Abstract
Misoprostol is a life-saving drug in many developing countries for women at risk of post-partum hemorrhaging due to its affordability, stability, ease of administration and clinical efficacy. However, misoprostol lacks receptor and tissue selectivities and thus its use is accompanied by a number of serious side-effects. The development of pharmacological agents combining the advantages of misoprostol with improved selectivity is hindered by the absence of atomic details of misoprostol action in labor induction. Here, we present the 2.5 Å resolution crystal structure of misoprostol free-acid form bound to the myometrium labor-inducing prostaglandin E2 receptor 3 (EP3). The active-state structure reveals a completely enclosed binding pocket containing a structured water molecule that coordinates misoprostol ring structure. Modelling of selective agonists in EP3 structure reveals rationales for selectivity. These findings will provide the basis for the next generation of uterotonic drugs that will be suitable for administration in low resource settings.
INTRODUCTION
Misoprostol is a very close analogue of the natural bioactive lipids prostaglandin E1 and prostaglandin E2 (PGE1 and PGE2) that are crucial autocrine and paracrine mediators in many physiological and pathophysiological conditions such as labor, inflammation, pain, fever, atherosclerosis, cardiac ischemia, asthma and cancer1–5. They are synthesized locally and are metabolites of the cyclooxygenases, the target of non-steroidal anti-inflammatory drugs such as aspirin. PGE2 is the most abundant subtype of prostanoids in the human body and its physiological actions are mediated by four G Protein-Coupled Receptors (GPCR), E-Prostanoid (EP) type 1 to 4 receptors6. As such, they are the target of many drugs and clinical trials7.
Misoprostol activates myometrium contraction involved in all stages of parturition through the binding and activation of the G protein-coupled receptor (GPCR) prostaglandin E2 receptor 3 (EP3), mimicking the action of PGE28,9. Although it is officially approved by the FDA for the treatment of gastric ulcers, it is used off-label in many aspects of labor management and been proven to display good efficacy at term to induce the expulsion of placenta, a critical step in avoiding clinical complication such as postpartum bleeding, the leading cause of women’s death in childbirth worldwide10,11. It is affordable, stable at room temperature and can be auto-administered orally. As such, it is on the World Health Organization’s list of essential medicines12. Misoprostol is used in medical settings as a prodrug methyl ester derivative (misoprostol-ME) that is rapidly metabolized by cellular esterases yielding the biologically-active free acid (misoprostol-FA) (Supplementary Fig. 1)13. However, like PGE2, misoprostol-FA also activates other prostaglandin receptors while displaying poor tissue selectivity, thus activating EP receptors in other tissues such as gastrointestinal track, cardiovascular and central nervous systems, resulting in deleterious side-effects and potential risks depending on the individual’s health, dose required, and experience of the caregiver12. Medicinal chemistry efforts have yielded selective drugs but their widespread clinical use is hindered by lack of stability, oral availability, and the need to be administered by trained healthcare professionals. Notwithstanding the high quality of Western health care systems, this combination of unfavorable properties also potentially limits clinical investigations of EP3 agonists on other relevant indications.
Unfortunately, annually 45 million women in the least developed countries do not have access to trained medical staff and thus experience unacceptable rates of maternal mortality during childbirth10. In these countries, clinically relevant EP3 selective agonists or other uterotonics drugs such as oxytocin are generally not available due to their cost, instability and their mode of administration, which requires a hospital setting. Therefore, misoprostol started to be distributed in the third world communities as the only available therapeutic for labor management14, despite the fact that this indication is not approved by the FDA due to lack of evidence for safety associated with its use during pregnancy12. Thus, new potent, safe and selective EP3 agonists that display better tissue selectivity toward myometrium and that retain the advantages of misoprostol are required to fully exploit the clinical advantages of EP3 activation in women reproductive health. Indeed, expanding the diversity of chemical scaffolds that act as activators of EP3 are essential for the development of EP3 agonists with improved pharmaco-economical properties as the few agonists available share very similar structures as the natural prostaglandin lipids7. Also, it would be beneficial for the evaluation of EP3 as a clinical target on a wider range of indications even in developed countries.
Recent breakthroughs in membrane protein crystallography methodology fostered the elucidation of GPCR structures15. These structures have enabled successful structure-based rational approaches using computer-assisted drug design to explore new ligand chemical space for some of these receptors16. Unfortunately, there is no crystal structure of active-state prostaglandin receptor bound to an agonist available to date. Moreover, the understanding of the precise molecular mechanism of binding and selectivity of agonists on EP3 is very limited but essential for further development of alternative agonists scaffold selectively targeting this receptor.
Here, we present the high-resolution crystal structure of the misoprostol in its free acid form bound to the human EP3 receptor. Using the combination of site-directed mutagenesis, molecular docking of selective and non-selective agonists, receptor signaling and binding assays, the key structural elements of EP3 agonists binding, selectivity and agonist-mediated receptor activation are revealed. This structure provides a first atomic description of the action of prostaglandins and their close derivatives on a prostanoid receptor and rational grounds as the basis for the design of a cost-effective, next-generation pharmacological tools aimed at reducing maternal mortality across the world.
RESULTS
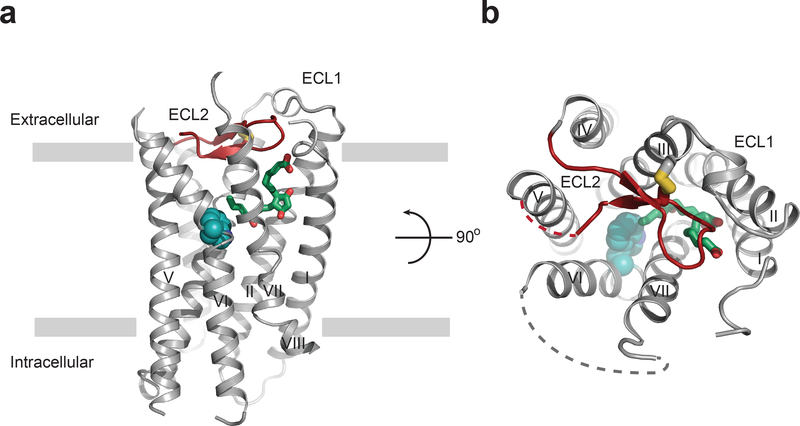
Active-like conformation of EP3 receptor structure
To obtain the structure of misoprostol-FA in complex with the EP3 receptor (EP3-miso), we modified the native receptor sequence by removing the initial methionine, replacing the third intracellular loop (ICL3) with T4-lysosyme, introducing a Gly2866.39Ala mutation17, and truncating the C-terminus after Leu3538.50 (see Methods; Supplementary Fig. 2). Then we crystallized EP3-miso in lipidic cubic phase, collected diffraction data using an X-ray free electron laser at room temperature, and determined the structure at 2.5 Å resolution (Fig. 1; Supplementary Fig. 2; Supplementary Table 1). The EP3-miso structure exhibited classic GPCR architecture, including a seven-transmembrane (7TM) helical bundle (helices I-VII) linked by three extracellular (ECL1–3) and three intracellular (ICL1–3) loops and a truncated helix VIII (Fig. 1; Supplementary Fig. 2). Interestingly, the partial helix VIII displays non-classical orientation relative to the 7TM bundle. Indeed, modelling of the rest of the helix VIII and C-terminus in a classical position in our structure would clash with the contact of an adjacent EP3 monomer in the crystal and support the notion that the truncation of the C-terminus was necessary for EP3 crystallization (Supplementary Fig. 2d). This truncation also removes the C-terminal sequence that discriminates between the human EP3 isoforms18. Overall, the receptor modifications did not affect the binding affinity of misoprostol-FA or PGE2, and neither the Gly2866.39Ala mutation nor the C-terminal truncation affected agonist-induced EP3 signaling (Supplementary Fig. 3a-c; Supplementary Table 2–4). We compared the EP3 receptor’s intracellular helical arrangements and micro-switches (conserved sub-structures in class A GPCRs that undergo rearrangement upon receptor activation) with the inactive- and active-state structures of another lipid-binding GPCR, the cannabinoid receptor Cb1 (37% sequence similarity, 20% identity in the 7TM domain)19,20. This analysis revealed that the EP3-miso structure shares the conformational features of an active-like structure, such as the intracellular opening at helix VI and N/DPxxY motif active-state configuration (Supplementary Fig. 4), consistent with the agonist efficacy of misoprostol-FA.
