Abstract
Objectives
Scoring laboratory polysomnography (PSG) data remains a manual task of visually annotating 3 primary categories: sleep stages, sleep disordered breathing, and limb movements. Attempts to automate this process have been hampered by the complexity of PSG signals and physiological heterogeneity between patients. Deep neural networks, which have recently achieved expert-level performance for other complex medical tasks, are ideally suited to PSG scoring, given sufficient training data.
Methods
We used a combination of deep recurrent and convolutional neural networks (RCNN) for supervised learning of clinical labels designating sleep stages, sleep apnea events, and limb movements. The data for testing and training were derived from 10 000 clinical PSGs and 5804 research PSGs.
Results
When trained on the clinical dataset, the RCNN reproduces PSG diagnostic scoring for sleep staging, sleep apnea, and limb movements with accuracies of 87.6%, 88.2% and 84.7% on held-out test data, a level of performance comparable to human experts. The RCNN model performs equally well when tested on the independent research PSG database. Only small reductions in accuracy were noted when training on limited channels to mimic at-home monitoring devices: frontal leads only for sleep staging, and thoracic belt signals only for the apnea-hypopnea index.
Conclusions
By creating accurate deep learning models for sleep scoring, our work opens the path toward broader and more timely access to sleep diagnostics. Accurate scoring automation can improve the utility and efficiency of in-lab and at-home approaches to sleep diagnostics, potentially extending the reach of sleep expertise beyond specialty clinics.
Keywords: deep learning, sleep scoring, neural network, EEG analysis
Introduction
Common sleep disorders such as sleep apnea, insomnia, and restless legs syndrome impact tens of millions of adults and are significant risk factors for cardiometabolic and neurodegenerative diseases, impaired performance, and decreased quality of life.1–7 The population health impact is enormous, including medical and psychiatric morbidity, motor vehicle accidents, decreased work productivity and quality of life, and increased mortality.7,8 Timely and accurate diagnosis of sleep disorders is critical to pursue appropriate treatment and improve health outcomes,9 yet most sleep disorders remain undiagnosed.10,11 Recent advances in portable monitoring technology have increased access to sleep diagnostics, yet both at-home and the gold-standard in-lab polysomnography (PSG) still require manual scoring.
Previous attempts to automate diagnosis of sleep disorders have generally relied on fewer than 100 PSGs from relatively homogeneous groups of healthy individuals.12 Models trained on such datasets are not likely to generalize well, because PSG signals vary widely due to differences in demographics, medication effects, sleep conditions, and medical conditions. We address this variability using a data-driven approach based on 79 456 hours of clinical data from 10 000 nights of PSG recording. This real-world data, recorded over 8 years in a clinical sleep laboratory, makes our PSG analysis system robust to physiologic variability between patients. Most prior approaches involve preprocessing and extraction of carefully engineered features before classification.12 Our system is trained end-to-end, directly from labeled signals.
Deep neural networks, fueled by increases in computing power and availability of large labeled datasets, have recently matched the performance of medical experts in complex medical pattern recognition tasks such as visual diagnosis of dermatologic lesions13 and diabetic retinopathy.14 In this paper, we outline the development of a Recurrent Convolutional Neural Network (RCNN) that matches the performance of sleep experts in annotating overnight PSGs. No prior study has simultaneously addressed all 3 key types of PSG information extracted by expert scorers: sleep stages, respiratory events, and limb movements. Our system uses a unified deep network architecture (RCNN) to accomplish all 3 tasks. Prior work, for comparison, uses mainly small datasets, and mainly of healthy adults.15–30
Methods
Description of deep neural network development
Deep learning algorithms such as multi-layer perceptrons, convolutional neural networks (CNN), and recurrent neural networks (RNN) have been successfully applied to many domains to solve challenging tasks. The most basic computation unit in neural networks is a perceptron which performs linear combinations of input features followed by a nonlinear transformation. The standard deep neural networks (DNN) consist of multiple layers of perceptrons, which all fully connected across consecutive layers. To avoid dense connections in DNN, CNNs introduces local connections and parameter sharing through convolution operations, which demonstrated numerous successes in computer vision application such as object recognition. RNNs are another extension of DNN that are suitable for modeling sequential data such as natural language text and time series. A detailed overview of various deep learning models for analyzing medical data can be found at Xiao et al.31 Here, we briefly describe the rationale of designing and developing deep neural networks for analysis of clinical sleep data. We initially used classical machine learning algorithms such as logistic regression and random forest directly on expert defined features. However, the resulting performance is not very high as shown in Supplementary Table S2. Also, it often takes a lot of time and effort to carefully develop expert defined features, since it requires domain expertise. On the other hand, deep neural networks, such as a convolutional neural network, can extract better features and then pass those learned features in a recurrent neural network to detect sleep stages over time.
