Abstract
The purpose of this study was to determine the distribution of and potential significance of laminin 332 (LM332) in breast cancer. Specimens from a population based cohort (N=297) from 1994–1995 were stained for estrogen receptor (ER), progesterone receptor (PgR), HER2 and the LM332 β3 chain. Seventy five tumors were LM332 positive and 222 were negative. LM332 β3 stained 16.0% of ER and/or PgR positive tumors and 73.2% of triple negative breast cancers (TNBC). Immunoblotting revealed LM332 in TNBC and HER2 positive samples, but not in an ER positive breast carcinoma or a phyllodes tumor. After 20 years, 172 patients were alive, 43 had died of breast cancer and 82 of other causes. Patients with LM332 positive tumors had significantly worse 5 (p < 0.0001) and 10 (p < 0.05) year overall and breast cancer specific survival. Among patients with LM332 β3 expressing and ER/PgR negative carcinomas, 10 year survival was significantly reduced (p < 0.0450). In a multivariate analysis LM332 positive patients had significant Hazard Ratios of 3.9 with 95% confidence intervals (CI) of (2.0–7.7) and 2.2 with 95% CI of (1.3–3.8) for 5 and 10 year overall survival respectively. Because tumor cell motility is required for metastasis, the effect of LM332 on MDA-MB-231 migration was determined using siRNA. Knock down of LM332-specific β3 and γ2 chains reduced motility without affecting viability. Our observation that LM332 in breast carcinoma is associated with decreased survival provides evidence that LM332 may have a role in the aggressive phenotype of some breast cancers.
Keywords: Laminin 332, Breast carcinoma, Triple negative, Immunohistochemistry, Prognosis, siRNA, Tumor Cell Migration
1. INTRODUCTION
Numerous prognostic markers have been identified in breast carcinoma and among these, estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2), have resulted in successful targeted therapies. Prognostic markers and targeted therapies are needed for triple negative breast carcinomas (TNBC), which do not express ER, progesterone receptor (PgR) or HER2. We examined whether laminin 332 (LM332) might provide a prognostic marker for breast carcinoma and TNBC in particular. LM332 is expressed by myoepithelium and is a heterotrimer of α3, β3 and γ2 chains (1–3). The α3 chain is a component of other laminins, but γ2 is LM332 –specific, and the □3 chain is specific with only rare exceptions (4). LM332 stimulates the migration of breast and other carcinoma cells (5) and correlates with tumor invasiveness (1, 6).
Previously we demonstrated LM332 expression in over 70% of TNBC (7), but the cases did not have long term follow-up. We now examine a population based cohort of breast cancer cases from 1994–1995 to better define the expression of LM332 among breast carcinoma subtypes with over 20 years of follow-up. We used a β3 chain antibody, which is sensitive and stains in the same pattern as γ2 antibodies (8). We provide additional validation of this antibody using an immunoblot of breast cancer samples and immunohistochemistry (IHC). Finally, we provide data to suggest a mechanism to explain the more aggressive phenotype of LM332-expressing breast cancers.
2. MATERIALS AND METHODS
2.1. Tissue micro array construction
The cohort was a subset of patients from the Orange County Breast and Ovarian Cancer Study, during the 1-year period beginning March 1, 1994 (9, 10). The study was approved by the Institutional Review Board of the University of California, Irvine, with informed consent obtained from all subjects. Demographic data were obtained from surveys and from the California Cancer Registry (CCR). Either paraffin blocks or unstained slides of breast cancer samples were procured from the facilities where surgeries were performed. All tissues were from excision specimens. The slides were reviewed, and cases of invasive carcinoma with either sufficient unstained slides or paraffin blocks for a tissue micro array (TMA) were used for histopathologic studies. TMA construction used a Tissue-Tek Quick Ray kit (Sakura Finetek USA, Torrance, CA) with 1 mm punches. Sufficient tumor was available in all paraffin blocks except one for at least two punches from the block to be placed on the array. Thirteen cases with ductal carcinoma in situ (DCIS) only and 23 cases without tumor in any of the punches were excluded. 314 specimens from 313 subjects were stained. IHC data was collected from 297 subjects after excluding cases without staining for all markers. Of these, 137 specimens were from unstained slides and 160 specimens were from the TMA.
2.2. Immunohistochemistry
Slides were stained with hematoxylin and eosin (H&E) and IHC for ER, PgR, HER2 and LM332 as previously described using a BenchMark Ultra (Ventana Medical Systems, Inc. Tucson AZ) immunostainer (7). The anti-HER2 antibody 4B5 (PATHWAY anti-HER2/neu Rabbit Monoclonal Primary Antibody), mouse ER (clone SP1) and mouse PgR (clone 1E2) antibodies were prediluted (Ventana). The LM332 β3 chain clone 17 (kalinin B1, BD Transduction Laboratories, Lexington KY) was diluted at 1:250. The appropriate breast tissue provided the positive controls. Microscopy and photography were performed using an Olympus (Center Valley, PA) BX 46 microscope with a DP71 camera and DP Controller and DP Manager Software.