Figure 1|. Overall structure of EP3 receptor bound to misoprostol-FA.
(a) EP3 receptor viewed parallel to the plasma membrane, and (b) from the extracellular side looking down into the misoprostol-FA binding pocket. Misoprostol-FA (green carbon) is shown as sticks. Disordered parts of loops are represented as dashed lines. ECL2 is shown in red. The canonical ‘toggle-switch’ Trp2956.48 is shown with cyan atom spheres for reference.
Misoprostol-FA binds EP3 receptor using 3 subpockets
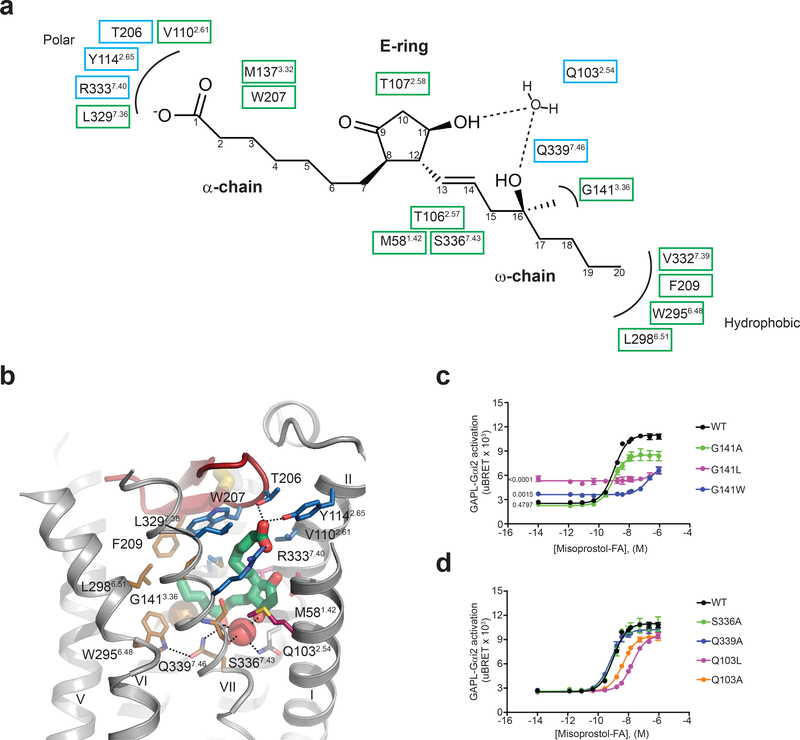
The misoprostol-FA binding pocket in EP3 has a volume of 778 Å3, which is one of the smallest, behind 11-cis-retinal-bound in rhodopsin (604 Å3) (PDB code 1u19). Seventeen EP3 residues from helices I, II, III, VI, and VII and ECL2 are within 4 Å of misoprostol-FA, clustering in three different sub-pockets that coordinate its α-chain, ω-chain, and E-ring (Fig 2a and b). The polar sub-pocket consisting of residues Arg3337.40, Tyr1142.65, and Thr206 forms a network of hydrogen bonds and ionic interactions with the acidic group of the α-chain, consistent with the observed effect of residue mutation on agonist binding21,22. In addition, Leu3297.36, Val1102.61, Met1373.32, and Trp207 form hydrophobic interactions with the α-chain. The ω-chain protrudes into a hydrophobic sub-pocket consisting of residues Gly1413.36, Trp2956.48, Leu2986.51, Val3327.39, Ser3367.43, and Phe209. There is a single polar interaction between Gln3397.46 and the hydroxyl group at carbon 16. The E-ring of misoprostol-FA interacts with residues Thr1072.58, Thr1062.57, and Met581.42 through hydrophobic interactions.
Figure 2|. Detailed structure of the misoprostol-FA binding pocket and interactions of the ligand with EP3 receptor.
(a) Two-dimensional view of receptor interactions with misoprostol-FA. Polar and hydrophobic interactions are shown in blue and green boxes, respectively. (b) Detailed view of misoprostol-FA interactions. Polar interactions are shown as dotted lines. Misoprostol-FA is shown as green sticks. Water and Gly1413.36 are shown as red and gold spheres, respectively. Side chains coordinating the α-chain, ω-chain, and E-ring regions are shown as blue, gold, and maroon carbons, respectively. (c,d) Misoprostol-FA Gi signaling on wild-type (WT) and EP3 receptor mutants. Values are mean ± SEM of 6 and 3 independent experiments for the WT and mutants, respectively. Significance of the basal value was determined using one-way ANOVA analysis followed by a Dunnett post-hoc test against the mean basal WT signaling. For the ANOVA, p < 0.0001, F is 62.07, among and within group degrees of freedom are 3 and 11, respectively. Post-hoc test p values are indicated on the graph. Mean pEC50 ± SEM are shown in Supplementary Table 3.
Prostaglandin-induced signaling mechanism
Fifteen of the seventeen residues forming the misoprostol-FA binding pocket were mutated and tested for misoprostol-FA binding affinity and in all but two cases, a significant reduction was observed (p < 0.0001 for all 15 residues) (Supplementary Fig. 3b, d-f; Supplementary Table 2). The two mutations that did not affect binding affinity (Leu3297.36Ala and Val1102.61Ala) decreased misoprostol-FA signaling potency by ~100- and 5-fold, respectively (Supplementary Table 3). Gly1413.36 reduced misoprostol-FA potency by three orders of magnitude after mutation of the side chain to progressively larger amino acids (leucine and tryptophan) while increasing EP3’s basal signaling by as much as 2.5 times that of wild-type. This suggests that occupancy of the ω-chain binding region is important for activation of EP3 (Fig. 2c; Supplementary Table 3). Interestingly, we found no direct interaction between the protein and the E-ring hydroxyl group despite its importance in PGE2-induced receptor activity23 and its presence in most synthetic EP3 agonists7 (Supplementary Fig. 1). The nearby residue Ser3367.43 forms a hydrogen-bond with the main chain carbonyl of Val3327.39, avoiding interactions with the E-ring (Fig. 2a, b). Instead, a structured water molecule at the center of a hydrogen bond network bridges both hydroxyl groups of misoprostol-FA with the main chain carbonyl of Ser3367.43 and the side chain of Gln1032.54 (Fig. 2a, b). Supporting this observation, alanine mutation of Gln1032.54 causes a significant reduction in affinity and potency (p < 0.0001 for both) of misoprostol-FA even without direct interactions with the ligand, while Ser3367.43Ala mutation does not affect signaling (Fig. 2d; Supplementary Table 3).
