Abstract
Human APOBEC3H is a single-stranded (ss)DNA deoxycytidine deaminase that inhibits replication of retroelements and HIV-1 in CD4+ T cells. When aberrantly expressed in lung or breast tissue APOBEC3H can contribute to cancer mutagenesis. These different activities are carried out by different haplotypes of APOBEC3H. Here we studied APOBEC3H haplotype II, which is able to restrict HIV-1 replication and retroelements. We determined how the dimerization mechanism, which is mediated by a double-stranded RNA molecule, influenced interactions with and activity on ssDNA. The data demonstrate that the cellular RNA bound by APOBEC3H does not completely inhibit enzyme activity, in contrast to other APOBEC family members. Despite degradation of the cellular RNA, an approximately 12 nt RNA remains bound to the enzyme, even in the presence of ssDNA. The RNA-mediated dimer is disrupted by mutating W115 on loop 7 or R175 and R176 on helix 6, but this also disrupts protein stability. In contrast, mutation of Y112 and Y113 on loop 7 also destabilizes RNA-mediated dimerization but results in a stable enzyme. Mutants unable to bind cellular RNA are unable to bind RNA oligonucleotides, oligomerize, deaminate ssDNA in vitro, but ssDNA binding is retained. Comparison of A3H wild type and Y112A/Y113A by fluorescence polarization, single molecule optical tweezer, and atomic force microscopy experiments demonstrates that RNA mediated dimerization alters the interactions of A3H with ssDNA and other RNA molecules. Altogether, the biochemical analysis demonstrates that RNA binding is integral to APOBEC3H function.
Keywords: HIV-1, APOBEC3H, restriction factor, enzyme-DNA interactions, enzyme-RNA interactions
INTRODUCTION
The APOBEC family of enzymes are cytidine deaminases and are important for a variety of cellular processes [1, 2]. The namesake family member, APOBEC1, deaminates cytidine in various mRNAs, with its deamination activity on the APOB mRNA being the best characterized in terms of function [3]. APOBEC3A can also deaminate mRNAs in macrophages and monocytes [4]. All the other active APOBEC enzymes, including APOBEC1 and APOBEC3A, deaminate deoxycytidine in single-stranded (ss)1 DNA [1, 2]. The functions of the APOBEC3 enzymes (A-H, excluding E) are to use cytosine deamination to inactivate retroviruses and retroelements and destroy inflammation inducing or foreign DNA [5–8]. This occurs through the formation of uracil, which has many downstream effects leading to either mutagenesis or DNA degradation [5–8]. APOBEC3A is specifically expressed in monocytes and macrophages [9, 10]. The other APOBEC3 enzymes are expressed in CD4+ T cells [9, 10]. Another family member, Activation Induced Cytidine Deaminase (AID) is expressed in B-cells and initiates somatic hypermutation and class switch recombination through the formation of uracils in genomic DNA [11, 12].
Despite these essential roles in immunity and metabolism, the APOBEC family must be tightly regulated since aberrant expression in cells where they are normally expressed or other tissue types can lead to genomic mutagenesis and cancer progression [13–18]. Regulatory mechanisms to protect the genome from “off-target” APOBEC activity include cellular localization, protein stability, and RNA binding, the latter of which has been the least well characterized [19–24]. It was discovered in 2003 that in order to obtain catalytically active GST-AID in vitro, when purified from Sf9 insect cells, that the enzyme had to be treated with RNase A [23]. The RNase A treatment resulted in the degradation of cellular RNA that was bound to AID and inhibiting catalytic activity on ssDNA [23]. However, GST-AID produced from E. coli does not require RNase A treatment and AID activity is not enhanced by RNase A treatment [25]. Nonetheless, for many APOBEC enzymes degradation of bound cellular RNA disrupts formation of large molecular weight ribonucleoprotein complexes and is required for enzyme activity [24, 26, 27]. Recent studies have also suggested that the RNA may be required to inhibit APOBEC3 activity in human genomic DNA and promote cytoplasmic localization [21, 22]. Conversely, RNA binding is required for virion encapsidation of APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H (haplotypes II, V, and VII), as a necessary first step for their restriction of the retrovirus, HIV-1 [28, 29]. Although it is not yet known the applicability to other APOBEC3 family members, recent crystal structures of APOBEC3H have shown that a double-stranded (ds) RNA remains associated with the enzyme in the presence of RNase A and mediates APOBEC3H dimerization [21, 22, 30, 31].
APOBEC3H is the most variable APOBEC3 gene with five major single-nucleotide polymorphisms, that results in seven major haplotypes [20, 32]. Of the seven, only haplotypes II, V, and VII are stable and active against HIV-1 [20, 32]. APOBEC3H haplotypes III, IV, and VI are unstable in cells and become rapidly degraded [20, 32]. APOBEC3H haplotype I has an intermediate phenotype where it has a short half-life in cells, cannot restrict HIV-1, but is still catalytically active in cells and in vitro [13, 20, 32, 33]. The stable haplotypes can restrict the replication of HIV-1 by becoming virion encapsidated in infected CD4+ T cells and deaminating cytosines in HIV-1 (−) DNA when it becomes single-stranded during reverse transcription [6]. The resulting uracils template the addition of adenine upon (+) DNA synthesis, resulting in C/G to T/A mutations that can functionally inactivate the provirus [6]. This occurs if the APOBEC3 enzymes can avoid proteasomal degradation induced by HIV-1 Vif, which becomes the substrate receptor of a Cullin 5 E3 ubiquitin ligase complex in infected cells [6]. However, there are also examples where APOBEC3 enzymes do not fully inactivate HIV-1 and can contribute to viral evolution in the form of drug resistance or CTL escape [34–40]. Due to the variability of APOBEC3H in the human population, the Vif proteins of many HIV-1 strains cannot induce degradation of APOBEC3H and it can effectively suppress viral replication or act as an infection barrier, until viral adaptation occurs [41, 42]. In contrast, APOBEC3H haplotype I has no known beneficial function, but can deaminate genomic DNA in breast and lung cancers, leading to increased genetic diversity of tumors [13]. APOBEC3 enzymes are Zinc co-ordinating deaminases and APOBEC3H also has a unique Zinc co-ordinating domain from the other APOBEC3 enzymes, based on amino acid sequence similarity [43].