Data regarding tissue collection, including differences in cold ischemic time and time in formalin was not available for these cases, since the importance of documenting these variables has only recently been appreciated (11). We chose an alternate method of determining whether the tissues were affected by pre-analytic variables, including not only cold ischemic time and fixation time, but also variability that might have occurred as a consequence of prolonged time in paraffin, transfer of tissue to microarrays or a long interval between sectioning and staining. To examine whether staining quality was affected by pre-analytic variables, sections were reviewed for staining of ER and PgR in low grade tumors and/or normal glandular elements as internal controls. In all cases where either or both these elements were present, ER, PgR or both showed a reaction, indicating that the quality of the sections was sufficient to allow adequate staining by IHC.
2.3. Immunoblot
For a second method of examining LM332 β3 expression in breast carcinoma, an immunoblot was performed using lysates of 5 frozen breast cancer samples obtained from the UCI Chao Family Comprehensive Cancer Center Experimental Tissue Core Facility. Frozen sections of tumors were performed to confirm that the tissue had at least 80% tumor cells. Approximately 30 mg from each of 2 metaplastic carcinomas, an ER positive ductal carcinoma, a HER2 positive DCIS and one malignant phyllodes tumor was homogenized in 0.5 ml of Laemmli buffer, boiled for 10 minutes, and centrifuged for 10 minutes. The breast carcinomas and phyllodes tumor were not from members of the cohort since specimens collected for that study did not include frozen tumor tissue, and thus the results from the immunoblot are not included among the results listed in Table 1. Supernatants normalized for an equal number of tumor cells per lane underwent sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), transfer to nitrocellulose and incubation with anti-LM332 β3 chain clone 17, (BD Transduction Laboratories) diluted 1: 2000 and mouse anti-glyceraldehyde-3 phosphate dehydrogenase (GADPH) (Millipore, Temecula CA) diluted 1:50,000 in blotto buffer. For tissue culture experiments, β actin Abcam8226 (Abcam, Cambridge, MA) was diluted 1:1000 to 23 μg/mL and γ2 laminin chain (clone D4B5 Millipore) was diluted 1:800 to 2 μg/mL. The biotinylated anti-mouse secondary antibody (Pierce, Rockford IL) was diluted 1:10,000, and the bands were developed with the Pico Super Signal West chemoluminescent detection system (Pierce) according to the manufacturer’s instructions and visualized by exposure to XAR film (Kodak, Rochester NY) or visualized with a C-Digit scanner (Licor, Lincoln, NE).
Table 1.
Comparison of laminin 332 β3 chain staining among epidemiologic, clinical and pathological subsets.
| Laminin β chain <5% | Laminin β chain ≥5% | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Count | Column% | Row% | Count | Column% | Row% | p_value |
| Age at diagnosis | |||||||
| <45 | 22 | 9.91 | 44.90 | 27 | 36.00 | 55.10 | 8.06E-07 |
| 45–54 | 53 | 23.87 | 77.94 | 15 | 20.00 | 22.06 | |
| >=55 | 147 | 66.22 | 81.67 | 33 | 44.00 | 18.33 | |
| Race/Ethnicity | |||||||
| White | 175 | 78.83 | 76.42 | 54 | 72.00 | 23.58 | 0.4505 |
| African American | 7 | 3.15 | 77.78 | 2 | 2.67 | 22.22 | |
| Hispanic | 23 | 10.36 | 63.89 | 13 | 17.33 | 36.11 | |
| Asian | 17 | 7.66 | 73.91 | 6 | 8.00 | 26.09 | |
| Grade | |||||||
| I | 100 | 45.05 | 88.50 | 13 | 17.33 | 11.50 | 7.34E-09 |
| II | 90 | 40.54 | 76.92 | 27 | 36.00 | 23.08 | |
| III | 32 | 14.41 | 47.76 | 35 | 46.67 | 52.24 | |
| Histology | |||||||
| Ductal | 181 | 81.53 | 71.54 | 72 | 96.00 | 28.46 | 0.0187 |
| Lobular | 16 | 7.21 | 88.89 | 2 | 2.67 | 11.11 | |
| Tubular | 8 | 3.60 | 88.89 | 1 | 1.33 | 11.11 | |
| Other | 17 | 7.66 | 100.00 | 0 | 0.00 | 0.00 | |
| Stage | |||||||
| Localized | 136 | 61.26 | 76.84 | 41 | 54.67 | 23.16 | 0.4680 |
| Regional/Metastatic | 85 | 38.29 | 72.03 | 33 | 44.00 | 27.97 | |
| Unknown | 1 | 0.45 | 50.00 | 1 | 1.33 | 50.00 | |
| ER and PgR combinations | |||||||
| ER+ or PgR+ | 200 | 90.09 | 84.03 | 38 | 50.67 | 15.97 | 1.38E-13 |
| ER− and PgR− | 22 | 9.91 | 37.29 | 37 | 49.33 | 62.71 | |
| Chemotherapy | |||||||
| No | 141 | 63.51 | 83.43 | 28 | 37.33 | 16.57 | 0.0003 |
| Yes | 72 | 32.43 | 62.61 | 43 | 57.33 | 37.39 | |
| 9 | 4.05 | 69.23 | 4 | 5.33 | 30.77 | ||
| Hormone Therapy | |||||||