The binding pocket of misoprostol-FA is totally enclosed
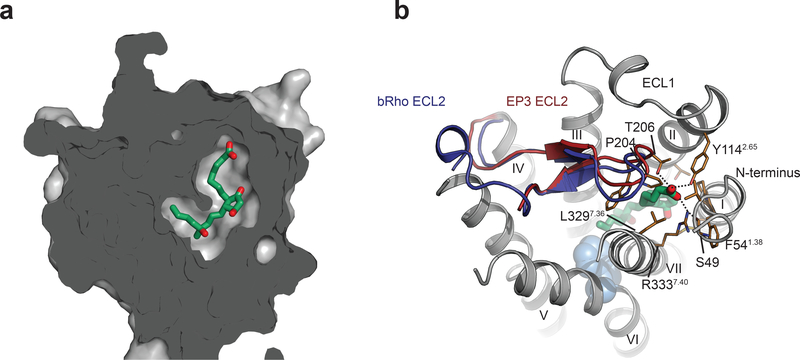
Close inspection of the EP3-miso structure reveals several key features distinguishing the receptor from other lipid-binding GPCRs. In contrast to previously solved structures of lipid-binding receptors, the binding site of EP3 is completely occluded (Fig. 3a). ECL2 blocks ligand access from the extracellular side similarly to that observed in bovine rhodopsin, and unlike other lipid receptors, the remaining extracellular loops do not appear to directly participate in the extracellular lid (Fig. 3b). Interactions formed by the negatively charged carboxyl of misoprostol-FA bring together residues from the N-terminus with the extracellular tips of helices II and VII, as well as ECL2 over the top of the ligand, closing the lid after its entrance into the pocket. It is not clear however if this is the result of an induced fit occlusion of the binding pocket or that the misoprostol enters the binding pocket through a putative channel between two helices of the transmembrane domain. The latter mechanism has already been reported previously for other lipid receptors24. Nevertheless, the small agonist bound in an enclosed state likely contributes to the long misoprostol residence time (Supplementary Fig. 5). The Arg3337.40 side chain, conserved among the prostaglandin receptor subfamily, plays the key role in this interaction network by forming a salt bridge to the ligand’s carboxyl, while making additional stacking interactions with Phe541.38, and a hydrogen bond to the main chain carbonyl of the N-terminal residue Ser49 (Fig. 3b). Interestingly, alanine mutation of all residues in the interacting network, affects signaling with the most pronounced effect for the Arg3337.40Ala mutant (p < 0.0001) (Supplementary Table 3). Overall, the configuration of prostaglandin binding pocket on EP3 is different from any other GPCR solved to date and was not predicted by previous molecular model25. This further confirm the validity of our approach and suggest the importance of our structural finding for a predictive rationalization of new EP3 ligands.
Figure 3|. The binding pocket of misoprostol-FA in EP3 receptor is totally enclosed.
(a) Sliced surface representation of the misoprostol binding pocket. (b) Extracellular view of EP3 receptor with its ECL2 (red) overlaid with the one of bovine rhodopsin (dark blue) (PDB code 1gzm). Trp2956.48 is shown with cyan spheres for reference. Misoprostol-FA is shown as green sticks. EP3 residues are shown as gold stick. Polar interactions are shown as dotted lines.
Prostaglandins-derivatives selectivity on EP3 receptor
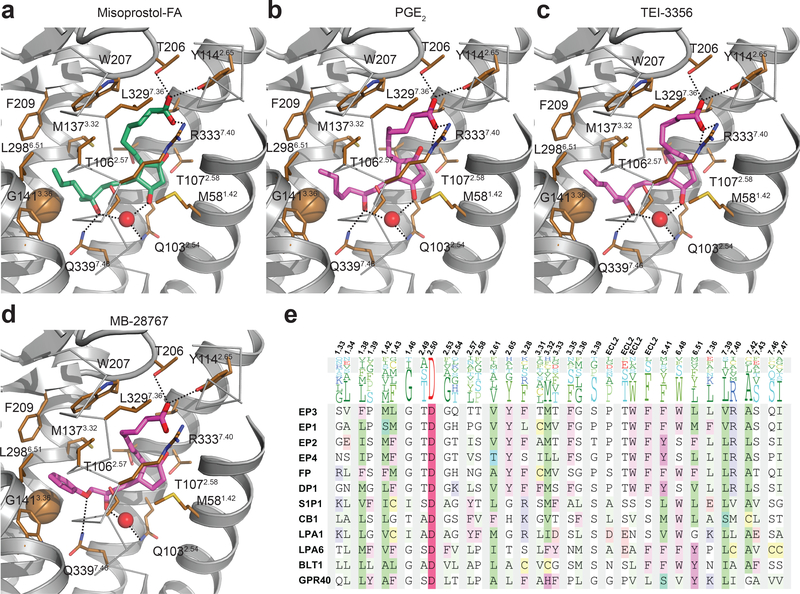
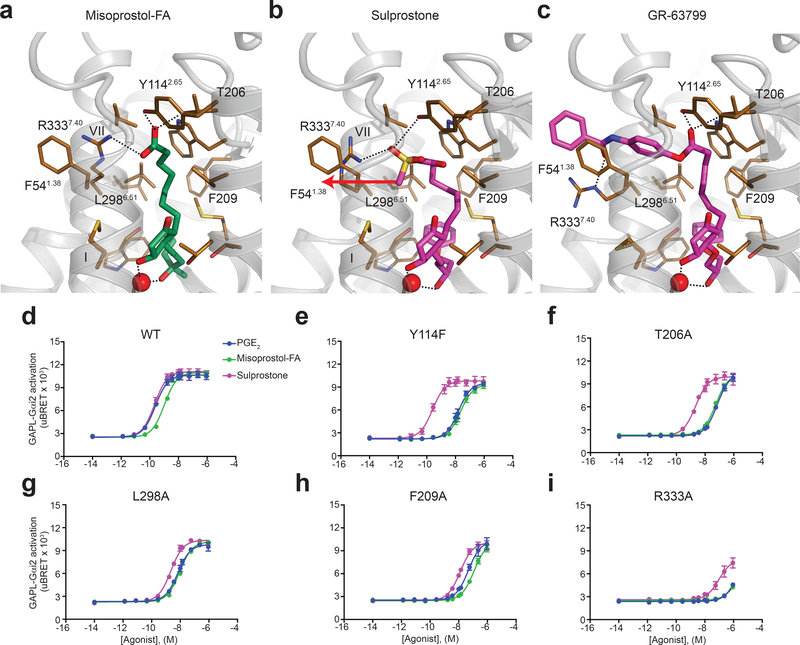
Achieving a specific drug-induced activation of EP3 is important for reducing adverse side-effects and improving safety and clinical efficacy. As such, there are a few EP3-specific agonists that have been developed that are lipid-like and very similar to prostaglandins. The effect of chemical substitutions on EP receptor selectivity has been described previously26–28. In contrast to the natural lipids PGE1 and PGE2, prostaglandin derivatives can achieve strong selectivity toward EP3 by adding a branched carbon 16 (misoprostol-FA, TEI-3356, 16,16-dimethyl-PGE2), a bulky group at the extremity of the ω-chain (MB-28767, sulprostone, GR-63799), and by the elongation of the α-chain beyond the acid functionality (sulprostone, GR-63799) (Supplementary Fig. 1)26–28. We took advantage of our crystal structure to understand these chemical selectivity features at the receptor level. Docking of these prostaglandin derivatives on EP3-miso structure shows that the methyl group of the branched carbon on the ω-chain contacts Gly1413.36 constraining the orientation of the rest of the hydrophobic tail in the pocket (Fig. 4a-c), and that the additional bulky group at the end of the ω-chain forms additional hydrophobic interactions (Fig. 4d). Alignment of prostaglandin E2 receptors reveals the presence of methionine and phenylalanine at position 6.51 of the EP1 and EP2 receptors, respectively (Fig. 4e). This additional size of these side chains in the hydrophobic pocket could clash with the ω-chain, when it is constrained or modified with a bulky chemical group, conferring selectivity toward EP3 and EP4. Interestingly, the enclosed receptor’s sub-pocket does not accommodate α-chain extensions as found in some EP3-selective ligands such as sulprostone and GR-63799 (Supplementary Fig. 1). Consequently, the binding pocket must have some degree of plasticity in this region. In order to dock sulprostone and GR-63799, we simulated the induced-fit in the pocket by allowing side chain flexibility of the contact residues. The docking simulations suggest that in the absence of a charged carboxyl group, the side chain of Arg3337.40 is more dynamic, opening a channel for the bulky extensions (Fig. 5a-c). Unlike small carboxyl-containing ligands, sulprostone no longer interacts with Thr206 and has suboptimal interaction with Tyr1142.65 (Fig. 5b), correlating with the reduced effects of mutating these residues on sulprostone potency (Fig. 5d-i; Supplementary Table 5). The larger 4-benzamidophenyl extension in GR-63799 can protrude outside of the pocket through a channel between helices I and VII, opened by repositioning of the Arg3337.40 side chain (Fig. 5c). Protrusion of ligands between transmembrane helices has precedence in other structures of lipid-binding receptors, such as the crystal structure of GPR40 in complex with the agonist TAK-875 29. Although GR-63799 does not form polar contacts with Arg3337.40, it still maintains the common hydrogen bond network with Thr206 and Tyr1142.65 side chains (Fig. 5c). Generally, the protruding bulky groups could increase EP3 selectivity by forming new selective interactions with non-conserved residues outside the orthosteric binding pocket, e.g. stacking with Phe541.38 (Fig. 4e; Fig. 5c). The binding pocket flexibility required for binding these ligands strengthens the hypothesis that this opening may provide dynamic access for endogenous and synthetic ligands to the orthosteric pocket. Taken together, analysis of EP3 selective prostaglandin-derivative agonists provides the fundamental information needed to guide the selective design of novel ligand scaffolds at the EP3 receptor level.
