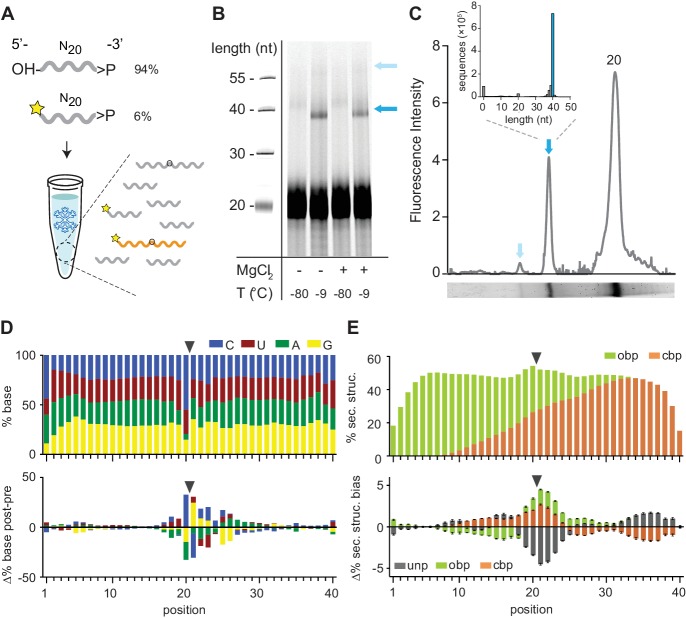
Figure 1. Reactivity of 2’, 3’ cyclic phosphate (>p) activated N20 RNA pools.
(A) N20>p pools are incubated in eutectic ice phases, spiked with 6% 5’FAM (Carboxyfluorescein)-labelled N20>p to facilitate detection of ligation products (orange) by Urea-PAGE. (B) Scan of a denaturing gel for FAM-labelled products after 57 days of incubation either in eutectic ice (−9 °C) or at −80 °C, and +/- MgCl2. Note that ligation of FAM-N20>p is inhibited due to the blocked 5’-OH. (C) Densitogram of a SYBR-gold stained Urea-PAGE gel trace (-MgCl2, −9 °C) (bottom panel) showing total ligation (both FAM-labelled and unlabelled ligation products (indicated by arrows)). The size distribution and size of the main ligation product (40mer) from deep sequencing indicates direct ligation of two eicosamers (inset). (D) Upper panel: Average nucleotide distribution profile of the 40mer ligation products. Lower panel: Changes in nucleotide composition compared to the unligated input N20>p pool. Black arrows indicate the ligation site. (E) Upper panel: Average base-pairing frequencies of sequenced 40mers (as predicted by RNAfold (Lorenz et al., 2011): ‘Opening’ base pairs (obp, green; defined as hybridization to a downstream nucleotide) and ‘closing’ base pairs (cpb, hybridization to an upstream nucleotide, orange). Lower panel: Average difference of predicted base pairing between experimental pool and synthetic sequence pools (N = 3; 300,000 sequences each) generated in silico using experimental nucleotide frequencies shown in panel D. Grey indicates predicted unpaired bases (unp), black standard deviations.