Dataset
The datasets used in this paper are from 2 sources: The Massachusetts General Hospital (MGH) sleep laboratory and the Sleep Heart Health Study (SHHS), summarized in Supplementary Table S1. Permissions for the SHHS were obtained via the online portal: www.sleepdata.org. The MGH Institutional Review Board approved retrospective analysis of clinically acquired PSG data without requiring additional consent. These 2 datasets consist of in-lab (MGH) and at-home (SHHS) PSG recordings which include combinations of electroencephalogram (EEG), respiratory signals, and electromyogram signals (EMG). The MGH dataset was scored as part of routine clinical practice by certified sleep technicians using the American Academy of Sleep Medicine (AASM) guidelines. The SHHS dataset was scored using the Rechtschaffen and Kales (R&K) guidelines. R&K scores are converted to AASM scores by combining stages NREM 3 and 4, designated in AASM as the single stages N3. The MGH dataset consists of a mixture of diagnostic, split night, and titration protocols. The SHHS PSGs are all diagnostic. EEG data is used for sleep staging, respiratory channels are used for apnea detection, and, for the MGH set, and the bilateral leg EMG channels are used for limb movement detection. The MGH dataset and SHHS dataset have 2 EEG channels in common (central). All 4 respiratory channels are present in both datasets. Pressure transducer airflow (PTAF) and EMG channels are available in the MGH dataset only.
Classification targets
Different target labels are modeled for the 3 scoring tasks.
For sleep staging, EEG signals are scored in non-overlapping 30-second epochs according to AASM standards as one of 5 stages: wake (W), rapid eye movement (REM) – R, non-REM stage 1 (N1), non- REM stage 2 (N2), and non-REM stage 3 (N3). Thus, sleep staging is formulated as a 5-class classification problem.
For respiratory event detection, we consider the following classes: obstructive apnea, central apnea, mixed apnea, and hypopnea (defined using the 4% desaturation rule). We combine these different respiratory event class labels into a single class (apnea event), and; thus, we perform a binary classification, ie presence or absence of apnea respiratory event, to mimic the clinical use of the composite apnea-hypopnea index (AHI). Event detections are performed in consecutive, non-overlapping 1-second intervals. Event detections in consecutive time windows are merged into a single “apnea event” in order to calculate the AHI, defined as the total number of apneas during sleep, divided by the number of hours of sleep (ie in sleep stage N1, N2, N3, or R). Calculation of AHI depends on the results of automated sleep staging, needed to calculate the total sleep time (the sum of N1-N3 and R).
For limb movement detection, EMG signals are marked for presence or absence of limb movement events. The majority (>90%) are periodic, and, because from a signal standpoint isolated limb movements have similar properties, we combine them into a single label. Limb movement detection is, therefore, formulated as a binary classification problem when we detect the presence or absence of limb movement events. Limb movement detections are performed in consecutive, non-overlapping 1-second intervals. Like AHI detection, limb movements detected in consecutive seconds are merged into a single event. Limb movement burden is quantified by the limb movement index (LMI), the number of limb movements per hour of sleep. Calculation of LMI for AHI depends on the results of automated sleep staging.
Data preparation
EEG data in PSG consists of signals from 6 channels, ie F3, F4, C3, C4, O1 and O2, each referenced to the contralateral mastoid. In Supplementary Figure S1, we show a schematic of the locations of the electrodes. While the MGH dataset has 6 electrodes, the SHHS dataset has only 2 EEG electrodes (C3, C4). Both MGH and SHHS datasets contain the following respiratory signals: chest belt, abdomen belt, SaO2 (oximetry), and airflow. The pressure transducer airflow (PTAF) present in the diagnosis phase of MGH set is not used in the final model, since including it yields no significant performance improvement (data not shown). The left and right anterior tibialis (LAT and RAT) EMG channels for limb movement detection are present in MGH dataset only. The sampling frequency of the data is 200Hz.
We use both raw waveform and spectrogram representations of the data as inputs for our models. For the spectrogram representation of EEG and EMG data, we segment each 30-second epoch into 29 subepochs of 2 seconds duration with 1-second overlap. For each 2-second subepoch, we use Thomson’s multitaper method to estimate the power spectral density (PSD), with the following parameters: window length, T = 2s, time-bandwidth product, TW = 3, number of tapers K = 5.32–34 For respiratory signals, the parameters are T = 30s, TW = 1.5, K = 2.
We split our datasets into train and test sets using 90/10 percentage splits of the original cohorts. Model performance is evaluated on the test sets. There is no overlap between test and training sets. As the MGH dataset has 10 000 PSGs, the train set consists of 9000 cases, and the test set consists of 1000 cases. The SHHS dataset has 5804 PSGs, so the train set has 5224 and the test set has 580 cases.
Sample selection
The sleep staging task has five different target classes: N1, N2, N3, R and W. These classes have approximately 19 million, 75 million, 22 million, 21 million, and 18 million 30 second epochs from the MGH dataset for N1-N3, R and W, respectively. Similarly, the SHHS dataset consists of approximately 11 million, 46 million, 9 million, 8 million, and 11 million, 30 second epochs for N1-N3, R and W, respectively. For sleep apnea detection, the MGH dataset contains approximately 2 million respiratory events. Similarly, the SHHS dataset has about 650 000 apnea events. For limb movement detection, the MGH data has approximately 2.7 million limb movement events.