Figure 4|. Docking of small agonists on the EP3 receptor.
(a) Misoprostol-FA in the EP3-miso structure. (b-d) Docking of the indicated agonists on EP3-miso crystal structure. EP3 receptor is show as cartoon except helix VII shown as ribbon. Misoprostol-FA and modelled agonists are in green and magenta sticks, respectively. Water and Gly1413.36 are shown as red and gold spheres, respectively. Dotted lines represent polar interactions. (e) Sequence alignment of the EP3 receptor binding pocket with other lipid receptors.
Figure 5|. Modelling of sulprostone and GR-63799 on EP3 receptor.
(a) Misoprostol-FA in the EP3-miso structure. (b,c) Docking of sulprostone (b) and GR-63799 (c) in the EP3-miso crystal structure. Misoprostol-FA and sulprostone/GR-63799 are in green and magenta sticks, respectively. The arrow represents the direction of displacement of the end of sulprostone’s α-chain compared to misoprostol-FA’s α-chain location in the EP3-miso structure. Dotted lines represent polar interactions. (d-i) Agonist-mediated signaling on EP3 receptor WT and mutants. Values represent the mean ± SEM of 3 independent experiments. PGE2 and misoprostol-FA mean pEC50 ± SEM are shown in Supplementary Table 3. The mean pEC50 ± SEM (M) for sulprostone is shown in Supplementary Table 5.
DISCUSSION
Overall, the EP3-miso structure presented here is the first atomic description of a prostanoid receptor bound to a close prostaglandin analogue and provides a high-resolution template for understanding ligand binding to prostaglandin receptors. The small, encapsulated binding site may be a special feature of misoprostol, PGE2, and similar prostaglandins resulting from induced formation of the lid occluding access after ligand binding. Alternatively, a putative ligand channel similar to what was found in other lipid receptors may participate in the ligand binding. The prostaglandin-coordinated lid likely impacts the kinetics of binding, and explains our observation that misoprostol displays a long residence time on the EP3-T4L crystallization construct (Supplementary Fig. 5). This is consistent with early reports of prostaglandin binding kinetics which have shown that prostaglandins would not dissociate completely from cells or tissues expressing EP3 receptor after more than 1 hour of equilibrium, including studies of membrane preparations of human myometrium30–32. Moreover, kinetics of prostaglandin dissociation from the EP3 receptor are prolonged by the coupling of the heterotrimeric Gi protein, supporting the notion that the induced-fit or coordinated-lid could be a feature of the stimulated receptor’s activated-state 30,31. Side-effects associated with the clinical use of misoprostol range from diarrhea to uterine hyper stimulation, fetal heart abnormalities, fetal death and uterine rupture; the more serious side-effects generally occur after high dose regimens used in obstetrics and pregnancy, both were contra-indicated for misoprostol use until 200212. Some of the side-effects associated with hyper-stimulation could be inferred from the structure, as the pharmacokinetic properties of the compound may not be predictive of its receptor occupancy due to the prostaglandin-coordinated lid on the receptor.
The presence of a water molecule appears to be an important feature for the E-ring coordination and induction of signal transduction in the binding site of EP3. To our knowledge, this has not been previously reported. Generally, water molecules can only be resolved and identified in crystal structures at resolutions at and below that of our study. The detection of this important feature using our approach could have been a challenge as most GPCR structures using x-ray crystallography and electron microscopy are solved at resolution above water detection15,33. In our study, the ability to detect the water associated to the E-ring revealed to be important to understand misoprostol action on EP3. Indeed, the ring substructures is the only difference between natural prostanoids and PGE2 E-ring hydroxyl is only found at analogous position in the F-ring of the prostaglandin F2α (PGF2α). Interestingly, in addition to acting on its receptor (FP), PGF2α still displays high affinity for EP3 and not the other EP receptors26. High-resolution prostaglandins-bound crystal structure of the other EPs and especially FP receptor would be required to validate the more general role of the water molecule at coordinating the ring structure across the prostanoid receptors subfamily and its role in prostanoid receptors’ selectivity.
The small and enclosed prostaglandin orthostheric binding pocket of EP3 suggests that alternative drug design strategies should be envisioned that allow a more diverse array of chemical scaffold to modulate the receptor. As such, an allosteric ligand has already been developed for the FP receptor34 and few structural elements on the EP3-miso structure suggest it could be a successful avenue for EP3. First, the sidechains of the binding site residues Arg3337.40 and Met581.42 are uniquely exposed to outside space of the transmembrane bundle, while interacting with misoprostol-FA (Fig. 2a; Fig. 3b; Fig. 4a-d; Fig. 5a-c). Additionally, other residues such as Ile3407.47 and Phe541.38 not interacting with misoprostol and located in the same interface of the transmembrane bundle are involved in the agonist-induced activation of EP3, as alanine mutations of these positions reduce the potency of misoprostol-FA and PGE2 (Fig. 3b; Fig. 5a-c; Supplementary Table 3).
Identifying new scaffolds that retain the benefits associated with misoprostol’s usage in the management of pregnancy in low-resource countries, particularly its affordability, stability, ease of administration, but also reducing associated health risks should be within reach utilizing the structure of the EP3 receptor as a starting point for further discovery. As such, the misoprostol-FA EP3 receptor complex structure provides the first essential breakthrough for future medicinal chemistry efforts to tackle life-threatening complications during childbirth for millions of women in developing countries.