| No | 125 | 56.31 | 70.22 | 53 | 70.67 | 29.78 | 0.09 |
| Yes | 88 | 39.64 | 81.48 | 20 | 26.67 | 18.52 | |
| Unknown | 9 | 4.05 | 81.82 | 2 | 2.67 | 18.18 | |
| Radiation | |||||||
| No | 127 | 57.21 | 75.60 | 41 | 54.67 | 24.40 | 0.701 |
| Yes | 95 | 42.79 | 73.64 | 34 | 45.33 | 26.36 | |
| Total | 222 | 75 | |||||
2.4. Fluorescence In Situ Hybridization
HER2 gene amplification using the PathVysion fluorescence in situ hybridization (FISH) assays (Abbott Molecular, Inc.) is described elsewhere (12). In brief, rehydration of paraffin sections and protease digestion was followed by hybridizing with fluorescent-labeled probes for HER2 gene and alpha-satellite DNA for chromosome 17, and counterstaining with 4’−6’-diamidino-2’-phenylindole. Staining was visualized with an Axioskop 20 fluorescence microscope (Carl Zeiss, Inc., Oberkochen, Germany). For each tumor, at least 20 nuclei were scored for both HER2 gene copy number and chromosome 17 centromere numbers. The presence or absence of HER2 gene amplification was determined according to standard guidelines (12).
2.5. IHC grading of microarray
IHC and H&E were evaluated for tumor subtype and Nottingham grades by two reviewers, including one breast pathologist (PMC), with differences resolved over a two-headed microscope. ER, PgR (13) and HER2 IHC were scored using standard guidelines (13, 14). Cytoplasmic staining of 5% or more of tumor cells was considered a positive result for LM332. Staining of normal elements was not scored. For ER and PgR negative, HER2 equivocal cases (2+ by IHC), slides were evaluated for HER2 gene amplification by FISH, to determine whether they were TNBC or HER2 over-expressing carcinomas.
2.6. Analysis of LM332 expression
We determined the relationship between LM332 expression and patient demographics, survival, and tumor characteristics. For comparison between LM332 and traditional biomarkers, cases were divided into two groups. The first group was tumors that were ER or PgR positive, with any value of HER2. The second group of tumors was negative for ER and PR, and thus included both TNBC and HER2 expressing carcinomas. Previous studies have shown that this grouping correlates with LM332 expression (7) and having two subgroups provided greater statistical power for evaluating the prognostic value of LM332 expression.
2.7. Follow-up and statistics
Patients were linked with the CCR between 1996 and 2012. Cause of death was recorded according to International Classification of Disease criteria. Last date of follow-up was either the date of death or the last date of contact. The CCR is the population-based surveillance system for all cancers diagnosed in California since 1988, with standardized data collection and quality control procedures (15). Explanatory variables included patient and tumor characteristics (Table 1). Demographic and clinical characteristics, such as age, race/ethnicity, stage, grade, tumor markers, and treatment were analyzed with χ2 Test or Fisher’s Exact Test for categorical variables. Kaplan–Meier estimation was used to generate group specific survival curves for 5, 10 and 20 year overall and breast cancer specific survival. Log-rank test was conducted to evaluate the relationship between categorical predictor variables and survival. After verifying proportionality assumption, Cox proportional hazards regression model was performed for multivariate analysis to produce hazard ratios (HR) and 95% confidence intervals. Cox regression using forward stepwise selection of covariates was accomplished to assess the association of LM332 with survival accounting for covariates including race, age group, stage, tumor markers and tumor characteristics. Two tailed P value of less than0.05 was considered significant.
2.8. Tissue culture of breast cell lines
Tissue culture components and other chemicals were obtained from Sigma Chemicals (St. Louis MO) except where otherwise indicated. Verification of cell lines by short tandem repeat sequence analysis was performed by the Molecular Pathology Research Core at the University of Southern California. MDA-MB-231 was maintained in RPMI 1640 with 4 mM glutamine, 10 U/mL penicillin, 10 μg/mL streptomycin and 10% fetal bovine serum (FBS) at 37° C in 5% CO2. To demonstrate LM332 expression, cells were scraped from tissue culture plates into 10% neutral buffered formalin, pelleted by centrifugation, paraffin embedded, cut and stained for the LM332 β3 chain as described.
