Abstract

A promising anticancer therapeutic strategy is the stabilization of telomeric G-quadruplexes using G-quadruplex-binding small molecules. Although many G-quadruplex-specific ligands have been developed, their low potency and selectivity to G-quadruplexes over duplex remains unsolved. Recently, a crystal structure of a telomeric 3′ quadruplex–duplex hybrid was reported and the quadruplex–duplex interface was suggested to a good target to address the issues. However, there are no high-resolution complex structures reported for G-quadruplex ligands except for a docked BSU6037. In this study, molecular dynamic (MD) binding simulations with a free ligand were used to study binding poses and dynamics of four representative ligands: telomestatin, TMPyP4, BSU6037, and BRACO19. The MD data showed that BSU6037 was able to fully intercalate into the interface whereas TMPyP4 and BRACO19 could only maintain partial intercalation into the interface and telomestatin only binds at the quadruplex and duplex ends. Both linear ligands, BSU6037 and BRACO19, were able to interact with the interface, yet they were not selective over duplex DNA. The DNA geometry, binding modes, and binding pathways were systematically characterized, and the binding energy was calculated and compared for each system. The interaction of the ligands to the interface was by the means of an induced-fit binding mechanism rather than a lock–key mechanism, consisting of the DNA unfolding at the interface to allow entrance of the drug and then the refolding and repacking of the DNA and the ligand to further stabilize the G-quadruplex. On the basis of the findings in this study, modifications were suggested to optimize the interface binding for TMPyp4 and telomestatin.
Introduction
A promising anticancer therapeutic strategy is the stabilization of telomeric G-quadruplexes using quadruplex-binding small molecules.1−4 G-quadruplexes are formed from the stacking of guanine (G) tetrads via Hoogsteen base pairing in G-rich sequences.3−12 Human telomeres appearing at the end of chromosomes consist of a 3′ single-stranded TTAGGG repeat overhang.2,10−18 This TTAGGG repeat overhang has the ability to form G-quadruplexes. During cell division, the (TTAGGG)n sequence is progressively shortened until a growth-arrest state and eventual senescence, the complete stopping of cell division, is reached.4,12,16,18−22 When senescence is bypassed due to the absence of cell cycle checkpoints, a cell then reaches a state of crisis where it undergoes uncontrolled growth.2,4,9,12,14,19,22,23 The overexpression of telomerase, a reverse transcriptase that lengthens telomeres,2−4,9,12,18,19,24 results in the bypass of senescence and thus indefinite growth of cancerous cells.9,10,12,13,17−19,25 G-quadruplexes have been shown to inhibit telomerase by making the 3′ overhang inaccessible to the telomerase RNA template.9,16,26−29 It has also been shown that G-quadruplex specific ligands compete with the protection of telomere protein 1 binding, where the binding of the ligands leads to a series of events: the uncapping of telomeres, DNA damage response signaling, and eventual apoptosis of the mutated cells.23,30−32 Consequently, the stabilization of telomeric G-quadruplexes is a viable therapeutic strategy to treat cancers.12 Furthermore, the over-representation of G-quadruplexes in cancer cells21,33−37 makes them cancer-specific targets.31 Because of this, many studies have been performed on the telomeric and promoter G-quadruplexes with various G-quadruplex specific ligands to inhibit cancer cell growth.1,2,9,15,16,22,27,38−47
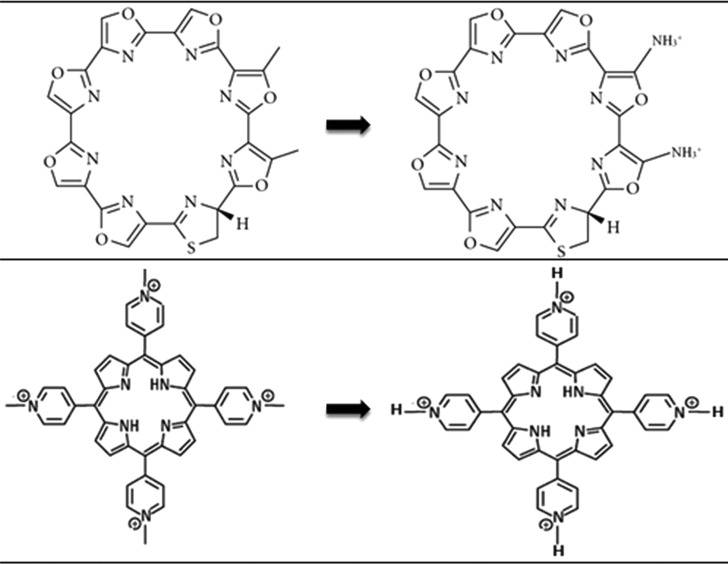
The class of anticancer drugs in this paper is widely represented by two structural characterizations: cyclical and linear. Four ligands (telomestatin, TMPyP4, BSU6037, and BRACO19 in Figure 1B–E) are used to exemplify ligands of both structural families within this class of drugs. Telomestatin and TMPyP4 are representative of the cyclical ligands; BSU6037 and BRACO19 are representative of the linear ligands. Telomestatin,48−51 TMPyP4,52−55 and BRACO1956−71 were chosen explicitly because they are the most studied ligands that are potent against various cancer cell lines. The ligand BSU6037 was previously docked to the quadruplex–duplex interface by Krauss et al.46
Figure 1.
Structures of the telomeric DNA quadruplex–duplex hybrid (A), telomestatin (B), TMPyP4 (C), BSU6037 (D), and BRACO19 (E).
A large variety of G-quadruplex specific ligands have been developed on the following criteria: an aromatic core that allows π–π stacking to the G-quartet and a side chain that allows electrostatic interactions with the sugar-phosphate backbone and ensures solubility.3,4,10−12,17,18,29,31,72 For a ligand to target G-quadruplexes successfully, its binding equilibrium constant must be at least 106 M–1 and it also must be selective over duplex binding by at least 2 orders of magnitude. The binding properties of three leading ligands (telomestatin, TMPyP4, and BRCO19) is listed in Table S1 of the Supporting Information. Telomestatin, a natural product, shows the highest potency and telomerase inhibition, and it is also 70 times more selective toward G-quadruplexes than duplex DNA.26,73 Despite this, telomestatin’s hydrophobicity and low water solubility impedes its bioavailability, which is an integral feature in successful drug design.3,31,74 TMPyP4 is a synthetic ligand designed for G-quadruplex stabilization with an affinity of 20 × 106 M–1 and a TRAP-LIG EC50 (TRAP: the telomere repeat amplification protocol for telomerase, which is used for determination of telomerase activity) of 8.9 μM but is only 2 times more selective to the G-quadruplex than duplex DNA.21,75 The synthetic acridine, BRACO19, has been reported to have promising potency, KG4 = 30 × 106 M–1, and telomeric inhibition, TRAP-LIG EC50 = 6.3 μM, with 10 times more selectivity over duplex DNA.31,32,75 However, BRACO19 has low membrane permeability and decomposes quickly, which decreases the binding of BRACO19 to G-quadruplexes.76,77 It is clear that potency, selectivity, and other properties need to be further improved to convert these compounds to therapeutic agents.
The detailed binding poses of multiple G-quadruplex ligands have been experimentally and computationally characterized; these studies suggested two well-known binding poses: end-to-end stacking (top or bottom stacking) and groove binding.9,10,21,22,31,33,41,43−45,78−80 The main binding interactions have been shown to be π–π stacking of the aromatic core of small ligands with the G-quartet, which is best achieved in end-to-end stacking.74 However, since terminal G-quartets are present in the majority of G-quadruplexes, achieving selectivity to a distinct G-quadruplex has become another issue.
A recent work by Krauss and others involving G-quadruplexes has begun to focus on targeting the telomeric 3′ junction formed between a G-quadruplex and duplex DNA as a solution to increase the binding selectivity and potency to G-quadruplexes over duplexes for G-quadruplex ligands.46 In their work, a crystal structure of a telomeric 3′ quadruplex–duplex hybrid formed from putative parallel G-quadruplex and duplex DNA was reported (Figure 1A) and the junction was suggested as a more specific G-quadruplex site that can be targeted by G-quadruplex ligands.46 To probe the ligand binding at the quadruplex–duplex interface, a diverse range of G-quadruplex ligands (BSU6037, BSU6039, BRAC-19, TMPyP4, and FC4ND10) were selected for ligand soaking experiments with preformed crystals of the quadruplex–duplex hybrid. Ligands that did not significantly degrade crystal quality but appeared to bind to the construct were TMPyP4, Cu-porphyrin, BSU6037, and FC4ND10. However, the binding pose at the interfaces was not obtained due to different reasons. Binding of TMPyP4 caused a large decrease in diffraction quality (from about 3.0 to 4.3 Å resolution) and a slight change in cell dimensions. Analysis of different Fourier maps did not show the presence of BSU6037 or Cu-porphyrin bound to the quadruplex–duplex hybrid, and FC4ND10 was observed to bind in the duplex region. Since no structural data is available on ligand binding at the quadruplex–duplex interface, BSU6037 was docked to a pseudo-ligand-binding site at the interface that was generated by rotating the phosphodiester backbone of T17, while maintaining strand polarity of the G-rich strand. Although the obtained binding pose of BSU6037 is closely analogous to that observed on a crystal complex structure formed between BSU6039 and a bimolecular quadruplex (1L1H),81 lack of structural flexibility in the docking may limit induced-fit binding modes.
This study aims at furthering molecular modeling investigations with four representative ligands (telomestatin, TMPyP4, BSU6037, and BRACO19 in Figure 1B–E) binding to the telomeric 3′ quadruplex–duplex construct in hopes of characterizing binding poses and their binding mechanisms using molecular dynamics (MD) binding simulation with a free ligand. The MD simulations probed the structure, dynamics, and interaction of the G-quadruplex–duplex construct with the ligands in high spatial and temporal resolution.82 Detailed process information was used to decipher the binding mechanism. Specifically, this study utilized the latest AMBER DNA force field (OL15) and drug (GAFF2) force fields to simulate the binding process between the ligands and the human telomeric G-quadruplex–duplex construct from an initially unbound state (where the ligand is 10 Å away from the DNA). The binding modes and pathways of each ligand to the G-quadruplex–duplex DNA were characterized, and then the molecular mechanics generalized born/surface area (MM-GBSA) binding energies of each binding pose were compared. Three major modes of binding were observed for TMPyP4, BSU6037, and BRACO19: interface intercalation, binding to the quadruplex, and binding to the duplex. No intercalation binding mode was observed for telomestatin, but suggestions on how to alter the telomestatin scaffold to allow for interface intercalation were given. The MM-GBSA binding energy analysis indicates that the charged aromatic TMPyP4 scaffold was better than both the uncharged aromatic structures telomestatin and linear structures (BSU6037 and BRACO19). Additionally, an implication on designing a better TMPyP4 analog was also discussed (Table 1).
Table 1. Molecular Dynamics Simulation Runs.
| system IDa | DNA | no. of ligands | no. of runs | no. of water molecules | ions | box size (Å)b | drug initial pose | NPT eq. (ns) | NVT (ns) | total time (μs) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 (5dww) | 0 | 3 | 6242 | 30 K+ | 67.7 | N/A | 1 | 1999 | 6 |
| 2 | 0 | 1 | 5 | 1491 | 0 | 31.8 | free | 1 | 499 | 2.5 |
| 3 | 0 | 1 | 5 | 1043 | 4 Cl– | 37.4 | free | 1 | 499 | 2.5 |
| 4 | 0 | 1 | 5 | 2065 | 2 Cl– | 45.8 | free | 1 | 499 | 2.5 |
| 5 | 0 | 1 | 5 | 1487 | 3 Cl– | 41.5 | free | 1 | 499 | 2.5 |
| 6 | 1 (5dww) | 1 | 19 + 1 | 18 926 | 30 K+ | 94.1 | free | 1 | 499/1999 | 11.5 |
| 7 | 1 (5dww) | 1 | 19 + 1 | 11 298 | 26 K+ | 80.2 | free | 1 | 499/1999 | 11.5 |
| 8 | 1 (5dww) | 1 | 19 + 1 | 12 131 | 28 K+ | 82.0 | free | 1 | 499/1999 | 11.5 |
| 9 | 1 (5dww) | 1 | 19 + 1 | 15 541 | 27 K+ | 88.4 | free | 1 | 499 1999 | 11.5 |
System 1 refers to the free DNA-only and systems 2–5 refer to the free ligand-only simulations (2–5: telomestatin, TMPyP4, BSU6037, and BRACO19, respectively). Systems 6–9 refer to the free DNA plus free ligand simulations (6–9: telomestatin, TMPyP4, BSU6037, and BRACO19, respectively).