We sought to characterize the RNA mediated dimerization of APOBEC3H to better understand how it influences the catalytic activity and association of the enzyme with ssDNA. The APOBEC3 enzymes have a common structure with a five-stranded β-sheet core that is surrounded by six α-helices [44–46]. The dsRNA mediated dimer is formed in the absence of protein-protein contacts in the dimer and loop 7 and helix 6 contact the RNA [21, 22, 30, 31]. For A3G, loop 7 has been shown to mediate dimerization [47, 48]. For all APOBEC3 enzymes, the loop 7 and/or helix 6 structures have been shown to be important for processivity on and binding to ssDNA and loop 7 is required for recognition of the cytosine containing motif for deamination [44, 49–54]. The loop 7 can be transferred between APOBEC enzymes to directly transfer motif specificities (5′CC for APOBEC3G and 5′TC for all other APOBEC3s) [50, 51]. These motifs can be further specified, e.g., 5′TTC for APOBEC3F and 5′CTC for APOBEC3H, and some characterizations have found preferences up to a total 9 nt preferred recognition sequence, although all the enzymes are active on the basic dinucleotide motif of their preference [24, 41, 49, 55–57].
We studied APOBEC3H haplotype II (referred to as A3H) and derived mutants that were purified from Sf9 insect cells, to determine how a dsRNA mediated dimer influenced interactions with and activity on ssDNA. We identify extended residues on loop 7 that stabilize RNA binding and affect oligomerization on RNA and DNA oligonucleotides. We also show that in contrast to A3G [24], bound cellular RNA in wild type (WT) A3H does not completely inhibit enzyme activity, although the RNA does alter ssDNA binding kinetics and affinity. However, degradation of cellular RNA with RNase A leaves an approximately 12 nt protected RNA that can remain bound in the presence of ssDNA. Mutants unable to bind cellular RNA and oligomerize have a loss of enzyme activity and protein stability. These data support a model in which the RNA-mediated dimer of A3H is integral for enzyme function. If A3H uniquely uses this dimerization mechanism, the data may also explain why A3H is less stable than other APOBEC3 enzymes since it relies on a noncovalently linked counterpart for structure and function.
Results
RNA bound to A3H is protected from RNase A degradation and mediates oligomerization
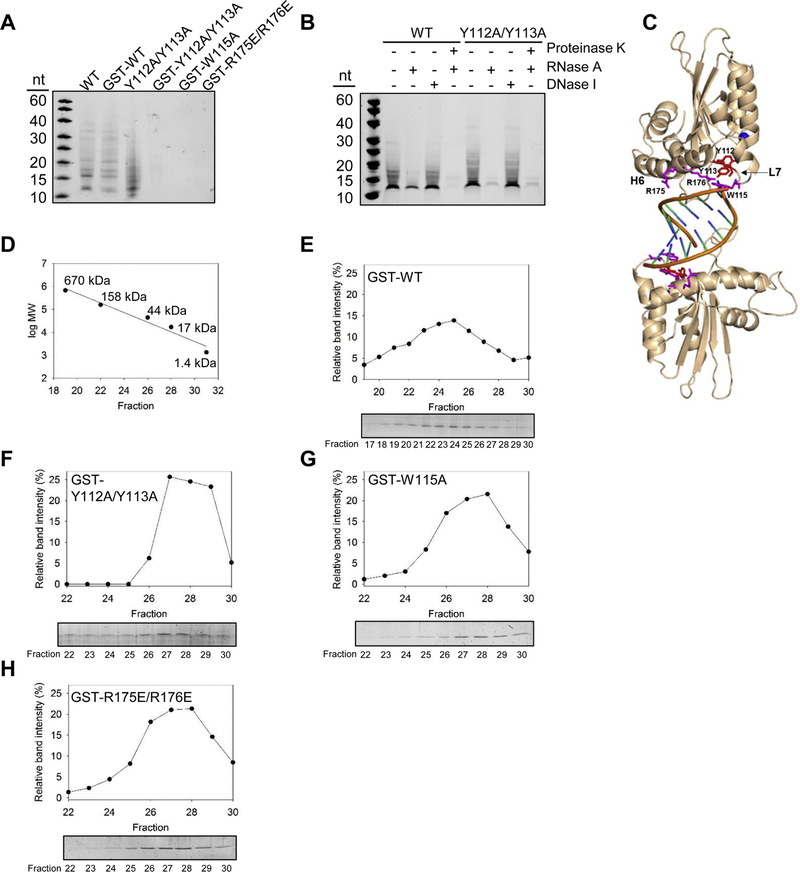
Similar to other APOBEC3 enzymes, we found that A3H WT purifies bound to nucleic acids (Figure 1A, WT) [47, 48]. The nucleic acid was identified to be RNA after comparison of treatments of the enzyme preparation with RNase A and DNase I (Figure 1B). Consistent with A3H crystal structures, we found that there was a small RNA bound to the A3H that could not be fully degraded without Proteinase K treatment of the protein preparation, indicating that A3H protects the RNA from nuclease treatment (Figure 1B). Crystal structures that resulted from A3H produced from E. coli recovered a 9 nt RNA [22, 30, 31] and we find that A3H from Sf9 insect cells is bound to a slightly larger, approximately 12 nt RNA (Figure 1B).