2.9. LM332 β3 and γ2 chain knock down
MDA-MB-231 cells were transfected with siRNA, (Qiagen Germantown, MD) targeting the LM332-specific, LAMB3 and LAMC2 genes. The RNA sequences were GCTTCAATGGTCTCCTTACTA and CCAGTGCAAAGCAGGCTACTT respectively. A random negative control, ACACTAAGTACGTCGTATTAC, was used at the same concentration as the total concentration for the laminin RNAs. MDA-MB-231 was transfected with each of the siRNAs or a combination of β3 and γ2 chain siRNA for 48 hours. For each condition, 1.0 μL of Surefect transfection reagent (Qiagen) was pre-incubated for 20 min with 0.13 nmole siRNA, in 625 μl of Optimem medium with 2% FBS. Subsequently, 50,000 MDA-MB-231 cells in 0.5 ml of Optimem with 2% FBS were added to the siRNA complex with 0.6 μl Alamar Blue (Thermo Fisher Scientific, Waltham, MA) and incubated overnight. The cells were divided into triplicate Millicell inserts (Millipore) with 12 μm pores for Boyden chamber assays, with 0.5 ml Optimem on the opposite side of the membrane. After 24 hours, the medium was removed for Alamar blue viability assays by measuring fluorescence using an excitation wavelength of 530 nm and an emission wavelength of 590 nm on a Synergy 2 Spectrophotometer (Biotek, Winooski, VT). To quantify cell migration, non-migrating cells were removed from the upper membrane with cotton-tipped swabs and cells that migrated to the other side of the membrane were stained with 2% crystal violet in 40% ethanol. The average number of cells per mm2 was counted in triplicate for each of the membranes, resulting in a total of 9 fields for each condition. The standard error and unpaired Student’s t-test were calculated for each condition.
Transfections were repeated using 7.5 μL of Surefect or 20 μl HiPerfect transfection reagent pre-incubated for 20 min with 0.5 nmole siRNA, in 1 ml of Optimem without serum, and then the mixture was added to 4mL of Optimem medium with 2% FBS and 400,000–600,000 MDA-MB-231 for each condition. After 48 hours, cells were lysed with 200 μL of SDS gel loading buffer for SDS PAGE and immunoblotting using equal amounts of lysate for each condition.Immunoblotting was performed as described above.
3. RESULTS
3.1. Demographics
The 297 subjects from this study showed similar demographics as the original population based cohort from which these cases were taken with respect to age and stage (9,10). The current study included 293 female and 4 male subjects, who ranged in age from 26 to 91 years with mean±std and median ages of 58.2±14.0 and 57 years respectively. Invasive carcinoma subtypes included 256 ductal, 18 lobular, 9 tubular, 3 cribriform, 2 pleomorphic, 1 papillary and 4 each of micropapillary and mucinous carcinomas. One patient had a lobular carcinoma and a separate ductal carcinoma. ER and/or PgR was found in 238 tumors (2 tumors were ER negative, PgR positive), 41 tumors were TNBC and 22 were ER and PgR negative, HER2 positive. All patients were treated with surgery. Adjuvant chemotherapy was administered to 115 patients, hormonal therapy to 108 patients and radiation therapy to 129 patients, but exact agents or doses were not specified in the CCR database. No patients received neoadjuvant therapy. After 20 years, 172 patients were alive, 43 had died of breast cancer and 82 from other causes.
3.2. LM332 expression in breast carcinoma
Breast carcinomas were stained with the laminin β3 chain, which in breast is only a component of LM332 and is present in the same distribution as the γ2 chain (8). Thus, positive staining for laminin β3 chain is consistent with staining for LM332. Staining was present in normal myoepithelium (Figure 1A), and in tumor cell cytoplasm (Figure 1B). Seventy five tumors were LM332 positive and 222 were LM332 negative. LM332 expression was significantly associated with age at diagnosis, tumor grade, treatment and steroid receptor expression (Table 1). Similar to our previous study on a different cohort (7), LM332 expression was found in 16.0 % of ER and/or PgR positive tumors and 62.7% of ER and PgR negative tumors. LM332 was positive in 73.2% of TNBC, similar to our previous observation that 70% of TNBC were LM332 positive (7). Between the two studies of 540 total breast carcinoma samples including 121 TN cases, 71% of the TN tumors were LM332 positive.
Figure 1.
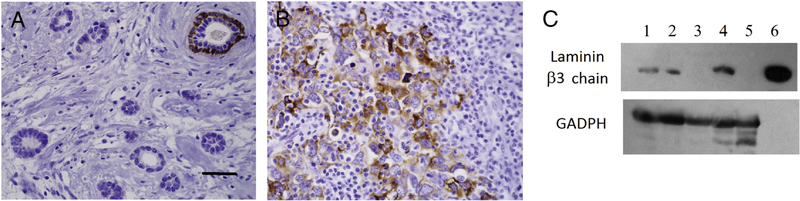
Immunohistochemistry for LM332 β3 chain in breast carcinoma. A. ER positive tumor with positive staining in the basal layer an entrapped normal duct. B. Diffuse staining of high grade tumor cells of a triple negative carcinoma. Original magnification 400 X, scale bar = 100μm C. Immunoblot of breast carcinoma lysates, developed using antibodies against the β3 chain of LM332 and GADPH as a loading control. Lanes 1 and 2 – metaplastic TNBC, Lane 3 - ER and PgR positive breast carcinoma, Lane 4 - HER2 over expressing breast carcinoma, Lane 5 malignant phyllodes tumor, Lane 6 - LM332 positive control.