Triclinic box equivalent to the true truncated octahedral box.
Results
Scaffold of the Quadruplex–Duplex Hybrid Was Maintained in the DNA-Only Simulations
To validate the force field for DNA, the DNA-only system was constructed from the crystal structure (Protein Data Bank (PDB) ID: 5DWW) for stability simulations. Three independent runs for this DNA-only system were carried for 2.0 μs. DNA backbone root-mean-square deviation (RMSD) plots and the last snapshots are shown in Figures S3 and S6, respectively. Clearly, the DNA hybrid was stable in all three runs, as indicated by the small root-mean square deviation of the backbone from the starting crystal structure (RMSD = 2–3 Å). Neither of the two K+ ions inside the G-quadruplex moved out, and all of them maintained the initial positions in three runs.
To show more structural details, a representative trajectory and its order parameters are shown in Figure 2. First, the G-quadruplex was stable, indicated by maintaining ∼8, ∼9, and ∼8 hydrogen bonds for the top, middle, and bottom G4 layers. Second, the interface was stable, indicated by ∼3 and ∼1 hydrogen bonds for the tribase layer and the A7–T19 pair. Third, the duplex part was stable, indicated by maintaining ∼2 hydrogen bonds for each base pair in the duplex part. Fourth, the DNA backbone was stable, indicated by a small RMSD of ∼2.5 Å. Fifth, the two K+ ions inside the quadruplex were stable, indicated by maintaining the K+ to K+ distance of ∼3.8 Å, with a small fluctuation. Sixth, the calculated relative conformation energy using MM-GBSA showed fluctuation but was flat overall.
Figure 2.
Representative trajectory of the DNA-only simulation and the order parameter plot illustrating the breaking and reforming of hydrogen bonds (at the quadruplex (1), interface (2), and duplex (3)), RMSD (Å) with reference to the final structure (4), K+ to K+ distance (5), and the MM-GBSA binding energy (ΔE in kcal/mol) (6). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by a yellow ball.
Geometric Characterization of the G-Quadruplex Part Showed a Right-Handed Helical Twist of the Three G4 Layers, the Ligand Binding Can Make Subtle Changes
Although the scaffold of a G-quadruplex has been extensively studied and classified into parallel, antiparallel, and hybrid class, the inter-G4 layer geometry from the MD simulations has not been thoroughly examined. From the top view of the G-quadruplex part (Figure S1a), the right-handed helical rotation of G4 layers was obviously identified. Analogous to the right-handed helical rise and twist in B-form DNA, this work has defined and calculated the rise, H-rise, and H-twist, as detailed in the method section for the crystal structure and five simulated systems (Table S6). For the DNA-only simulated system, the rise, H-rise, and H-twist averaged over the two layer steps (i.e., bottom to middle and middle to top) are ∼3.44 Å, ∼2.09 Å, and ∼17.6°, respectively, which are close to the values of the crystal structures (∼3.42 Å, ∼1.13 Å, and ∼10.7°). The slightly larger differences for H-rise and H-twist may be attributed to the crystal packing in the condense phase or the force-field artifact. Further comparison to the solution NMR structure will clarify this issue. This is supported by a recent bioinformatics study on 74 G-quadruplex structures in the PDB, suggesting that the distribution of the twist angle between the two adjacent G-quartets for biomolecular parallel G-quadruplexes is ranged from 10 to 35°, with a maximum at 31 ± 3°.83 Nonetheless, H-rise and H-twist in G-quadruplex are smaller than those in standard B-DNA (3.32 Å and 34.3° for H-rise and H-twist, respectively). Interestingly, the stacking of telomestatin on the top of the G-quadruplex (Figure 5) did not change these parameters much (∼3.37 Å, ∼1.79 Å, and ∼15.8°) neither did the binding of BSU6037 (Figure 7) at the G-quadruplex–duplex interface (∼3.44 Å, ∼1.92 Å, and ∼16.9°) but the interaction from TMPyP4 and BRACO19 changed H-rise to from ∼2.09 to ∼3 Å. Thus, ligand binding can slightly modulate the G-quadruplex interlayer geometry.
Figure 5.
Representative trajectory of the top stacking mode of telomestatin (run 9) and the order parameter plot, illustrating the breaking and reforming of hydrogen bonds (at the quadruplex (1), interface (2), and duplex (3)), drug-base dihedral angle (4), ligand (black)/DNA (red) RMSD (Å) with reference to the final structure (5), ligand center-to-DNA center distance (black) and K+-to-K+ distance (red) (6), and the MM-GBSA binding energy (ΔE in kcal/mol) (7). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by yellow balls.
Figure 7.
BSU6037, run 19, trajectory snapshots and the order parameter plot, illustrating the breaking and reforming of hydrogen bonds (at the quadruplex (1), interface (2), and duplex (3)), drug-base dihedral angle (4), ligand (black)/DNA (red) RMSD (Å) with reference to the final structure (5), ligand center-to-DNA center distance (black) and K+-to-K+ distance (red) (6), and the MM-GBSA binding energy (ΔE in kcal/mol) (7). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by yellow balls.
Ligand Binding Did Not Change the Interaction between K+ and the G-Quadruplex in Cases When K+ Did Not Move Out
Potassium ions play an important role in stabilizing G-quadruplexes. The distances between a potassium ion and eight neighboring oxygen atoms and the oxygen–oxygen distances were calculated, and these values are shown in Figure S1c,d for the DNA-only system. The values for the crystal structure and other four DNA–ligand systems are shown in Tables S2 and S3. The average potassium–oxygen distance difference between the crystal structure and the simulated DNA-only system was within 0.16 Å, which is comparable to the structural fluctuation (±0.18 Å) observed in the MD simulations. The average oxygen–oxygen distance difference between the crystal structure and simulated DNA-only system was within 0.34 Å, which is slightly larger than the structural fluctuation (±0.21 Å). Thus, the simulation well reproduced these distances. The average potassium–oxygen distance difference between the simulated DNA-only system and DNA–ligand systems was within 0.05 Å, which is much smaller than the structural fluctuation (±0.18 Å) observed in the MD simulations. The average oxygen–oxygen distance difference between the simulated DNA-only system and DNA–ligand systems was within 0.03 Å, which is slightly larger than the structural fluctuation (±0.21 Å). Thus, ligand binding did not change the interaction between K+ and the G-quadruplex when K+ did not move out.
Geometric Characterization of the Duplex Part Showed That a Normal B-DNA Geometry for Most Base Pairs Was Maintained, the Ligand Binding on the Top of the G-Quadruplex or at the G-Quadruplex–Duplex Interface Did Not Change the Duplex Geometry
Various geometry parameters including base pair-axis, base pair-step, and paired base–base parameters were calculated, as detailed in the method section, to characterize the duplex part for the crystal structure and the five simulated systems (Table S4). The values from the five simulated systems generally agree with the values from the crystal structure. For example, the crystal structure has average values of ∼3.3 Å and ∼31° for H-rise and H-twist, respectively. The H-rise for the DNA-only system, DNA–telomestatin system (top binding), DNA–TMPyP4 (interface interacting), DNA–BSU6037 (interface binding), and DNA–BRACO19 (interface interacting) is ∼2.8, ∼3.4, ∼3.0, ∼3.0, and ∼3.1 Å. The H-twists for the DNA-only system, DNA–telomestatin system (top binding), DNA–TMPyP4 (interface interacting), DNA–BSU6037 (interface binding), and DNA–BRACO19 (interface interacting) is ∼30, ∼33, ∼30, ∼30, and ∼32°. All of these values are close to the values of 3.3 Å and 34° for H-rise and H-twist of B-DNA helix, respectively. Thus, the helical structure of the duplex was maintained throughout the MD simulations. This is also supported from the DNA backbone dihedral angle and pucker calculations (Table S5): although there are some subtle differences, most values are close to each other.
Root-Mean-Square Fluctuation (RMSF) Data Showed That the Three loops (T4, T8, and T12–T13) in the G-Quadruplex Part (1–16) and Interface Region (T17–A19, T7–A8) and the Two Termini of the Duplex Part (A25 and A2) had Larger Local Fluctuations Than Those of the Four G Strands (G1–G3, G5–G7, G9–G11, and G14–G16) for the DNA-Only System
Figure 3 shows RMSF plot for the five systems. As to the DNA-only system (black line), five peaks were identified, corresponding to T4, T8, T12–T13, T18, and A25–A2. In contrast, the other regions (G1–G3, G5–G7, G9–G11, G14–G16, G20–T24, and A3–C6) show lower RMSF values. Thus, although the four G4 strands and two helical strands had lower structural fluctuation, the three loops in the G-quadruplex part, the interface region, and the termini of the duplex part had larger structural fluctuation. The G-quadruplex top binding by telomestatin (red line) slightly reduced the RMSF values at the five peaks, in particular, the T12–T13 loop peak and the A24–A2 terminal peak, indicating the telomestatin might slightly stabilize the overall structure. The interface interaction by TMPyP4 (green line) slightly reduced the first three peaks but significantly reduced the fourth peak in the interface region. The interface interaction by BRACO19 (blue line) slightly reduced the first three peaks but significantly reduced the fourth and fifth peaks. Thus, the binding by TMPyP4 and BRACO19 stabilized the interface region. The interface binding by BSU6037 (purple line) slightly increased the first, second, and fifth peaks but significantly reduced the third peak and shifted the fourth peak to G20–C21 region. These changes can be attributed to the change of the interface by the BSU6037 interface binding (Figure 7).
Figure 3.
Root-mean-square fluctuation (RMSF) plot of the five systems (DNA-only, top binding of telomestatin, interface intercalation mode of BSU6037, and the interface interacting mode of TMPyP4 and BRACO19). The x axis refers to the residue number of the DNA quadruplex hybrid structure (Figure 1a); the residue 1 of the second chain was missed in the crystal structure.
Three Major Binding Modes Were Observed for TMPyP4, BSU6037, and BRACO19: Interface Interaction, Binding to the Quadruplex, and Binding to the Duplex, but No Interface Interaction Mode Was Observed for Telomestatin
Starting from the unbound state, 20 binding simulation runs (500 ns of each) were carried out using each ligand. The RMSD of the DNA backbone heavy showed that a majority of the trajectories for each complex reached and remained in a stable form (Figure S4). Those that experienced a larger fluctuation in RMSD values were those where the DNA backbone started to slightly unravel and unfold. The convergence of the binding simulations was confirmed, as described in the Methods section, with the simulation systems reaching a relatively steady state where the DNA–ligand complex retained 20 or more atom contacts (Figure S5). A table to summarize the number of trajectories in each final binding pose as well as a figure of the final binding poses of each trajectory is available in Figures S7 and S8–S11, respectively.
The most stable complexes (Figure S13) of the 20 trajectories were extracted and categorized into structural families on the basis of a clustering analysis for each ligand, as described in the methods section. By setting the threshold of 1% population, 6 structural families were identified for telomestatin (Figure S14), 9 were for TMPyP4 (Figure S15), 10 were for BSU6037 (Figure S16), and 12 were for BRACO19 (Figure S17). Then, these families were further merged into three binding modes: interface interacting, quadruplex side binding, and duplex side binding. After analyzing each of the modes, it was discovered that only TMPyP4, BSU6037, and BRACO19 were observed in interface interacting, quadruplex binding, and duplex binding modes: telomestatin was not able to bind to or interact with the interface (Figure 5), and BSU6037 was the only ligand able to fully intercalate into the interface (Figure 7). The results, including percentages, of the most abundant clusters are reported in Figure 4. For telomestatin, overbinding to ends of the DNA duplex could be a simulation artifact due to the missing T1 in the crystal structure. The hydrogen bond networks of the interface interacting modes as well as detailed two-dimensional ligand–DNA interactions of the major binding modes are presented in Figures S18 and S19, respectively.
Figure 4.
Major binding modes between ligands and human telomeric quadruplex–duplex (PDB ID: 5DWW). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by yellow balls. Overall population abundance (MM-GBSA binding energy) of each binding mode is annotated.
From the trajectory and clustering analysis, it is clear that unlike BSU6037, the ligands TMPyP4 and BRACO19 did not fully intercalate into interface. Instead, TMPyP4 and BRACO19’s side chains extended into the interface to make contact. As the interface opened up, the drugs were able to move further into the interface and the interactions with the interface were enhanced as the DNA deformed, which suggests an induced fit binding mechanism. However, despite the utilization of an induced fit binding mechanism, the final binding mode remained a partial interaction and not full intercalation for both TMPyP4 and BRACO19.