ONLINE METHODS
Cloning of EP3 Receptor
Isoform A of the human EP3 receptor was codon-optimized for Spodoptera frugiperda (Sf9) expression, synthesized (Genewiz), and subcloned in pFastBac1 and pcDNA3.1 vectors. For the pFastBac1 and the pcDNA3.1 derived constructs, the coding sequence harbours an HA signal peptide and an affinity FLAG tag at the N-terminus, and a precision protease sequence followed by a 10xHis affinity tag at the C-terminus. Several modifications were introduced to the receptor in the crystallization construct to improve its expression, stability, and crystallization. The receptor’s initial methionine was removed, the C-terminus was truncated after residue Leu3538.50, Gly2866.39 was mutated to an alanine, residues 260–272 from ICL3 were replaced by a cysteine-free T4-lysosyme (T4L) harboring an Ala73Thr mutation, and a glycine-serine linker was included on each side of the junction sites yielding the final EP3-T4L crystallization construct (Supplementary Fig. 2). The mutation Gly2866.39Ala was one of the mutations designed by the machine learning-based software CompoMug35 and rationalized to favor a helical conformation at the bottom of helix VI. Although we did not observe improved receptor thermostability for the Gly2866.39Ala mutation, it improved protein expression and crystal resolution from previously low-resolution diffracting crystals obtained without the mutation. For the radioligand binding assays and signaling assays, mutations were derived from the pcDNA3.1-EP3 isoform A construct by site-directed mutagenesis.
Expression and Purification of EP3 Receptor
We expressed the EP3-T4L protein in Sf9 insect cells (Invitrogen) using the Bac-to-Bac Baculovirus Expression System (Invitrogen) in the presence of 1 μM misoprostol methyl ester (Cayman Chemical). Membranes from cells expressing EP3-T4L were prepared using 3 rounds of washing and ultracentrifugation at 250,000 g, first in the presence of lysis buffer containing 10 mM HEPES, pH 7.5, 10 mM MgCl2, 20 mM KCl, and EDTA-free protease inhibitor cocktail tablets (Roche), and then twice with a washing buffer containing 10 mM HEPES, pH 7.5, 1 M NaCl, 10 mM MgCl2 and 20 mM KCl.. We resuspended the purified membranes using 25 ml lysis buffer supplemented with 20% glycerol and then incubated with EDTA-free protease inhibitor cocktail (Roche), 2 mg/ml iodoacetamide (Sigma) at room temperature for 1 h, then 30 mins at 4 °C. The receptor was then solubilized in 50 mM HEPES, pH 7.5, 800 mM NaCl, 1% (w/v) n-dodecyl-b-D-maltopyranoside (DDM, Anatrace), and 0.2% (w/v) cholesteryl hemisuccinate (CHS, Sigma) at 4 °C for 2 h. The supernatant was isolated by ultracentrifugation at 350,000 g for 60 min and then incubated with TALON resin (Clontech) overnight at 4 °C. The TALON resin was washed with 20 column volumes of wash buffer 1 containing 50 mM HEPES, pH 7.2, 150 mM NaCl, 10 mM MgCl2, 8 mM ATP, 0.1% (w/v) DDM, 0.01% (w/v) CHS, 10% (v/v) glycerol, and 20 mM imidazole and followed by 10 column volumes of wash buffer 2 containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 10% (v/v) glycerol, and 20 mM imidazole. We eluted EP3-T4L protein using 2.5 column volumes of elution buffer containing 50 mM HEPES, pH 7.5, 200 mM imidazole, 10% (v/v) glycerol, 150 mM NaCl and 0.01% (w/v) DDM, 0.002% (w/v) CHS. His-tagged PNGase F (custom-made) was added to the eluted sample to deglycosylate the receptor. Finally, we used a 100 kDa molecular weight cut-off concentrator (Sartorius) to concentrate EP3-T4L protein at a concentration of 20–30 mg/ml. The de-esterification of misoprostol ester by cellular esterase in the cell culture step yielding to the sole presence of the free acid form in the purified sample was confirmed by mass spectrometry (see Methods; Supplementary Fig. 5). No additional ligand was used during receptor extraction, purification, and crystallization.
Lipidic Cubic Phase Crystallization and Data Collection
We used a lipid syringe mixer to reconstitute EP3-T4L protein in a mixture of monoolein (9.9 MAG) (Sigma) lipid supplemented by 10% (w/w) cholesterol (Avanti Polar Lipids) at a protein-lipid ratio of 2:3 (v/v)36. We then performed crystallization trials by dispensing 40 nl of protein-laden LCP overlaid with 800 nl of precipitant per well in a 96-well glass sandwich plates (Marienfeld) using a NT8-LCP robot (Formulatrix). Plates were stored at 20 °C and imaged using a RockImager 1000 (Formulatrix). The EP3-miso crystals grew to a maximum size of 250 × 150 × 20 μm after 10 d at 20 °C in the presence of 100 mM sodium citrate pH 3.8–4.2, 10–35 mM magnesium sulfate, 20–23% (v/v) PEG 400, 2.5% Jeffamine M-600. Preliminary screening of these crystals at the Advanced Photon Source synchrotron showed diffraction to 2.6 Å resolution. However, a poor electron density in the binding pocket did not allow for modeling of the ligand. Then, microcrystals of EP3-miso for serial femtosecond crystallography (SFX) were grown in syringes37 by injecting 5 μl of LCP-containing receptor into 100 μl gas-tight syringes (Hamilton) filled with 60 μl of precipitant solution (100 mM sodium citrate pH 4.2, 45 mM magnesium sulfate, 21% (v/v) PEG 400, 2.5% Jeffamine M-600). The crystals grew to an average size of 20 × 10 × 5 μm within 10 d at 20 °C. The excess precipitant was then removed from the syringes and the remaining LCP containing crystals was consolidated. Finally, the micro-crystals were loaded into an LCP injector38, Approximately 5% (v/v) 9.9 MAG and 5% (v/v) 7.9 MAG were added and mixed with the LCP to absorb the residual precipitant solution and prevent the formation of a lamellar crystalline phase due to rapid evaporative cooling when injecting LCP into vacuum38.
Diffraction Data Collection Using X-ray Free-Electron Laser
CXI instrument at the Linac Coherent Light Source (LCLS) at SLAC National Accelerator Laboratory was used to collect SFX diffraction data with individual X-ray pulses of 40 fs durations at a wavelength of 1.302 Å (9.52 keV). A pair of Kirkpatrick–Baez mirrors was used to focus approximately 1011 photons per pulse onto a spot size of approximately 1.5 μm in diameter. Microcrystals of EP3-miso in the LCP media were extruded in the sample vacuum chamber at room temperature using a 50 μm nozzle LCP injector running at a flow rate of approximately 220 nl/min38. Diffraction images were recorded at a rate of 120 frames s−1 with the 2.3 Megapixel Cornell-SLAC Pixel Array Detector (CSPAD). Crystal hit frames were identified with the Cheetah program39, and diffraction patterns were indexed, integrated, and merged using the version 0.6.2 of the CrystFEL software suite40 with the pushres parameter set to 1.6 nm−1.
