed Res Int. 2016;2016:8079372. doi: 10.1155/2016/8079372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mentz RJ, O'Connor CM. Pathophysiology and clinical evaluation of acute heart failure. Nat Rev Cardiol. 2016;13:28–35. doi: 10.1038/nrcardio.2015.134. [DOI] [PubMed] [Google Scholar]
- 79.Olivari MT. Behavioral and environmental factors contributing to the development and progression of congestive heart failure. J Heart Lung Transplant. 2000;19:S12–20. doi: 10.1016/s1053-2498(99)00106-0. [DOI] [PubMed] [Google Scholar]
- 80.O'Donoghue M, Chen A, Baggish AL, Anwaruddin S, Krauser DG, Tung R, et al. The effects of ejection fraction on N-terminal ProBNP and BNP levels in patients with acute CHF: analysis from the ProBNP Investigation of Dyspnea in the Emergency Department (PRIDE) study. J Card Fail. 2005;11:S9–14. doi: 10.1016/j.cardfail.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 81.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 82.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 83.Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci U S A. 2014;111:11151–6. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15:650–9. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 85.Li H, Fan J, Yin Z, Wang F, Chen C, Wang DW. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ikitimur B, Cakmak HA, Coskunpinar E, Barman HA, Vural VA. The relationship between circulating microRNAs and left ventricular mass in symptomatic heart failure patients with systolic dysfunction. Kardiol Pol. 2015;73:740–6. doi: 10.5603/KP.a2015.0082. [DOI] [PubMed] [Google Scholar]
- 87.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015;17:393–404. doi: 10.1002/ejhf.223. [DOI] [PubMed] [Google Scholar]
- 88.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–75. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 89.Xuan L, Sun L, Zhang Y, Huang Y, Hou Y, Li Q, et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J Cell Mol Med. 2017;21:1803–14. doi: 10.1111/jcmm.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–16. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 91.Coats CJ, Heywood WE, Mills K, Elliott PM. Current applications of biomarkers in cardiomyopathies. Expert Rev Cardiovasc Ther. 2015;13:825–37. doi: 10.1586/14779072.2015.1053873. [DOI] [PubMed] [Google Scholar]
- 92.Fan KL, Zhang HF, Shen J, Zhang Q, Li XL. Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy. Indian Heart J. 2013;65:12–6. doi: 10.1016/j.ihj.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roncarati R, Viviani Anselmi C, Losi MA, Papa L, Cavarretta E, Da Costa Martins P, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2014;63:920–7. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 94.Miyamoto SD, Karimpour-Fard A, Peterson V, Auerbach SR, Stenmark KR, Stauffer BL, et al. Circulating microRNA as a biomarker for recovery in pediatric dilated cardiomyopathy. J Heart Lung Transplant. 2015;34:724–33. doi: 10.1016/j.healun.2015.01.979. [DOI] [PubMed] [Google Scholar]
- 95.Fang L, Ellims AH, Moore XL, White DA, Taylor AJ, Chin-Dusting J, et al. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J Transl Med. 2015;13:314. doi: 10.1186/s12967-015-0672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feinberg MW, Moore KJ. MicroRNA regulation of atherosclerosis. Circ Res. 2016;118:703–20. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Markus B, Grote K, Worsch M, Parviz B, Boening A, Schieffer B, et al. Differential expression of MicroRNAs in endarterectomy specimens taken from patients with asymptomatic and symptomatic carotid plaques. PLoS One. 2016;11:e0161632. doi: 10.1371/journal.pone.0161632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao W, He HW, Wang ZM, Zhao H, Lian XQ, Wang YS, et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012;11:55. doi: 10.1186/1476-511X-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y, Li HH, Yang R, Yang BJ, Gao ZY. Association between circulating microRNA-208a and severity of coronary heart disease. Scand J Clin Lab Invest. 2017;77:379–84. doi: 10.1080/00365513.2017.1328740. [DOI] [PubMed] [Google Scholar]