MM-GBSA Binding Energy Data Showed That the Interface Interaction Mode of TMPyP4 Was the Most Energetically Favorable, the G-Quadruplex Binding Mode of Telomestatin Was Much More Favorable Than Its Duplex Binding Mode, and the Interface Interactions of BSU6037 and BRACO19 Were Comparable to the Duplex Binding Mode
To examine the relative stability for the three binding modes observed, MM-GBSA binding energy calculations were conducted on each binding mode for each complex (Table 2). TMPyP4 had the highest binding energy for interface interaction, −96.2 ± 6.8 kcal/mol, which was 13.3 kcal/mol higher than that for G-quadruplex binding and 17.7 kcal/mol higher than that for duplex binding. These values indicate that TMPyP4 strongly interacts with the interface and is selective over the duplex DNA. Telomestatin did not interact with the interface but had a much stronger binding energy and selectivity to the G-quadruplex (−70.4 ± 5.1 kcal/mol) over the duplex (−30.3 ± 5.0 kcal/mol). BSU6037 and BRACO19 both had similar interface interaction and duplex binding energies and a lower comparative G-quadruplex binding energy. To further understand the nature of binding, the binding energy was broken down into van der Waals (VDW) interactions, hydrophobic interactions (SUR), electrostatic interactions (GBELE), and the conformational energy change induced from the complex forming (CONF) (Table 2). As shown in Table 2, a majority of the binding energy is contributed by the VDW energy and the conformational energy change. As expected, the top stacking for telomestatin had a higher VDW energy than the duplex binding; this is likely due to the uniplanar structure of telomestatin, making stacking a more favorable binding choice. TMPyP4 had the largest VDW energy for the interface interaction mode, which could be tied to its charged planar structure. Both linear structures, BSU6037 and BRACO19, had higher VDW energies for the duplex and interface than the G-quadruplex, which could be explained by the fact that the linear structures can better bind to the duplex groove or to the interface pocket than to the quadruplex loops or top (stacked). Both planar structures, telomestatin and TMPyP4, had larger conformational energies for the G-quadruplex than either the interface or duplex, likely due to their ability to stack on top of the quadruplex to further stabilize the complex. These large conformational binding energies further support the induced fit binding mechanism of the ligands to the quadruplex–duplex DNA. Each of the four ligands varied in their respective charged state; telomestatin had no charge (charge state of 0), TMPyP4 carried the largest charge (+4), BSU6037 had a charge state of +2, and BRACO19 had a charge state of +3. The MM-GBSA binding energy data revealed that BRACO19 exhibited the strongest electrostatic interactions (ΔEGBELE) with the hybrid quadruplex–duplex DNA, indicating that a charge state of +3 is the best.
Table 2. MM-GBSA Binding Energy of Ligands to the Quadruplex–Duplex DNAa,b,c,d,e,f.
| ligands (charge state) | binding pose | ΔVDW | ΔSUR | ΔGBELE | ΔCONF | ΔTOT | ΔΔE |
|---|---|---|---|---|---|---|---|
| telomestatin (0) | G-quadruplex | –50.1 ± 2.0 | –2.7 ± 0.1 | 18.3 ± 1.4 | –35.9 ± 4.5 | –70.4 ± 5.1 | 25.8 |
| duplex | –41.4 ± 5.4 | –2.9 ± 0.3 | 16.4 ± 2.8 | –2.4 ± 3.5 | –30.3 ± 5.0 | 65.9 | |
| TMPyP4 (+4) | interface | –68.2 ± 3.7 | –5.1 ± 0.3 | –11.4 ± 1.6 | –11.5 ± 5.8 | –96.2 ± 6.8 | 0.0 |
| G-quadruplex | –51.8 ± 3.1 | –3.4 ± 0.2 | –12.6 ± 1.8 | –15.1 ± 2.9 | –82.9 ± 3.7 | 13.3 | |
| duplex | –68.4 ± 3.9 | –4.8 ± 0.2 | –8.0 ± 2.0 | 3.5 ± 6.3 | –78.5 ± 6.8 | 17.7 | |
| BSU6037 (+2) | interface | –52.9 ± 5.6 | –4.2 ± 0.6 | –6.0 ± 4.0 | –29.2 ± 0.8 | –92.3 ± 5.6 | 3.9 |
| G-quadruplex | –15.8 ± 6.7 | –1.6 ± 0.7 | –8.9 ± 3.6 | –33.2 ± 1.0 | –59.5 ± 6.8 | 36.7 | |
| duplex | –52.9 ± 3.9 | –4.7 ± 0.3 | –7.3 ± 2.8 | –20.0 ± 0.3 | –90.6 ± 4.5 | 5.6 | |
| BRACO19 (+3) | interface | –37.7 ± 3.1 | –4.4 ± 0.3 | –19.1 ± 3.3 | –26.1 ± 2.8 | –87.3 ± 3.4 | 8.9 |
| G-quadruplex | –44.8 ± 3.5 | –3.3 ± 0.4 | –10.7 ± 3.2 | –24.5 ± 5.9 | –77.7 ± 6.1 | 18.5 | |
| duplex | –42.2 ± 8.9 | –4.4 ± 0.7 | –14.5 ± 5.3 | –23.2 ± 4.3 | –84.3 ± 9.0 | 11.9 |
ΔVDW = change of VDW energy in gas phase upon complex formation (unit: kcal/mol).
ΔSUR = change of energy due to surface area change upon complex formation (unit: kcal/mol).
ΔEBELE = change of GB reaction field energy + gas phase elec. energy upon complex formation (unit: kcal/mol).
ΔCONF = change of conformational energy upon complex formation (unit: kcal/mol).
ΔTOT = ΔVDW +ΔSUR + ΔEBELE + ΔCONF change of potential energy in water upon complex formation (unit: kcal/mol).
ΔΔE = ΔTOT– (−96.2).
Statistics of the interface interacting trajectories are presented in Table 3. This table provides insight on the overall binding pathways for the trajectories that maintained a final interface interaction binding pose. Some trajectories were not included from this analysis, despite making contact with the interface at some point during the trajectory. For TMPyP4 runs 3 (Figure S22) and 5 (Figure 6), the initial contact with the DNA is with a residue on a side loop of the quadruplex. In both cases, within 40 ns after the initial contact, the ligand relocates closer to the interface where its aromatic side chains first make contact with interface residues. The ligand makes minor adjustments to maximize interactions with the interface but remains in the same binding mode for the remainder of the trajectory. BSU6037 system runs 7 (Figure S25) and 19 (Figure 7) make initial contact with the loop residues on the quadruplex region before interacting with residues of the duplex region. In both trajectories, the ligand proceeds to interact with terminal residue of the interface, which induces conformational changes to the DNA, moving the ligand inside of the binding pocket, increasing its interaction with the interface until ligand stacking is achieved. BRACO19 made initial contact with the duplex in runs 6 (Figure 9), 8 (Figure S28), and 10 (Figure S29). After initial contact, the side chains of BRACO19 made contact with the interface while still bound to the duplex until the final pose was achieved.
Table 3. Statistics of the Interface Interacting Trajectoriesa.
| system | total interface interactions | quadruplex to interface | duplex to interface | quadruplex + duplex to interface |
|---|---|---|---|---|
| TMPyP4 | 2 | 2 | ||
| BSU6037 | 2 | 2 | ||
| BRACO19 | 3 | 3 |
Provided is the overall binding pathway taken by each ligand in its interface interacting mode.
Figure 6.
Representative trajectory of the interface interacting mode of TMPyP4 (run 5) and the order parameter plot, illustrating the breaking and reforming of hydrogen bonds (at the quadruplex (1), interface (2), and duplex (3)), drug-base dihedral angle (4), ligand (black)/DNA (red) RMSD (Å) with reference to the final structure (5), ligand center-to-DNA center distance (black) and K+-to-K+ distance (red) (6), and the MM-GBSA binding energy (ΔE in kcal/mol) (7). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by yellow balls.
Figure 9.
Suggested modifications for telomestatin and TMPyP4.
To Characterize the Binding Pathway of the Two/Three Modes of Each Ligand, Seven Order Parameters Were Calculated for Representative Trajectories, As Described in the Methods Section (Figures 2, Figures 5–8, and S20–S31)
Figure 8.
BRACO19, run 06, trajectory snapshots and the order parameter plot, illustrating the breaking and reforming of hydrogen bonds (at the quadruplex (1), interface (2), and duplex (3)), drug-base dihedral angle (4), ligand (black)/DNA (red) RMSD (Å) with reference to the final structure (5), ligand center-to-DNA center distance (black) and K+-to-K+ distance (red) (6), and the MM-GBSA binding energy (ΔE in kcal/mol) (7). 5′ and 3′ of the DNA chain are indicated by a red and blue ball, respectively. K+ ions are indicated by yellow balls.
The plots are included in the supporting document for telomestatin (the quadruplex binding mode (Figure S20) and the duplex binding mode (Figure S21)), TMPyP4 (the interface binding mode (Figure S22), the G-quadruplex binding mode (Figure S23), and the duplex binding mode (Figure S24)), BSU6037 (the interface binding mode (Figure S25), the G-quadruplex binding mode (Figure S26), and the duplex binding mode (Figure S27)), and BRACO19 (the interface binding mode (Figures S28 and S29), the G-quadruplex binding mode (Figure S30), and the duplex binding mode (Figure S31)). A representative trajectory for each ligand (the quadruplex binding mode for telomestatin and the interface interacting mode for TMPyP4, BSU6037, and BRACO19) was selected to be extended from 500 to 2000 ns for checking the stability of these binding modes. The information gathered from the 2000 ns simulations is reported in Figures 5–8. The occupancy time for the final stable binding mode was identified from the RMSD plot with reference to the final snapshot: ∼1800 ns for telomestatin, ∼1750 ns for TMPyP4, ∼1200 ns for BSU6037, and ∼1200 ns for BRACO19.
Top Stacking Binding Mode of Telomestatin Provides Support for the Selectivity of This Ligand to the G-Quadruplex DNA over Duplex DNA
Telomestatin was unable to bind to or interact with the interface in any of the simulation runs, but it did bind very selectively to the top of the quadruplex over the duplex, which is qualitatively consistent with the experimentally observed 70-fold selectivity (Table S1). In the representative trajectory (Figure 5), telomestatin initially contacted the DNA at 14 ns, binding to the side loop of the quadruplex. It released from this position at 67 ns and randomly searched for another binding pose until 105 ns, where it slightly moved up and rebound to the quadruplex side loop. It moved out and stacked on top of the quadruplex at 165 ns and remained in this top stacking binding mode for the remaining time. No significant changes in the number of hydrogen bonds were observed in either the G-quadruplex, duplex, or interface, demonstrating that it was not an induced fit binding mechanism. The two potassium cations maintained their position and distance throughout the trajectory. The MM-GBSA calculated binding energy revealed that telomestatin top stacking to the quadruplex (∼−70 kcal/mol) was more favorable than binding to the side loop of the quadruplex (∼−30 kcal/mol), which can help explain why this interconversion occurred.
Binding Mechanism of the Ligands to the Interface Seems to Be via an Induced Fit Binding Mechanism Rather Than a “Lock–Key” Binding Mechanism
The interface interaction modes for TMPyP4, BSU6037, and BRACO19 are presented in Figures 6–8. The pattern changes in the number of hydrogen bonds for the interaction of the three ligands at the interface are consistent with the trajectories (Figures 6–8). A decrease in the number of hydrogen bonds among the base triad (red) and base pair (green) was observed during initial ligand–interface interaction. The hydrogen bonds for the A7–T19 base pair were completely disrupted for each interface interaction. The energetic cost of breaking these hydrogen bonds was likely compensated by the favorable ligand–interface interaction energy gain. During the interactions, a portion of the ligands were also extended to group bind to the sides of the DNA backbone adjacent to the interface. The stacking between the bases and the interacting portion of the ligands in addition to the group binding between the extended ligand portion and the DNA is likely further stabilizing the complex.
TMPyP4 Interface Interaction Follows a Base “Flip-Insertion” Binding Mechanism
TMPyP4 demonstrates a subtle induced fit binding mechanism. In the representative trajectory (Figure 6), TMPyP4 made initial contact with the DNA at the side loop of the quadruplex at 4 ns. At 37 ns, TMPyP4 shifted down toward the top of the duplex, with a region of the ligand making contact with the A7 and T19 base pair of the interface, breaking the two hydrogen bonds shared between them. By examining the trajectory, we discovered that the initial interface contact is concurrent with the flipping out of base T19, creating a binding pocket to allow entrance of the ligand into the interface region. One hydrogen bond between the base triad (T8, A18, and T17) also gets disrupted by this initial interface contact. TMPyP4 moves further into the interface at 255 ns, completely disrupting all hydrogen bonds within the base triad. These bases that made up the base triad in the interface flip out and bond to the aromatic regions of TMPyP4, such as the methylpiperidine side chains positioned around the central aromatic ring, rather than to each other. After this interaction, the top layer of the quadruplex has an increase in the number of hydrogen bonds but it also exhibits a higher fluctuation in them; these large fluctuations indicate that when TMPyP4 moves further into the interface and maintains a partial intercalation, it does not stabilize the G-quadruplex as much as the initial interface interaction does. The middle layer of the quadruplex experiences a loss of a hydrogen bond during this same period. No significant change in hydrogen bonds is present within the duplex, as the conformation of this region does not undergo any major changes. The top potassium ion moved out at 339 ns, whereas the bottom potassium ion remained stable throughout the course of the trajectory.
