Abstract
Notch signalling regulates a plethora of developmental processes and is also essential for the maintenance of tissue homeostasis in adults. Therefore, fine-tuning of Notch signalling strength needs to be tightly regulated. Of key importance for the regulation of Notch signaling are O-fucose, O-GlcNAc and O-glucose glycans attached to the extracellular domain of Notch receptors. The EGF repeats of the Notch receptor extracellular domain harbor consensus sites for addition of the different types of O-glycan to Ser or Thr, which takes place in the endoplasmic reticulum. Studies from Drosophila to mammals have demonstrated the multifaceted roles of O-glycosylation in regulating Notch signalling. O-glycosylation modulates different aspects of Notch signalling including recognition by Notch ligands, the strength of ligand binding, Notch receptor trafficking, stability and activation at the cell surface. Defects in O-glycosylation of Notch receptors give rise to pathologies in humans. This Review summarizes the nature of the O-glycans on Notch receptors and their differential effects on Notch signalling.
Keywords: Notch signalling, Glycosylation, Glycosyltransferases, O-Glucose, O-Fucose, O-GlcNAc
Introduction:
Notch signalling is a well-characterized, evolutionarily-conserved pathway that plays multiple roles in the regulation of embryonic development, and in the maintenance of tissue homeostasis. Defective Notch signalling leads to numerous pathologies in development, and to different adult diseases [1]. In mammals, there are four different Notch receptors (NOTCH1 to NOTCH4), whereas Drosophila has only one homologue which is most similar to NOTCH1. Notch receptors are single transmembrane glycoproteins, comprising an extracellular domain (NECD), a transmembrane region, and an intracellular domain (NICD)(reviewed in [2–4]). The NECD comprises 29–36 epidermal growth factor-like (EGF) repeats, which include Notch ligand-binding domains. The EGF repeats are followed by the negative regulatory region (NRR), which is composed of three cysteine-rich Lin12 Notch repeats and a heterodimerization domain (HD). NECD is non-covalently linked at the HD to the N-terminal 12 amino acids of NICD that are external to the transmembrane domain (termed NEXT for Notch extracellular truncation; [5]). NICD and NECD are generated by furin cleavage at the S1 site during passage through the Golgi. NICD is characterized by an RBP-Jκ-associated module (RAM) and ankyrin (ANK) repeats, both of which are required for interactions with the DNA-binding complex CBF1–Suppressor of Hairless–LAG1 (CSL). Near the C-terminus of Notch receptors is a PEST domain, which regulates NICD degradation by the proteasome. Between the ANK repeats and the PEST domain, NICD also contains several nuclear localization signals, and a domain that confers transactivation of transcriptional repressors (TAD).
Notch signalling is mediated by short-range, cell-cell interactions between signal-sending cells expressing Notch ligands, and signal-receiving cells expressing Notch receptors. Signalling strength is fine-tuned by numerous factors, including the expression of Notch ligands that cause cis-inhibition of Notch receptors in signal-receiving cells, molecules involved in secretory pathway trafficking, and the O-glycans attached to NECD. The canonical Notch signalling pathway involves Notch ligands Delta or Serrate (in Drosophila) and Delta-like or Jagged (in mammals), binding to NECD of Notch receptors and initiating two, sequential proteolytic cleavages. The first is caused by a disintegrin and metalloprotease (ADAM) and occurs at the S2 site adjacent to the Notch transmembrane domain [6, 7]. This generates soluble NECD bound to Notch ligand that is endocytosed into the signal-sending, ligand-expressing cell [8], and the membrane-bound NEXT fragment described above. The second cleavage occurs within the transmembrane domain of Notch receptors at the S3 site, and is catalyzed by a complex that includes presenilins and has γ-secretase activity [9]. Released NICD complexes with CSL/RBP-Jκ, recruits the co-activator Mastermind (MAML) and other factors, and the complex activates Notch target genes [10–12]. Ligand-induced Notch receptor cleavage (activation) alters the expression of many Notch target genes which regulate diverse signalling outcomes, ranging from cell proliferation to cell fate determination, and cell death. Aberrant changes in Notch signalling cause disorders of development and adult diseases. Therefore, precise temporal and spatial regulation of Notch signalling at appropriate levels is critical for optimal Notch signalling [13].
The EGF repeats in NECD are post-translationally modified by distinct O-glycans. Glycosyltransferases catalyze the addition of O-glycans to Notch EGF repeats by transferring fucose from GDP-fucose, glucose from UDP-glucose or N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to a Ser or Thr residue in a specific consensus sequence. Each sugar may subsequently be extended by the addition of 1–3 sugar residues added sequentially (Fig. 1). The different O-glycans on NECD can regulate similar or different aspects of Notch signalling (reviewed in [14–18]). Roles for O-glycosylation of Notch have been performed in vivo in different tissues, and in culture using cell-based assays with various cell lines. More recently, with the advent of exome sequencing, several human pathologies have been associated with defects in the O-glycosylation of Notch receptors (Fig. 2). This Review will describe the O-glycans of NECD and the different functions of each type of O-glycan in Notch signalling in cells and organisms. It must be noted that Notch ligands also contain EGF repeats that may be O-glycosylated, and a limited subset of other secretory pathway proteins contain EGF repeats with appropriate consensus sequence(s) for modification with O-fucose, O-glucose or O-GlcNAc glycans [19–21]. Functions of O-glycans attributed to Notch signalling arising from mutations in glycosyltransferase genes are therefore based on evidence of Notch signalling dependence using Notch signalling reporter constructs, gamma-secretase inhibitors (GSI) that prevent S3 cleavage, effects on the expression of Notch pathway genes, and also the similarity of observed phenotypes to mutants with defects in other Notch pathway members. Mutation of O-glycan attachment sites (Ser/Thr) to investigate functions of a specific O-glycan is also an important strategy. However, such experiments require probing of several amino acid changes that abolish O-glycosylation, as well as the exchange of Ser to Thr or Thr to Ser respectively, to establish that it is the missing O-glycan, and not the altered amino acid, that is required at a given attachment site [22].
Figure 1.
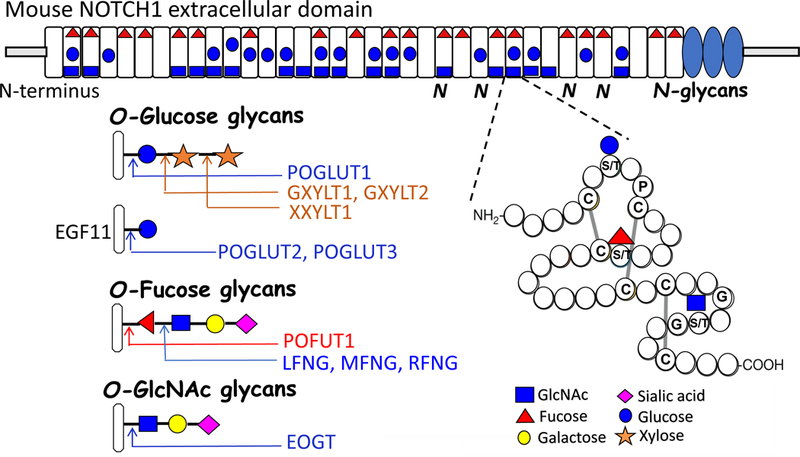
Representation of mouse NOTCH1 extracellular domain depicting EGF repeats with different O-glycan consensus sites that may be modified with the O-glycans shown. One of the EGF domains is magnified to show the consensus site for each type of O-glycan. Different O-glycans, their respective differential extension with sugars (+/−), and the glycosyltransferases responsible for the transfer of each sugar are shown below the diagram. The transfer of O-glucose by POGLUT2 or POGLUT3 occurs only on EGF11 in NOTCH1. The consensus site is between Cys3 and Cys4, indicated by the different location of the glucose symbol in EGF11 in the diagram.
Figure 2.
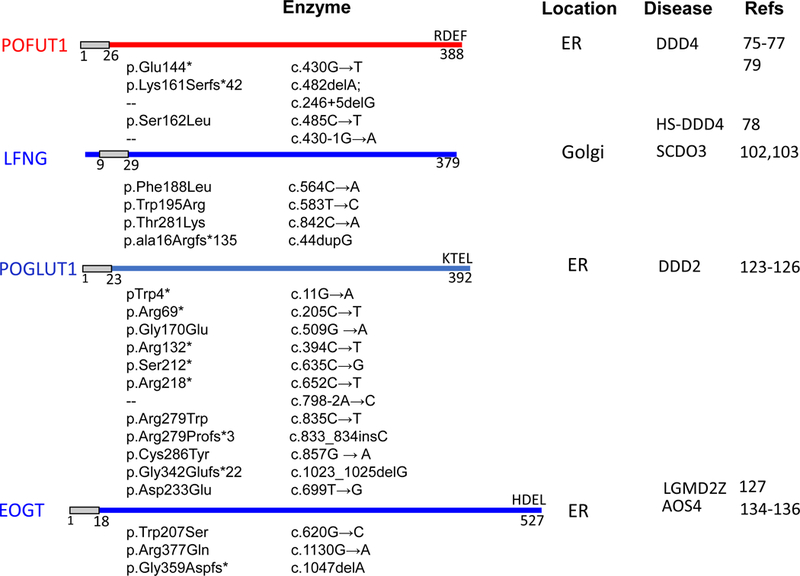
The human mutations and pathologies associated with glycosyltransferases that modify Notch receptors. The diagram of each glycosyltransferase represents protein size, with the signal peptide or transmembrane domain identified by a grey box and each ER retention sequence given at the C-terminus. The human mutations identified so far are mentioned below each protein diagram, with the corresponding pathologies referred to in the disease column.