Training algorithms
We combine two different primary types of neural networks in all experiments. We use a convolutional neural network (CNN) and recurrent neural network (RNN). We refer to the combination of these models as RCNN. The combination of CNN with RNN enables us to extract features from raw data using the CNN and to model long-range temporal dependencies present in the data with the RNN. The CNN module contains 2 filter sizes (100 and 200 dimensions) to capture patterns across different time scales, which we empirically find to have better performance than just a single filter size.
The details of the RCNN architecture used in our experiments are presented in Supplementary Figure S1. For sleep staging, the input for the CNN is the spectrogram representation of the EEG signal. Similarly, for AHI detection, we provide 60 second blocks of respiratory signal data or spectrogram representation of these channels. For the limb movement detection task, we provide 60 second blocks of EMG (LAT, RAT) raw waveform or spectrogram representation to the CNN.
Our models are trained using backpropagation. We use cross-entropy as the loss function to train the models. The categorical cross-entropy loss is given by
where y is the predicted probability distribution and y’ is the true distribution.
We adopt batch normalization (BN) after each convolution and before activation.35 We initialize the weights as in36 and train all neural networks from scratch. We use stochastic gradient descent (SGD) with a mini-batch size of 100. The learning rate starts from 0.1 and is divided by 10 when the error plateaus. We use a weight decay of 0.0001 and a momentum of 0.9. We perform 50 iterations of random search over a set of parameter choices for hyper-parameter tuning. All models are implemented using PyTorch (http://pytorch.org/). All experiments are conducted on a server with Intel Xeon E5-2640, 256GB RAM, four NVidia Titan X GPU, and CUDA 8.0.
Scoring algorithms
Given an input sample, the trained model outputs a probability distribution over the possible target classes. In the sleep stage detection model, the model provides the probability distribution over the 5 AASM sleep stages. Similarly, in the AHI and LMI detection tasks, the model provides a probability that the sample is an apnea or a limb movement event. We use a sliding window to combine adjacent one-second output decisions to define individual apnea or limb-movement events. By merging adjacent one second outputs, we combine them into a single detected apnea or limb movement event. This allows us to compare annotations from the RCNN directly with those from experts, since experts label entire events (eg by marking the beginning and ending of an apnea) rather than independently labeling 2 second intervals. Expressing detections as single merged events also allows us to calculate the clinically relevant measures of apnea and limb movement abnormality, AHI (apnea hypopnea index) and LMI (limb movement index), which are the number of apneas or limb movements, respectively, per hour of sleep.
Evaluation
To measure performance on sleep stage classification, we use the overall classification accuracy, and classification accuracy broken down by stage, shown as a confusion matrix. Element (i, j) of each confusion matrix represents the empirical probability of predicting class j given that the ground truth (expert label) is class i.
To measure performance on apnea classification, we use the correlation value (r2) between the algorithm-predicted AHI and the AHI computed from expert-scored PSGs, where AHI = (Apnea + Hypopnea events)/hours of sleep. To measure performance in the limb movement detection task, we calculate the correlation value (r2) between the algorithm-predicted LMI and the LMI based on expert scoring of PSGs, where LMI = (number of leg movement events)/hours of sleep.
Cross dataset experiments
We evaluate our models for sleep stage detection and apnea detection in both the MGH and the SHHS datasets in the supplemental material (Supplementary Tables S2–S4). In Supplementary Table S2 we present the accuracy of models trained using MGH data and tested on SHHS, trained using SHHS and tested using MGH, and trained using the combination of MGH and SHHS and tested on MGH or on SHHS. We also show the test performance of the MGH model using only frontal channels to simulate sleep monitoring using home monitoring devices. Supplementary Table S3 shows the AHI estimation in different test-train and limited-channel contexts. Unlike multi-channel PSG data, home monitoring sensors often come from a single channel such as abdomen or chest belt. To simulate home monitoring, we assess how well models trained on a single channel (either abdomen or chest belt) perform in comparison to models given access to multi-channel PSG data. Finally, Supplementary Table S4 shows model performance for limb movement detection on MGH data.
Results
Our data consisted of 10 000 clinical PSGs performed at the Massachusetts General Hospital Sleep Laboratory (MGH data), split into 9000 training and validation PSGs and 1000 PSGs held out for testing.
We utilized a convolutional neural network (CNN) to model the local spatiotemporal characteristics of 30-second PSGs, combined with a recurrent neural network (RNN) to model long-range temporal dependencies. Figure 1 shows the RCNN system architecture. Our dataset was composed of PSGs labeled by certified sleep technologists, following the American Academy of Sleep Medicine (AASM) standards.37 The RCNN was trained to use 6 EEG channels to assign to each 30-second PSG to one of 5 sleep stages: awake (W), rapid eye movement (REM) sleep (R), and non-REM stages 1-3 (N1-N3). In addition, the RCNN was trained to use 5 respiratory channels to detect apnea events, quantified as the apnea-hypopnea index (AHI; events/hour of sleep), and limb movement events using the leg EMG channels, quantified as the limb movement index (LMI; events/hour of sleep).