Interface Intercalation of BSU6037 Requires the DNA To Open Up at the Interface Region To Provide the Ligand Access to the Binding Site
The binding mechanism of BSU6037 to the interface is particularly interesting as it clearly demonstrates an induced fit binding mechanism (Figure 7). In the representative trajectory, BSU6037 initially binds to the side of the duplex until 82 ns where it makes the first contact with the interface residues. BSU6037 binds to terminal residue A8 that flips into the core of the interface, moving BSU6037 closer into the interface where it is able to interact with interface residue T19 where it remains until 300 ns. At 301 ns, the duplex begins to unravel until 483 ns where it is approximately 90° from the starting position. At 458 ns, the top potassium cation leaves the quadruplex. With BSU6037 in the interface, the duplex reforms itself until complete at 584 ns where BSU6037 remains stacking in the interface for the remainder of the trajectory. Under further examination, it was discovered that when the BSU6037 ligand bonded to the interface of the DNA, it forced the bonds in the base pair and base triad to break, unfolding and opening the DNA up, which allowed the ligand into the binding pocket. The binding energy of the interface (−92.3 ± 5.6 kcal/mol) was much larger than that of the G-quadruplex (−59.5 ± 6.8 kcal/mol), which could explain why after BSU6037 made an initial contact with the quadruplex it inserted itself into the interface instead. These conformational changes that take place during binding demonstrate the induced fit binding mechanism that the ligand (BSU6037) produces.
BRACO19 Also Interacted with the Interface Region
BRACO19 demonstrates another subtle induced fit binding mechanism. In the representative trajectory (Figure 8), BRACO19 bound to the major groove of the duplex part at 3 ns where the N,N-dimethylcyclohexylamine side chain made the initial contact with the side of the quadruplex where it remained until 107 ns. At 107 ns, a side chain of BRACO19 began making contact with the interface residues T17, A18, and T19. This side chain binding to the interface region forced base A7 to flip out, eliminating the hydrogen bonds formed by the A7–T19 base pair so that the drug moved up and interacted with the interface region until 767 ns. At 767 ns, the interface region bends open, allowing the N,N-dimethylcyclohexylamine side chain of BRACO19 to reposition slightly deeper into the interface region. BRACO19 maintained this interaction with the interface region throughout the simulation; the G-quadruplex part was stable through the trajectory. The top potassium cation moved out of the quadruplex at 1636 ns. MM-GBSA calculations determined that the binding energy fluctuated between −80 and −90 kcal/mol.
Discussion
G-quadruplex DNA has become an important therapeutic target recently as it has been found to be over-represented in a wide variety of cancers.84 Many ligands targeting G-quadruplexes have been developed; however, selectivity to a specific G-quadruplex over DNA duplex is yet to be achieved at a level acceptable for therapeutic use. Recently, the junction between the G-quadruplex and duplex hybrid DNA (PDB: 5DWW) in the 3′ end of telomeric overhang was targeted as a promising opportunity to improve upon G-quadruplex selectivity and ligand potency.46 Yet, there are no detailed molecular interactions or high-resolution complex structures reported for most of the ligands except for BSU6037,46 which has been docked to the crystal structure of the G-quadruplex–duplex hybrid.
This study was done to gain molecular insights into the binding of telomestatin, TMPyP4, BSU6037, and BRACO19 to the quadruplex–duplex DNA by using binding molecular dynamic simulations with a free ligand. The free ligand simulation allowed for more dynamics and flexibility within both the DNA and ligand, which in turn showed that ligand interaction with the interface was by means of an induced fit binding mechanism rather than a lock–key mechanism. The MM-GBSA binding energy calculations (Table 2) showed that the ligands binding to the DNA induced a large change in each complex’s conformational energy, which was a major contributor to the high total energies. Large conformational binding energies are concurrent with an induced fit binding mechanism occurring. The DNA subtly opening up at the interface region makes room for the drug interactions (Figures 6–8). Once the insertion is completed, the interface closes and the complex is stabilized. Energetically, the loss of hydrogen bonds at the interface is compensated by the initial ligand–DNA interaction and final π–π interactions once the induced fit mechanism is completed. Of course, more experimental evidence is required to support this interface open-close mechanism.
Out of multiple major binding poses for each system, our MM-GBSA binding energy calculations indicate that interface interactions were the most stable binding pose. The large magnitude of the binding energy in combination with the long time (2000 ns) stability suggests that the binding is likely enthalpically driven, with a minor contribution in regard to the entropic binding energy. The binding energy decomposition analysis further suggests that the VDW and CONF terms make the largest contributions to the total binding energy. This indicates that drug specific packing optimization is a possible way to further stabilize G-quadruplexes.
MM-GBSA binding energy calculations indicate that the most stable binding pose for telomestatin was top stacking to the G-quadruplex; its large uncharged planar structure was better suited to stacking on top of the G-quadruplex, as top stacking results in the most favorable packing interaction. However, telomestatin did not bind to or interact with the interface. Because the other three charged ligands were found to be able to interact with the interface, it is speculated that lack of charge for telomestatin might hinder its ability to interact with the DNA backbone for opening the binding pocket and interacting with the interface. If this is true, the findings of this work suggest that to allow telomestatin to bind at the interface region and maintain its high selectivity over duplex DNA, the planar scaffold needs to incorporate some small charged side chains (Figure 9 top). These charged side chains can disrupt the DNA backbone interactions, thereby destabilizing the DNA scaffold enough to allow a binding pocket to open up and permit ligand insertion into the interface.
To test this, induced fit docking was performed on the original ligand, telomestatin, as well as a modified ligand containing two charged side chains (Figure S32a–d). The results of the induced fit docking revealed there was a −3.1 kcal/mol difference between the original ligand (−7 kcal/mol) and the modified ligand (−10.1 kcal/mol) binding energies. This supports the proposed modification that adding a small charged side chain would increase the ability of the ligand to insert into the interface. Nonetheless, further simulation and experimentation are required to verify the proposed modification.
TMPyP4 exhibited the highest interface MM-GBSA binding energy (−96.2 kcal/mol), which was the strongest binding energy of all binding modes among the four ligands. In fact, the MM-GBSA binding energy shows that the interface interaction mode is significantly more stable for TMPyP4 than the quadruplex and duplex binding modes by 13.3 and 17.7 kcal/mol, respectively, indicating that the interface mode may significantly contribute to the higher affinity as well as a higher selectivity to the 3′ telomeric quadruplex–duplex junction. However, TMPyP4 has a preference to bind to diad base over quartet due to its size and structure and it only interacts with the interface partially when the interface is completely distorted. It is clear that the size and charge of TMPyP4 hindered the ligands ability to fully intercalate into the interface so that the central chromophore could interact with both the bottom quartet of the G4 as well as the top diad of the duplex inside of the interface simultaneously; this was also the case for BRACO19. Therefore, it is speculated that the methyl groups of TMPyP4 should be removed from the aromatic side chains to increase stability and allow the ligand to move deeper into the interface to increase ligand–DNA interactions. The proposed structure (Figure 9 bottom) is designed to keep TMPyP4 intact and to increase packing interactions with a deeper insertion of the drug into to the interface (Figure S32E–H). This could potentially create more potent and selective ligands to bind to the interface of the G-quadruplex–duplex hybrid DNA. As a result of induced fit docking, the proposed structure had a docking score of −12.81 kcal/mol, which is 0.37 kcal/mol stronger than that of the original structure. The proposed TMPyP4 analog showed an increase in its ability to remain planar and increase the interactions with the interface (Figure 32H). This supports the proposed modification to the original ligand to increase the ligands ability to move deeper into the interface. Nonetheless, further simulation and experimentation are required to verify the proposed modification.
The MM-GBSA binding energy analysis revealed that BSU6037 bound to the interface and duplex at comparable binding energies (∼−91.5 kcal/mol), which was about 30 kcal/mol stronger than binding to the quadruplex part. Similarly, BRACO19 interacted with the interface and bound at the duplex at comparable binding energies (∼−86.6 kcal/mol), which was around 10 kcal/mol stronger than binding to the quadruplex. Not only do these ligands lack selectivity over duplex DNA, they also bind to the interface weaker than TMPyP4 between 3.9 and 8.9 kcal/mol. Interestingly, despite BSU6037’s ability to intercalate into the interface, the partial interaction between TMPyP4 and the interface remains more energetically favorable. These findings indicate that linear structures, even those that contain charged side chains, lack the selectivity and potency needed to be a viable G-quadruplex stabilizer. Therefore, future ligand development should focus on charged aromatic scaffolds that optimize packing and π–π interactions within the interface and include charged side chains that destabilize the DNA backbone to allow the ligand entrance to the interface binding pocket.
The free binding simulations also revealed that BSU6037 had a different binding pose than the one described by Krauss et al. in 2016. Krauss et al. found that a pseudo binding site for BSU6037 could be constructed at the interface region by disrupting T17 of the TAT base triad, which consists of bases T17(A)–A18(A)–T8(B), to expose the surface of the bottom G-quartet; specifically, they thought that if T17 of the base triad flipped out, the ligand would have access to the bottom of the quadruplex.46 When they docked BSU6037 into this pseudo binding site, the ligand fit directly below the bottom G-quartet, with the ligand oriented so that it laid in-plane with the A7–T19 base pair. However, the free ligand molecular dynamic simulations in this work did not show BSU6037 binding in this position. BSU6037 destabilized DNA at the interface region, causing bases T7 and A8 to break apart from their respective partners and flip out, which in turn unfolded the DNA at the interface. BSU6037 fit into the opened binding pocket, and the DNA refolded around it (Figure 7). Furthermore, the simulations resulted in BSU6037 experiencing slightly different interactions then what was observed in the docked experiments. In the previous docking experiment, the central planar regions of BSU6037 exhibited π–π stacking interactions with bases G11, G16, and T17, the center protonated nitrogen of BSU6037 had a hydrogen bond with the N1 atom of base T19, the side chain amide nitrogen had a hydrogen bond with the carbonyl oxygen atom (O4) of base A8, and the other side-chain amide nitrogen had a hydrogen bond with the N3 atom of base A18.46 In this work, π–π stacking interactions with the central planar regions of BSU6037 and bases G11 and A8 on the 3′ end of the shorter DNA strand were observed, as well as a hydrogen bond was formed between the side-chain amide nitrogen and base T17. This change in ligand–DNA interactions is likely brought on by the different binding location of BSU6037.
Coordination of O6 carbonyls by cations within the ion pore is known to be a major factor in stabilizing the quadruplex structure. Successful stabilization from the cations has reportedly maintained an average interquartet distance of 3.3 Å between the bipyramidal antiprism that is formed.83 Zheng and co-workers85 have shown that single-strand G-rich sequence can fold into a G-quadruplex with very low K+-ion concentration (0.01–01 mM), indicating strong binding of K+. To look into this feature, distance calculations were performed to define some parameters of the K+ cations relative to the surrounding G4 DNA. Analyzing the oxygen–potassium and oxygen–oxygen distance calculations (Figure S1; Tables S2, S3, and S6) revealed that each residue was able to maintain an average oxygen–oxygen distance close to 3.3 Å during the simulations. The similar mean values and very low standard deviations for both calculations indicate that the simulation parameters for the K+ cations are appropriately set despite the K+ cation moving out of the ion pore in some ligand binding trajectories. Because none of the K+ moved out in three 2.0 μs simulations of the DNA-only system, the ligand binding might lead to the increase of the dynamic fluctuation of the quadruplex and the K+-ion pore. In fact, the trajectory analysis indicates that the moving out of a K+ ion occurred at the induced fit binding process with a large deformation of the DNA structure. It is suspected that further extending the simulation period may show the reversible binding of the K+ cation into the ion pore as the DNA structure is stabilized from the ligand binding. In future work, a more detailed analysis can be performed to elucidate the dynamic properties of the reversible binding of K+ cations.