O-Fucose Glycans
• Discovery and cell-based assays
In a Drosophila screen for modifiers of Notch signalling, a gene named Fringe was shown to be required for Notch signalling at the dorsal/ventral boundary of the wing disc in third instar larvae [23, 24]. Fringe was subsequently shown to be a glycosyltransferase that transfers GlcNAc in β1,3-linkage to O-fucose on certain Notch EGF repeats [25, 26]. These findings established a new paradigm of glycan regulation of a cell fate-determining signalling pathway. Amongst potential substrates of Fringe, Notch receptors contain a high number of putative consensus sites for the O-fucose modification [27], and all are indeed modified [28, 29]. Based on in silico and structural studies, the consensus site for the addition of O-fucose is C2xxxxS/TC3 (where Ser (S) or Thr (T) accepts the fucose, and C2 and C3 are the second and third cysteines of the EGF repeat; x is any amino acid) [19, 21]. The enzyme responsible for the addition of O-fucose to appropriate EGF repeats, protein O-fucosyltransferase 1 (POFUT1), is encoded by Ofut1 in Drosophila and Pofut1 in mammals [30]. Pofut2 encodes a distinct protein O-fucosyltransferase that does not act on EGF repeats, but instead transfers O-fucose to thrombospondin type 1 repeats [31].
POFUT1 activity was first identified in Chinese hamster ovary (CHO) cells [32], and cloning revealed high sequence conservation from C. elegans to mammals [33]. The OFUT1/POFUT1 enzyme is a resident of the endoplasmic reticulum (ER) with an ER-retention signal at the C-terminus. OFUT1/POFUT1 catalyzes transfer of fucose most efficiently to properly-folded EGF repeats [25, 34, 35]. Loss of Ofut1 in Drososphila S2 cells results in the loss of Notch ligand binding [36, 37]. CHO cells deficient in fucosylation exhibit reduced Notch signalling stimulated by Jagged 1 (JAG1) [25, 38]. Initial studies indicated that knockdown or loss of Ofut1 did not alter Notch receptor expression at the cell surface [37, 39]. However, subsequent investigations suggested a more complex picture. One group provided evidence that Drosophila OFUT1 is a chaperone for Notch required for its trafficking out of the endoplasmic reticulum (ER) [40], while others proposed that OFUT1 was required later in the secretory pathway to maintain Notch stability at the cell surface [41]. To distinguish roles for the O-fucose on Notch EGF repeats versus O-fucosyltransferase activity in Notch receptor trafficking, experiments were performed with a mutant Ofut1R245A which has little or no O-fucosyltransferase activity. Ofut1-null Drosophila expressing Ofut1R245A were partially rescued in development, and Notch cell surface expression was increased in embryos, consistent with enzyme-dead OFUT1 acting as a Notch chaperone [42]. Further analysis in other Drosophila mutants with intact Ofut1, but lacking the ability to synthesize GDP-fucose, showed that O-fucose has a role in Notch signalling that is not merely to provide a substrate for Fringe [43]. In Pofut1-null mouse embryonic stem (ES) cells [44], hematopoietic stem cells (HSC) [45] and CHO cells [46], Notch receptor cell surface expression is essentially unaltered or somewhat reduced [47], but Notch-ligand binding and ligand-induced Notch signalling are greatly reduced. Although Pofut1R245A partially rescues Notch signalling in Pofut1-null ES cells, so does an enzyme-dead, ER glucosidase [44], suggesting that up-regulation of general chaperones may be responsible. Interestingly, overexpression of the human equivalent POFUT1R240A, in POFUT1-null human osteosarcaoma (U2OS) cells partially rescues Notch ligand binding and Notch signalling [48]. However, mouse embryos that are homozygous for Pofut1R245A die at mid-gestation, with a phenotype indistinguishable from Pofut1-null embryos [49]. This is presumably because of the unexpected finding that POFUT1(R245A) is degraded in embryos, making homozygous mutant embryos effectively null. Accumulation of NOTCH1 intracellularly is observed in Pofut1-null somites [49, 50]. By contrast, Pofut1-null inner ear cells [51], endocardium [46], and HSC [52] exhibit substantially reduced Notch signalling, but were shown in the latter two cases to express NOTCH1 at the cell surface equivalently to wild type.
Several groups have addressed the requirement for O-fucose versus POFUT1 by examining mutants unable to synthesize GDP-fucose (GDP-Fuc), the nucleotide sugar substrate of POFUT1. In the mouse, homozygous embryos that cannot make GDP-fucose are rescued by maternal GDP-Fuc to varying degrees [53, 54]. Homozygous mutants that are born have greatly reduced myeloid and lymphoid cells showing that POFUT1, which is present at normal levels, cannot rescue Notch signalling by chaperone activity. In Lec13 CHO cells which have very low levels of GDP-fucose, Notch receptors are well expressed at the cell surface, but Notch ligand binding and ligand-induced Notch signalling are markedly reduced, while POFUT1 levels are unaltered [44]. In Drosophila, embryos unable to synthesize GDP-Fuc do not show typical Notch neurogenic defects as observed if Ofut1 is mutated, but die as first instar larvae, suggesting that O-fucose is required as a substrate of Fringe [42]. However, loss of GDP-Fuc synthesis in large areas of the wing reveal Fringe-independent Notch signalling defects [55]. Similar experiments reveal a temperature-sensitive loss of Notch signaling during neurogenesis in Drosophila mutants that cannot make GDP-Fuc [43]. At 30ºC, Notch lacking O-fucose is unable to signal whereas signaling is normal at 25ºC. Loss of GDP-Fuc from mouse HSC leads to a reduction in self renewal, and an altered ability to occupy the bone marrow niche [56]. Thus, it is apparent that OFUT1/POFUT1 present at normal levels is unable to rescue Notch signalling in several in vivo contexts in which Notch does not carry O-fucose glycans, but is nevertheless expressed at the cell surface. Therefore, broad generalizations about OFUT1/POFUT1 as a Notch chaperone required for cell surface expression of Notch cannot be made, since cellular context is clearly of utmost importance in determining effects on Notch receptor trafficking when OFUT1/POFUT1 is enzymatically inactive or absent.
While ligand-induced Notch signalling assays reveal that O-fucose glycans regulate the strength of Notch signalling, and flow cytometry and surface plasmon resonance assays reveal effects on the binding of different Notch ligands, such assays do not address whether O-fucose glycans on Notch receptors physically interact with Notch ligands. However, recent X-ray studies have begun to elucidate mechanistic roles for O-fucose in Notch receptor-ligand binding. Thus, co-crystals of NOTCH1 EGF11–13 with a Delta-like 4 (DLL4) N-terminal ECD fragment identified precisely how the O-fucose on EGF12 of NOTCH1 interacts directly with the module at the N-terminus of Notch ligands (C2 domain) of DLL4 [57]. The authors propose that the fucose functions as a twenty-first amino acid. Subsequently, they solved co-crystals of a larger NOTCH1 fragment (EGF8-EGF12) and a JAG1 N-terminal ECD fragment [58]. In this case, they observed not only interactions with the O-fucose in NOTCH1 EGF12 with the C2 domain of JAG1, but also between EGF3 of JAG1 and the O-Fuc in EGF8 of NOTCH1, thereby extending the ligand binding domain of NOTCH1. This finding was consistent with the effects of a mutation in Drosophila Notch EGF8 that affects Serrate-induced, but not Delta-induced, Notch signalling [59]. Co-culture assays using engineered forms of Notch receptors and ligands have further defined mechanisms by which DLL1, DLL4 and JAG1 regulate Notch signalling [60, 61]. Single cell reporter assays showed that DLL1 stimulates Notch signalling in a pulsatile fashion, while DLL4 stimulates in a sustained manner [61]. These differences lead to different cell fate outcomes – DLL1-induced Notch signalling promotes myogenesis, whereas DLL4-induced signalling inhibits myogenesis [61, 62]. Such assays elegantly dissect the Notch signalling pathway and reveal the critical necessity of optimal regulation of Notch signalling strength. Too much, or too little Notch signalling may each have deleterious consequences.