Immunoblot analysis revealed the expected bands at 140 kDa for the β3 chain in the metaplastic and the HER2 over-expressing carcinomas (Figure 1C). The ER positive carcinoma and the phyllodes tumor did not reveal bands. The findings are consistent with our observation that TNBC and a few HER2 expressing tumors react with LM332 antibodies by IHC (7, 8). The absence of LM332 in the phyllodes tumor is consistent with previous IHC observations (8, 16).
3.3. LM332 expression and prognosis
Using Kaplan-Meyer analysis, LM332 expression was significantly associated with worse 5 and 10 year survival (Figure 2A), with p values of <0.0001 and 0.050 respectively, although by 20 years, survival was similar for patients with tumors with or without LM332 over expression, p< 0.246. Similarly, there was also an increase in breast cancer specific mortality in the LM332-expressing group at 5 and 10 years (Figure 2B, p< 0.0078 and 0.0437 respectively), but not at 20 years (p=0.4603).
Figure 2.
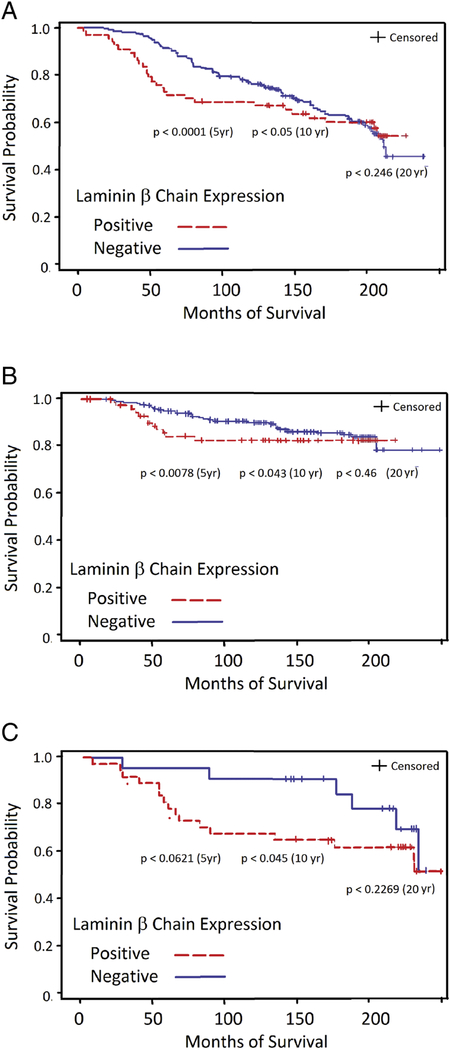
Kaplan-Meyer analysis of survival of patients whose breast cancers are positive (dashed red line) or negative (solid blue line) for LM332 β3 chain expression. A. Overall survival. B. Breast cancer specific survival. C. Survival of patients with ER-negative and PgR-negative breast carcinomas.
The observation that TNBC and HER2 positive tumors have a worse prognosis (17) raises the possibility that the decreased survival of patients with LM332 expressing tumors was simply due to the fact that most of these cases were TNBC and HER2 expressing tumors. To investigate this possibility, we examined LM332 expression in the subgroup with ER and PgR negative tumors. LM332 expression in tumors from this group of patients also was associated with a trend toward a decreased survival at 5 years (p<0.0621) and significantly decreased survival at 10 years (p < 0.0450) (Figure 2C). Thus, LM332 expression is independent of subtype as a prognostic marker in breast cancer.
In a multivariate cox regression analysis of 5 year survival after adjusting for age, race/ethnicity and social economic status, patients with LM332 positive tumors were associated with an increased HR of 3.9 with 95% confidence interval (CI) (2.0–7.7). No other marker, mainly ER, PgR or HER2, was associated with increased risk of death. Similar results observed also in a multivariate cox regression analysis of 10 year survival, where after adjusting for age, race/ethnicity and social economic status, patients with LM332 positive tumors were associated with an increased HR of 2.2 with 95% CI (1.3–3.8).
3.4. LM332 function in breast carcinoma in vitro
A functional study of LM332 in breast carcinoma cells was performed to determine if its expression has a role in imparting aggressive properties to the tumor. Tumor migratory ability is a determinant of metastatic potential (18), the primary cause of almost all breast cancer deaths, thus we sought to determine whether LM332 had a role in the motility of breast cancer cell lines in vitro. IHC on pellets from cultures of the TNBC line MDA-MB-231 revealed LM332 β3 chain expression (Figure 3A). The siRNA against the LM332-specific chains β3, γ2 or both together showed knock down of β3 and γ2 chain protein expression, respectively (Figure 3b, lanes 1 and 2), and as expected, the combination of the two siRNAs knocked down both chains. Motility of MDA-231-MB cells was significantly reduced by siRNA against the laminin chains, although cell viability was not affected. The observation that 2 different sequences of siRNA both contributed to a similar reduction of motility, but the control did not is evidence that the siRNA effect was targeted to LM332 and not an off target effect. This decrease in motility is evidence that LM332 contributes to the migratory ability of this breast cancer cell line. Inhibition of motility was not complete, so it is likely that there are other determinants of motility other than LM332 in this cell line and in breast cancer in general.