Figure 1.
A3H dimerization is mediated by RNA. (A) Purified A3H was denatured in formamide buffer and samples were resolved by urea denaturing PAGE. The gel was stained with SYBR Gold to detect nucleic acids. The A3H WT, GST-WT, and Y112A/Y113A purify with associated nucleic acids, but the GST-Y112A/Y113A, GST-W115A, and GST-R175E/R176E do not. Samples in Figure 1A were also resolved by SDS-PAGE and Commassie stained to detect proteins (Supplementary Figure S1A). (B) The nucleic acids that purify with the A3H WT and Y112A/Y113A are RNA and the A3H protects the RNA from full degradation, leaving an approximately 12 nt bound RNA. Samples in Figure 1B were also resolved by SDS-PAGE and Commassie stained to detect proteins and confirm Proteinase K degradation of the proteins (Supplementary Figure S1B). (C) The human A3H haplotype II dimer (wheat) is mediated by a dsRNA (orange). There are no protein-protein contacts in the dimer. The RNA makes key contacts with helix 6 amino acids R175 and R176 and loop 7 amino acid W115 (magenta). These amino acids and Y112 and Y113 (red) were mutated to study A3H dimerization. The catalytic center contains Zn2+ (blue). The structure is from PDB entry 6BBO [22]. (D) The oligomerization state of A3H was calculated from a standard calibration curve. (E) SEC profile for 10 µg of A3H GST-WT, (F) A3H GST-Y112A/Y113A, (G) A3H GST-W115A, and (H) A3H GST-R175E/R176E from a 10 mL Superdex 200 column. Due to the GST tag (26.9 kDa) fused to the A3H (21.5 kDa) the monomer fraction had an apparent molecular weight of ~48 kDa and the dimer fraction had an apparent molecular weight of 96 kDa. GST-A3H formed dimers. The A3H GST-Y112A/Y113A, A3H GST-W115A, and A3H GST-R175E/R176E were monomeric. (A-B and E-H) Representative images are shown from two independent experiments.
To biochemically investigate A3H dimerization and its relationship with RNA, we made mutants on loop 7 (W115A) and helix 6 (R175E/R176E) according to published structures [21, 22, 30, 31] and an additional loop 7 mutant (Figure 1C, Y112A/Y113A) that was predicted to be involved in A3H tetramerization on ssDNA through biochemical analysis [49]. We examined the A3H mutants for their ability to bind cellular nucleic acids. The loop 7 mutant that was previously characterized to mediate A3H tetramerization (Y112A/Y113A) retained the ability to bind the RNA molecule [49], although the nuclease protection appeared to be compromised since the RNA molecules after RNase A treatment were almost completely degraded (Figure 1A-B). This suggests that the Y112A/Y113A mutant did not bind the dsRNA in the same conformation as A3H WT. Importantly, the A3H Y112A/Y113A mutant was still stable and could be cleaved from the GST tag. However, when the residues W115 and R175/R176, which according to crystal structures more directly interact with the RNA were mutated (Figure 1C) [22, 30, 31], the A3H expression levels decreased and the GST tag needed to be retained for protein stability, consistent with the findings of Matsuoka et al. [31] (data not shown). The GST-tagged W115A and R175E/R176E mutants did not bind any nucleic acids during purification (Figure 1A). This was unlikely due to the GST tag since A3H WT also bound RNA when the GST tag was not cleaved (Figure 1A). Rather, these data were in agreement with A3H dimerization being mediated by residues on loop 7 and helix 6 and through an RNA molecule (Figure 1C) [22, 30]. However, the GST-tagged A3H Y112A/Y113A was not able to retain binding to RNA in contrast to the untagged enzyme (Figure 1A). This data and the almost complete lack of ability to protect the RNA from RNase A treatment suggests that extended residues on loop 7, which were shown to form an aromatic cage to stabilize unpaired RNA bases, can also disrupt dsRNAmediated dimerization (Figure 1B-C) [30, 31].
To confirm the relationship between RNA binding of A3H and oligomerization we used size exclusion chromatography (SEC). To directly compare SEC profiles, we used GST-tagged A3H for all experiments, which has an expected molecular weight of 48 kDa. We have previously shown that GST cannot dimerize when fused to A3H [49]. Based on the standard curve and consistent with previous data, GST-A3H WT forms a dimer [21, 22, 30, 49] (Figure 1D-E, peak fractions 23–24 have an apparent molecular weight of 102–96 kDa). Previous SEC data that used 100 μg (44 μM) of the A3H Y112A/Y113A mutant found that A3H Y112A/Y113A was a dimer [49]. In our SEC experiments, we used only 10 μg (1.5 μM) enzyme to maintain an equal amount of enzyme to the A3H mutants that expressed 10-fold (W115A) to 20-fold (R175E/R176E) less well than A3H WT. Using these lower enzyme amounts, we found that GST-A3H Y112A/Y113A was a monomer (Figure 1F, apparent molecular weight of 44 kDa), consistent with a compromised ability to bind RNA with or without a GST tag (Figure 1AB). The GST-W115A and GST-R175E/R176E mutants were also monomeric, consistent with previous analyses (Figure 1G-H) [21, 22, 30]. Altogether, these data demonstrate that both helix 6 and loop 7 are involved in A3H’s RNA mediated dimerization.