The O-fucose modification on Notch may be elongated to form a disaccharide, trisaccharide or tetrasaccharide by the sequential addition of GlcNAc, galactose (Gal) and N-acetylneuraminic acid (NeuAc), respectively [25, 27]. Fringe adds GlcNAc to O-fucose on properly-folded EGF repeats at highest efficiency [25, 63]. Drosophila has a single Fringe, but there are three homologues in mammals - Lunatic (LFNG), Manic (MFNG) and Radical (RFNG) [64]. In vitro assays of recombinant enzymes show mouse LFNG to be the most active [65]. Co-culture Notch reporter assays and ligand binding assays revealed the important role of GlcNAc addition to O-fucose in promoting Delta binding to Notch receptors and Delta-induced Notch signalling, while simultaneously inhibiting Jagged binding to Notch and reducing Jagged-induced Notch signalling [25, 36, 38, 44, 66–69]. Mosaic experiments in Drosophila showed that removal of Fringe from Notch ligand-expressing cells of the wing disc did not alter ligand ability to induce Notch signalling [24]. Co-culture assays with CHO cells expressing a Notch reporter with or without LFNG, MFNG or RFNG, or inducible DLL1 or JAG1, showed that Fringe modification in cis (i.e. in the Notch expressing cell) promotes cis inhibition of Notch signalling by DLL1, but weakens cis inhibition by JAG1 in the cases of LFNG and MFNG, whereas RFNG promotes cis inhibition by DLL1 and JAG1 alike [60]. These effects are similar to the effects of Fringe on Notch signalling in trans. It will be most interesting to see co-crystals of NOTCH1 and Notch ligand fragments with O-fucose plus GlcNAc to gain molecular insights into the different effects of Fringe on Delta versus Jagged binding. Modeling GlcNAc into the DLL4/NOTCH1 fragment crystal structure predicts specific amino acid interactions [57]. Binding to both DLL1 and JAG1 is increased when the O-fucose in EGF12 of the NOTCH1 EGF11–13 fragment is elongated by Fringe [70]. A Fringe code has been proposed based on the differential modification of NOTCH1 EGF repeats by LFNG versus MFNG or RFNG, and the consequences for Notch ligand binding and Notch signalling in human embryonic kidney (HEK)-293T cells [29]. However, these results come from overexpressed transfected Fringe genes in cultured cells. It will be interesting to determine whether a Fringe code leads to functional consequences in vivo in mice expressing only a single Fringe [71]. Elongation of O-Fuc-GlcNAc by Gal was shown to be necessary for optimal Notch signalling in CHO cell reporter assays, whereas the further elongation of Gal by NeuAc appears to be dispensable for Notch signalling [38, 72].
• In vivo consequences of defective O-fucose glycan synthesis or loss of O-fucose sites
In Drosophila, expression of Ofut1 is regulated during embryonic development and Ofut1 is differentially expressed in adult tissues. Loss or suppression of Ofut1 in Drosophila results in phenotypes similar to those of Notch pathway mutants such as lateral inhibition in the nervous system, and cell lineage decisions in sensory organ precursor cells [37, 39]. Expression of Ofut1 with no or low fucosyltransferase activity (Ofut1R245A) rescues fringe-dependent neurogenesis in Drosophila embryos [42]. Loss of Pofut1 in mice is embryonic lethal [50, 73], as are Pofut1R245A homozygotes due to degradation of the mutant enzyme [49]. These mice show the characteristic phenotype associated with defective Notch signalling, including growth retardation, due in part to disrupted somitogenesis, vascularization defects, and defects in neural tube formation. Pofut1 null embryos share phenotypes not only with Notch receptor null mice, but also with Notch downstream effector deficient mice. Therefore, only conditional deletion of Pofut1 can be used to study requirements for O-fucose glycans in Notch signalling in different cell types. Alternatively, mutation of O-fucose sites in the EGF repeats of Notch receptors or Notch ligands has been used.
Conditional deletion of Pofut1 in bone marrow cells and stroma with Mx1-Cre causes cell-fate defects in lymphoid and myeloid cell differentiation. Cells lacking POFUT1 exhibit no Notch ligand binding but only a slight decrease in cell surface expression of NOTCH1 and NOTCH2 receptors [45]. Residual Notch signalling occurs in these bone marrow cells, since deletion of the Notch downstream effector RBP-Jκ via the same method gave a more severe phenotype [52, 74]. Similarly, deletion of Pofut1 in the endocardium via Nfatc1-Cre is less severe than deletion of Notch1 by the same strategy [46]. Residual DLL4-induced Notch signalling in this case allowed the identification of angiogenic precursor cells involved in coronary arteriogenesis. Yet another example in which conditional deletion of Pofut1 gives a milder phenotype than deletion of Notch1 is deletion via Pax2-Cre in the inner ear [51]. These examples were unexpected given that the Pofut1-null embryonic phenotype is similar to that of a Notch1-null. Deletion of Pofut1 in intestinal epithelium by Villin-Cre [75] or in bone marrow by Mx1-Cre [45] has milder consequences than deleting RBP-Jκ in the same manners [52, 76]. By contrast, deletion of Pofut1 in lung [77] or skin [78] gives severe Notch signalling-defective phenotypes.
In humans, several heterozygous autosomal dominant mutations, and a homozygous recessive mutation in the POFUT1 gene, have been associated with disease (Fig. 2). Pigmentation defects are characteristic of the autosomal dominant mutations which give rise to a syndrome termed Dowling-Degos Disease 4 (DDD4) [79–81]. A recently identified heterozygous mutation in POFUT1, associated with DDD4 is also accompanied by Hidradenitis Suppurativa (HS) which is marked by recurrent painful nodules and abscesses [82]. A homozygous recessive mutation in POFUT1 ablates a N-glycan site and is correlated with more severe developmental defects [83]. However, the loss of the N-glycan is not the basis of the reduced activity of POFUT1. Rather, it seems that Ser162 in the Asn-Lys-Ser N-glycan sequon cannot be replaced by Leu, though it can be replaced by Gln, a change that apparently enhances POFUT1 activity. Enhanced POFUT1 activity has been associated with tumor progression and increased Notch signalling in liver cancer [84].
Since POFUT1 may modify ~100 different proteins that contain appropriate EGF repeat(s), it is also important to determine functions of O-fucose by mutating O-fucose sites in POFUT1 substrates. Removal of the O-fucose site in EGF12 of Drosophila Notch (N-EGF12f) revealed that the O-fucose glycan is important for inhibiting Serrate-induced Notch functions in the wing disc, and binding of Delta and Serrate to N-EGF12f was enhanced [85]. This is surprising considering the crystal structures showing the key role of NOTCH1 EGF12 O-fucose in mammalian DLL4 and JAG1 binding [57, 58]. The Notch1[12f] mutation in mouse causes reduced ligand binding to thymocytes, but no apparent effects on viability or fertility [86]. However, Notch signalling is compromised in Notch1[12f/12f] mice, as reflected by the cell-autonomous reduction in T cell development. Point mutations have also been introduced into Notch ligands DLL1 [87] and DLL3 [88]. Only in the case of DLL3 did elimination of two O-fucose sites in EGF2 and EGF5 have a functional effect, in that a mutant transgene could not rescue somitogenesis in Dll3 null embryos. It will be important to perform this experiment by mutating the endogenous Dll3 gene and replacing the O-fucose Ser/Thr with the alternative (Thr/Ser) that could receive a fucose, as well as with amino acids other than Ala that cannot be O-fucosylated. DLL1 has four EGF repeats that receive O-fucose but DLL1 expressed in Pofut1-null presomitic mesoderm or mouse embryo fibroblasts was localized to the cell surface and stimulated Notch signalling [87]. By contrast, experiments with intestinal cells showed that Paneth cells have slightly reduced cell surface expression of DLL1 and DLL4 when RFNG is absent, and cells from cultured intestinal organoids have reduced DLL1 on the cell surface after knockdown of Lfng [89]. Lfng knockout mice have reduced DLL1 and DLL4 on the surface of goblet cells, whose numbers are increased due to a reduction in Notch signalling in the absence of Lfng [89]. Thus, depending on cellular context, cell surface expression of Notch ligands may be promoted by Fringe modification.
Elongation of O-fucose with a GlcNAc transferred by Fringe is critical for development in Drosophila and in mammals [90]. While there are three Fringe homologues in mammals, numerous studies reveal a dominant role for Lfng [91–93]. Mice lacking Lfng display severe defects in somitogenesis [91, 94, 95], reproduction [96, 97], and T cell and B cell development [71, 98, 99]. However, genetic background affects survival and longevity of Lfng-null mice [71, 91]. The dominance of Lfng is clearly observed in retinal angiogenesis, in which all three Fringe homologues are expressed by tip cells of the growing angiogenic front [100]. Deletion of Lfng causes excess vessel sprouting, despite the continued expression of Mfng and Rfng. Lfng is proposed to promote DLL4-induced Notch signalling in tip cells, and to inhibit JAG1-induced Notch signalling in stalk cells, thereby promoting selection of tip cells [101]. An additive role of Lfng and Mfng occurs in marginal zone B cell development and in T cell development [71, 102, 103]. In fact, each Fringe gene expressed in the absence of the other two can rescue altered T and B cell development compared to triple Fringe knockout mice [71]. Fringe regulates Notch signalling based on differential interactions between Notch modified by Fringe with Delta-like versus Jagged Notch ligands. Fringe may also promote the cell surface expression of Delta-like ligands [89]. A role for the addition of Gal to Fringe-modified Notch receptors was observed in B4galt1 null embryos which exhibit reduced expression of several Notch target genes during somitogenesis [104]. A patient with a mutation in B4GALT1 had severe neurological defects and other pathologies consistent with reduced Notch signalling [105].
Human mutations in LFNG give rise to spondylocostal dysostosis [106, 107], but no mutations in MFNG or RFNG have yet been associated with any human pathology. However, upregulation of MFNG has been correlated with tumor progression in claudin-low breast cancer, due to increased Notch signalling and the induction of PI3KCG [108]. By contrast, loss of Lfng which suppresses JAG1-induced NOTCH1 signalling in mammary epithelium, in cooperation with MET/CAVEOLIN gene amplification, promotes basal-like breast cancer [109]. LFNG also functions as a tumor suppressor in melanoma metastasis [110], and in mouse models of pancreatic [111] and prostate cancer [112]. These results reveal fundamental roles of Fringe in regulating Notch signalling. Further studies will help better elucidate different functions of the three fringe homologues in mammals.