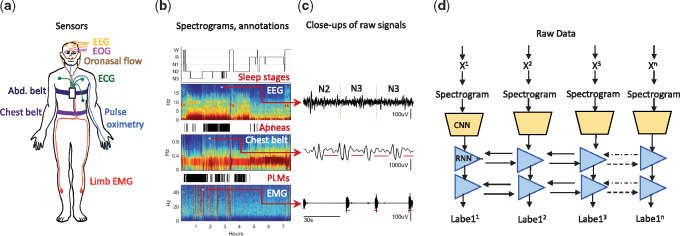
Figure 1.
Deep RCNN layout for automated polysomnography analysis. a. Data are recorded during sleep by sensors that measure brain activity (electroencephalography, EEG), eye movements (electrooculogram, and EOG), oronasal airflow, heart rhythm (electrocardiography, ECG), blood oxygenation (pulse oximetry), respiration (chest and abdominal belts), and limb movements (limb electromyography (EMG), placed over the anterior tibialis muscles. b. Examples of some of the signals and event labels provided by experts. Top: hypnogram showing sleep stages, and the corresponding spectrogram for one of the 6 EEG channels. Middle: Apnea events (black bars) and corresponding spectrogram for the chest best signal. Bottom: limb movement events (black bars) and corresponding spectrogram for one of the limb EMG signals. c. Close ups, showing details of the selected signals and labeled events. d. Architecture of the RCNN model. Signals consecutive epochs (xi) are sequentially fed into a convolutional neural network module (CNN). The CNN output is fed into a bidirectional recurrent neural network, which a sequence of inferred labels: sleep stages, apnea detections, and PLM detections. Details of the CNN architecture are provided in the supplemental material.
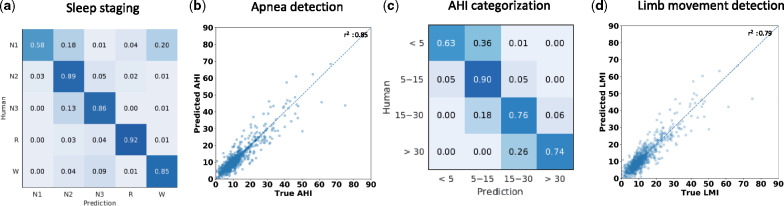
For sleep staging, the RCNN achieved an overall accuracy of 87.5% [84.2, 90.9], which compares favorably to human expert performance38,39 (Figure 2a). Also RCNN significantly outperformed classical machine learning methods such as logistic regression (accuracy 69.34%) and random forest (accuracy 74.52%) as shown in Supplementary Table S2. Besides lower performance in terms of accuracy, classical machine learning methods require expert-defined features, which are not always available such as for AHI and LMI prediction. AHI inferred by the RCNN strongly correlated with expert scoring (r2= 0.85) (Figure 2b). Converting AHI values into standard clinical categories of mild, moderate and severe disease, the RCNN achieved an overall diagnostic accuracy of 88.2% [84.7, 91.4] (Table 1). Importantly, when the apnea severity inferred by the RCNN disagreed with experts, misclassification was mainly to an adjacent severity category (Figure 2c). We used the desaturation criteria (rather than arousal criteria) for calculating AHI events, as inter-rater reliability is higher for desaturation criteria.40 The predicted LMI correlated strongly with expert scoring (r2 =0.79; Figure 2d). This level of performance is comparable with expert performance, though annotation performance of limb movements is less well studied, particularly in subjects with concurrent sleep apnea.41
Figure 2.
Classification performance of the RCNN for polysomnography scoring. The labels inferred by the RCNN are tested against the annotations of medical experts. a. Confusion matrix for sleep staging, showing RCNN agreement with expert scores. Sleep experts score each 30 second EEG epoch as 1 of 5 sleep stages: awake (W), non-REM stage 1, 2, or 3 (N1, N2, and N3), or rapid eye movement sleep (R). The RCNN outputs a probability for each stage, and we compare the highest probability class against the expert’s score for each epoch. The RCNN’s labels show >80% agreement for all classes except N1, comparable to levels of agreement between human experts. b. Sleep apnea events are detected by the RCNN in 1 second epochs, and the AHI (apnea hypopnea index: number of RCNN-detected apnea events per hour of sleep) is plotted against the AHI estimated from expert PSG scores. The correlation between expert and RCNN AHI scores is shown. c. Confusion matrix for the classification of AHI severity (none, 5; mild, 5-15; moderate, 15-30; severe, >30 per hour), comparing AHI scores inferred by the RCNN against expert scores. d. Limb movement index (LMI) are detected in consecutive one second intervals, and the total burden of lime movements, summarized as the limb movement index (LMI, number of lime movements per hour of sleep). The LMI inferred by the RCNN is compared with scores from sleep experts.
Table 1.