RNA-mediated oligomerization required for A3H catalytic activity
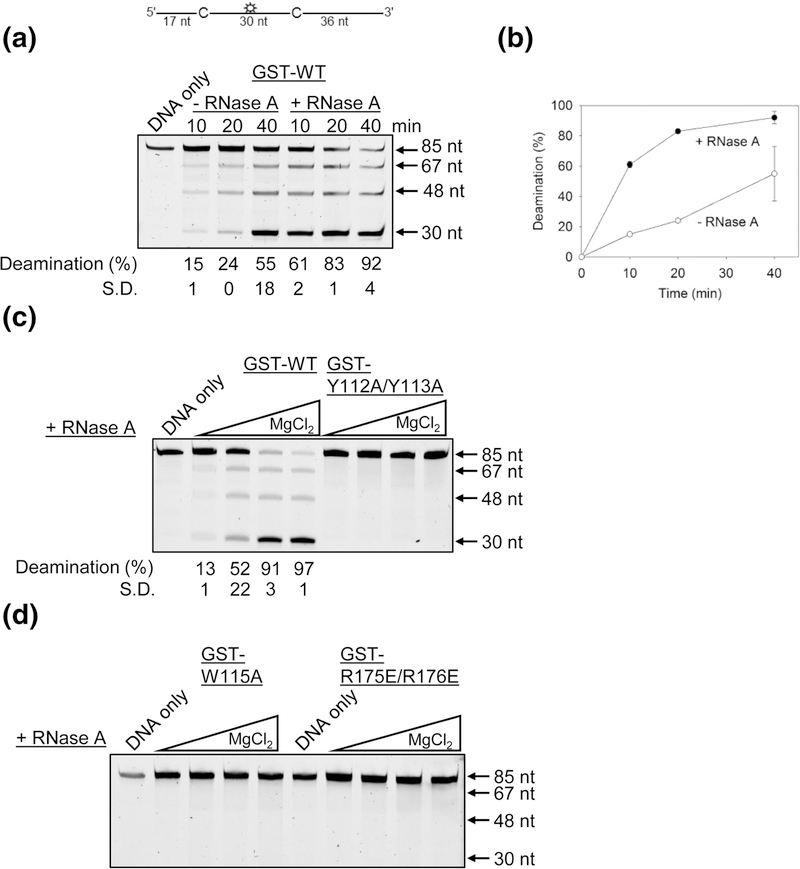
We tested if the A3H purified from Sf9 cells and bound to cellular RNA was catalytically active on ssDNA. We used an 85 nt fluorescein labeled ssDNA substrate that contained two deamination sites that were 30 nt apart (Figure 2A, sketch). The two deamination sites were used to avoid potential differences in specific activities of A3H across the length of an ssDNA confounding the results. The ssDNA was incubated with an excess amount of A3H for 40 min (ssDNA: A3H ratio of 1:8). After stopping the reaction, the deaminations were visualized through PAGE using a standard Uracil DNA glycosylase-based assay. We found that the GSTA3H bound to RNA (-RNase A) was catalytically active, but that the catalytic activity was approximately 4-fold less than the catalytic activity of A3H treated with RNase A when the enzyme activity was in the linear range (Figure 2A-B). This 4-fold difference in A3H WT activity occurred when the RNase A was added directly to the deamination reaction (Figure 2AB) or the A3H WT was purified in the presence of RNase A (data not shown). A3H that has been treated with RNase A has previously been characterized to be a processive enzyme that is able to deaminate multiple cytosines in a single enzyme-substrate encounter [49]. Processive deaminations are measured under single-hit conditions (<15% substrate usage) to ensure that each ssDNA substrate was acted upon by only one enzyme during the course of the reaction [58]. Under these conditions a processivity factor is calculated as the ratio of the processive deaminations occurring during the experiment, i.e., deamination of both 5’CTC motifs in a single enzyme-substrate encounter, in comparison to the calculated theoretical number of deaminations that would occur independently at both 5’CTC motifs if the enzyme were non-processive (see Materials and Methods) [24]. The A3H that was not treated with RNase A had a processivity factor of 4.6 ± 0.3 (Figure 2A, 10 min, 15% deamination, -RNase A), which is comparable to the processivity of A3H in the presence of RNase A (Figure 2C, processivity factor 6.4 ± 0.6, 13% deamination, +RNase A).
Figure 2.
Deamination of ssDNA by GST-A3H. Deamination activity was tested on an 85 nt ssDNA with two 5′CTC deamination motifs separated by 30 nt. (A-B) A3H WT can deaminate ssDNA in the presence and absence of bound cellular RNA. (C-D) Monomeric A3H mutants have disrupted ssDNA deaminase activity. Using different MgCl2 concentrations that were found to enhance GST-A3H WT activity, the deamination activity of A3H mutants were tested in the presence of RNase A. (C-D) The A3H GST-Y112A/, GST-W115A and GST-R175E/R176E were not active on ssDNA. A representative image is shown from three independent experiments. The S.D. was calculated from three independent experiments and is shown below the gel or for panel (B) is represented by error bars. Some error bars in (B) are obscured by the symbol.
The Y112A/Y113A A3H has previously been shown to not be active on ssDNA [49, 52]. To ensure that these results were not buffer specific, we tested the GST-Y112A/Y113A activity alongside A3H WT in a range of MgCl2 concentrations, since different MgCl2 concentrations can influence the interaction with and activity on ssDNA of other APOBEC3 enzymes, such as A3G [59]. Although titration of the MgCl2 from 0–10 mM enhances WT GST-A3H activity, there was no observed activity for GST-Y112A/Y113A, consistent with previous results [49, 52] (Figure 2C). We also found that in a range of MgCl2 concentrations the GST-tagged mutants W115A and R175E/R176E were not active (Figure 2D). Altogether, the data demonstrate that deamination activity of A3H requires dimerization through an RNA molecule, but not degradation of bound RNA.