Figure 3.
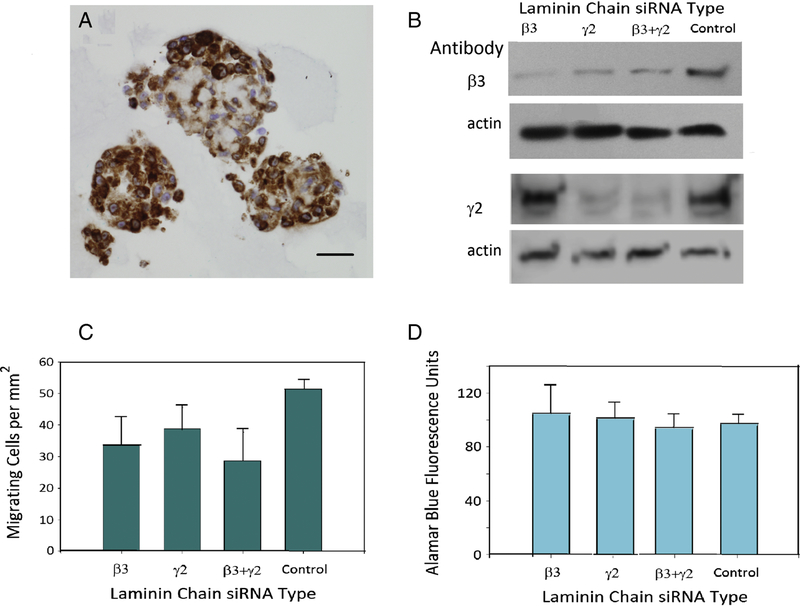
Laminin chain knockdown in breast carcinoma cells. A. LM332 β3 chain immunohistochemistry in MDA-MB-231. Original magnification 400 X, scale bar = 100μm. B. Immunoblot showing knockdown of LM332 β3 and γ2 chains in MDA-MB-231 by siRNA against each of the two chans, the two combined, and a negative control. C. Migration of siRNA tranfected MDA-MB-231 cells in a Boyden chamber assay. The values indicate the number of migrating cells per mm2. D. Viability of the cells in the Boyden chamber measured using an Alamar blue cell assay. Error bars for all graphs represent standard error of the mean.
4. DISCUSSION
The current study extended our previous work (7) using a different data set by showing that LM332-expressing carcinomas had a worse prognosis, including among patients with TNBC and ER negative, HER2 positive carcinomas. A few studies have examined LM332 in breast cancer (7, 8, 16, 19–21). Henning et al. (19) argued that LM332 was lost during progression, but did not subtype breast carcinomas, so it is possible that TNBC was underrepresented. Kim et al. (20) detected LM332 in breast stroma and suggested that this location may exert a microenvironmental signal that induces carcinoma invasion in vivo, consistent with our earlier in vitro observations (5). Diaz et al. noted a trend toward a worse prognosis for 10 patients whose tumors co-expressed peritumoral LM332 γ2 chain and β4 integrin (21), but they were unable to demonstrate a significant difference in prognosis between cancers with and without LM332. In contrast, we have shown that LM332 in the breast tumor cells themselves were associated with a worse prognosis after up to 10 years of follow-up. Finally, breast cancers from 109 Nigerian women with tumors positive for laminin of unspecified type, also had a worse prognosis (22). Because the specificity of the antibody in that study was not provided, it is unclear whether their findings reflect the expression of LM332, or another laminin. Our study provides more cases stained with a specific subunit of LM332 with longer follow-up. Our examination of cytoplasmic LM332 provides better evidence that LM332 is synthesized by the tumor cells themselves. Other cancers in which LM332 has been implicated in a worse prognosis include colorectal, esophageal, pancreas, tongue, vaginal, ovarian, head and neck, bladder and lung (6, 23–25).
A major goal of this study was to examine a cohort with long term follow-up. The disadvantage of studying cases from 1994 is that significant changes in breast cancer treatment have occurred since then, particularly the introduction of targeted therapies. The potential prognostic value of LM332 in patients treated with modern therapies can be explored as databases of patients with 10 or more years of follow-up become available.
We also explored a possible mechanism for the aggressive phenotype of breast carcinomas with LM332 expression. Exogenous LM332 increases the migration and invasion of breast carcinoma cells (5), but there is less known about whether endogenous expression of LM332 in breast carcinoma cells might contribute to their migratory ability. Knock down of specific LM332 chains in the TNBC line MDA-MB-231 reduced their migration, suggesting that LM332 may induce the migration and subsequent metastasis of carcinoma cells, in turn leading to breast cancer mortality.