Generalization experiments when applying models trained on clinical data (MGH) to the MGH and SHHS test sets
| Task | Experiment setup | Accuracy | Kappa |
|---|---|---|---|
| Sleep staging | Train and test on MGH (6 channels) | 87.5% | 80.5 |
| Train on MGH and test on MGH (2 channels) | 81.9% | 76.4 | |
| Train on MGH and test on SHHS (2 channels) | 77.7% | 73.2 | |
| Sleep apnea detection | Accuracy | r2 (AHI) | |
| Train and test on MGH | 88.2% | 0.85 | |
| Train on MGH and test on SHHS | 80.2% | 0.77 | |
| Limb movement detection | Accuracy | r2 (LMI) | |
| Train and test on MGH data | 84.7% | 0.79 |
Note: Accuracy is measured as the percent agreement between labels inferred by the algorithm and expert labels. For apnea and limb movement detection, accuracy is measured both by the correlation () with expert scores of the algorithm’s estimate of the number of events per hour of sleep (apnea-hypopnea index (AHI), or limb movement index (LMI)), and by the expert-algorithm agreement of regarding categorization of the event burden as mild, moderate or severe. Cohen’s Kappa is provided as a complementary measure of accuracy which takes into account the probability of agreement occurring by chance.
To further validate the RCNN’s generalization capability, we evaluated the performance of sleep staging and sleep apnea detection on an independent set of publicly available PSGs (SHHS data; n = 5804; www.sleepdata.org). The SHHS utilizes limited-channel EEG data (2 central channels), and respiratory effort, airflow and oximetry channels, but does not include limb electromyogram signals (EMG). First, we tested the MGH-trained RCNN on 1000 randomly selected PSGs from SHHS. To enable testing on SHHS, we first retrained the sleep staging RCNN on the MGH training data while allowing access to only 2 central EEG channels to mimic the SHHS EEG configuration. For sleep staging the MGH-trained RCNN, when tested on the SHHS testing PSGs, achieved an accuracy of 77.7% [74.3, 79.7]. Next, we applied the MGH-trained AHI prediction model to the SHHS test set, which also demonstrated a strong correlation with expert labels (r2 =0.77) [0.72, 0.79]. By comparison, on the MGH test data, the limited-channel RCNN classified sleep stages with 81.9% [78.2, 84.9] overall accuracy, and AHI with r2 = 0.85 [0.83, 0.87].
To compare generalization capabilities of RCNNs trained with real-world clinical PSG data (MGH) vs standardized clinical trial data (SHHS), we evaluated sleep staging and apnea detection in cross-training experiments: train on MGH data, test on both MGH and SHHS data; then train on SHHS data, and test on MGH and SHHS data. In all experiments, the PSG sets used for training and testing are kept constant. Results are shown in Table 1 (with additional experiments shown in Supplementary Tables S2–S4). In all cases, models trained with MGH data performed well on test sets from both MGH and SHHS, confirming the importance and sufficiency of large heterogeneous datasets, even when they derive from routine clinical practice settings, for robust model training.
We next investigated the features learned by the RCNN for scoring sleep stages (Figure 3a), AHI (Figure 3b), and LMI (Figure 3c), using t-SNE (t-distributed Stochastic Neighbor Embedding).42 Each point represented a signal segment projected from the 124-dimensional high-dimensional output of the RCNN’s last hidden layer onto a plane. The RCNN has learned to form well-separated clusters of points from signals belonging to the same annotation classes.
Figure 3.
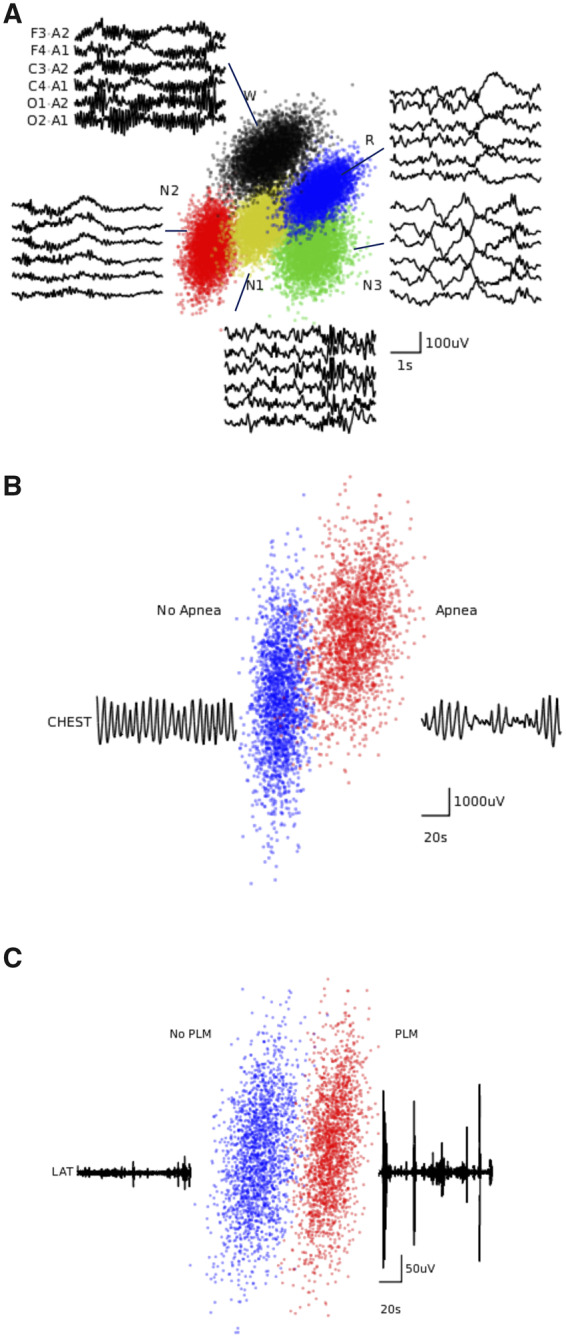
t-SNE visualization of the last hidden layer representations in the CNN. Here we show the CNN’s internal representation of a) sleep stages, b) apnea events, and c) limb movements. Points are obtained by applying t-SNE, a method for visualizing high-dimensional data, to the last hidden layer representation in the RCNN for each model. Colored points represent the different event types, showing how the algorithm learns to cluster the signals. Waveforms near show typical examples from each cluster.