O-Glucose glycans
• Discovery and cell-based assays
Bovine blood coagulation factors VII and IX, were the first proteins identified with an O-glucose modification on EGF repeats [113]. O-glucose was subsequently identified on NOTCH1 in CHO cells [27]. O-glucose was also found extended by xylose in α1,3-linkage, or by two xylose moieties to form a trisaccharide [114]. In Drosophila, only one enzyme has been identified as a glucoside xylosyltransferase (GXYLT) termed Shams [115], whereas in mammals, the addition of the first xylose is mediated by GXYLT1 or GXYLT2, and the second xylose is added in α1,3-linkage by xylose xylosyltransferase 1 (XXYLT1) [114, 116]. A Drosophila Xxylt1 was recently identified and shown to repress Delta-Notch signaling [117]. Thus, loss of Drosophila Xxylt1 was found to promote Delta-Notch signaling in an appropriately sensitized genetic background. The consensus site for O-glucose addition to most EGF repeats of Notch receptors and ligands is C1xSxA/PC2, where C1 and C2 are the first and second cysteines of the EGF repeat, S is the Ser that accepts glucose, x is any amino acid, P is Pro and A is Ala [118, 119] (Fig. 1). Mass spectrometric analysis of Drosophila NOTCH1 ECD from S2 cells and NOTCH isolated from Drosophila embryos demonstrated the presence of O-glucose on all predicted 18 sites. However, O-glucose-xylose disaccharide was found only on EGF13–20 and EGF25. Similarly, the trisaccharide was found on a restricted subset of the EGF repeats with O-glucose [28]. Both the addition of O-glucose and its elongation by xylose is dependent on the amino acids in the consensus site and the proper folding of the EGF repeat [120]. The O-glucosyltransferase is encoded by Rumi in Drosophila [121] and the protein O-glucosyltransferase 1 gene Poglut1 in mammals [122]. POGLUT1 is an ER resident enzyme [121]. In vitro knockdown of Rumi in Drosophila S2 cells and mammalian cell lines reduces Notch signalling due to defects in Notch receptor cleavage upon ligand binding. However, loss of O-glucose glycans does not reduce Notch ligand binding. Thus, O-glucose glycans appear to promote a conformational change in Notch receptors after ligand interaction, and are required for S2 cleavage by ADAM proteases [123, 124]. Further, crystal structures of a NOTCH1 ligand-binding fragment bound to a N-terminal fragment of DLL4 demonstrate that O-glucose on EGF12 and EGF13 are located away from the DLL4 binding face, and cover hydrophobic residues Pro and Phe in these EGF repeats [57], hence supporting the notion that O-glucose is not required for Notch-ligand interactions. The presence of a novel, O-linked hexose attached to Ser in EGF11 at a site which does not match the consensus of an O-glucose site was revealed in crystal structures [57, 125]. This modification is conserved in other Notch receptors, except NOTCH2, at the consensus site C3xNTxGSFxC4. The hexose was recently identified as a glucose residue which is added by POGLUT2 or POGLUT3, homologues of POGLUT1 [126]. Mutations of single O-glucose sites in NECD do not affect the cell surface expression of Notch, nor impair Notch activation, except for mutation in EGF28 [15, 127]. Though, the O-glucose consensus site in EGF28 is present in mammalian NOTCH1, and not present in Drosophila Notch or other mammalian Notch receptors, mutation of Ser to Ala in EGF28 of NOTCH1 causes a decrease of Delta-induced Notch signalling, but does not affect signalling mediated by Jagged ligands. Deletion of Poglut1 in HEK-293T cells causes a mild reduction in cell surface expression of NOTCH1, and enhanced secretion of soluble NECD, suggesting that O-glucose glycans contribute to trafficking or stability of Notch receptors [47]. Mutation of the O-glucose site in EGF11 to Ala has no effects on cell surface expression of NOTCH1, DLL1 binding or DLL1-induced NOTCH1 signalling [126]. However, the combined mutation of EGF11 and the O-fucose site in EGF8 or EGF12 of NOTCH1 has somewhat greater effects than the single O-fucose mutations. It would be interesting to see if cells lacking both POGLUT2 and POGLUT3, but retaining Ser in EGF11, give the same results with NOTCH1 carrying the EGF8 or EGF12 O-fucose mutations.
• In vivo consequences of defects in O-glucose glycan synthesis or loss of O-glucose sites
Loss of Rumi from Drosophila results in temperature-sensitive Notch signalling defects [121]. Rumi-null flies are viable at 180C but exhibit a slight Delta wing vein phenotype. However, at 280C lethality occurs at the larval stage, and reduced Notch signalling is observed in all contexts studied. O-glucose on Notch in flies is critical for S2 cleavage of Notch but ligand binding remains unaffected. Further, rumi mutant G189E lacking transferase activity did not rescue defective Notch signalling in rumi null flies, suggesting the importance of O-glucose glycans to Notch receptor trafficking and stability at the cell surface. Multiple mutations in O-glucosylation sites on NECD were essential for temperature-sensitive Notch signalling defects to arise, whereas single site mutations did not affect Notch signalling [127]. Poglut1−/− mice are embryonic lethal and die before E9.5 with severe defects in neural tube development, cardiogenesis and somitogenesis [122]. Poglut1 null mutants die earlier than Notch pathway null mutants, apparently due to the loss of O-glucose from CRUMBS2 [122]. Haploinsufficiency of Poglut1 on a Jag1 heterozygous background results in decreased O-glucosylation of NOTCH1 and severe defects in bile duct morphogenesis, suggesting a genetic interaction between Poglut1 and members of the Notch signalling pathway [123]. Additionally, mutations in human POGLUT1 cause an autosomal dominant form of Dowling-Degos Disease termed DDD2 [128–131], or a recessive limb-girdle muscular dystrophy [132].
O-GlcNAc glycans
• Discovery and cell-based assays
The presence of O-GlcNAc on EGF repeats of Notch receptors was first identified in S2 cells on a Drosophila NECD fragment containing EGF20 [133]. O-GlcNAc is added to Ser/Thr between the fifth and sixth cysteines of an EGF repeat with the consensus site C5xxG(Y/F)(T/S)Gx2–3C6 [20, 133–135]. Of the 36 EGF mouse NOTCH1 repeats, 17 have a consensus site for O-GlcNAc and Drosophila Notch has 18 consensus sites. However, mass spectrometry on Notch purified from S2 cells and Drosophila larvae identified O-GlcNAc on only 5 sites [28]. In mammalian cells, O-GlcNAc can be further elongated by Gal [134] and probably sialic acid. The enzyme responsible for the addition of O-GlcNAc on EGF repeats was identified in Drosophila as EGF-domain specific O-GlcNAc-transferase (EOGT). EOGT is conserved across species and is localized to the ER by a signal peptide at the N terminus and a C-terminal KDEL sequence [134]. Studies using knockdown and knockout of Eogt in mammalian cell lines suggest that O-GlcNAc on Notch receptors promotes Delta-mediated Notch signalling, but does not significantly affect JAG1-induced Notch signalling [136]. Loss of EOGT inhibits binding of Delta-like ligands but not JAG1, suggesting that O-GlcNAc on Notch plays specific roles in Notch ligand binding and Notch signalling [136].
• In vivo consequences of defects in O-GlcNAc glycans and loss of O-GlcNAc sites
Loss of eogt in flies results in lethality, mostly during second instar larval development, with a few survivors at the early third-instar stage [134, 135]. However, larvae lacking eogt do not show a phenotype similar to flies with Notch-deficient signalling. Knockdown of eogt in the fly wing results in blistering that may arise from roles for O-GlcNAc on Dumpy, an extracellular matrix protein with a large number of EGF repeats. Ligand-induced Notch signaling promotes blistering because the phenotype is partially rescued with the loss of one allele of Notch or Notch pathway members such as Delta or Serrate that reduces Notch signalling, suppressor of hairless (Su(H) or maml [135]. Such genetic interaction studies provided the first link between Notch signalling and Eogt which has now been validated in mammalian cells and mice. In the mouse, Eogt expression is enhanced in the presomitic mesoderm at E9.5, and limited to the digits of developing limbs by E12.5 [137]. Eogt null mice are viable, fertile and do not show a typical Notch phenotype [136]. Using retinal angiogenesis as a sensitive assay for ligand-induced Notch signalling [100], defective angiogenesis with leaky blood vessels is observed in mice lacking Eogt [136]. Loss of Eogt results in increased blood vessel branching and increased tip cell numbers, which is characteristic of disrupted DLL4-NOTCH1 signalling Thus, in retinal angiogenesis, loss of Eogt recapitulates results in cell lines and reveals the importance of O-GlcNAc in promoting optimal Delta-induced Notch signalling. In humans, mutations in EOGT cause a rare, congenital disorder termed Adams-Oliver Syndrome 4 (AOS4) [137–139]. Symptoms include cutis aplasia of the scalp, defects in the development of digits, vascular defects and, in some cases, cardiac defects. Autosomal dominant mutations in NOTCH1, DLL4 and RBPJ genes have also been identified in patients diagnosed with AOS [140–143].