Structure Determination
The structure was solved initially by molecular replacement implemented in Phaser41 using dataset collected at LCLS, an active-state model of EP3, and a model of T4L where all side chains were trimmed to alanine. The EP3 model was created by aligning the receptor sequence with the nanobody-bound β2-adrenergic receptor structure (PDB code 3p0g). The T4L model was derived from (PDB code 2rh1). A molecular replacement search identified one receptor and one T4L in the asymmetric unit with a TFZ > 9. Refinement and model completion were done by repetitive cycling between PHENIX42 and BUSTER43, followed by manual examination and rebuilding of the coordinates in Coot44, using both |2Fo| - |Fc| and |Fo| - |Fc| maps. Ramachandran plot analysis of the final structure with Molprobity45 indicates that 100% of the residues are in either favored (95.4%) or allowed regions (4.6%), respectively (no outliers). The final data collection and refinement statistics are shown in Supplementary Table 1. Fourteen percent of the residues in the final structure represent residues that are RSRZ (real space R-factor Z-score) outliers in the PDB validation report. However, manual inspection of 2Fo-Fc maps of these residues confirms the correct fit of these residues in the electron density. The overall B-factor of the structure is 128 Å2, which is higher than average for a 2.5 Å resolution structure. The T4L fusion protein displays the highest B-factor values of the protein domain (146.8 Å2), probably because of the insertion of flexible Glycine-Serine linkers between the T4L and the receptor, and contributing to the high overall B-factor. The linkers were essential for the crystallization. In contrast, the EP3 receptor displays a lower overall B-factor of 117 Å2, with the binding pocket and most of the transmembrane domain having B-factors in the 80–100 Å2 range (Supplementary Fig. 7, Supplementary table 1). B-factor is a measure of the static and dynamic atomic disorder of a structure, but a high B-factor could also indicate a poor fit of the model in the electron density. In our case, the model fits the electron density very well with overall real space correlation coefficients (RSCCs) of 0.956 and 0.856 for the receptor main chain and side chains, respectively, and 0.952 and 0.857 for the T4L main chain and side chains, respectively. Importantly, the B-factor distribution is consistent with the structure without any anomalies. Further, membrane protein structures solved by serial crystallography have typically shown higher B-factors than structures solved by regular synchrotron data collection (Supplementary Fig. 8)46, and is currently the subject of investigation and discussion among experts in the field. Multiple factors could contribute to this, such as data collection temperature (room vs cryogenic temperatures), the merge of thousands of crystals in serial crystallography vs one or few crystals for traditional synchrotron data, anisotropy (which was not observed in the EP3 structure) and differences in the data processing.
Final 2Fo-Fc and simulated annealing Fo-Fc omit maps were created using Phenix to validate the fit of the electron density to the protein residues and misoprostol-FA in the binding site (Supplementary Fig. 6). Residues 1–45, 213–223 and 309–319 of the N-terminus, ECL2 and ECL3, respectively, were disordered and were not included in the final EP3-miso structure model (Fig. 1; Fig. 2b). Immediately C-terminal to the truncation at residue Leu3538.50, we were able to model a continuous 1.5-turn α-helical structure consisting of a non-native sequence of the precision protease cleavage site that is used as a linker to the polyhistidine affinity tag (Supplementary Fig. 2)17. Some residues have a missing sidechain: Cys47, Tyr77, Arg80, Glu81, Ser82, Arg84, Phe88, Arg119, His122, Leu179, Asn 224, Arg275, Lys304, Glu320, Lys321, and Lys323 in the receptor, and Arg 2001, Glu2004, Val2005, and Leu2006 in the C-terminal non-native linker. The presence of the precision protease site does not appear to affect the overall structure of the receptor. The closest tertiary interaction near this site (between side chains of Glu2796.32 and Leu4 of the linker) is approximately 3.5 Å away, and no direct interaction could be observed between the linker, the receptor or other receptor monomers in the crystal lattice (Supplementary Fig. 2c, d). The final EP3-miso complex contains 273 residues of EP3, 160 residues of T4L, and 7 residues of a C-terminal precision protease sequence.
Molecular Modelling
Membrane position was predicted by OPM database47. Selective EP3 agonists were docked into the orthosteric pocket of the EP3 receptor. Ligand structures were generated from 2D representations and their 3D geometry was optimized using MMFF-94 force field. The energy-based docking was performed using a protocol from ICM-Pro software (Molsoft) in the rectangular box that comprised the EP3 orthosteric pocket. Docking simulations used biased probability Monte Carlo (BPMC) optimization of the compound’s internal coordinates in the pre-calculated grid energy potentials of the receptor. The exhaustive sampling of the compound conformational space was done with the thoroughness parameter set to 10 and performing at least three independent docking runs for each compound. Docking poses from binding simulation of misoprostol-FA largely overlap with misoprostol-FA in the crystal structure (RMSD of 0.30 Å).All ligands except sulprostone and GR-63799, afforded optimal docking into the grid potential EP3 receptor model. For sulprostone and GR-63799, the bulkier chemical groups at the extremity of the α-chain required an induced-fit docking simulation by setting the side chains of the pocket lining residues in helices I, VII, and ECL2 as “explicitly flexible group” during the docking. The volumes of EP3-miso and Rhodopsin 11-cis-retinal (PDB code 1u19) binding pockets as well as the interaction of the residues in the binding site with misoprostol-FA were calculated in ICM-Pro software (Fig. 2a). All receptor alignments, molecular representations and distance measurements in the article were created using PyMol version 2.048.
Radioligand Binding Assay
The radioligand binding assay was adapted from a previously published protocol26. pcDNA3.1-EP3 receptor constructs were transfected in HEK293s cells, cultured in FreeStyle™ 293 expression medium (Thermo Fisher) at 37 °C with 5% CO2 cells, using 293fectin™ (Thermo Fisher). Forty-eight hours after transfection, cells were incubated with anti-FLAG FITC-labelled antibody (Sigma) in the absence and presence of 0.15% Triton X-100, and receptor surface and total cell expression were determined by fluorescence measurement using a Guava® flow cytometer (Millipore). Cellular membranes were prepared as described previously26 and radioligand binding was performed. Briefly, 125 μl of HEK293 membranes expressing WT or mutant EP3 receptors were incubated for 60 min at 30 °C shaking with 0.3 – 0.5 nM [3H]-PGE2 radioligand (Perkin-Elmer) and a range of competing ligand (Cayman Chemical) concentrations in a buffer containing 10 mM MES/KOH (pH 6.0), 1 mM EDTA, 10 mM MgCl2, 0.1% bovine serum albumin, 1% dimethylsulfoxide. Membrane concentration was adjusted to have approximately 2000 cpm at radioligand saturation. We then harvested the incubated membranes on 96-well filter mats treated with 0.3% polyethyleneimine using a 96-well Filtermate harvester (Perkin Elmer), and washed 3 times with a buffer containing 10 mM MES/KOH (pH 6.0) and 10 mM MgCl2. The filters were dried, heat sealed with a wax scintillant, and the radioactivity signal was determined using a MicroBeta2 Trilux scintillation counter (Perkin Elmer). Homologous competition binding was used to determine PGE2 dissociation constant (KD) values for WT and mutant receptors. Ki values were determined using [3H]-PGE2 competition with 12 concentrations of competing ligand. The highest Pearson correlation R coefficient was used to determine the best fit for 1 or 2 binding sites. The data were analysed by Prism 7 to give pKd and pKi values and reported as the mean ± SEM of independent replicated experiments as detailed in Supplementary Tables 2 and 4.