Discussion
Our results demonstrate human-level performance of deep learning algorithms trained on large PSG datasets to replicate the primary categories of scoring: stages, sleep apnea, and limb movements. Our large PSG sample size allows cross-validation steps during training (to minimize the risk of model overfitting), and testing on an independent, held-out set of 1000 PSGs to obtain unbiased estimates of performance. The clinical heterogeneity and lack of special selection or exclusion of cases supports the generalization of performance when trained on the MGH set and tested on the independent research cohort of the SHHS. External validation of this kind is crucial to address a common criticism of scoring automation: will the algorithm performance be robust when applied broadly?
Our results also address one form of conventional wisdom that careful standardization of PSG recording conditions and homogeneity of patient characteristics are critical to obtaining generalizable algorithms. Our results suggest that, given sufficiently large datasets, training on real-world data can yield human level performance and generalize to standardized data sets (such as SHHS). The capacity to generalizability is a pre-requisite for algorithm deployment in real-world settings, especially in medical diagnostics that routinely encounter heterogeneous pathophysiology. Further, the availability of clinical datasets obtained in routine practice in principle far exceeds that of research studies, and our results provide motivation to utilize such “in-hand” data to develop predictive algorithms.
Feature selection is another key problem in the application of supervised machine learning. Although it is natural to assume that features informed by experts with domain knowledge, sometimes described as “feature engineering,” ought to be an important component in developing machine learning algorithms, our results shows that deep learning models can learn better features than human in this specific task. Specifically, we train our deep learning algorithms using generic features, as well as direct time series data, and obtain human-level scoring accuracy. The advantages to this approach include the minimization of bias, as well as reduced burden on human capital, which can be spent more efficiently on preparation and interpretation.
In addition to automation of in-lab scoring, the accuracy of portable sleep recording systems stand to directly benefit from reliable and robust algorithms. Because portable systems reach a far larger audience, whether clinical or consumer is in nature, robust and scalable scoring is necessary, if only to accommodate the increased scale. The minor reduction in accuracy when moving from 6- to 2-channel EEG for staging, and from 5 respiratory channels to 1 for sleep apnea, is still on par with the level of accuracy attained by experts. These results suggest that accurate automated analysis of sleep stages and apnea is attainable with limited-channel devices such as those available for at-home use. Improvement of classification with limited channels has important implications for clinical diagnostics such as home sleep apnea testing kits,43 as well as consumer facing devices,44 which the Food and Drug Administration is showing increasing willingness to consider for some medical uses (for example, arrhythmia detection).45
In summary, our deep network is accurate and scalable, and can be deployed on multi-channel (eg in-lab PSG) or limited channel (eg portable) acquisition systems. The potential for substantial clinical impact includes broadening the reach of clinical sleep medicine, augmenting clinical decision-making for sleep specialists, and improving the accuracy and reliability of at-home portable systems. Further work should focus on integrating this new technology into specific monitoring devices and optimizing performance in real-world clinical settings. The ability to automate overnight PSG scoring with the accuracy of a sleep specialist has the potential to expand access to essential medical care.
Funding
Dr Bianchi has received funding from, the Center for Integration of Medicine and Innovative Technology, the Milton Family Foundation, the MGH-MIT Grand Challenge, and the American Sleep Medicine Foundation, and the Department of Neurology. Dr Westover has received funding from NIH-NINDS (1K23NS090900). Dr Sun received funding from the National Science Foundation (IIS-1418511, CCF-1533768), NIH (1R01MD011682-01, R56HL138415), Children’s Healthcare of Atlanta, and UCB. This was not an industry supported study.
Contributors
Biswal implemented the algorithms and conducted the experiments. Biswal, Westover, J. Sun and Bianchi developed methods. H. Sun and Goparaju extracted and provided the data. All authors were involved in drafting the paper.