Ability to bind cellular RNA dictates ability to bind RNA oligonucleotides, but not DNA oligonucleotides
To determine if there was a relationship between enzyme activity and ssDNA binding and to further understand the relationship of A3H with RNA, we quantitatively analyzed A3H binding to ssDNA and RNA. We used fluorescence polarization to measure the rotational anisotropy and determine an apparent dissociation constant (Kd). Rotational anisotropy is a measurement of the apparent change in rotation speed of a fluorescently labeled molecule due to a change in shape and/or molar mass of the molecule upon binding an unlabeled partner [60]. We fluorescently labeled ssDNA and RNA and titrated A3H into the binding reaction. The binding of the fluorescently labeled RNA or ssDNA to A3H will result in a change in rotation speed (anisotropy) until the fluorescently labeled RNA or ssDNA is saturated with A3H. Usually upon a fluorescently labeled molecule interacting with a binding partner, the increase in size of the complex causes the rotation speed to decrease, which results in an increase in rotational anisotropy [60]. From these measurements, an apparent Kd can be calculated.
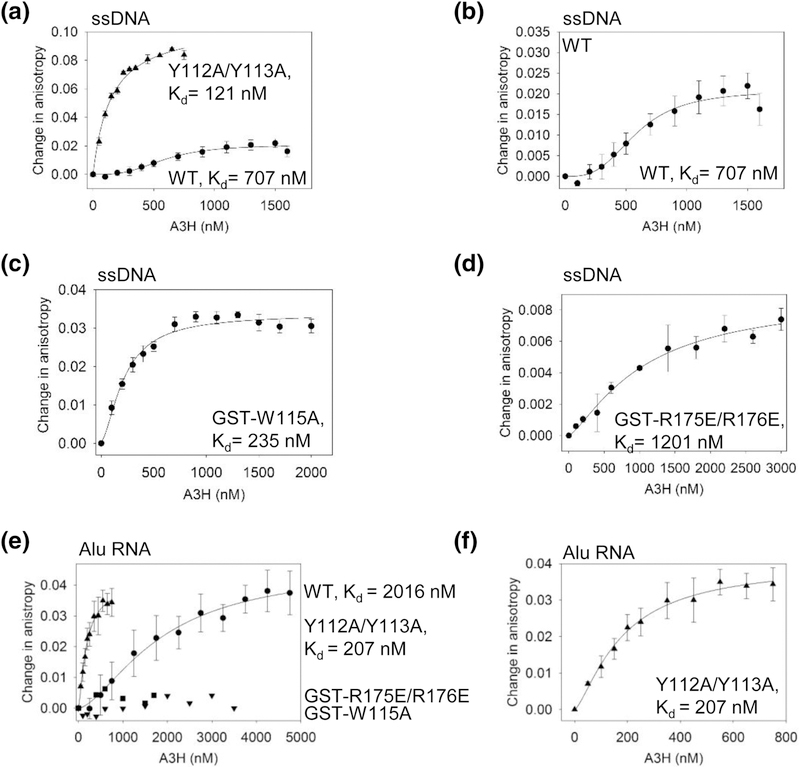
Using A3H that had been treated with RNase A during purification to remove any cellular RNA not protected by dimerization, we determined the apparent Kd values of WT and mutant A3H for a 118 nt linear ssDNA. We used the A3H Y112A/Y113A, which was a monomer in solution and could be cleaved from the GST tag (Figure 1), as a tool to interpret the effect of RNA-mediated dimerization on the nucleic acid binding properties of A3H WT. The A3H WT bound the ssDNA cooperatively, but with 6-fold less affinity than the A3H Y112A/Y113A (Figure 3A and Table 1, WT Kd of 707 ± 29 nM, Hill coefficient of 1.5, Y112A/Y113A Kd of 121 ± 43 nM). The A3H Y112A/Y113A bound the ssDNA noncooperatively, as determined by regression analysis. Notably, we observed that the change in anisotropy for the A3H Y112A/Y113A mutant was 4-fold higher than the A3H WT, suggesting that more molecules of the A3H Y112A/Y113A mutant were able to bind ssDNA than A3H WT (Figure 3A-B). The higher affinity binding of the A3H Y112A/Y113A mutant suggests that the weaker binding to the dsRNA enables more residues to contact and stabilize binding to ssDNA, suggesting that the A3H-bound RNA interferes with the ssDNA interaction. Since the A3H WT, but not the A3H Y112A/Y113A, bound DNA cooperatively, the data suggest either that A3H WT dimers can promote the binding of other A3H dimers on ssDNA or that each A3H subunit of the dimer cooperates to enhance binding. The RNA binding deficient mutants, GST-W115A and GST-R175E/R176E could both bind ssDNA. The binding of A3H W115A to ssDNA was comparable to the A3H Y112A/Y113A mutant in that there was no cooperativity and the apparent Kd was only 2-fold higher than that of A3H Y112A/Y113A (Figure 3C and Table 1, Kd of 235 ± 30 nM). This is consistent with Ito et al. that found the A3H W115A mutant bound ssDNA with higher affinity than A3H WT [21]. The binding of GST-R175E/R176E was of lower affinity, but still non-cooperative (Figure 3D and Table 1, Kd of 1201 ± 466 nM).
Figure 3.
A3H WT and mutants have different abilities to bind RNA and ssDNA. The apparent Kd of A3H enzymes from a 118 nt ssDNA was analyzed by steady-state rotational anisotropy. (A) The WT (circles, 707 ± 29 nM, Hill coefficient of 1.5) and Y112A/Y113A (triangles, 121 ± 43 nM) bind the ssDNA with different affinities and cause a different change in anisotropy, which is (B) shown alone for A3H WT for comparison to the (A) Y112A/Y113A mutant. The (C) GST-W115A (235 ± 30 nM) and (D) GST- R175E/R176E (1201 ± 466 nM) mutants bind the ssDNA, but induce a smaller change in anisotropy than the Y112A/Y113A mutant, similar to A3H WT. (E-F) The apparent Kd of A3H enzymes from a 91 nt Alu RNA was analyzed by steady-state rotational anisotropy for (E-F) WT (circles, 2016 ± 883 nM, Hill coefficient 1.5), Y112A/Y113A (triangles, 207 ± 60 nM, Hill coefficient 1.5), GST-W115A (upside down triangles), and GST-R175E/R176E (squares). The mutants GST-W115A and GST-R175E/R176E did not bind the RNA. (F) The Y112A/Y113A binds the Alu RNA with a higher affinity than A3H WT and is shown alone for comparison to (A). Error bars represent the S.D. from three independent experiments.