Although prognostic information by itself may help in determining which patients need more aggressive therapy, a biomarker is potentially more useful if it identifies a therapeutic target. Molecular subtyping (26) and biomarkers such as epidermal growth factor receptor (27) and androgen receptor (28) have been proposed as prognostic and predictive biomarkers for TNBC. Because of its role in tumor progression, LM332 (29) and its receptor, α3β1 integrin (30) have been proposed as anti-metastatic targets. In experimental models, inhibition of either LM332 or its receptor, α3β1 integrin, has resulted in a decrease in metastases in various tumor types (23, 31–35). IHC, such as used in our study, is a technique that is routine in pathology laboratories worldwide, whereas molecular tests require specialized laboratories.
In summary, our observation that LM332 expression in breast carcinoma is associated with decreased survival provides additional evidence that LM332 has a role in the aggressive phenotype of breast cancers, particularly TNBC. Additional studies will be needed to determine if inhibition of LM332 can be used for breast cancer therapy.
Highlights.
Laminin 332, which is implicated in metastasis, stains 73% of triple negative breast carcinomas.
Laminin 332 is associated with worse 5 and 10 year overall and breast cancer specific survival.
Laminin 332 is associated with worse 10 year overall survival for triple negative breast cancer.
Knock down of laminin 332 in triple negative breast cancer cells decreased cell migration in vitro.
ACKNOWLEDGEMENTS
The authors thank Jeffrey Kim and the UCI Chao Family Comprehensive Cancer Center Experimental Tissue Core Facility, directed by Dr. Robert Edwards, for performance of immunoperoxidase reactions. We thank Ivonne Villalobos, Roberta Guzman, Angela Santiago and Dr. Michael Press for their assistance with HER2 FISH testing.
Funding support: The study was supported by the National Cancer Institute Support Grant CA-62203 to the Chao Family Cancer Center, Dr. Frank Meyskens, Principal Investigator, National Cancer Institute Grant CA58860 and the Lon V. Smith Foundation Grant LVS-18840.
Abbreviations:
- LM332
laminin 332
- TNBC
Triple negative breast carcinoma
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- PgR
progesterone receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
PMC is a supplier of purified laminin 332 to Kerafast, Inc. The other authors have no disclosures.
REFERENCES
- 1.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer 2007; 7:370–380. [DOI] [PubMed] [Google Scholar]
- 2.Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol 2002; 12:197–207. [DOI] [PubMed] [Google Scholar]
- 3.Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol 2012; 28:523–53. [DOI] [PubMed] [Google Scholar]
- 4.Yan HHN, Mruk DD, Wong EWP, Lee WM, Cheng CY. An autocrine axis in the testis that coordinates spermiation and blood–testis barrier restructuring during spermatogenesis. PNAS 2008; 105:8950–8955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter PM, Dao AV, Arain ZS, Chang MK, Nguyen HP, Arain S, et al. Motility induction in breast carcinoma by mammary epithelial laminin 332 (laminin 5). Mol Cancer Res 2009; 7:462–75. [DOI] [PubMed] [Google Scholar]
- 6.Guess CM, Quaranta V. Defining the role of laminin-332 in carcinoma. Matrix Biology 2009; 28:445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon S-Y, Chae S-W, Wilczynski SP, Arain A, Carpenter PM. Laminin 332 expression in breast carcinoma. Appl Immunohistochem Mol Morphol 2012; 20: 159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter PM, Wang-Rodriguez J, Chan OTM, Wilczynski SP. Laminin 5 Expression in metaplastic breast carcinomas. Am J Surg Pathol 2007; 32:345–53. [DOI] [PubMed] [Google Scholar]
- 9.Ziogas A, Gildea M, Cohen P, Bringman D, Taylor TH, Seminara D, et al. Cancer risk estimates for family members of a population-based family registry for breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev 2000;9:103–11. [PubMed] [Google Scholar]
- 10.Anton-Culver H, Cohen PF, Gildea ME, Ziogas A. Characteristics of BRCA1 mutations in a population-based case series of breast and ovarian cancer. Eur J Cancer 2000;36:1200–8. [DOI] [PubMed] [Google Scholar]
- 11.Khoury T, Sait S, Hwang H, Chandrasekhar R, Wilding G, Tan D, et al. Delay to formalin fixation effect on breast biomarkers. Mod. Pathol 2009; 22: 1457–1467. [DOI] [PubMed] [Google Scholar]