Measurement of EP3 Receptor Activation on the Gi2 Signaling Pathway
GAPL-Gi2 bioSensAll® is a bioluminescence resonance energy transfer (BRET)-based assay that monitors the activation of the heterotrimeric Gi2 protein specifically at the plasma membrane. Heterotrimeric Gi2 protein activation following receptor stimulation increases the BRET signal. GAPL-Gi2 biosensor coding plasmid and related information are the property of Domain Therapeutics NA Inc. (Cat # DTNA A29). All the BRET assays were done at Domain Therapeutics NA Inc. (Montreal, QC, Canada). Briefly, assays were performed in HEK-293T cells, cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Wisent) supplemented with 1% penicillin-streptomycin (Wisent) and 10% fetal bovine serum (Wisent) and maintained at 37 °C with 5% CO2. GAPL-Gi2 and pcDNA3.1-EP3 receptor constructs were co-transfected using 25-kDa linear polyethyleneimine (PEI) (Polysciences, Warrington, PA) at a mass PEI / DNA ratio of 3:1. Briefly, separate solution of diluted DNA and PEI in 150 mM NaCl were mixed and incubated at room temperature for more than 20 min. Then, DNA/PEI complexes were added to HEK-293T cells resuspended into cell culture medium at a density of 350,000 cells/mL followed by gentle mixing. Thirty-five thousand transfected cells were then distributed per well of a cell culture-treated 96-well plates (Greiner) and incubated 48 h at 37 °C. Then, cells were washed once with 100 μL of Tyrode-HEPES buffer (Sigma) and incubated with 100 μL of fresh Tyrode-HEPES buffer at room temperature for 1h. Then, e-Coelenterazine Prolume Purple (Methoxy e-CTZ; Nanolight) was added to a final concentration of 2 μM followed immediately by the addition of increasing agonist concentrations using the HP D300 digital dispenser (Tecan). After 10 min of incubation at room temperature, the BRET signal was subsequently detected at 0.2 sec integration time using a Synergy NEO plate reader (BioTek) equipped with emission filters centered at 400 ± 35 nm and 515 ± 10 nm for the BRET donor and acceptor channels, respectively. The BRET signal is the ratio of acceptor emission to donor emission. All BRET ratios were standardized using the equation below with pre-established BRET values for positive and negative BRET controls. The standardized BRET ratio is referred to as the uBRET = ((BRET ratio – A)/(B-A)) * 10,000 where A is the BRET ratio obtained from transfection of negative BRET control and B is the BRET ratio obtained from transfection of positive BRET control. The standardized BRET ratio is used to normalize and compare BRET signal obtained from different sets of filters and detectors.
Detection of Misoprostol Bound to EP3 by Mass Spectrometry
Purified EP3-T4L proteins that were expressed with misoprostol methyl ester or vehicle supplemented media were analyzed using mass spectrometry. Each sample was buffer exchanged into 150 mM ammonium acetate with 0.01% (w/v) DDM, 0.002% (w/v) CHS using a 100-kDa MW cut-off ultrafiltration device (Sartorius, Germany) by centrifugation at 13,000 × g for 10 min at 4 °C. After exchanging the buffer twice, the solution of protein complexes retained on the ultrafiltration membrane was transferred into a new centrifugal tube and treated with 100% methanol to denature the protein-ligand complexes. Released compounds were then separated from protein precipitates by centrifugation at 13,000 g for 20 min at 4 °C. The supernatant was dried out in a speed vacuum, and re-dissolved in 50% methanol before LC-MS analysis. Purified human A2A adenosine receptor served as negative control and underwent the same treatment as the specific EP3-T4L samples. Three experimental replicates were prepared and analysed for each pair of the EP3-T4L samples and the control.
All samples were analyzed on a TripleTOF 6600 mass spectrometer (AB SCIEX, USA) coupled to a Shimazu L30A UPLC system (Shimazu, Japan). Compounds were eluted from an Eclipse Plus C18 column (2.1 mm × 100 mm, 3.5 μm, Agilent, USA) at a flow rate of 0.4 mL/min, with the mobile phases of water/0.1% formic acid (A) and acetonitrile/0.1% formic acid (B). The LC gradient was as follows: 0–2 min, 5% B; 2–2.1 min, 5–20% B; 2.1–4 min, 20–35% B; 4–6 min, 35–60% B; 6–6.5 min, 60–90% B; 6.5–9.0 min, 90% B and re-equilibration for 3 min. Full-scan mass spectra were acquired in the range of 100–1000 m/z with ESI source settings: voltage 5.0–5.5 kV; gas temperature 500 °C; curtain gas 35 psi; nebulizer gas 55 psi; and heater gas 55 psi. Compound identification was based on accurate mass measurement (mass error <10 ppm) and retention time matching with the pure standard (RT shift <0.1 min). LC-MS chromatograms for specific compounds were extracted using Peakview 2.2 (AB SCIEX) according to the aforementioned criteria. The MS response of each compound is represented by the integrated peak area of the corresponding extracted LC-MS chromatogram. The S/C ratio refers to the ratio of the MS response of a specific compound detected in the EP3 receptor sample relative to that of the control. S/C >2 in all replicates indicate positive binding of the compound to the protein target49. The S/C ratios for misoprostol detection in EP3-T4L sample treated with misoprostol methyl ester during cell expression are 8.33, 9.62 and 6.63, confirming the presence of misoprostol acid. No MS signal of misoprostol could be detected from vehicle treated. No MS signal of misoprostol methyl ester could be detected in any samples (Supplementary Fig. 5).
Statistical analysis
One-way ANOVA statistical analysis was performed using GraphPad 7.0 software (Prims). Dunnett post-hoc test was used for comparison multiple samples with a single control. Tukey post-hoc test was used to compare multiple sample with multiple control. The replicate, error bars and P value, F value and degrees of freedom are indicated in the relevant figure, legends and table footnotes.
Data availability
The misoprostol-FA EP3 receptor complex structure coordinates and structure factors are available via the Protein DataBank (PDB) accession code 6m9t.
Supplementary Material
Acknowledgments
This research was supported by NIH R01 GM108635 (V.C.), R21 DA042298 (W.L.), R01 GM124152 (W.L.), the STC Program of the National Science Foundation through BioXFEL (No. 1231306) (U.W. and W.L.), the Russian Science Foundation (project no. 16–14-10273), and the GPCR Consortium. MA was supported by a Canadian Institute of Health and Research (CIHR) Postdoctoral Fellowship Award. CG acknowledges the Panofsky Fellowship from SLAC National Accelerator Laboratory and Stanford University for financial support. PP acknowledges the Russian Foundation for Basic Research (RFBR No.18–34-00990). Parts of this research were carried out at the LCLS, a National User Facility operated by Stanford University on behalf of the U.S. Department of Energy and is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC0276SF00515, and at the GM/CA CAT of the Argonne Photon Source, Argonne National Laboratory. We thank A. Walker for assistance in manuscript preparation; M. Chu, K. Villiers and C. Hanson for baculovirus expression and mammalian cell culture, and N. Sawyer for helpful suggestions. We are grateful to F. Badeaux and E. Audet-Badeaux for their encouragement and support.
Footnotes
Competing financial interests
B.B is an employee of Domain Therapeutics NA, a company focused on GPCR drug discovery that sells and licenses the cell signaling kit used in this study. All other authors declare no competing interests.