Table 1.
Apparent dissociation constants (Kd) of A3H WT and mutants for ssDNA and RNA.
| Kd | ||
|---|---|---|
| A3H | ssDNA (nM) | RNA (nM) |
| WT | 707 ± 29 (1.5)1 | 2016 ± 883 (1.5) |
| Y112A/Y113A | 121 ± 43 | 207 ± 60 (1.5) |
| GST-W115A | 235 ± 30 | No binding detected |
| GST-R175E/R176E | 1201 ± 446 | No binding detected |
Hill Coefficient for cooperative binding.
We used a 91 nt Alu RNA to test A3H binding affinity for RNA since this retrotransposon RNA is bound by A3H and other APOBEC3 enzymes [27, 61–64]. The A3H WT could bind the Alu RNA with an apparent Kd of 2016 ± 883 nM (Figure 3E and Table 1). This binding was cooperative with a Hill coefficient of 1.5. Although the A3H Y112A/Y113A mutant also bound the Alu RNA cooperatively, it bound with an approximately 10-fold higher affinity (Figure 3E-F and Table 1, apparent Kd of 207 ± 6 nM, Hill coefficient of 1.5). The two A3H mutants with no cellular RNA binding ability (Figure 1A) were unable to bind the Alu RNA in vitro (Figure 3E and Table 1, GST-W115A and GST-R175E/R176E). Since the main RNA binding residues, W115, R175, and R176, were still present, we reasoned that the A3H Y112A/Y113A was still able to bind and oligomerize on longer RNAs, but perhaps with a different conformation than A3H WT since the Y112A/Y113A had an impaired ability to protect the cellular RNA from RNase A treatment (Figure 1A-B). The increased affinity of A3H Y112A/Y113A for RNA compared to A3H WT may be due to the small RNA being outcompeted by the Alu RNA for A3H Y112A/Y113A, whereas for A3H WT the Alu RNA needs to bind in addition to the dsRNA mediating dimerization. Altogether, the binding data suggest that the determinants for RNA and DNA binding were different for A3H and that RNAmediated dimerization influenced the interaction of A3H WT with ssDNA and Alu RNA.
RNA-mediated oligomerization of A3H inhibits saturated binding of ssDNA
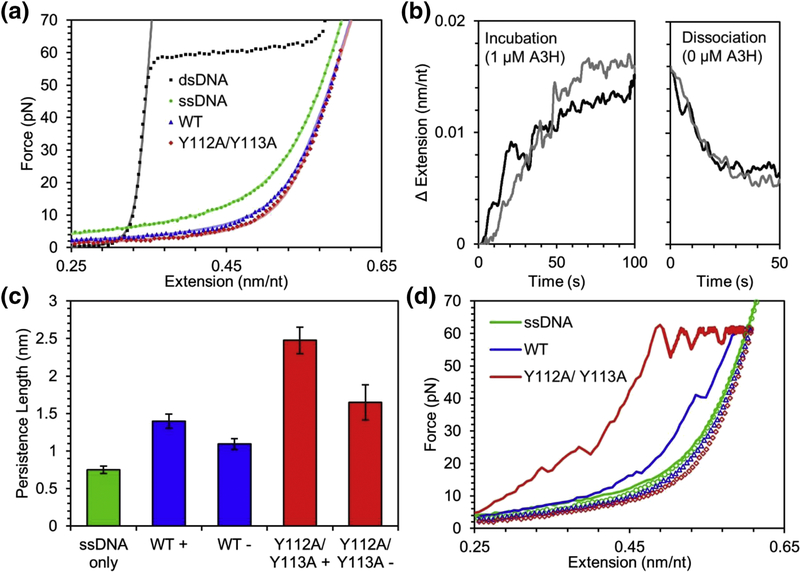
To understand how the bound dsRNA mediating dimerization influences the ability of A3H WT to bind ssDNA, we used optical tweezers to isolate a single ssDNA template and observe its interactions with A3H (see Materials and Methods). The A3H used for these experiments was treated with RNase A during purification. The physical characteristics of ssDNA can be well described by the freely jointed chain (FJC) polymer model [65], with a contour length (end to end length without bending or elastic stretching) of 0.56 nm/nt, a persistence length (length over which the ssDNA appears approximately straight) of 0.75 nm, and a stretch modulus (elasticity of the ssDNA) of 600 pN (Figure 4A). The very short persistence length (about the size of 1.5 nt), indicates the ssDNA backbone is very flexible and easily bends in the absence of applied force. When incubated with 1 μM A3H WT while held at a constant force of 30 pN, the ssDNA elongates by ~0.015 nm/nt over the timescale of 100 s (Figure 4B). By comparing the force extension curve of the A3H WT bound ssDNA with bare ssDNA, we observed that significantly less force is required to stretch to a given extension, consistent with an increased persistence length induced by A3H WT, which we confirm by fitting the FJC model to our data. The persistence length of the ssDNA doubles to 1.40 ± 0.09 nm while the contour length slightly decreases to 0.551 ± 0.003 nm/nt (Figure 3C, WT+). These data indicate that upon A3H WT saturated binding to the ssDNA, the ssDNA becomes just 2% shorter, when completely extended, but twice as rigid. Once the free protein is removed from the sample, the extended length of the ssDNA while held at 30 pN decreases relatively quickly, on the timescale of ~10 s, but not back to its original length, indicating some A3H remains bound to the ssDNA on a longer timescale (Figure 4B). Similarly, the FJC fit of the force extension curves of ssDNA with residual bound A3H in protein free buffer returns a persistence length between that of bare ssDNA and A3H saturated ssDNA (Figure 4C, WT-). This semi-irreversible binding has been previously observed in similar experiments with A3G and has been associated with the formation of larger oligomers [66]. We repeated this experiment with the A3H Y112A/Y113A mutant and found qualitatively similar results, consistent with rotational anisotropy data (Figure 3). Binding of 1 μM A3H Y112A/Y113A triples the ssDNA persistence length to 2.48 ± 0.18 nm and causes a larger reduction in contour length to 0.537 ± 0.002 nm/nt, indicating an enhanced ability of A3H Y112A/Y113A to stiffen the ssDNA compared to A3H WT (Figure 4C-D). These data are consistent with the A3H Y112A/Y113A more stably interacting with ssDNA than A3H WT (Figure 3A). Although less than A3H WT, the A3H Y112A/Y113A was able to stay associated with the ssDNA in protein free buffer (Figure 4C, Y112A/Y113A -). This supports the conclusion that A3H Y112A/Y113A oligomers can form at high local concentration, consistent with previous data [49].