Synergistic and redundant roles for O-fucose and O-glucose glycans
While the majority of studies to date have focused on understanding the functions of each type of O-glycan independently, a few studies have investigated roles for O-glucose and O-fucose glycans together. In Drosophila, loss of O-fucose or O-glucose separately causes temperature-sensitive Notch signalling defects that manifest at 30ºC [43]. However, at 25ºC each mutant behaves essentially like wild type. When, however, both O-fucose and O-glucose glycans are not transferred to Notch, Notch signalling is lost at 25ºC. This correlates with accumulation of Notch in the ER, whereas loss of O-fucose or O-glucose alone allowed exit of Notch from the ER. Thus O-glucose and O-fucose glycans function synergistically to support Notch trafficking out of the ER in Drosophila. Consistent with this, in mammalian cells in which both POFUT1 and POGLUT1 were deleted to give NOTCH1 lacking both O-fucose and O-glucose glycans, NOTCH1 was not expressed well at the cell surface, whereas loss of either O-glycan alone allowed cell surface expression of NOTCH1 [47]. In another study, the presence of xylose in O-glucose glycans was also found to contribute to Notch trafficking and Notch signalling in Drosophila but only in the context of a double mutant [144]. Thus, Notch lacking both O-fucose and the dixylose on O-glucose was mislocalized from the apical plasma membrane to adherens junctions, and had reduced Notch signalling [144]. Notch lacking O-glucose (including dixylose) and O-fucose was not exported from the endoplasmic reticulum. Thus, some functions of sugars (e. g. dixylose) may only be observed in the absence of a compensatory sugar (e. g. O-fucose), and this may in turn depend on cellular context.
Conclusions
It is apparent from studies published over the past 18 years that distinct O-glycans on Notch receptors are essential for regulating and optimizing different aspects of Notch signalling. O-glucose glycans on Notch positively regulate the cleavage of Notch receptors upon ligand binding, and promote receptor trafficking to the cell surface, but do not directly mediate interactions with Notch ligands. However, extension of O-glucose by xylose negatively regulates Notch signalling, in a context dependent manner. O-fucose glycans influence Notch signalling by differentially regulating Notch ligand binding to Delta and Jagged ligands. In Drosophila and certain numerous mammalian cell types, POFUT1 is important for promoting Notch receptor trafficking to the cell surface. However, in mammalian cells lacking POFUT1 NOTCH1 and other Notch receptors are well expressed at the cell surface but do not bind Notch ligands or exhibit ligand-induced Notch signalling. The addition of GlcNAc to O-fucose by Fringe differentially modulates Notch receptor interactions with the various ligands. LFNG and MFNG generally promote Delta ligand binding and inhibit Jagged ligand binding, whereas RFNG promotes both Delta and Jagged ligand binding. The more recently identified O-GlcNAc modification on Notch appears to mediate Notch signalling via Delta but not Jagged ligands based on both cell-based and in vivo studies. Mutations in several of the glycosyltransferases that synthesize the O-glycans on Notch receptors cause a variety of defects in Notch signalling, establishing the biological importance of O-glycans in regulating and optimizing the strength of Notch signalling. Human mutations in several glycosyltransferase are associated with different pathologies, which are also associated with mutations in Notch and Notch pathway members. The ExAc browser [145] describes exon sequencing data from 60,706 unrelated individuals, not including people with known congenital mutations, and reports single nucleotide mutations including missense and nonsense mutations in the different glycosyltransferase genes described above. A few healthy people with homozygous missense mutations have been reported, indicating that these mutations are not important for the activity of the relevant glycosyltransferase. While the majority of studies to date have focused on understanding the roles of each type of glycan independently, a few studies have revealed synergistic and redundant roles of O-glucose and O-fucose glycans. Future efforts should continue along this line of inquiry to reveal how all the O-glycans on Notch receptors and ligands work separately and together to optimize Notch signalling. These studies will reveal synergistic, redundant and non-overlapping functions of the glycans which will further help to elucidate how O-glycans on Notch regulate diverse cell fate decisions. Insights from these studies will help to design potential targets for therapeutic purposes.
Acknowledgments
This work was supported by funding from National Institutes of Health grant RO1 GM106417 to PS.
Abbreviations
- NECD
Notch extracellular domain
- NICD
Notch intracellular domain
- EGF
Epidermal growth factor-like
- NRR
Negative regulatory region
- HD
Heterodimerization domain
- NEXT
Notch extracellular truncation
- RAM
RBP-Jκ-associated module
- ANK
Ankyrin repeats
- TAD
transcriptional activation domain
- CSL
CBF-1 Suppressor of Hairless-LAG1
- ADAM
A disintegrin and metalloprotease
- MAML
Mastermind-like
- GlcNAc
N-acetylglucosamine
- GSI
gamma-secretase inhibitor
- POFUT1
Protein O-fucosyltransferase 1
- CHO
Chinese hamster ovary
- ER
Endoplasmic reticulum
- ES
Embryonic stem cells
- HSC
Hematopoietic stem cells
- U2OS
Human osteosarcoma cell line
- GDP-Fuc
GDP-Fucose
- Gal
Galactose
- NeuAc
N-acetylneuraminic acid
- LFNG
Lunatic fringe
- MFNG
Manic fringe
- RFNG
Radical fringe
- HEK
Human embryonic kidney
- DLL
Delta-like
- JAG1
Jagged 1
- C2 domain
module at the N-terminus of Notch ligands
- POGLUT1
Protein O-glucosyltransferase 1
- EOGT
EGF-domain specific O-GlcNAc
- Su(H)
Suppressor of hairless
- AOS4
Adams-Oliver Syndrome 4
- DDD
Dowling-Degos Disease
- SCDO3
Spondylocostal dysostosis 3
- LGMD2Z
Limb-girdle muscular dystrophy type 2Z
- HS-DDD4
Hidradenitis Suppurativa-Dowling-Degos Disease 4
- GXYLT1
Glucoside xylosyltransferase 1
- GXYLT2
Glucoside xylosyltransferase 2
- XXYLT1
Xylose xylosyltransferase 1
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Siebel C & Lendahl U (2017) Notch Signaling in Development, Tissue Homeostasis, and Disease, Physiol Rev 97, 1235–1294. [DOI] [PubMed] [Google Scholar]
- 2.Fortini ME (2009) Notch signaling: the core pathway and its posttranslational regulation, Dev Cell 16, 633–47. [DOI] [PubMed] [Google Scholar]
- 3.Kopan R & Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism, Cell 137, 216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovall RA, Gebelein B, Sprinzak D & Kopan R (2017) The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force, Dev Cell 41, 228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ & Kopan R (2000) A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1, Mol Cell 5, 197–206. [DOI] [PubMed] [Google Scholar]
- 6.Bozkulak EC & Weinmaster G (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling, Mol Cell Biol 29, 5679–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R & Vooijs M (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage, J Biol Chem 284, 31018–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musse AA, Meloty-Kapella L & Weinmaster G (2012) Notch ligand endocytosis: mechanistic basis of signaling activity, Semin Cell Dev Biol 23, 429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struhl G & Greenwald I (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila, Nature 398, 522–5. [DOI] [PubMed] [Google Scholar]
- 10.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T & Honjo T (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H), Curr Biol 5, 1416–23. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R & Israel A (1995) Signalling downstream of activated mammalian Notch, Nature 377, 355–8. [DOI] [PubMed] [Google Scholar]
- 12.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S & Griffin JD (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors, Nat Genet 26, 484–9. [DOI] [PubMed] [Google Scholar]
- 13.Bray SJ (2016) Notch signalling in context, Nat Rev Mol Cell Biol 17, 722–735. [DOI] [PubMed] [Google Scholar]
- 14.Stanley P & Okajima T (2010) Roles of glycosylation in Notch signaling, Curr Top Dev Biol 92, 131–64. [DOI] [PubMed] [Google Scholar]
- 15.Jafar-Nejad H, Leonardi J & Fernandez-Valdivia R (2010) Role of glycans and glycosyltransferases in the regulation of Notch signaling, Glycobiology 20, 931–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi H & Haltiwanger RS (2014) Significance of glycosylation in Notch signaling, Biochem Biophys Res Commun 453, 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haltom AR & Jafar-Nejad H (2015) The multiple roles of epidermal growth factor repeat O-glycans in animal development, Glycobiology 25, 1027–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshney S & Stanley P (2017) EOGT and O-GlcNAc on secreted and membrane proteins, Biochem Soc Trans 45, 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rampal R, Luther KB & Haltiwanger RS (2007) Notch signaling in normal and disease States: possible therapies related to glycosylation, Curr Mol Med 7, 427–45. [DOI] [PubMed] [Google Scholar]
- 20.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F & Smith RD (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets, Proc Natl Acad Sci U S A 109, 7280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider M, Al-Shareffi E & Haltiwanger RS (2017) Biological functions of fucose in mammals, Glycobiology 27, 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi S, Ge C, Luo Y, Hou X, Haltiwanger RS & Stanley P (2007) The threonine that carries fucose, but not fucose, is required for Cripto to facilitate Nodal signaling, J Biol Chem 282, 20133–41. [DOI] [PubMed] [Google Scholar]