References (for main text only)
- 1.Ricciotti E & FitzGerald GA Prostaglandins and Inflammation. Arteriosclerosis, thrombosis, and vascular biology 31, 986–1000, doi:10.1161/atvbaha.110.207449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodward DF, Jones RL & Narumiya S International Union of Basic and Clinical Pharmacology. LXXXIII: Classification of Prostanoid Receptors, Updating 15 Years of Progress. Pharmacol Rev 63, 471 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Wang D & DuBois RN Eicosanoids and cancer. Nat Rev Cancer 10, 181, doi:10.1038/nrc2809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelson AD Antiplatelet therapies for the treatment of cardiovascular disease. Nat Rev Drug Discov 9, 154, doi:10.1038/nrd2957 (2010). [DOI] [PubMed] [Google Scholar]
- 5.O’Callaghan G & Houston A Prostaglandin E2 and the EP receptors in malignancy: possible therapeutic targets? Br J Pharmacol 172, 5239–5250, doi:10.1111/bph.13331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao C-M & Breyer MD Physiological Regulation of Prostaglandins in the Kidney. Annu Rev Physiol 70, 357–377, doi:10.1146/annurev.physiol.70.113006.100614 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Markovič T, Jakopin Ž, Dolenc MS & Mlinarič-Raščan I Structural features of subtype-selective EP receptor modulators. Drug Discov Today 22, 57–71, doi:https://doi.org/10.1016/j.drudis.2016.08.003 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Arulkumaran S et al. The roles of prostaglandin EP 1 and 3 receptors in the control of human myometrial contractility. J Clin Endocrinol Metab 97, 489–498, doi:10.1210/jc.2011-1991 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Kandola MK et al. EP2 Receptor Activates Dual G Protein Signaling Pathways that Mediate Contrasting Proinflammatory and Relaxatory Responses in Term Pregnant Human Myometrium. Endocrinology 155, 605–617, doi:10.1210/en.2013-1761 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Potts M, Prata N & Sahin-Hodoglugil NN Maternal mortality: one death every 7 min. Lancet 375, 1762–1763, doi:https://doi.org/10.1016/S0140-6736(10)60750-7 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Widmer M et al. Misoprostol as an adjunct to standard uterotonics for treatment of post-partum haemorrhage: a multicentre, double-blind randomised trial. Lancet 375, 1808–1813, doi:https://doi.org/10.1016/S0140-6736(10)60348-0 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Allen R & O’Brien BM Uses of Misoprostol in Obstetrics and Gynecology. Rev Obstet Gynecol 2, 159–168 (2009). [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai BS, Kessler LK, Stolzenbach J, Schoenhard G & Bauer RF Expression of gastric antisecretory and prostaglandin E receptor binding activity of misoprostol by misoprostol free acid. Dig Dis Sci 36, 588–593 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Orobaton N et al. Implementing at-scale, community-based distribution of misoprostol tablets to mothers in the third stage of labor for the prevention of postpartum haemorrhage in Sokoto State, Nigeria: Early results and lessons learned. PLOS ONE 12, e0170739, doi:10.1371/journal.pone.0170739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang J et al. Successful Strategies to Determine High-Resolution Structures of GPCRs. Trends Pharmacol Sci 37, 1055–1069, doi:https://doi.org/10.1016/j.tips.2016.09.009 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ngo T et al. Identifying ligands at orphan GPCRs: current status using structure‐based approaches. Br J Pharmacol 173, 2934–2951, doi:10.1111/bph.13452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballesteros JA & Weinstein H in Meth Neurosci Vol. 25 (ed Sealfon Stuart C.) 366–428 (Academic Press, 1995). [Google Scholar]
- 18.Schmid A, Thierauch KH, Schleuning WD & Dinter H Splice variants of the human EP3 receptor for prostaglandin E2. Eur J Biochem 228, 23–30 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Hua T et al. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 547, 468–471, doi:10.1038/nature23272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua T et al. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 167, 750–762.e714, doi:https://doi.org/10.1016/j.cell.2016.10.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Audoly L & Breyer RM The second extracellular loop of the prostaglandin EP3 receptor is an essential determinant of ligand selectivity. J Biol Chem 272, 13475–13478 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Audoly L & Breyer RM Substitution of charged amino acid residues in transmembrane regions 6 and 7 affect ligand binding and signal transduction of the prostaglandin EP3 receptor. Mol Pharmacol 51, 61–68 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Ungrin MD et al. Key structural features of prostaglandin E(2) and prostanoid analogs involved in binding and activation of the human EP(1) prostanoid receptor. Mol Pharmacol 59, 1446–1456 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Hanson MA et al. Crystal structure of a lipid G protein-coupled receptor. Science (New York, N.Y.) 335, 851–855, doi:10.1126/science.1215904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akasaka H et al. The key residue within the second extracellular loop of human EP3 involved in selectively turning down PGE2- and retaining PGE1-mediated signaling in live cells. Arch Biochem Biophys 616, 20–29, doi:https://doi.org/10.1016/j.abb.2016.12.001 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Abramovitz M et al. The utilization of recombinant prostanoid receptors to determine the affinities and selectivities of prostaglandins and related analogs. Biochim Biophys Acta 1483, 285–293 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Kiriyama M et al. Ligand binding specificities of the eight types and subtypes of the mouse prostanoid receptors expressed in Chinese hamster ovary cells. Br J Pharmacol 122, 217–224, doi:10.1038/sj.bjp.0701367 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negishi M et al. TEI-3356, a highly selective agonist for the prostaglandin EP3 receptor. Prostaglandins 48, 275–283, doi:https://doi.org/10.1016/0090-6980(94)90028-0 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Srivastava A et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature 513, 124, doi:10.1038/nature13494 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Negishi M et al. Functional interaction of prostaglandin E receptor EP3 subtype with guanine nucleotide-binding proteins, showing low-affinity ligand binding. Biochim Biophys Acta 1175, 343–350 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Hamon M et al. Modulation of human myometrial PGE2 receptor by GTP characterization of receptor subtype. Prostaglandins 46, 251–268 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Tsai BS, Kessler LK, Schoenhard G, Collins PW & Bauer RF Demonstration of specific E-type prostaglandin receptors using enriched preparations of canine parietal cells and [3H]misoprostol free acid. Am J Med 83, 9–14 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Liang YL et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature 555, 121–125, doi:10.1038/nature25773 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Goupil E et al. A novel biased allosteric compound inhibitor of parturition selectively impedes the prostaglandin F2alpha-mediated Rho/ROCK signaling pathway. J Biol Chem 285, 25624–25636, doi:10.1074/jbc.M110.115196 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popov P et al. Computational design of thermostabilizing point mutations for G protein-coupled receptors. eLife 7, e34729, doi:10.7554/eLife.34729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caffrey M & Cherezov V Crystallizing membrane proteins using lipidic mesophases. Nature protocols 4, 706–731, doi:10.1038/nprot.2009.31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Ishchenko A & Cherezov V Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nature protocols 9, 2123, doi:10.1038/nprot.2014.141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weierstall U et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature communications 5, 3309, doi:10.1038/ncomms4309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barty A et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. Journal of applied crystallography 47, 1118–1131, doi:10.1107/s1600576714007626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TA et al. CrystFEL: a software suite for snapshot serial crystallography. Journal of applied crystallography 45, 335–341, doi:doi:10.1107/S0021889812002312 (2012). [Google Scholar]
- 41.McCoy AJ et al. Phaser crystallographic software. Journal of applied crystallography 40, 658–674, doi:10.1107/s0021889807021206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography 68, 352–367, doi:10.1107/s0907444912001308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smart OS et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta crystallographica. Section D, Biological crystallography 68, 368–380, doi:10.1107/s0907444911056058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501, doi:10.1107/s0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 66, 12–21, doi:10.1107/s0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawaya MR et al. Protein crystal structure obtained at 2.9 A resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proc Natl Acad Sci U S A 111, 12769–12774, doi:10.1073/pnas.1413456111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomize MA, Pogozheva ID, Joo H, Mosberg HI & Lomize AL OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic acids research 40, D370–376, doi:10.1093/nar/gkr703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 2.0 (2015).
- 49.Chen X et al. Identification of inhibitors of the antibiotic-resistance target New Delhi metallo-beta-lactamase 1 by both nanoelectrospray ionization mass spectrometry and ultrafiltration liquid chromatography/mass spectrometry approaches. Analytical chemistry 85, 7957–7965, doi:10.1021/ac401732d (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The misoprostol-FA EP3 receptor complex structure coordinates and structure factors are available via the Protein DataBank (PDB) accession code 6m9t.