Competing interests
Dr Bianchi has a patent pending on a home sleep monitoring device, has research agreements with MC10 and Insomnisolv, and consulting agreements with McKesson, International Flavors and Fragrances, and Apple Inc., serves as a medical monitor for Pfizer, and has provided expert testimony in sleep medicine. This was not an industry supported study, and none of these entities had any role in the study.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Abbreviations
AASM: American Academy of Sleep Medicine
AHI: apnea-hypopnea index
CNN: convolutional neural network
EEG: electroencephalogram
EMG: electromyogram
EOG: electrooculogram
LMI: limb movement index
MGH: Massachusetts General Hospital
NREM: non-rapid eye movement
PSG: polysomnogram
R&K: Rechtschaffen and Kales
REM: rapid eye movement
RNN: recurrent neural network
SHHS: Sleep Heart Health Study
Supplementary Material
References
- 1. Buysse DJ. Insomnia. JAMA 2013; 309 (7): 706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iranzo A. Sleep in neurodegenerative diseases. Sleep Med Clin 2016; 11 (1): 1–18. [DOI] [PubMed] [Google Scholar]
- 3. Kapur VK. Obstructive sleep apnea: diagnosis, epidemiology, and economics. Respir Care 2010; 55 (9): 1155–67. [PubMed] [Google Scholar]
- 4. Budhiraja R, Budhiraja P, Quan SF.. Sleep-disordered breathing and cardiovascular disorders. Respir Care 2010; 55 (10): 1322–32; discussion 30–2. [PMC free article] [PubMed] [Google Scholar]
- 5. Tregear S, Reston J, Schoelles K, Phillips B.. Obstructive sleep apnea and risk of motor vehicle crash: systematic review and meta-analysis. J Clin Sleep Med 2009; 5 (6): 573–81. [PMC free article] [PubMed] [Google Scholar]
- 6. Smolensky MH, Di Milia L, Ohayon MM, Philip P.. Sleep disorders, medical conditions, and road accident risk. Accid Anal Prev 2011; 43 (2): 533–48. [DOI] [PubMed] [Google Scholar]
- 7. Skaer TL, Sclar DA.. Economic implications of sleep disorders. Pharmacoeconomics 2010; 28 (11): 1015–23. [DOI] [PubMed] [Google Scholar]
- 8. Pietzsch JB, Garner A, Cipriano LE, Linehan JH.. An integrated health-economic analysis of diagnostic and therapeutic strategies in the treatment of moderate-to-severe obstructive sleep apnea. Sleep 2011; 34 (6): 695–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDaid C, Duree KH, Griffin SC, et al. A systematic review of continuous positive airway pressure for obstructive sleep apnoea-hypopnoea syndrome. Sleep Med Rev 2009; 13 (6): 427–36. [DOI] [PubMed] [Google Scholar]
- 10. Usmani ZA, Chai-Coetzer CL, Antic NA, McEvoy RD.. Obstructive sleep apnoea in adults. Postgrad Med J 2013; 89 (1049): 148–56. [DOI] [PubMed] [Google Scholar]
- 11. Leger D, Bayon V.. Societal costs of insomnia. Sleep Med Rev 2010; 14 (6): 379–89. [DOI] [PubMed] [Google Scholar]
- 12. Sun H, Jia J, Goparaju B, et al. Large-scale automated sleep staging. Sleep 2017; 40 (10): doi:10.1093/sleep/zsx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542 (7639): 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316 (22): 2402–10. [DOI] [PubMed] [Google Scholar]
- 15. Fraiwan L, Lweesy K, Khasawneh N, Fraiwan M, Wenz H, Dickhaus H.. Classification of sleep stages using multi-wavelet time frequency entropy and LDA. Methods Inf Med 2010; 49 (3): 230–7. [DOI] [PubMed] [Google Scholar]
- 16. Lajnef T, Chaibi S, Ruby P, et al. Learning machines and sleeping brains: automatic sleep stage classification using decision-tree multi-class support vector machines. J Neurosci Methods 2015; 250: 94–105. [DOI] [PubMed] [Google Scholar]
- 17. Liang SF, Kuo CE, Hu YH, Cheng YS.. A rule-based automatic sleep staging method. J Neurosci Methods 2012; 205 (1): 169–76. [DOI] [PubMed] [Google Scholar]
- 18. Anderer P, Gruber G, Parapatics S, et al. health solution for automatic sleep classification according to Rechtschaffen and Kales: validation study of the Somnolyzer 24 x 7 utilizing the Siesta database. Neuropsychobiology 2005; 51 (3): 115–33. [DOI] [PubMed] [Google Scholar]
- 19. Berthomier C, Drouot X, Herman-Stoica M, et al. Automatic analysis of single-channel sleep EEG: validation in healthy individuals. Sleep 2007; 30 (11): 1587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Loparo KA, Kelly MR, Kaplan RF.. Evaluation of an automated single-channel sleep staging algorithm. Nat Sci Sleep 2015; 7: 101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hassan AR, Bhuiyan MI.. A decision support system for automatic sleep staging from EEG signals using tunable Q-factor wavelet transform and spectral features. J Neurosci Methods 2016; 271: 107.. [DOI] [PubMed] [Google Scholar]