- 23.Irvine KD & Wieschaus E (1994) fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development, Cell 79, 595–606. [DOI] [PubMed] [Google Scholar]
- 24.Panin VM, Papayannopoulos V, Wilson R & Irvine KD (1997) Fringe modulates Notch-ligand interactions, Nature 387, 908–12. [DOI] [PubMed] [Google Scholar]
- 25.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine KD, Haltiwanger RS & Vogt TF (2000) Fringe is a glycosyltransferase that modifies Notch, Nature 406, 369–75. [DOI] [PubMed] [Google Scholar]
- 26.Bruckner K, Perez L, Clausen H & Cohen S (2000) Glycosyltransferase activity of Fringe modulates Notch-Delta interactions, Nature 406, 411–5. [DOI] [PubMed] [Google Scholar]
- 27.Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL & Haltiwanger RS (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules, J Biol Chem 275, 9604–11. [DOI] [PubMed] [Google Scholar]
- 28.Harvey BM, Rana NA, Moss H, Leonardi J, Jafar-Nejad H & Haltiwanger RS (2016) Mapping Sites of O-Glycosylation and Fringe Elongation on Drosophila Notch, J Biol Chem 291, 16348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakuda S & Haltiwanger RS (2017) Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands, Dev Cell 40, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haines N & Irvine KD (2003) Glycosylation regulates Notch signalling, Nat Rev Mol Cell Biol 4, 786–97. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Koles K, Vorndam W, Haltiwanger RS & Panin VM (2006) Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats, J Biol Chem 281, 9393–9. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Lee GF, Kelley RF & Spellman MW (1996) Identification of a GDP-L-fucose:polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains, Glycobiology 6, 837–42. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P & Haltiwanger RS (2001) Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase, J Biol Chem 276, 40338–45. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y & Spellman MW (1998) Purification and characterization of a GDP-fucose:polypeptide fucosyltransferase from Chinese hamster ovary cells, J Biol Chem 273, 8112–8. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y & Haltiwanger RS (2005) O-fucosylation of notch occurs in the endoplasmic reticulum, J Biol Chem 280, 11289–94. [DOI] [PubMed] [Google Scholar]
- 36.Okajima T, Xu A & Irvine KD (2003) Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe, J Biol Chem 278, 42340–5. [DOI] [PubMed] [Google Scholar]
- 37.Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, Perrimon N & Matsuno K (2003) neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions, Development 130, 4785–95. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Moloney DJ & Stanley P (2001) Fringe modulation of Jagged1-induced Notch signaling requires the action of beta 4galactosyltransferase-1, Proc Natl Acad Sci U S A 98, 13716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okajima T & Irvine KD (2002) Regulation of notch signaling by o-linked fucose, Cell 111, 893–904. [DOI] [PubMed] [Google Scholar]
- 40.Okajima T, Xu A, Lei L & Irvine KD (2005) Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding, Science 307, 1599–603. [DOI] [PubMed] [Google Scholar]
- 41.Sasamura T, Ishikawa HO, Sasaki N, Higashi S, Kanai M, Nakao S, Ayukawa T, Aigaki T, Noda K, Miyoshi E, Taniguchi N & Matsuno K (2007) The O-fucosyltransferase O-fut1 is an extracellular component that is essential for the constitutive endocytic trafficking of Notch in Drosophila, Development 134, 1347–56. [DOI] [PubMed] [Google Scholar]
- 42.Okajima T, Reddy B, Matsuda T & Irvine KD (2008) Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling, BMC Biol 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishio A, Sasamura T, Ayukawa T, Kuroda J, Ishikawa HO, Aoyama N, Matsumoto K, Gushiken T, Okajima T, Yamakawa T & Matsuno K (2015) O-fucose monosaccharide of Drosophila Notch has a temperature-sensitive function and cooperates with O-glucose glycan in Notch transport and Notch signaling activation, J Biol Chem 290, 505–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl M, Uemura K, Ge C, Shi S, Tashima Y & Stanley P (2008) Roles of Pofut1 and O-fucose in mammalian Notch signaling, J Biol Chem 283, 13638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB & Zhou L (2011) Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions, Blood 117, 5652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Wu B, Lu P, Zhang D, Wu B, Varshney S, Del Monte-Nieto G, Zhuang Z, Charafeddine R, Kramer AH, Sibinga NE, Frangogiannis NG, Kitsis RN, Adams RH, Alitalo K, Sharp DJ, Harvey RP, Stanley P & Zhou B (2017) Uncontrolled angiogenic precursor expansion causes coronary artery anomalies in mice lacking Pofut1, Nat Commun 8, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H & Haltiwanger RS (2017) O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking, J Biol Chem 292, 15964–15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMillan BJ, Zimmerman B, Egan ED, Lofgren M, Xu X, Hesser A & Blacklow SC (2017) Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations, Glycobiology 27, 777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ajima R, Suzuki E & Saga Y (2017) Pofut1 point-mutations that disrupt O-fucosyltransferase activity destabilize the protein and abolish Notch1 signaling during mouse somitogenesis, PLoS One 12, e0187248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamura Y & Saga Y (2008) Pofut1 is required for the proper localization of the Notch receptor during mouse development, Mech Dev 125, 663–73. [DOI] [PubMed] [Google Scholar]
- 51.Basch ML, Brown RM, Jen HI, Semerci F, Depreux F, Edlund RK, Zhang H, Norton CR, Gridley T, Cole SE, Doetzlhofer A, Maletic-Savatic M, Segil N & Groves AK (2016) Fine-tuning of Notch signaling sets the boundary of the organ of Corti and establishes sensory cell fates, Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu VW, Saez B, Cook C, Lotinun S, Pardo-Saganta A, Wang YH, Lymperi S, Ferraro F, Raaijmakers MH, Wu JY, Zhou L, Rajagopal J, Kronenberg HM, Baron R & Scadden DT (2015) Specific bone cells produce DLL4 to generate thymus-seeding progenitors from bone marrow, J Exp Med 212, 759–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith PL, Myers JT, Rogers CE, Zhou L, Petryniak B, Becker DJ, Homeister JW & Lowe JB (2002) Conditional control of selectin ligand expression and global fucosylation events in mice with a targeted mutation at the FX locus, J Cell Biol 158, 801–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker DJ, Myers JT, Ruff MM, Smith PL, Gillespie BW, Ginsburg DW & Lowe JB (2003) Strain-specific modification of lethality in fucose-deficient mice, Mamm Genome 14, 130–9. [DOI] [PubMed] [Google Scholar]
- 55.Glavic A, Lopez-Varea A & de Celis JF (2011) The balance between GMD and OFUT1 regulates Notch signaling pathway activity by modulating Notch stability, Biol Res 44, 25–34. [DOI] [PubMed] [Google Scholar]
- 56.Myers J, Huang Y, Wei L, Yan Q, Huang A & Zhou L (2010) Fucose-deficient hematopoietic stem cells have decreased self-renewal and aberrant marrow niche occupancy, Transfusion 50, 2660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S & Garcia KC (2015) Structural basis for Notch1 engagement of Delta-like 4, Science 347, 847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, Haltiwanger RS, Zhu C, Ha T & Garcia KC (2017) Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity, Science 355, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto S, Charng WL, Rana NA, Kakuda S, Jaiswal M, Bayat V, Xiong B, Zhang K, Sandoval H, David G, Wang H, Haltiwanger RS & Bellen HJ (2012) A mutation in EGF repeat-8 of Notch discriminates between Serrate/Jagged and Delta family ligands, Science 338, 1229–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LeBon L, Lee TV, Sprinzak D, Jafar-Nejad H & Elowitz MB (2014) Fringe proteins modulate Notch-ligand cis and trans interactions to specify signaling states, Elife 3, e02950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME & Elowitz MB (2018) Dynamic Ligand Discrimination in the Notch Signaling Pathway, Cell 172, 869–880 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buas MF, Kabak S & Kadesch T (2009) Inhibition of myogenesis by Notch: evidence for multiple pathways, J Cell Physiol 218, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luther KB, Schindelin H & Haltiwanger RS (2009) Structural and mechanistic insights into lunatic fringe from a kinetic analysis of enzyme mutants, J Biol Chem 284, 3294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD & Vogt TF (1997) A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway, Development 124, 2245–54. [DOI] [PubMed] [Google Scholar]
- 65.Rampal R, Li AS, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A & Haltiwanger RS (2005) Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats, J Biol Chem 280, 42454–63. [DOI] [PubMed] [Google Scholar]
- 66.Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF & Weinmaster G (2000) Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2, Nat Cell Biol 2, 515–20. [DOI] [PubMed] [Google Scholar]