Literature suggested the H-rise and H-twist geometry parameters were the most important parameters to analyze the overall helical structure. Despite a number of works defining duplex DNA, there has been little research into the helical rise and rotation of the quadruplex structure from the simulations. This research was able to identify average values for the H-rise (2.4 Å) and H-twist (20 Å) of the bases within the three quartets of the DNA G-quadruplex, as well as reproduce an average value for the rise parameter (3.5 Å). A common feature defining helical B-DNA is maintaining average values of 3.32 Å and 34.3° for H-rise and H-twist, respectively. The results of the H-rise and H-twist helical geometry parameter analysis revealed comparable values for each of the systems being analyzed, providing quantitative support that the helical structure was maintained throughout the simulations.
In addition to the interface interacting mode, a top stacking mode on the parallel G-quadruplex part for TMPyP4 and BRACO19 was also observed (Figure 4). Interestingly, their complex crystal structure with a two-stranded parallel human telomeric G-quadruplex are available in the PDB databank.32,86 (Figure 10). Although the number of strands forming the parallel G-quadruplex is different, the binding mode from the simulations shows a good similarity to the crystal structure. This supports the simulation methodology in this study.
Figure 10.

Comparison of the G-quadruplex end binding mode from the ligand binding simulations and from the crystallography studies. 5′ and 3′ are represented by red and blue balls, respectively. Potassium cations are represented as yellow balls.
Conclusions
Computational methods have become essential in drug discovery as they provide the detailed structural information that experimental results may lack. Although many G-quadruplex ligands have been designed, they still lack the potency and selectivity to a specific G-quadruplex over duplex DNA that is required for therapeutic use. A promising opportunity to improve upon both selectivity and potency is by targeting the recently solved junction between G-quadruplex–duplex hybrid DNA in the 3′ end of telomeric overhang. In this study, free ligand molecular dynamic simulations were utilized in addition to MM-GBSA binding energy calculations to evaluate the binding modes and mechanisms of telomestatin, TMPyP4, BSU6037, and BRACO19, which have had little research previously done. Three different major binding modes were identified: interface interacting, quadruplex side binding, and duplex side binding. The MM-GBSA binding energy analysis revealed that of the three binding modes, the interactions with the interface region of the quadruplex–duplex hybrid DNA would result in the most stable structure. The MM-GBSA binding energy analysis also revealed that TMPyP4 has the strongest binding energy to the interface region. The trajectory analysis revealed that TMPyP4, BSU6037, and BRACO19 were able to interact with the interface via an induced-fit mechanism, consistent with subtly opening near the interface and repacking after ligand insertion. However, BSU6037 was the only ligand able to remain fully intercalated into the interface region. Both linear ligands BSU6037 and BRACO19 were not selective over duplex DNA. The results of the induced fit docking suggest that removal of the methyl groups from the aromatic side chains of TMPyP4 could allow the central ring to interact deeper within the interface region; the docking also suggests that adding some small charged side chains to telomestatin’s aromatic scaffold may allow this ligand to interact with the interface region. With these findings, it is believed that further research in increasing the binding stability could result in promising telomeric G-quadruplex–duplex stabilizers.
Methods
Simulation System
Three types of systems were constructed: a DNA-only, a ligand-only, and four DNA + ligand (Table 1). The unbound DNA–ligand system was constructed using the X-ray crystallography solved human telomeric G-quadruplex–duplex structure (PDB ID: 5DWW(46)), including the K+ ions inside the G-quadruplex and a ligand molecule that was at least 10 Å away from the DNA. The 10 Å distance makes sure that there are at least three layers of water molecules separating the ligand from the DNA; therefore, there is not much interaction between the ligand and the DNA (i.e., an unbound state). This setup allowed the probing of the binding pathways and binding mechanisms. This unbound system (DNA plus ligand) was solvated in a water box of truncated octahedron with 10 Å water buffer plus K+ as counter ions to neutralize the system, leading to ∼120 mM of K+ cation concentration. The latest version of the AMBER DNA OL15 ff99 (i.e., parm99bsc087 + χOL488 + ε/ζOL189 + βOL190 updates) was applied to represent the DNA, TIP3P model91 was used to represent water, and the recently developed K+ model by Cheatham group was used for the system.92 The partial charges for each ligand molecule were obtained using standard AMBER protocol: the electrostatic potential of the ligand molecule was obtained at the HF/6-31G* level after geometry optimization at the same level; the electrostatic potential using the restrained electrostatic potential method determined the partial charges,93 and other force-field parameters were taken from the AMBER GAFF294 force field. The ligand force field in Mol2 format can be found in the supporting document (Figure S33). These AMBER force fields are the most common ones used in nucleic acid simulations including G-quadruplexes.80,82,88,95−99
Simulation Protocols
Three runs for the DNA-only, five runs for the ligand-only, and the 20 simulation runs for each unbound DNA–ligand system (Table 1) were conducted using the GPU version of the AMBER 14 simulation package.100 The detailed protocol followed this groups’ early studies.101,102 Each unbound DNA–ligand system underwent a 500 ps prerun at 500 K to ensure the position and orientation of the free ligand was randomized before a production run at 300 K; during this prerun, the receptors’ position remained fixed. After the energy minimization for the initial structure of a system, 3, 5, or 20 independent runs were performed by assigning different initial velocities to the atoms of the system; this was followed by a Maxwell–Boltzmann distribution using a different random seed. The multiple independent runs for the DNA–ligand system allow for a better sampling of binding poses and pathway. A production run of 500 ns at 300 K included a short 1.0 ns molecular dynamics run in the NPT ensemble mode (constant pressure and temperature) to equilibrate the system density in which the DNA and ligand were subjected to Cartesian restraints (1.0 kcal/mol Å) and 499.0 ns dynamics in the equivalent NVT ensemble mode (constant volume and temperature) (Table 1). SHAKE103 was applied to constrain all bonds connecting hydrogen atoms, enabling a 2.0 fs time step in the simulations. The particle-mesh Ewald method104 was used to treat long-range electrostatic interactions under periodic boundary conditions (charge grid spacing of ∼1.0 Å, the fourth order of the B-spline charge interpolation, and direct sum tolerance of 10–5). The cutoff distance for short-range nonbonded interactions was 10 Å, with the long-range van der Waals interactions based on a uniform density approximation. To reduce the computation, nonbonded forces were calculated using a two-stage RESPA approach105 where the short-range forces were updated every step and the long-range forces were updated every two steps. Temperature was controlled using the Langevin thermostat with a coupling constant of 2.0 ps. The trajectories were saved at 50.0 ps intervals for analysis.
Convergence of Simulations
The root-mean-square deviation (RMSD) of DNA/ligand backbone heavy was calculated against the starting structure. The flat and small average RMSDs (Figures S3 and S4) over all runs indicate that these systems were stable. Atom contacts between the DNA fragment and the drug molecule were calculated using an atom-to-atom distance cutoff of 3.0 Å. The simulation systems reached a steady state, as indicated by the stable average contact number (Figure S5) over all runs. A stable complex was defined as a complex with the number of atom contacts between the DNA fragment and the drug molecule greater than 20. The last snap shots of the apo form are shown (Figure S6).The free drugs bound to different sites, as shown in the last snapshots for the 20 runs (Figures S8–S11), indicate a good sampling of binding sites.
K+ Position
Distances were calculated to determine the position of the K+ cation relative to the surrounding DNA G-quadraplex (Figure S1 and Table S6) because the electrostatic interactions between the K+ cation and the partially negative oxygen atom on each residue of the surrounding G-quartet are critical in stabilizing the G-quadruplex structure. For the apo DNA form, the distance between the oxygen on each residue and the nearest K+ cation (Figure S1c), as well as the distance between the oxygen of each residue and the oxygen of the nearest residue (Figure S1d) was calculated. These oxygen–oxygen and oxygen–potassium distances per residue were also calculated for the four ligand binding simulations (Tables S2 and S3).
G-Quadruplex Parameters
To understand inter-G4 layer geometry, the rise, H-rise, and H-twist parameters were calculated. The rise parameter was defined as the distance (Å) between the centers of the guanine bases, excluding the sugar phosphate backbone, of the lower G4 layer to the center of the guanine bases on the G4 layer above. The H-rise parameter calculated the projection of the G4 layer rise with respect to the Z axis, which was defined as the vertical axis through the two K+ cations located inside the G4-ion channel. The H-twist parameter calculated the rotational angle of the bases with respect to the aforementioned Z axis. The rotation in the H-twist parameter was calculated from the bottom, meaning that the layer above would be used as a reference, measuring the degree of clockwise rotation required of the lower layer to align with the position of the reference layer (Table S6).
DNA Duplex Parameters
Various helical parameters were calculated using Curves+106 to characterize the base pairing in the helical portion of the DNA (Table S4). The base pair-axis parameters were used to quantify the rigid-body transformation of the base pair reference frame and the reference frame of the local helical axis. The base pair-axis parameters were calculated by measuring the movement of the bases toward the grooves (x-displacement), movement of the bases perpendicular to the grooves (y-displacement), rotation around the short axis of the base pairs (inclination), and rotation around the long axis of the base pairs (tip). Intrabase pair parameters were calculated using the rigid-body transformation from one base reference system to its respective paired base reference system. The three translational parameters calculated measured the change in distance with respect to the vector pointing to the major groove (shear), with respect to the vector pointing along the long axis of the base pair (stretch), and with respect to the vector of the base pair (stagger). Three rotational parameters were also calculated that measured the rotation with respect to the vector pointing into the major groove (buckle), pointing along the long axis of the base pair (propel), and with respect to the vector normal to the base pair (opening). Another set of parameters calculated was the interbase pairing parameters that calculated the rigid-body transformations between base pairing steps. The interbase pair parameters calculated included three translational parameters (shift, slide, and rise) and three translational parameters (tilt, roll, and twist), along with two additional parameters that considered the rigid body movement of each step relative to the overall helical axis (H-rise and H-twist).
DNA Backbone Parameters
Backbone torsional angles (α, β, γ, δ, ε, ζ, χ, Pucker) (Figure S12) were calculated to further characterize the conformational changes of the DNA in each system (Table S5). The standard backbone dihedral angles (α, β, γ, δ, ε, and ζ) were measured around the covalent bonds of the sugar ring and χ about the glycosidic bond. The sugar puckering was calculated by determining the position of the phosphorus atoms relative to the sugars.
Structural Fluctuation of the DNA
For each system, the root-mean-square fluctuations (RMSFs) of each individual residue of DNA were calculated to characterize the local structural fluctuation (Figure 3).
Binding Mode Identification
Because the DNA backbone was relatively stable in the binding process, we aligned the DNA backbone of the stable complexes from the trajectories by a least square fitting. The aligned complexes were clustered into different structural families on the basis of the 2 Å pair-wise RMSD cutoff of the drug molecule only using the Daura algorithm,107 in which the number of neighboring structures was calculated for every structure on the basis of the RMSD cutoff. The structure with the largest number of neighbors together with its neighboring structures was removed to form a structure family, and the process continued for the remaining structures until all structures were assigned to a structural family (Figure S13). The centroid structure (i.e., the structure having the largest number of neighbors in the structural family) was used to represent the family. The centroid structures of populated structural families (>1% of total structure population) are shown in Figures S14–S17. On the basis of visual inspection, the centroid structures were further merged into superfamilies corresponding to major binding modes (interface, quadruplex, and duplex binding).
Parameters for Characterizing DNA–Drug Binding Pathway
Seven order parameters were calculated to characterize the DNA–drug binding process: hydrogen bond analysis at the quadruplex, interface, and duplex, drug-base dihedral angle, ligand RMSD (Å), center-to-center distances (Å), and MM-GBSA binding energy (ΔE in kcal/mol). The distance cutoff between H-donor and H-acceptor was 3.5 Å, and the donor-H-acceptor angle cutoff was 120°. The hydrogen bonds for the G-quadruplex were calculated for the top (red; G1, G5, G9, and G14), middle (green; G2, G6, G10, and G15), and bottom (blue; G3, G7, G11, and G16) G-tetrads over the course of the trajectories (Figure 1A). The hydrogen bonds for the interface were calculated for the top/three-base layer (red, T8, T17, and A18) and bottom/two-base layer (green, A7 and T19) over the course of the trajectories. The hydrogen bonds for the duplex were calculated for the first (red, C6, and G20), second (green; G5–C21), and third (blue; C4–G22) base pairs below the interface, over the course of the trajectories. The ligand-base dihedral angle was defined as the dihedral angle between the plane of the DNA layer that is closest to the drug and the drug’s ring plane. Atom contact number between the DNA and the drug molecule was calculated using an atom-to-atom distance cutoff of 3.0 Å. Ligand–DNA distance was defined as the distance from the DNA center to the drug molecule center, indicated by a black line. Potassium-ion distance was defined as the distance from the center to center of the two potassium ions inside the G-quadruplex, indicated by a red line.