- 22. Punjabi NM, Shifa N, Dorffner G, Patil S, Pien G, Aurora RN.. Computer-assisted automated scoring of polysomnograms using the somnolyzer system. Sleep 2015; 38 (10): 1555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malhotra A, Younes M, Kuna ST, et al. Performance of an automated polysomnography scoring system versus computer-assisted manual scoring. Sleep 2013; 36 (4): 573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anderer P, Moreau A, Woertz M, et al. Computer-assisted sleep classification according to the standard of the American Academy of Sleep Medicine: validation study of the AASM version of the Somnolyzer 24 x 7. Neuropsychobiology 2010; 62 (4): 250–64. [DOI] [PubMed] [Google Scholar]
- 25. Schaltenbrand N, Lengelle R, Toussaint M, et al. Sleep stage scoring using the neural network model: comparison between visual and automatic analysis in normal subjects and patients. Sleep 1996; 19 (1): 26–35. [DOI] [PubMed] [Google Scholar]
- 26. Younes M, Younes M, Giannouli E.. Accuracy of automatic polysomnography scoring using frontal electrodes. J Clin Sleep Med 2016; 12 (05): 735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Younes M, Soiferman M, Thompson W, Giannouli E.. Performance of a new portable wireless sleep monitor. J Clin Sleep Med 2017; 13 (02): 245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shambroom JR, Fabregas SE, Johnstone J.. Validation of an automated wireless system to monitor sleep in healthy adults. J Sleep Res 2012; 21 (2): 221–30. [DOI] [PubMed] [Google Scholar]
- 29. Vilamala A, Madsen KH, Hansen LK.. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. arXiv:171000633 2017. [Google Scholar]
- 30. Zhang J, Wu Y, Bai J, Chen F.. Automatic sleep stage classification based on sparse deep belief net and combination of multiple classifiers. Trans Inst Meas Control 2016; 38 (4): 435–51. [Google Scholar]
- 31. Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc 2018; 25 (10): 1419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomson DJ. Spectrum estimation and harmonic analysis. Proc IEEE 1982; 70: 1055–96. [Google Scholar]
- 33. Bokil H, Andrews P, Kulkarni JE, Mehta S, Mitra PP.. Chronux: a platform for analyzing neural signals. J Neurosci Methods 2010; 192 (1): 146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bokil H, Purpura K, Schoffelen JM, Thomson D, Mitra P.. Comparing spectra and coherences for groups of unequal size. J Neurosci Methods 2007; 159 (2): 337–45. [DOI] [PubMed] [Google Scholar]
- 35. Ioffe S, Szegedy C.. Batch normalization: accelerating deep network training by reducing internal covariate shift. ArXiv 2015;1502.03167v3. [Google Scholar]
- 36. Glorot X, Bengio Y.. Understanding the difficulty of training deep feedforward neural networks. Proc Mach Learn Res 2010; 9: 249–56. [Google Scholar]
- 37. Silber MH, Ancoli-Israel S, Bonnet MH, et al. The visual scoring of sleep in adults. J Clin Sleep Med 2007; 3 (2): 121–31. [PubMed] [Google Scholar]
- 38. Danker-Hopfe H, Anderer P, Zeitlhofer J, et al. Interrater reliability for sleep scoring according to the Rechtschaffen & Kales and the new AASM standard. J Sleep Res 2009; 18 (1): 74–84. [DOI] [PubMed] [Google Scholar]
- 39. Magalang UJ, Chen NH, Cistulli PA, et al. Agreement in the scoring of respiratory events and sleep among international sleep centers. Sleep 2013; 36 (4): 591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Redline S, Budhiraja R, Kapur V, et al. The scoring of respiratory events in sleep: reliability and validity. J Clin Sleep Med 2007; 3 (2): 169–200. [PubMed] [Google Scholar]
- 41. Stefani A, Heidbreder A, Hackner H, Hogl B.. Validation of a leg movements count and periodic leg movements analysis in a custom polysomnography system. BMC Neurol 2017; 17 (1): 42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van der Maaten L, Hinton G.. Visualizing data using t-SNE. J Mach Learn Res 2008; 9: 2579–605. [Google Scholar]
- 43. Collop NA, Tracy SL, Kapur V, et al. Obstructive sleep apnea devices for out-of-center (OOC) testing: technology evaluation. J Clin Sleep Med 2011; 7 (5): 531–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bianchi MT. Sleep devices: wearables and nearables, informational and interventional, consumer and clinical. Metabolism 2017; doi: 10.1016/j.metabol.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 45. Gottlieb S. FDA Announces New Steps to Empower Consumers and Advance Digital Healthcare. Secondary FDA Announces New Steps to Empower Consumers and Advance Digital Healthcare 2017; https://www.fda.gov/NewsEvents/Newsroom/FDAVoices/ucm612014.htm. Accessed October 18, 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.