Figure 4.
A3H alters physical properties of ssDNA upon binding. (A) The binding of A3H WT (blue) or Y112A/Y113A (red) greatly increases the persistence length of ssDNA (green). Symbols are experimental data and lines are fits of the wormlike chain (dsDNA) or FJC model. (B) The extended length of ssDNA increases while held at a constant tension of 30 pN while incubated with 1 μM A3H as shown by two sample curves. Once the free protein is removed from the sample, some bound A3H dissociates resulting in a partial reversal of the extension change. (C) The measured persistence length of ssDNA is greatly increased both after incubating with 1 μM A3H for 5 min (+) and after allowing bound protein to dissociate for 5 min in the absence of free protein (−). (D) ssDNA forms hairpins and loops in the absence of applied force, resulting in a shorter apparent length when stretched to high force (solid line) versus when relaxed to low force (open symbols). For bare ssDNA (green), hairpins are completely removed above ~15 pN of applied force, but bound A3H WT (blue) or Y112A/Y113A (red) stabilizes these structures requiring higher forces and longer times to break apart. Both the ability to locally straighten ssDNA and stabilize loop formation is enhanced for A3H Y112A/Y113A as compared to A3H WT.
Interestingly, A3H WT and Y112A/Y113A bound to the ssDNA stabilizes the formation of ssDNA hairpins and loops that naturally form in the absence of extending force. The presence of loops and hairpins can be detected by comparing the force extension curves of ssDNA obtained by increasing its extension starting at zero force and by decreasing its extension starting at a high force (Figure 4D). The hairpins and loops formed at zero force result in shorter apparent extensions in the former curve. For bare ssDNA, these loops and hairpins are removed entirely when forces above ~15 pN are applied and the two extension curves overlap (Figure 4D, green). With A3H WT bound, the shortening effect is more pronounced and persists to higher forces (Figure 4D, blue). Large sudden drops in force or increases in extension indicate large protein mediated loops are suddenly pulled apart. For A3H Y112A/Y113A, this effect is enhanced as nearly all excess ssDNA length has been stabilized into hairpins/loops and the applied force increases immediately when the ssDNA is extended (Figure 4D, red). Even when applying the very high force of 60 pN, ~100 s are required to remove all ssDNA secondary structures stabilized by A3H Y112A/Y113A. These data suggest that A3H Y112A/Y113A can bind and stabilize double-stranded structures in DNA, suggesting an ability to remodel the DNA. Although A3H WT also has this property, the effect is less pronounced.
A3H WT oligomerization primarily occurs on ssDNA
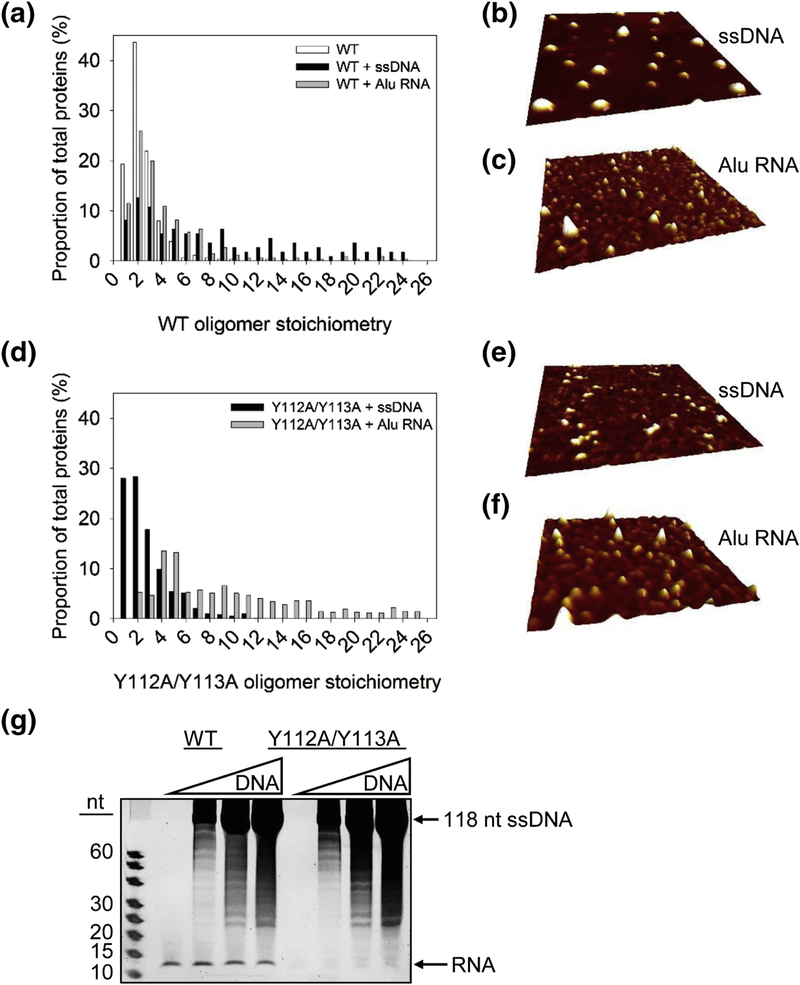
To determine the reason for these different dynamics on ssDNA, we examined the A3H WT and Y112A/Y113A using Atomic Force Microscopy (AFM). The A3H used for these experiments was treated with RNase A during purification. AFM can be used to generate topographic images of molecules deposited on a surface and is commonly used for examining the oligomerization states of proteins [47, 59, 66–68]. We reasoned that the differences in the dynamics of ssDNA binding would relate to the different oligomerization states of these proteins (Figure 3 and Figure 4). First, A3H WT alone was imaged to determine the size of a dimer deposited on the mica surface (see Materials and Methods). We determined the size of a dimer (stoichiometry of 2) and normalized all other conditions relative to this value (Figure 5A). Next, we examined the oligomerization states on the ssDNA that was used for the fluorescence polarization experiments. To prepare ssDNA bound A3H, we mixed A3H with a 10-fold excess of the ssDNA. The reaction was