The molecular mechanics generalized born/surface area (MM-GBSA)108 module in the AMBER package (GB1 model with Bondi radii set, salt of 0.2 M, and surface tension of 0.0072 kcal/mol/Å2) was used to analyze the energetics of the bound complexes to avoid the large energy fluctuation of explicit solvent. A recent evaluation study by Case and co-worker has concluded that realistic DNA modeling can be obtained with GB implicit solvation methods and the GB1 model performs better than GB2-OBC1 and GB5-OBC2.109 Note that the two K+ ions inside the G-quadruplex–duplex DNA were kept in MM-GBSA calculations because they were a part of the complex structure. The MM-GBSA binding energy for a system was calculated from three simulations:110 ligand only, DNA only, and DNA–ligand complex using eq 1. It has four components in the eq 2: van der Waals (VDW) interaction energy, hydrophobic interaction energy (SUR), electrostatic interaction (GBELE), and the change of the conformation energy for DNA and ligand. These terms were calculated using eqs 3 and 4.
| 1 |
| 2 |
| 3 |
| 4 |
Absolute binding free-energy calculations can be very useful in determining binding site selectivity in G-quadruplex DNA, which is not easily obtained through experimental methods.111 However, due to the high computation cost of the absolute free-energy calculation, the relative free-energy calculation with implicit solvent model can be useful for screening a large number of ligands. A recent study has shown that GB models make good prediction even for charged molecules when the relative solvation free energy is considered.112 Systematic benchmarking studies up to 1864 crystal complexes have shown that relative MM-GBSA binding energy is a powerful tool to rank ligand binding affinity.113−117 This is also supported by our previous studies, in which the binding energy of between anticancer drug doxorubicin and a B-DNA fragment (d(CGATCG)2),98 between anticancer drug daunomycin and a TGGGT G-quadruplex DNA,118 between RHPS4 and human telomeric G-quadruplexes/duplex,110 between telomestatin and a hybrid telomeric G-quadruplex,45 and between BRACO19 and a parallel-stranded telomeric G-quadruplex119 were successfully assessed by this MM-GBSA protocol.
Acknowledgments
We acknowledge Rowan SEED grant, the High Performance Computing Facility at Rowan funded by the National Science Foundation under Grants MRI-1429467 and XSEDE MCB160164/160173/170088.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01574.
RMSD and contact number plots for each trajectory, the last snapshots of quadruplex–duplex simulations for each ligand, representative structures of the most populated structural families, additional trajectories and order parameter plots of each ligand, the AMBER GAFF2 force field of the ligands (PDF)
Movies for the representative trajectory of each ligand (MPG)(MPG)(MPG)(MPG)(MPG)
The authors declare no competing financial interest.
Supplementary Material
References
- Gowan S. M.; Heald R.; Stevens M. F. G.; Kelland L. R. Potent Inhibition of Telomerase by Small-Molecule Pentacyclic Acridines Capable of Interacting with G-Quadruplexes. Mol. Pharmacol. 2001, 60, 981–988. 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- Gunaratnam M.; Greciano O.; Martins C.; Reszka A. P.; Schultes C. M.; Morjani H.; Riou J. F.; Neidle S. Mechanism of Acridine-Based Telomerase Inhibition and Telomere Shortening. Biochem. Pharmacol. 2007, 74, 679–689. 10.1016/j.bcp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Neidle S. Quadruplex Nucleic Acids as Novel Therapeutic Targets. J. Med. Chem. 2016, 59, 5987–6011. 10.1021/acs.jmedchem.5b01835. [DOI] [PubMed] [Google Scholar]
- Ali A.; Bhattacharya S. DNA binders in clinical trials and chemotherapy. Bioorg. Med. Chem. 2014, 22, 4506–4521. 10.1016/j.bmc.2014.05.030. [DOI] [PubMed] [Google Scholar]
- Simonsson T. G-Quadruplex DNA Structures - Variations on a Theme. Biol. Chem. 2001, 382, 621–628. 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoludis J. M.; Miller S. T.; Jeffrey P. D.; Barrett S. P.; Rablen P. R.; Lawton T. J.; Yatsunyk L. A. Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA. J. Am. Chem. Soc. 2012, 134, 20446–20456. 10.1021/ja3088746. [DOI] [PubMed] [Google Scholar]
- Ali A.; Bansal M.; Bhattacharya S. Ligand 5,10,15,20-Tetra(N-methyl-4-pyridyl)porphine (TMPyP4) Prefers the Parallel Propeller-Type Human Telomeric G-Quadruplex DNA over Its Other Polymorphs. J. Phys. Chem. B 2015, 119, 5–14. 10.1021/jp505792z. [DOI] [PubMed] [Google Scholar]
- Cao Q.; Li Y.; Freisinger E.; Qin P. Z.; Sigel R. K. O.; Mao Z. W. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs. Inorg. Chem. Front. 2017, 4, 10–32. 10.1039/C6QI00300A. [DOI] [Google Scholar]
- Jain A. K.; Bhattacharya S. Interaction of G-Quadruplexes with Nonintercalating Duplex-DNA Minor Groove Binding Ligands. Bioconjugate Chem. 2011, 22, 2355–2368. 10.1021/bc200268a. [DOI] [PubMed] [Google Scholar]
- Jain A. K.; Paul A.; Maji B.; Muniyappa K.; Bhattacharya S. Dimeric 1,3-Phenylene-bis(piperazinyl benzimidazole)s: Synthesis and Structure–Activity Investigations on their Binding with Human Telomeric G-Quadruplex DNA and Telomerase Inhibition Properties. J. Med. Chem. 2012, 55, 2981–2993. 10.1021/jm200860b. [DOI] [PubMed] [Google Scholar]
- Maji B.; Bhattacharya S. Advances in the molecular design of potential anticancer agents via targeting of human telomeric DNA. Chem. Commun. 2014, 50, 6422–6438. 10.1039/C4CC00611A. [DOI] [PubMed] [Google Scholar]
- Shay J. W.; Wright W. E. Role of telomeres and telomerase in cancer. Semin. Cancer Biol. 2011, 21, 349–353. 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W.; Wright W. E. Telomerase Therapeutics for Cancer: Challenges and New Directions. Nat. Rev. Drug Discovery 2006, 5, 577–584. 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- Alcaro S.; Costa G.; Distinto S.; Moraca F.; Ortuso F.; Parrotta L.; Artese A. The Polymorphisms of DNA G-quadruplex Investigated by Docking Experiments with Telomestatin Enantiomers. Curr. Pharm. Des. 2012, 18, 1873–1879. 10.2174/138161212799958495. [DOI] [PubMed] [Google Scholar]
- Ivancich M.; Schrank Z.; Wojdyla L.; Leviskas B.; Kuckovic A.; Sanjali A.; Puri N. Treating Cancer by Targeting Telomeres and Telomerase. Antioxidants 2017, 6, 15. 10.3390/antiox6010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S.; Chaudhuri P.; Jain A. K.; Paul A. Symmetrical Bisbenzimidazoles with Benzenediyl Spacer: The Role of the Shape of the Ligand on the Stabilization and Structural Alterations in Telomeric G-Quadruplex DNA and Telomerase Inhibition. Bioconjugate Chem. 2010, 21, 1148–1159. 10.1021/bc9003298. [DOI] [PubMed] [Google Scholar]
- Paul A.; Maji B.; Misra S. K.; Jain A. K.; Muniyappa K.; Bhattacharya S. Stabilization and Structural Alteration of the G-Quadruplex DNA Made from the Human Telomeric Repeat Mediated by Tröger’s Base Based Novel Benzimidazole Derivatives. J. Med. Chem. 2012, 55, 7460–7471. 10.1021/jm300442r. [DOI] [PubMed] [Google Scholar]
- Stewart S. A.; Weinberg R. A. Telomeres: Cancer to Human Aging. Annu. Rev. Cell Dev. Biol. 2006, 22, 531–557. 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- Shammas M. A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. 10.1097/MCO.0b013e32834121b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby T.; Fiaschetti G.; Nagasawa K.; Shin-ya K.; Baumgartner M.; Grotzer M. G-Quadruplexes as Potential Therapeutic Targets for Embryonal Tumors. Molecules 2013, 18, 12500–12537. 10.3390/molecules181012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E.; Heald R. A.; Stevens M. F. G.; Searle M. S. Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat. J. Mol. Biol. 2003, 334, 25–36. 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Gomez D.; O’Donohue M.-F.; Wenner T.; Douarre C.; Macadre J.; Koebel P.; Giraud-Panis M.-J.; Kaplan H.; Kolkes A.; Shin-ya K.; Riou J.-F. The G-quadruplex Ligand Telomestatin Inhibits POT1 Binding to Telomeric Sequences in Vitro and Induces GFP-POT1 Dissociation from Telomeres in Human Cells. Cancer Res. 2006, 66, 6908–6912. 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- Gray R. D.; Trent J. O.; Chaires J. B. Folding and Unfolding Pathways of the Human Telomeric G-Quadruplex. J. Mol. Biol. 2014, 426, 1629–1650. 10.1016/j.jmb.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. W.; Piatyszek M. A.; Prowse K. R.; Harley C. B.; West M. D.; Ho P. L. C.; Coviello G. M.; Wright W. E.; Weinrich S. L.; Shay J. W. Specific Association Of Human Telomerase Activity With Immortal Cells and Cancer. Science 1994, 266, 2011–2015. 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Tera M.; Ishizuka H.; Takagi M.; Suganuma M.; Shin-ya K.; Nagasawa K. Macrocyclic Hexaoxazoles as Sequence- and Mode-Selective G-Quadruplex Binders. Angew. Chem., Int. Ed. 2008, 47, 5557–5560. 10.1002/anie.200801235. [DOI] [PubMed] [Google Scholar]
- Uda R. M.; Matsui T.; Takei M. Binding of malachite green promotes stability and shows preference for a human telomere DNA G-quadruplex. Supramol. Chem. 2017, 29, 553–560. 10.1080/10610278.2017.1297447. [DOI] [Google Scholar]
- Collie G. W.; Promontorio R.; Hampel S. M.; Micco M.; Neidle S.; Parkinson G. N. Structural Basis for Telomeric G-Quadruplex Targeting by Naphthalene Diimide Ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. 10.1021/ja2102423. [DOI] [PubMed] [Google Scholar]
- Jain A. K.; Reddy V. V.; Paul A.; Muniyappa K.; Bhattacharya S. Synthesis and Evaluation of a Novel Class of G-Quadruplex-Stabilizing Small Molecules Based on the 1,3-Phenylene-Bis(piperazinyl benzimidazole) System. Biochemistry 2009, 48, 10693–10704. 10.1021/bi9003815. [DOI] [PubMed] [Google Scholar]
- Temime-Smaali N.; Guittat L.; Sidibe A.; Shin-ya K.; Trentesaux C.; Riou J.-F. The G-Quadruplex Ligand Telomestatin Impairs Binding of Topoisomerase III alpha to G-Quadruplex-Forming Oligonucleotides and Uncaps Telomeres in ALT Cells. PLoS One 2009, 4, e6919 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht S. A.; Neidle S. Small-Molecule Quadruplex-Targeted Drug Discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. 10.1016/j.bmcl.2014.04.029. [DOI] [PubMed] [Google Scholar]
- Campbell N. H.; Parkinson G. N.; Reszka A. P.; Neidle S. Structural Basis of DNA Quadruplex Recognition by an Acridine Drug. J. Am. Chem. Soc. 2008, 130, 6722–6734. 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- Müller S.; Rodriguez R. G-quadruplex interacting small molecules and drugs: from bench toward bedside. Expert Rev. Clin. Pharmacol. 2014, 7, 663–679. 10.1586/17512433.2014.945909. [DOI] [PubMed] [Google Scholar]
- Düchler M. G-Quadruplexes: Targets and Tools in Anticancer Drug Design. J. Drug Targeting 2012, 20, 389–400. 10.3109/1061186X.2012.669384. [DOI] [PubMed] [Google Scholar]