- 100.Zhang JY, Gong YL, Li CJ, Qi Q, Zhang QM, Yu DM. Circulating MiRNA biomarkers serve as a fingerprint for diabetic atherosclerosis. Am J Transl Res. 2016;8:2650–8. [PMC free article] [PubMed] [Google Scholar]
- 101.Jansen F, Yang X, Proebsting S, Hoelscher M, Przybilla D, Baumann K, et al. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc. 2014;3:e001249. doi: 10.1161/JAHA.114.001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394:792–7. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 103.Fan X, Wang E, Wang X, Cong X, Chen X. MicroRNA-21 is a unique signature associated with coronary plaque instability in humans by regulating matrix metalloproteinase-9 via reversion-inducing cysteine-rich protein with Kazal motifs. Exp Mol Pathol. 2014;96:242–9. doi: 10.1016/j.yexmp.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 104.Ren J, Zhang J, Xu N, Han G, Geng Q, Song J, et al. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS One. 2013;8:e80738. doi: 10.1371/journal.pone.0080738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Badrnya S, Baumgartner R, Assinger A. Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb Haemost. 2014;112:128–36. doi: 10.1160/TH13-11-0977. [DOI] [PubMed] [Google Scholar]
- 106.Bazan HA, Hatfield SA, Brug A, Brooks AJ, Lightell DJ Jr, Woods TC. Carotid plaque rupture is accompanied by an increase in the ratio of serum circR-284 to miR-221 levels. Circ Cardiovasc Genet 2017; 10. pii: e001720. doi: 10.1161/CIRCGENETICS.117.001720. [DOI] [PMC free article] [PubMed]
- 107.Ford TJ, Corcoran D, Berry C. Stable coronary syndromes: pathophysiology, diagnostic advances and therapeutic need. Heart. 2018;104:284–92. doi: 10.1136/heartjnl-2017-311446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.D'Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, et al. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One. 2013;8:e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeller T, Keller T, Ojeda F, Reichlin T, Twerenbold R, Tzikas S, et al. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J. 2014;35:2106–14. doi: 10.1093/eurheartj/ehu151. [DOI] [PubMed] [Google Scholar]
- 110.Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol. 2014;113:976–81. doi: 10.1016/j.amjcard.2013.11.060. [DOI] [PubMed] [Google Scholar]
- 111.Feldman A, Moreira DAR, Gun C, Wang HL, Hirata MH, de Freitas Germano J, et al. Analysis of circulating miR-1, miR-23a, and miR-26a in atrial fibrillation patients undergoing coronary bypass artery grafting surgery. Ann Hum Genet. 2017;81:99–105. doi: 10.1111/ahg.12188. [DOI] [PubMed] [Google Scholar]
- 112.Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, et al. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS One. 2012;7:e44906. doi: 10.1371/journal.pone.0044906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, et al. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study) Heart Rhythm. 2015;12:3–10. doi: 10.1016/j.hrthm.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu G, Cui Y, Jia Z, Yue Y, Yang S. The Values of Coronary Circulating miRNAs in Patients with Atrial Fibrillation. PLoS One. 2016;11:e0166235. doi: 10.1371/journal.pone.0166235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou X, Mao A, Wang X, Duan X, Yao Y, Zhang C. Urine and serum microRNA-1 as novel biomarkers for myocardial injury in open-heart surgeries with cardiopulmonary bypass. PLoS One. 2013;8:e62245. doi: 10.1371/journal.pone.0062245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Russo F, Di Bella S, Vannini F, Berti G, Scoyni F, Cook HV, et al. miRandola 2017: a curated knowledge base of non-invasive biomarkers. Nucleic Acids Res 2018; 46: D354–D359. doi: 10.1093/nar/gkx854. [DOI] [PMC free article] [PubMed]
- 117.Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, et al. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics. 2009;25:3327–9. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miRNAs in cardiovascular_pathologies.
Circulating ncRNAs as biomarkers in cardiac pathologies.