- 12.Press MF, Sauter G, Buyse M, Fourmanoir H, Quinaux E, Tsao-Wei DD, et al. HER2 Gene Amplification Testing by Fluorescent In Situ Hybridization (FISH): Comparison of the ASCO-College of American Pathologists Guidelines With FISH Scores Used for Enrollment in Breast Cancer International Research Group Clinical Trials. J Clin Oncol 2016;34:3518–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allred DC, Bustamante MA, Daniel CO, Gaskill HV, Cruz AB. Immunocytochemical analysis of estrogen receptors in human breast carcinomas. Evaluation of 130 cases and review of the literature regarding concordance with biochemical assay and clinical relevance. J Arch Surg 1990;125:107–13. [DOI] [PubMed] [Google Scholar]
- 14.Wolff AC, M. Hammond EH, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update Arch Pathol Lab Med 2014; 138: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.California Cancer Registry, Cancer Reporting, http://www.ccrcal.org/Cancer_Reporting/index.shtml [accessed 07 May 2018]
- 16.D’Alfonso TM, Ross DS, Liu YF, Shin SJ. Expression of p40 and laminin 332 in metaplastic spindle cell carcinoma of the breast compared with other malignant spindle cell tumours. J Clin Pathol 2015;68:516–21. [DOI] [PubMed] [Google Scholar]
- 17.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Cancer. Prognostic markers in triple-negative breast cancer 2007;109:25–32. [DOI] [PubMed] [Google Scholar]
- 18.Stracke ML, Aznavoorian SA, Beckner ME, Liotta LA, Schiffmann E. Cell motility, a principal requirement for metastasis. EXS 1991, 59: 147–62. [DOI] [PubMed] [Google Scholar]
- 19.Henning K, Berndt A, Katenkamp D, Kosmehl H. Loss of laminin-5 in the epithelium-stroma interface: an immunohistochemical marker of malignancy in epithelial lesions of the breast. Histopathology 1999; 34:305–309. [DOI] [PubMed] [Google Scholar]
- 20.Kim BG, An HJ, Kang S, Choi YP, Gao MQ, Park H, et al. Laminin-332-rich tumor microenvironment for tumor invasion in the interface zone of breast cancer. Am J Pathol 2011; 78:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz LK, Cristofanilli M, Zhou X, Welch KL, Smith TL, Yang Y, et al. β4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Mod Pathol 2005;18:1165–75. [DOI] [PubMed] [Google Scholar]
- 22.Agboola AOJ, Ebili HO, Iyawe VO, Banjo AAF, Salami BS, Rakha EA, et al. , Tumour cell membrane laminin expression is associated with basal-like phenotype and poor survival in Nigerian breast cancer. Malaysian J Pathol 2016; 38: 83–92. [PubMed] [Google Scholar]
- 23.Moon YW, Rao G, Kim JJ, Shim HS, Park KS, An SS, et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ 2015; 22:1341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Dang J, Chang KY, Yau E, Aza-Blanc P, Moscat J, et al. miR-1298 Inhibits Mutant KRAS-Driven Tumor Growth by Repressing FAK and LAMB3. Cancer Res 2016;76:5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sathyanarayana UG, Maruyama R, Padar A, Suzuki M, Bondaruk J, Sagalowsky A, et al. Molecular detection of noninvasive and invasive bladder tumor tissues and exfoliated cells by aberrant promoter methylation of laminin-5 encoding genes. Cancer Res 2004;64:1425–30. [DOI] [PubMed] [Google Scholar]
- 26.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016. 11(6):e0157368. doi:10.1371/journal.pone.0157368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol 2014; 27:1212e1222. [DOI] [PubMed] [Google Scholar]
- 28.Pietri E, Conteduca V, Andreis D, Massa I, Melegari E, Sarti S, et al. : Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr Relat Cancer 2016; 23:R485eR498. [DOI] [PubMed] [Google Scholar]
- 29.Tsuruta D, Kobayashi H, Imanishi H, Sugawara K, Ishii M, Jones JC. Laminin-332-integrin interaction: a target for cancer therapy? Curr Med Chem 2008; 15:1968–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med 2010;12:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Gibson-Corley KN, Herndon ME, Sun Y, Gustafson-Wagner E, Teoh-Fitzgerald M, et al. Integrin α3β1 can function to promote spontaneous metastasis and lung colonization of invasive breast carcinoma. Mol Cancer Res 2014;12:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salo S, Boutaud A, Hansen AJ, He L, Sun Y, Morales S, et al. Antibodies blocking adhesion and matrix binding domains of laminin-332 inhibit tumor growth and metastasis in vivo. Int J Cancer 2009; 125:1814–25. [DOI] [PubMed] [Google Scholar]
- 33.Wang X-M, Li J, Yan M-X, Liu L, Jia D-S, Geng Q, et al. Integrative Analyses Identify Osteopontin, LAMB3 and ITGB1 as Critical Pro-Metastatic Genes for Lung Cancer. PLoS One 2013. 8:e55714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Tran M, Rousselle P, Nokelaines P, Tallapragada S, Nguyen NT, Fincher EF, et al. Targeting a tumor-specific laminin domain critical for human carcinogenesis. Cancer Res 2008; 68:2885–94. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Fu W, Im JH, Zhou Z, Santoro SA, Iyer V, et al. Tumor cell α3β1 integrin and vascular laminin-5 mediate pulmonary arrest and metastasis J Cell Biol 2004; 164:935–41. [DOI] [PMC free article] [PubMed] [Google Scholar]