- Cree S. L.; Kennedy M. A. Relevance of G-Quadruplex Structures to Pharmacogenetics. Front. Pharmacol. 2014, 5, 160. 10.3389/fphar.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onel B.; Lin C.; Yang D. DNA G-Quadruplex and Its Potential as Anticancer Drug Target. Sci. China: Chem. 2014, 57, 1605–1614. 10.1007/s11426-014-5235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. Targeting G-Quadruplexes in Gene Promoters: A Novel Anticancer Strategy?. Nat. Rev. Drug Discovery 2011, 10, 261–275. 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaro S.; Musetti C.; Distinto S.; Casatti M.; Zagotto G.; Artese A.; Parrotta L.; Moraca F.; Costa G.; Ortuso F.; Maccioni E.; Sissi C. Identification and Characterization of New DNA G-Quadruplex Binders Selected by a Combination of Ligand and Structure-Based Virtual Screening Approaches. J. Med. Chem. 2013, 56, 843–855. 10.1021/jm3013486. [DOI] [PubMed] [Google Scholar]
- Dhamodharan V.; Harikrishna S.; Bhasikuttan A. C.; Pradeepkumar P. I. Topology Specific Stabilization of Promoter over Telomeric G-Quadruplex DNAs by Bisbenzimidazole Carboxamide Derivatives. ACS Chem. Biol. 2015, 10, 821–833. 10.1021/cb5008597. [DOI] [PubMed] [Google Scholar]
- Granotier C.; Pennarun G.; Riou L.; Hoffschir F.; Gauthier L. R.; De Cian A.; Gomez D.; Mandine E.; Riou J. F.; Mergny J. L.; Mailliet P.; Dutrillaux B.; Boussin F. D. Preferential Binding of a G-Quadruplex Ligand to Human Chromosome Ends. Nucleic Acids Res. 2005, 33, 4182–4190. 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J.-Q.; Chen S.-B.; Tan J.-H.; Ou T.-M.; Luo H.-B.; Li D.; Xu J.; Gu L.-Q.; Huang Z.-S. New Insights into the Structures of Ligand–Quadruplex Complexes from Molecular Dynamics Simulations. J. Phys. Chem. B 2010, 114, 15301–15310. 10.1021/jp106683n. [DOI] [PubMed] [Google Scholar]
- Hounsou C.; Guittat L.; Monchaud D.; Jourdan M.; Saettel N.; Mergny J. L.; Teulade-Fichou M. P. G-Quadruplex Recognition by Quinacridines: A SAR, NMR, and Biological Study. Chem. Med. Chem. 2007, 2, 655–666. 10.1002/cmdc.200600286. [DOI] [PubMed] [Google Scholar]
- Li M.-H.; Luo Q.; Li Z.-S. Molecular Dynamics Study on the Interactions of Porphyrin with Two Antiparallel Human Telomeric Quadruplexes. J. Phys. Chem. B 2010, 114, 6216–6224. 10.1021/jp101373p. [DOI] [PubMed] [Google Scholar]
- Luo D.; Mu Y. All-Atomic Simulations on Human Telomeric G-Quadruplex DNA Binding with Thioflavin T. J. Phys. Chem. B 2015, 119, 4955–4967. 10.1021/acs.jpcb.5b01107. [DOI] [PubMed] [Google Scholar]
- Mulholland K.; Wu C. Binding of Telomestatin to a Telomeric G-Quadruplex DNA Probed by All-Atom Molecular Dynamics Simulations with Explicit Solvent. J. Chem. Inf. Model. 2016, 56, 2093–2102. 10.1021/acs.jcim.6b00473. [DOI] [PubMed] [Google Scholar]
- Krauss I. R.; Ramaswamy S.; Neidle S.; Haider S.; Parkinson G. N. Structural Insights into the Quadruplex–Duplex 3′ Interface Formed from a Telomeric Repeat: A Potential Molecular Target. J. Am. Chem. Soc. 2016, 138, 1226–1233. 10.1021/jacs.5b10492. [DOI] [PubMed] [Google Scholar]
- Milelli A.; Tumiatti V.; Micco M.; Rosini M.; Zuccari G.; Raffaghello L.; Bianchi G.; Pistoia V.; Díaz J. F.; Pera B.; Trigili C.; Barasoain I.; Musetti C.; Toniolo M.; Sissi C.; Alcaro S.; Moraca F.; Zini M.; Stefanelli C.; Minarini A. Structure-Activity Relationships of Novel Substituted Naphthalene Diimides as Anticancer Agents. Eur. J. Med. Chem. 2012, 57, 417–428. 10.1016/j.ejmech.2012.06.045. [DOI] [PubMed] [Google Scholar]
- Binz N.; Shalaby T.; Rivera P.; Shin-Ya K.; Grotzer M. A. Telomerase Inhibition, Telomere Shortening, Cell Growth Suppression and Induction of Apoptosis by Telomestatin in Childhood Neuroblastoma Cells. Eur. J. Cancer 2005, 41, 2873–2881. 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Shammas M. A.; Reis R. J. S.; Li C.; Koley H.; Hurley L. H.; Anderson K. C.; Munshi N. C. Telomerase Inhibition and Cell Growth Arrest After Telomestatin Treatment in Multiple Myeloma. Clin. Cancer Res. 2004, 10, 770–776. 10.1158/1078-0432.CCR-0793-03. [DOI] [PubMed] [Google Scholar]
- Tahara H.; Shin-ya K.; Seimiya H.; Yamada H.; Tsuruo T.; Ide T. G-Quadruplex Stabilization by Telomestatin Induces TRF2 Protein Dissociation from Telomeres and Anaphase Bridge Formation Accompanied by Loss of the 3′ Telomeric Overhang in Cancer Cells. Oncogene 2006, 25, 1955–1966. 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- Tauchi T.; Shin-ya K.; Sashida G.; Sumi M.; Nakajima A.; Shimamoto T.; Ohyashiki J. H.; Ohyashiki K. Activity of a Novel G-Quadruplex-Interactive Telomerase Inhibitor, Telomestatin (SOT-095), Against Human Leukemia Cells: Involvement of ATM-Dependent DNA Damage Response Pathways. Oncogene 2003, 22, 5338–5347. 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- Siddiqui-Jain A.; Grand C. L.; Bearss D. J.; Hurley L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. Y.; Gleason-Guzman M.; Izbicka E.; Nishioka D.; Hurley L. H. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003, 63, 3247–3256. [PubMed] [Google Scholar]
- Mikami-Terao Y.; Akiyama M.; Yuza Y.; Yanagisawa T.; Yamada O.; Kawano T.; Agawa M.; Ida H.; Yamada H. Antitumor activity of TMPyP4 interacting G-quadruplex in retinoblastoma cell lines. Exp. Eye Res. 2009, 89, 200–208. 10.1016/j.exer.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Mikami-Terao Y.; Akiyama M.; Yuza Y.; Yanagisawa T.; Yamada O.; Yamada H. Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Lett. 2008, 261, 226–234. 10.1016/j.canlet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Gunaratnam M.; Greciano O.; Martins C.; Reszka A. P.; Schultes C. M.; Morjani H.; Riou J.-F.; Neidle S. Mechanism of Acridine-Based Telomerase Inhibition and Telomere Shortening. Biochem. Pharmacol. 2007, 74, 679–689. 10.1016/j.bcp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Chen T. R.; Drabkowski D.; Hay R. J.; Macy M.; Peterson W. Jr. WiDr is a Derivative of Another Colon Adenocarcinoma Cell Line, HT-29. Cancer Genet. Cytogenet. 1987, 27, 125–134. 10.1016/0165-4608(87)90267-6. [DOI] [PubMed] [Google Scholar]
- Fang R.; Aust A. E. Induction of Ferritin Synthesis in Human Lung Epithelial Cells Treated with Crocidolite Asbestos. Arch. Biochem. Biophys. 1997, 340, 369–375. 10.1006/abbi.1997.9892. [DOI] [PubMed] [Google Scholar]
- Mickey D. D.; Stone K. R.; Wunderli H.; Mickey G. H.; Vollmer R. T.; Paulson D. F. Heterotransplantation of a Human Prostatic Adenocarcinoma Cell Line in Nude Mice. Cancer Res. 1977, 37, 4049. [PubMed] [Google Scholar]
- Akagi T.; Kimoto T. Human Cell Line (Hgc-27) Derived from Metastatic Lymph-Node of Gastric Cancer. Acta Med. Okayama 1976, 30, 215–219. [PubMed] [Google Scholar]
- Brandes L. J.; Hermonat M. W. Receptor Status and Subsequent Sensitivity of Subclones of MCF-7 Human Breast Cancer Cells Surviving Exposure to Diethylstilbestrol. Cancer Res. 1983, 43, 2831–2835. [PubMed] [Google Scholar]
- Alizadehnohi M.; Nabiuni M.; Nazari Z.; Safaeinejad Z.; Irian S. The Synergistic Cytotoxic Effect of Cisplatin and Honey Bee Venom on Human Ovarian Cancer Cell Line A2780cp. J. Venom Res. 2012, 3, 22–27. [PMC free article] [PubMed] [Google Scholar]
- Nichols W. W.; Murphy D. G.; Cristofalo V. J.; Toji L. H.; Greene A. E.; Dwight S. A. Characterization of a New Human Diploid Cell Strain, IMR-90. Science 1977, 196, 60. 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- Landers J. E.; Cassel S. L.; George D. L. Translational Enhancement of Oncogene Expression in Human Tumor Cells Containing a Stabilized Wild-Type p53 Protein. Cancer Res. 1997, 57, 3562–3568. [PubMed] [Google Scholar]
- Olopade O. I.; Jenkins R. B.; Ransom D. T.; Malik K.; Pomykala H.; Nobori T.; Cowan J. M.; Rowley J. D.; Diaz M. O. Molecular Analysis of Deletions of the Short Arm of Chromosome 9 in Human Gliomas. Cancer Res. 1992, 52, 2523. [PubMed] [Google Scholar]
- Burger A. M.; Dai F. P.; Schultes C. M.; Reszka A. P.; Moore M. J.; Double J. A.; Neidle S. The G-Quadruplex-Interactive Molecule BRACO-19 Inhibits Tumor Growth, Consistent with Telomere Targeting and Interference with Telomerase Function. Cancer Res. 2005, 65, 1489–1496. 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- Zhou G.; Liu X.; Li Y.; Xu S.; Ma C.; Wu X.; Cheng Y.; Yu Z.; Zhao G.; Chen Y. Telomere Targeting with a Novel G-Quadruplex-Interactive Ligand BRACO-19 Induces T-Loop Disassembly and Telomerase Displacement in Human Glioblastoma Cells. Oncotarget 2016, 7, 14925–14939. 10.18632/oncotarget.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. J.; Reszka A. P.; Haider S. M.; Romagnoli B.; Morrell J.; Read M. A.; Gowan S. M.; Incles C. M.; Kelland L. R.; Neidle S. Evaluation of by Disubstituted Acridone Derivatives as Telomerase Inhibitors: The Importance of G-Quadruplex Binding. Bioorg. Med. Chem. Lett. 2004, 14, 5845–5849. 10.1016/j.bmcl.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Morimoto H.; Safrit J. T.; Bonavida B. Synergistic Effect of Tumor Necrosis Factor-Alpha- and Diphtheria Toxin-Mediated Cytotoxicity in Sensitive and Resistant Human Ovarian Tumor Cell Lines. J. Immunol. 1991, 147, 2609. [PubMed] [Google Scholar]
- Rankin A. M.; Faller D. V.; Spanjaard R. A. Telomerase Inhibitors and ‘T-oligo’ as Cancer Therapeutics: Contrasting Molecular Mechanisms of Cytotoxicity. Anti-Cancer Drugs 2008, 19, 329–338. 10.1097/CAD.0b013e3282f5d4c2. [DOI] [PubMed] [Google Scholar]
- Kellner J.; Wierda W.; Shpall E.; Keating M.; McNiece I. Isolation of a Novel Chronic Lymphocytic Leukemic (CLL) Cell Line and Development of an In Vivo Mouse Model of CLL. Leuk. Res. 2016, 40, 54–59. 10.1016/j.leukres.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyinsky N. S.; Varizhuk A. M.; Beniaminov A. D.; Puzanov M. A.; Shchyolkina A. K.; Kaluzhny D. N. G-quadruplex ligands: Mechanisms of anticancer action and target binding. Mol. Biol. 2014, 48, 778–794. 10.1134/S0026893314060077. [DOI] [PubMed] [Google Scholar]
- Miyazaki T.; Pan Y.; Joshi K.; Purohit D.; Hu B.; Demir H.; Mazumder S.; Okabe S.; Yamori T.; Viapiano M.; Shin-ya K.; Seimiya H.; Nakano I. Telomestatin Impairs Glioma Stem Cell Survival and Growth through the Disruption of Telomeric G-Quadruplex and Inhibition of the Proto-oncogene, c-Myb. Clin. Cancer Res. 2012, 18, 1268–1280. 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. J.; Heddi B.; Tera M.; Iida K.; Nagasawa K.; Phan A. T. Solution Structure of an Intramolecular (3+1) Human Telomeric G-Quadruplex Bound to a Telomestatin Derivative. J. Am. Chem. Soc. 2013, 135, 13495–13501. 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- Reed J.; Gunaratnam M.; Beltran M.; Reszka A. P.; Vilar R.; Neidle S. TRAP–LIG, a modified telomere repeat amplification protocol assay to quantitate telomerase inhibition by small molecules. Anal. Biochem. 2008, 380, 99–105. 10.1016/j.ab.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Taetz S.; Baldes C.; Mürdter T. E.; Kleideiter E.; Piotrowska K.; Bock U.; Haltner-Ukomadu E.; Mueller J.; Huwer H.; Schaefer U. F.; Klotz U.; Lehr C.-M. Biopharmaceutical Characterization of the Telomerase Inhibitor BRACO19. Pharm. Res. 2006, 23, 1031–1037. 10.1007/s11095-006-0026-y. [DOI] [PubMed] [Google Scholar]