- 67.Yang LT, Nichols JT, Yao C, Manilay JO, Robey EA & Weinmaster G (2005) Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1, Mol Biol Cell 16, 927–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C & Weinmaster G (2005) The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands, J Cell Biol 170, 983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS & Irvine KD (2007) In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe, J Biol Chem 282, 35153–62. [DOI] [PubMed] [Google Scholar]
- 70.Taylor P, Takeuchi H, Sheppard D, Chillakuri C, Lea SM, Haltiwanger RS & Handford PA (2014) Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands, Proc Natl Acad Sci U S A 111, 7290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song Y, Kumar V, Wei HX, Qiu J & Stanley P (2016) Lunatic, Manic, and Radical Fringe Each Promote T and B Cell Development, J Immunol 196, 232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou X, Tashima Y & Stanley P (2012) Galactose differentially modulates lunatic and manic fringe effects on Delta1-induced NOTCH signaling, J Biol Chem 287, 474–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi S & Stanley P (2003) Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways, Proc Natl Acad Sci U S A 100, 5234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W, Yu S, Zimmerman G, Wang Y, Myers J, Yu VW, Huang D, Huang X, Shim J, Huang Y, Xin W, Qiao P, Yan M, Xin W, Scadden DT, Stanley P, Lowe JB, Huang AY, Siebel CW & Zhou L (2015) Notch Receptor-Ligand Engagement Maintains Hematopoietic Stem Cell Quiescence and Niche Retention, Stem Cells 33, 2280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guilmeau S, Flandez M, Bancroft L, Sellers RS, Tear B, Stanley P & Augenlicht LH (2008) Intestinal deletion of Pofut1 in the mouse inactivates notch signaling and causes enterocolitis, Gastroenterology 135, 849–60, 860 e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F & Clevers H (2005) Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells, Nature 435, 959–63. [DOI] [PubMed] [Google Scholar]
- 77.Tsao PN, Chen F, Izvolsky KI, Walker J, Kukuruzinska MA, Lu J & Cardoso WV (2008) Gamma-secretase activation of notch signaling regulates the balance of proximal and distal fates in progenitor cells of the developing lung, J Biol Chem 283, 29532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin HY, Kao CH, Lin KM, Kaartinen V & Yang LT (2011) Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis, PLoS One 6, e15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen M, Li Y, Liu H, Fu X, Yu Y, Yu G, Wang C, Bao F, Liany H, Wang Z, Shi Z, Zhang D, Zhou G, Liu J & Zhang F (2014) Analysis of POFUT1 gene mutation in a Chinese family with Dowling-Degos disease, PLoS One 9, e104496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buket Basmanav F, Fritz G, Lestringant GG, Pachat D, Hoffjan S, Fischer J, Wehner M, Wolf S, Thiele H, Altmuller J, Pulimood SA, Rutten A, Kruse R, Hanneken S, Frank J, Danda S, Bygum A & Betz RC (2015) Pathogenicity of POFUT1 in Dowling-Degos disease: additional mutations and clinical overlap with reticulate acropigmentation of kitamura, J Invest Dermatol 135, 615–618. [DOI] [PubMed] [Google Scholar]
- 81.Li M, Cheng R, Liang J, Yan H, Zhang H, Yang L, Li C, Jiao Q, Lu Z, He J, Ji J, Shen Z, Li C, Hao F, Yu H & Yao Z (2013) Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease, Am J Hum Genet 92, 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gonzalez-Villanueva I, Gutierrez M, Hispan P, Betlloch I & Pascual JC (2018) Novel POFUT1 mutation associated with hidradenitis suppurativa-Dowling-Degos disease firm up a role for Notch signalling in the pathogenesis of this disorder, Br J Dermatol 178, 984–986. [DOI] [PubMed] [Google Scholar]
- 83.Takeuchi H, Wong D, Schneider M, Freeze HH, Takeuchi M, Berardinelli SJ, Ito A, Lee H, Nelson SF & Haltiwanger RS (2018) Variant in human POFUT1 reduces enzymatic activity and likely causes a recessive microcephaly, global developmental delay with cardiac and vascular features, Glycobiology [DOI] [PMC free article] [PubMed]
- 84.Ma L, Dong P, Liu L, Gao Q, Duan M, Zhang S, Chen S, Xue R & Wang X (2016) Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway, Biochem Biophys Res Commun 473, 503–10. [DOI] [PubMed] [Google Scholar]
- 85.Lei L, Xu A, Panin VM & Irvine KD (2003) An O-fucose site in the ligand binding domain inhibits Notch activation, Development 130, 6411–21. [DOI] [PubMed] [Google Scholar]
- 86.Ge C & Stanley P (2008) The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development, Proc Natl Acad Sci U S A 105, 1539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muller J, Rana NA, Serth K, Kakuda S, Haltiwanger RS & Gossler A (2014) O-fucosylation of the notch ligand mDLL1 by POFUT1 is dispensable for ligand function, PLoS One 9, e88571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Serth K, Schuster-Gossler K, Kremmer E, Hansen B, Marohn-Kohn B & Gossler A (2015) O-fucosylation of DLL3 is required for its function during somitogenesis, PLoS One 10, e0123776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kadur Lakshminarasimha Murthy P, Srinivasan T, Bochter MS, Xi R, Varanko AK, Tung KL, Semerci F, Xu K, Maletic-Savatic M, Cole SE & Shen X (2018) Radical and lunatic fringes modulate notch ligands to support mammalian intestinal homeostasis, Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rana NA & Haltiwanger RS (2011) Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors, Curr Opin Struct Biol 21, 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moran JL, Shifley ET, Levorse JM, Mani S, Ostmann K, Perez-Balaguer A, Walker DM, Vogt TF & Cole SE (2009) Manic fringe is not required for embryonic development, and fringe family members do not exhibit redundant functions in the axial skeleton, limb, or hindbrain, Dev Dyn 238, 1803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Svensson P, Bergqvist I, Norlin S & Edlund H (2009) MFng is dispensable for mouse pancreas development and function, Mol Cell Biol 29, 2129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moran JL, Levorse JM & Vogt TF (1999) Limbs move beyond the radical fringe, Nature 399, 742–3. [DOI] [PubMed] [Google Scholar]
- 94.Zhang N & Gridley T (1998) Defects in somite formation in lunatic fringe-deficient mice, Nature 394, 374–7. [DOI] [PubMed] [Google Scholar]
- 95.Evrard YA, Lun Y, Aulehla A, Gan L & Johnson RL (1998) lunatic fringe is an essential mediator of somite segmentation and patterning, Nature 394, 377–81. [DOI] [PubMed] [Google Scholar]
- 96.Hahn KL, Beres B, Rowton MJ, Skinner MK, Chang Y, Rawls A & Wilson-Rawls J (2009) A deficiency of lunatic fringe is associated with cystic dilation of the rete testis, Reproduction 137, 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hahn KL, Johnson J, Beres BJ, Howard S & Wilson-Rawls J (2005) Lunatic fringe null female mice are infertile due to defects in meiotic maturation, Development 132, 817–28. [DOI] [PubMed] [Google Scholar]
- 98.Visan I, Tan JB, Yuan JS, Harper JA, Koch U & Guidos CJ (2006) Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches, Nat Immunol 7, 634–43. [DOI] [PubMed] [Google Scholar]
- 99.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR & Aguet M (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1, Immunity 10, 547–58. [DOI] [PubMed] [Google Scholar]
- 100.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M & Adams RH (2009) The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis, Cell 137, 1124–35. [DOI] [PubMed] [Google Scholar]
- 101.Ehling M, Adams S, Benedito R & Adams RH (2013) Notch controls retinal blood vessel maturation and quiescence, Development 140, 3051–61. [DOI] [PubMed] [Google Scholar]
- 102.Tan JB, Xu K, Cretegny K, Visan I, Yuan JS, Egan SE & Guidos CJ (2009) Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches, Immunity 30, 254–63. [DOI] [PubMed] [Google Scholar]
- 103.Stanley P & Guidos CJ (2009) Regulation of Notch signaling during T- and B-cell development by O-fucose glycans, Immunol Rev 230, 201–15. [DOI] [PubMed] [Google Scholar]
- 104.Chen J, Lu L, Shi S & Stanley P (2006) Expression of Notch signaling pathway genes in mouse embryos lacking beta4galactosyltransferase-1, Gene Expr Patterns 6, 376–82. [DOI] [PubMed] [Google Scholar]
- 105.Hansske B, Thiel C, Lubke T, Hasilik M, Honing S, Peters V, Heidemann PH, Hoffmann GF, Berger EG, von Figura K & Korner C (2002) Deficiency of UDP-galactose:N-acetylglucosamine beta-1,4-galactosyltransferase I causes the congenital disorder of glycosylation type IId, J Clin Invest 109, 725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D & Dunwoodie SL (2006) Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype, Am J Hum Genet 78, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lefebvre M, Dieux-Coeslier A, Baujat G, Schaefer E, Judith SO, Bazin A, Pinson L, Attie-Bitach T, Baumann C, Fradin M, Pierquin G, Julia S, Quelin C, Doray B, Berg S, Vincent-Delorme C, Lambert L, Bachmann N, Lacombe D, Isidor B, Laurent N, Joelle R, Blanchet P, Odent S, Kervran D, Leporrier N, Abel C, Segers K, Guiliano F, Ginglinger-Fabre E, Selicorni A, Goldenberg A, El Chehadeh S, Francannet C, Demeer B, Duffourd Y, Thauvin-Robinet C, Verloes A, Cormier-Daire V, Riviere JB, Faivre L & Thevenon J (2018) Diagnostic strategy in segmentation defect of the vertebrae: a retrospective study of 73 patients, J Med Genet [DOI] [PubMed]