- Taetz S.; Mürdter T. E.; Zapp J.; Boettcher S.; Baldes C.; Kleideiter E.; Piotrowska K.; Schaefer U. F.; Klotz U.; Lehr C. M. Decomposition of the Telomere-Targeting agent BRACO19 in physiological media results in products with decreased inhibitory potential. Int. J. Pharm. 2008, 357, 6–14. 10.1016/j.ijpharm.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Patell D. I. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure 1993, 1, 263–282. 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Hurley L. H.; Yang D. Solution Structure of a 2:1 Quindoline-C-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design. J. Am. Chem. Soc. 2011, 133, 17673–17680. 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosconati S.; Marinelli L.; Trotta R.; Virno A.; De Tito S.; Romagnoli R.; Pagano B.; Limongelli V.; Giancola C.; Baraldi P. G.; Mayol L.; Novellino E.; Randazzo A. Structural and Conformational Requisites in DNA Quadruplex Groove Binding: Another Piece to the Puzzle. J. Am. Chem. Soc. 2010, 132, 6425–6433. 10.1021/ja1003872. [DOI] [PubMed] [Google Scholar]
- Haider S. M.; Parkinson G. N.; Neidle S. Structure of a G-quadruplex–Ligand Complex. J. Mol. Biol. 2003, 326, 117–125. 10.1016/S0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- Sponer J.; Cang X.; Cheatham T. E. III Molecular Dynamics Simulations of G-DNA and Perspectives on the Simulation of Nucleic Acid Structures. Methods 2012, 57, 25–39. 10.1016/j.ymeth.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnikov R. V.; Kopylov A. M.; Golovin A. V. Classification of G-Quadruplex DNA on the Basis of the Quadruplex Twist Angle and Planarity of G-Quartets. Acta Nat. 2010, 2, 72–81. [PMC free article] [PubMed] [Google Scholar]
- Biffi G.; Tannahill D.; Miller J.; Howat W. J.; Balasubramanian S. Elevated Levels of G-Quadruplex Formation in Human Stomach and Liver Cancer Tissues. PLoS One 2014, 9, e102711 10.1371/journal.pone.0102711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K. W.; Chen Z.; Hao Y. H.; Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010, 38, 327–338. 10.1093/nar/gkp898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G. N.; Ghosh R.; Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry 2007, 46, 2390–2397. 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- Pérez A.; Marchán I.; Svozil D.; Sponer J.; Cheatham T. E. III; Laughton C. A.; Orozco M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys. J. 2007, 92, 3817–3829. 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepl M.; Zgarbova M.; Stadlbauer P.; Otyepka M.; Banas P.; Koca J.; Cheatham T. E.; Jurecka P.; Sponer J. Reference Simulations of Noncanonical Nucleic Acids with Different chi Variants of the AMBER Force Field: Quadruplex DNA, Quadruplex RNA, and Z-DNA. J. Chem. Theory Comput. 2012, 8, 2506–2520. 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbová M.; Luque F. J.; Šponer J.; Cheatham T. E.; Otyepka M.; Jurečka P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J. Chem. Theory Comput. 2013, 9, 2339–2354. 10.1021/ct400154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbová M.; Šponer J.; Otyepka M.; Cheatham T. E.; Galindo-Murillo R.; Jurečka P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. 10.1021/acs.jctc.5b00716. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparisons of Simple Potential Functions for Simulating Liquid Qater. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Joung I. S.; Cheatham T. E. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112, 9020–9041. 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly C. I.; Cieplak P.; Cornell W. D.; Kollman P. A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the RESP Model. J. Phys. Chem. 1993, 97, 10269–10280. 10.1021/j100142a004. [DOI] [Google Scholar]
- Case D. A.; Betz R. M.; Botello-Smith W.; Cerutti D. S.; Cheatham T. E. III; Darden T. A.; Duke R. E.; Giese T. J.; Gohlke H.; Goetz A. W.; Homeyer N.; Izadi S.; Janowski P.; Kaus J.; Kovalenko A.; Lee T. S.; LeGrand S.; Li P.; Lin C.; Luchko T.; Luo R.; Madej B.. AMBER 2016; University of California: San Francisco, 2016. [Google Scholar]
- Lavery R.; Zakrzewska K.; Beveridge D.; Bishop T. C.; Case D. A.; Cheatham T. E. I.; Dixit S.; Jayaram B.; Lankas F.; Laughton C.; Maddocks J. H.; Michon A.; Osman R.; Orozco M.; Perez A.; Singh T.; Spackova N.; Sponer J. A Systematic Molecular Dynamics Study of Nearest-Neighbor Effects on Base Pair and Base Pair Step Conformations and Fluctuations in B-DNA. Nucleic Acids Res. 2010, 38, 299–313. 10.1093/nar/gkp834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrná E.; Špačková N.; Sarzyñska J.; Koča J.; Orozco M.; Cheatham T. E. III; Kulinski T.; Šponer J. Single Stranded Loops of Quadruplex DNA As Key Benchmark for Testing Nucleic Acids Force Fields. J. Chem. Theory Comput. 2009, 5, 2514–2530. 10.1021/ct900200k. [DOI] [PubMed] [Google Scholar]
- Mukherjee A.; Lavery R.; Bagchi B.; Hynes J. T. On the Molecular Mechanism of Drug Intercalation into DNA: A Simulation Study of the Intercalation Pathway, Free Energy, and DNA Structural Changes. J. Am. Chem. Soc. 2008, 130, 9747–9755. 10.1021/ja8001666. [DOI] [PubMed] [Google Scholar]
- Lei H.; Wang X.; Wu C. Early Stage Intercalation of Doxorubicin to DNA Fragments Observed in Molecular Dynamics Binding Simulations. J. Mol. Graph. Model. 2012, 38, 279–289. 10.1016/j.jmgm.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Islam B.; Stadlbauer P.; Krepl M.; Koca J.; Neidle S.; Haider S.; Sponer J. Extended molecular dynamics of a c-kit promoter quadruplex. Nucleic Acids Res. 2015, 43, 8673–8693. 10.1093/nar/gkv785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A.; Babin V.; Berryman J. T.; Betz R. M.; Cai Q.; Cerutti D. S.; Cheatham T. E. III; Darden T. A.; Duke R. E.; Gohlke H.; Goetz A. W.; Gusarov S.; Homeyer N.; Janowski P.; Kaus J.; Kolossváry I.; Kovalenko A.; Lee T. S.; LeGrand S.; Luchko T.; Luo R.; Madej B.; Merz K. M.; Paesani F.; Roe D. R.; Roitberg A.; Sagui C.; Salomon-Ferrer R.; Seabra G.; Simmerling C. L.; Smith W.; Swails J.; Walker R. C.; Wang J.; Wolf R. M.; Kollman P. A.; Wu X.. AMBER 14; University of California: San Francisco, 2014.
- Lei H.; Wang X.; Wu C. Early stage intercalation of doxorubicin to DNA fragments observed in molecular dynamics binding simulations. J. Mol. Graph. Model. 2012, 38, 279–89. 10.1016/j.jmgm.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Mulholland K.; Wu C. Binding of Telomestatin to a Telomeric G-Quadruplex DNA Probed by All-Atom Molecular Dynamics Simulations with Explicit Solvent. J. Chem. Inf. Model. 2016, 56, 2093–2102. 10.1021/acs.jcim.6b00473. [DOI] [PubMed] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T. A.; Lee H.; Pedersen L. G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. 10.1063/1.470117. [DOI] [Google Scholar]
- Procacci P.; Berne B. J. Multiple Time-Scale Methods For Constant-Pressure Molecular-Dynamics Simulations Of Molecular-Systems. Mol. Phys. 1994, 83, 255–272. 10.1080/00268979400101241. [DOI] [Google Scholar]
- Lavery R.; Moakher M.; Maddocks J. H.; Petkeviciute D.; Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves. Nucleic Acids Res. 2009, 37, 5917–5929. 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daura X.; Gademann K.; Jaun B.; Seebach D.; van Gunsteren W. F.; Mark A. E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem., Int. Ed. 1999, 38, 236–240. . [DOI] [Google Scholar]
- Kollman P. A.; Massova I.; Reyes C.; Kuhn B.; Huo S.; Chong L.; Lee M.; Lee T.; Duan Y.; Wang W.; Donini O.; Cieplak P.; Srinivasan J.; Case D. A.; Cheatham T. E. III Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Model. Acc. Chem. Res. 2000, 33, 889–897. 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Gaillard T.; Case D. A. Evaluation of DNA Force Fields in Implicit Solvation. J. Chem. Theory Comput. 2011, 7, 3181–3198. 10.1021/ct200384r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland K.; Siddiquei F.; Wu C. Binding Modes and Pathway of RHPS4 to Human Telomeric G-quadruplex and Duplex DNA Probed by All-Atom Molecular Dynamics Simulations with Explicit Solvent. Phys. Chem. Chem. Phys. 2017, 19, 18685–18694. 10.1039/C7CP03313C. [DOI] [PubMed] [Google Scholar]
- Deng N.; Wickstrom L.; Cieplak P.; Lin C.; Yang D. Resolving the Ligand-Binding Specificity in c-MYC G-Quadruplex DNA: Absolute Binding Free Energy Calculations and SPR Experiment. J. Phys. Chem. B 2017, 121, 10484–10497. 10.1021/acs.jpcb.7b09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsted J.; Soderhjelm P.; Ryde U. How Accurate are Continuum Solvation Models for Drug-Like Molecules?. J. Comput.-Aided Mol. Des. 2009, 23, 395–409. 10.1007/s10822-009-9271-6. [DOI] [PubMed] [Google Scholar]
- Kollman P. A.; Massova I.; Reyes C.; Kuhn B.; Huo S.; Chong L.; Lee M.; Lee T.; Duan Y.; Wang W.; Donini O.; Cieplak P.; Srinivasan J.; Case D. A.; Cheatham T. E. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Hou T.; Wang J.; Li Y.; Wang W. Assessing the Performance of the Molecular Mechanics/Poisson Boltzmann Surface Area and Molecular Mechanics/Generalized Born Surface Area Methods. II. The Accuracy of Ranking Poses Generated from Docking. J. Comput. Chem. 2011, 32, 866–877. 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T.; Wang J.; Li Y.; Wang W. Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. J. Chem. Inf. Model. 2011, 51, 69–82. 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Sun H.; Li Y.; Wang J.; Hou T. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 3. The Impact of Force Fields and Ligand Charge Models. J. Phys. Chem. B 2013, 117, 8408–8421. 10.1021/jp404160y. [DOI] [PubMed] [Google Scholar]
- Sun H.; Li Y.; Tian S.; Xub L.; Hou T. Assessing the Performance of MM/PBSA and MM/GBSA Methods. 4. Accuracies of MM/PBSA and MM/GBSA Methodologies Evaluated by Various Simulation Protocols Using PDBbind Data Set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. 10.1039/C4CP01388C. [DOI] [PubMed] [Google Scholar]
- Shen Z.; Mulholland K.; Zheng Y.; Wu C. Binding of Anticancer Drug Daunomycin to a TGGGGT G-Quadruplex DNA Probed by All-Atom Molecular Dynamics Simulations: Additional Pure Groove Binding Mode and Implications on Designing More Selective G-Quadruplex Ligands. J. Mol. Model. 2017, 23, 256. 10.1007/s00894-017-3417-6. [DOI] [PubMed] [Google Scholar]
- Machireddy B.; Kalra G.; Jonnalagadda S.; Ramanujachary K.; Wu C. Probing the Binding Pathway of BRACO19 to a Parallel-Stranded Human Telomeric G-Quadruplex Using Molecular Dynamics Binding Simulation with AMBER DNA OL15 and Ligand GAFF2 Force Fields. J. Chem. Inf. Model. 2017, 57, 2846–2864. 10.1021/acs.jcim.7b00287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.