- 108.Zhang S, Chung WC, Wu G, Egan SE, Miele L & Xu K (2015) Manic fringe promotes a claudin-low breast cancer phenotype through notch-mediated PIK3CG induction, Cancer Res 75, 1936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, Cardiff RD, Yoon K, Gaiano N, Ling V, Beyene J, Zacksenhaus E, Gridley T, Leong WL, Guidos CJ, Perou CM & Egan SE (2012) Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer, Cancer Cell 21, 626–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Del Castillo Velasco-Herrera M, van der Weyden L, Nsengimana J, Speak AO, Sjoberg MK, Bishop DT, Jonsson G, Newton-Bishop J & Adams DJ (2018) Comparative genomics reveals that loss of lunatic fringe (LFNG) promotes melanoma metastasis, Mol Oncol 12, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang S, Chung WC & Xu K (2016) Lunatic Fringe is a potent tumor suppressor in Kras-initiated pancreatic cancer, Oncogene 35, 2485–95. [DOI] [PubMed] [Google Scholar]
- 112.Zhang S, Chung WC, Wu G, Egan SE & Xu K (2014) Tumor-suppressive activity of Lunatic Fringe in prostate through differential modulation of Notch receptor activation, Neoplasia 16, 158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hase S, Kawabata S, Nishimura H, Takeya H, Sueyoshi T, Miyata T, Iwanaga S, Takao T, Shimonishi Y & Ikenaka T (1988) A new trisaccharide sugar chain linked to a serine residue in bovine blood coagulation factors VII and IX, J Biochem 104, 867–8. [DOI] [PubMed] [Google Scholar]
- 114.Sethi MK, Buettner FF, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R & Bakker H (2010) Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats, J Biol Chem 285, 1582–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee TV, Sethi MK, Leonardi J, Rana NA, Buettner FF, Haltiwanger RS, Bakker H & Jafar-Nejad H (2013) Negative regulation of notch signaling by xylose, PLoS Genet 9, e1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R & Bakker H (2012) Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch, J Biol Chem 287, 2739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pandey A, Li-Kroeger D, Sethi MK, Lee TV, Buettner FFR, Bakker H & Jafar-Nejad H (2018) Sensitized genetic backgrounds reveal differential roles for EGF repeat xylosyltransferases in Drosophila Notch signaling, Glycobiology, in press. [DOI] [PMC free article] [PubMed]
- 118.Nishimura H, Kawabata S, Kisiel W, Hase S, Ikenaka T, Takao T, Shimonishi Y & Iwanaga S (1989) Identification of a disaccharide (Xyl-Glc) and a trisaccharide (Xyl2-Glc) O-glycosidically linked to a serine residue in the first epidermal growth factor-like domain of human factors VII and IX and protein Z and bovine protein Z, J Biol Chem 264, 20320–5. [PubMed] [Google Scholar]
- 119.Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB & Haltiwanger RS (2011) O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1, J Biol Chem 286, 31623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takeuchi H, Kantharia J, Sethi MK, Bakker H & Haltiwanger RS (2012) Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats, J Biol Chem 287, 33934–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS & Bellen HJ (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling, Cell 132, 247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ramkumar N, Harvey BM, Lee JD, Alcorn HL, Silva-Gagliardi NF, McGlade CJ, Bestor TH, Wijnholds J, Haltiwanger RS & Anderson KV (2015) Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2, PLoS Genet 11, e1005551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS & Jafar-Nejad H (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi, Development 138, 1925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ma W, Du J, Chu Q, Wang Y, Liu L, Song M & Wang W (2011) hCLP46 regulates U937 cell proliferation via Notch signaling pathway, Biochem Biophys Res Commun 408, 84–8. [DOI] [PubMed] [Google Scholar]
- 125.Andrawes MB, Xu X, Liu H, Ficarro SB, Marto JA, Aster JC & Blacklow SC (2013) Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1, J Biol Chem 288, 25477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takeuchi H, Schneider M, Williamson DB, Ito A, Takeuchi M, Handford PA & Haltiwanger RS (2018) Two novel protein O-glucosyltransferases that modify sites distinct from POGLUT1 and affect Notch trafficking and signaling, Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed]
- 127.Leonardi J, Fernandez-Valdivia R, Li YD, Simcox AA & Jafar-Nejad H (2011) Multiple O-glucosylation sites on Notch function as a buffer against temperature-dependent loss of signaling, Development 138, 3569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Basmanav FB, Oprisoreanu AM, Pasternack SM, Thiele H, Fritz G, Wenzel J, Grosser L, Wehner M, Wolf S, Fagerberg C, Bygum A, Altmuller J, Rutten A, Parmentier L, El Shabrawi-Caelen L, Hafner C, Nurnberg P, Kruse R, Schoch S, Hanneken S & Betz RC (2014) Mutations in POGLUT1, encoding protein O-glucosyltransferase 1, cause autosomal-dominant Dowling-Degos disease, Am J Hum Genet 94, 135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hanneken S, Rutten A, Eigelshoven S, Braun-Falco M, Pasternack SM, Ruzicka T, Nothen MM, Betz RC & Kruse R (2011) [Galli-Galli disease. Clinical and histopathological investigation using a case series of 18 patients], Hautarzt 62, 842–51. [DOI] [PubMed] [Google Scholar]
- 130.Duchatelet S, Clerc H, Machet L, Gaboriaud P, Miskinyte S, Kervarrec T & Hovnanian A (2018) A new nonsense mutation in the POGLUT1 gene in two sisters with Dowling-Degos disease, J Eur Acad Dermatol Venereol [DOI] [PubMed]
- 131.Wilson NJ, Cole C, Kroboth K, Hunter WN, Mann JA, McLean WH, Kernland Lang K, Beltraminelli H, Sabroe RA, Tiffin N, Sobey GJ, Borradori L, Simpson E & Smith FJ (2017) Mutations in POGLUT1 in Galli-Galli/Dowling-Degos disease, Br J Dermatol 176, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Servian-Morilla E, Takeuchi H, Lee TV, Clarimon J, Mavillard F, Area-Gomez E, Rivas E, Nieto-Gonzalez JL, Rivero MC, Cabrera-Serrano M, Gomez-Sanchez L, Martinez-Lopez JA, Estrada B, Marquez C, Morgado Y, Suarez-Calvet X, Pita G, Bigot A, Gallardo E, Fernandez-Chacon R, Hirano M, Haltiwanger RS, Jafar-Nejad H & Paradas C (2016) A POGLUT1 mutation causes a muscular dystrophy with reduced Notch signaling and satellite cell loss, EMBO Mol Med 8, 1289–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T & Okajima T (2008) O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors, J Biol Chem 283, 35486–95. [DOI] [PubMed] [Google Scholar]
- 134.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K & Okajima T (2011) O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions, Nat Commun 2, 583. [DOI] [PubMed] [Google Scholar]
- 135.Muller R, Jenny A & Stanley P (2013) The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila, PLoS One 8, e62835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sawaguchi S, Varshney S, Ogawa M, Sakaidani Y, Yagi H, Takeshita K, Murohara T, Kato K, Sundaram S, Stanley P & Okajima T (2017) O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals, Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shaheen R, Aglan M, Keppler-Noreuil K, Faqeih E, Ansari S, Horton K, Ashour A, Zaki MS, Al-Zahrani F, Cueto-Gonzalez AM, Abdel-Salam G, Temtamy S & Alkuraya FS (2013) Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome, Am J Hum Genet 92, 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ogawa M, Sawaguchi S, Kawai T, Nadano D, Matsuda T, Yagi H, Kato K, Furukawa K & Okajima T (2015) Impaired O-linked N-acetylglucosaminylation in the endoplasmic reticulum by mutated epidermal growth factor (EGF) domain-specific O-linked N-acetylglucosamine transferase found in Adams-Oliver syndrome, J Biol Chem 290, 2137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cohen I, Silberstein E, Perez Y, Landau D, Elbedour K, Langer Y, Kadir R, Volodarsky M, Sivan S, Narkis G & Birk OS (2014) Autosomal recessive Adams-Oliver syndrome caused by homozygous mutation in EOGT, encoding an EGF domain-specific O-GlcNAc transferase, Eur J Hum Genet 22, 374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stittrich AB, Lehman A, Bodian DL, Ashworth J, Zong Z, Li H, Lam P, Khromykh A, Iyer RK, Vockley JG, Baveja R, Silva ES, Dixon J, Leon EL, Solomon BD, Glusman G, Niederhuber JE, Roach JC & Patel MS (2014) Mutations in NOTCH1 cause Adams-Oliver syndrome, Am J Hum Genet 95, 275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Southgate L, Sukalo M, Karountzos ASV, Taylor EJ, Collinson CS, Ruddy D, Snape KM, Dallapiccola B, Tolmie JL, Joss S, Brancati F, Digilio MC, Graul-Neumann LM, Salviati L, Coerdt W, Jacquemin E, Wuyts W, Zenker M, Machado RD & Trembath RC (2015) Haploinsufficiency of the NOTCH1 Receptor as a Cause of Adams-Oliver Syndrome With Variable Cardiac Anomalies, Circ Cardiovasc Genet 8, 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meester JA, Southgate L, Stittrich AB, Venselaar H, Beekmans SJ, den Hollander N, Bijlsma EK, Helderman-van den Enden A, Verheij JB, Glusman G, Roach JC, Lehman A, Patel MS, de Vries BB, Ruivenkamp C, Itin P, Prescott K, Clarke S, Trembath R, Zenker M, Sukalo M, Van Laer L, Loeys B & Wuyts W (2015) Heterozygous Loss-of-Function Mutations in DLL4 Cause Adams-Oliver Syndrome, Am J Hum Genet 97, 475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hassed SJ, Wiley GB, Wang S, Lee JY, Li S, Xu W, Zhao ZJ, Mulvihill JJ, Robertson J, Warner J & Gaffney PM (2012) RBPJ mutations identified in two families affected by Adams-Oliver syndrome, Am J Hum Genet 91, 391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsumoto K, Ayukawa T, Ishio A, Sasamura T, Yamakawa T & Matsuno K (2016) Dual Roles of O-Glucose Glycans Redundant with Monosaccharide O-Fucose on Notch in Notch Trafficking, J Biol Chem 291, 13743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, Birnbaum D, The Exome Aggregation C, Daly MJ & MacArthur DG (2017) The ExAC browser: displaying reference data information from over 60 000 exomes, Nucleic Acids Res 45, D840–D845. [DOI] [PMC free article] [PubMed] [Google Scholar]