set up in this manner to ensure distribution of individual A3H molecules on different nucleic acid molecules, such that there would be only single molecules of A3H per ssDNA, unless there were pre-existing oligomers in solution or there was a cooperative binding relationship with ssDNA. In contrast to the A3H WT alone, which had a strong peak as a dimer (Figure 5A, ~43%, oligomer stoichiometry of 2), the peak of the WT bound to ssDNA was much more disperse (Figure 5A-B). Only ~12% of the particles were in the main dimer peak (Figure 5A, oligomer stoichiometry of 2). The remaining particles were much larger, reaching sizes that were 10-fold more than the main peak. Since the A3H WT bound ssDNA cooperatively, the larger peaks likely represented larger oligomeric forms induced by cooperative binding of ssDNA (Figure 5A). However, the A3H WT had a much more defined peak particle size when binding to the Alu RNA (Figure 5A, C). These data indicated that the RNA-mediated dimer of A3H WT decreased the ability to further oligomerize on RNA, in comparison to ssDNA. The decreased ability of A3H WT to oligomerize on RNA, but not ssDNA suggests that the RNA may be displaced upon binding to ssDNA or that the different binding domains for RNA and DNA affect oligomerization.
Figure 5.
Oligomerization states of A3H on ssDNA and Alu RNA. (A-F) AFM imaging was done in the presence of 5 mM MgCl2. (A-C) The oligomeric state of A3H WT alone (WT) and bound to ssDNA (WT + ssDNA) or Alu RNA (WT + Alu RNA) demonstrated that oligomerization predominantly occurred on ssDNA. (D-F) A3H Y112A/Y113A bound to ssDNA (Y112A/Y113A + ssDNA) or Alu RNA (Y112A/Y113A + Alu RNA) demonstrated that oligomerization predominantly occurred on Alu RNA. Volume distributions of A3H were converted to stoichiometries based on analysis of dimeric A3H WT (WT). Stoichiometries were plotted against percentage of total proteins. The total proteins counted were: WT, 337; WT + ssDNA, 111; Y112A/Y113A + ssDNA, 625; WT + Alu RNA, 331; Y112A/Y113A + Alu RNA 1004. Representative AFM images of A3H WT and Y112A/Y113A are shown for (B, E) ssDNA and (C, F) Alu RNA. Images are 500 × 500 nm with a height scale of 3 nm. (G) The A3H WT or Y112A/Y113A were incubated with increasing amounts of a 118 nt ssDNA and treated with RNase A. The presence of ssDNA did not enable degradation of the bound RNA, which would occur if DNA binding caused release of the RNA by A3H. These data suggest that A3H can bind both short RNAs and ssDNA simultaneously. A representative image is shown from three independent experiments.
To investigate further the relationship between A3H dimerization and nucleic acid binding, we also determined the oligomerization states of A3H Y112A/Y113A using AFM. The A3H Y112A/Y113A mutant had two peaks on ssDNA, each accounting for ~28% of the particles and had stoichiometric values of 1 to 2, consistent with impaired RNA binding and dimerization and non-cooperative interaction with ssDNA (Figure 5D-E). However, that A3H Y112A/Y113A could still form some dimers was in agreement with partial ability to retain cellular RNA binding, the single molecule optical tweezer experiments, and previous results showing that SEC with A3H Y112A/Y113A at high concentration results in a dimeric form (Figure 1 and Figure 4) [49]. We also examined using AFM how the A3H Y112A/Y113A bound to Alu RNA. In contrast to the ssDNA binding, the AFM particle sizes for A3H Y112A/Y113A bound to Alu RNA were 2- to 4-fold larger than A3H Y112A/Y113A bound to ssDNA and had a wide peak indicating that this mutant could oligomerize on RNA (Figure 5D, F). These data are consistent with diminished, but not completely abrogated binding to cellular RNA (Figure 1A-B) and the ability to cooperatively bind Alu RNA in vitro (Figure 3E-F). These data support the model that the RNA and ssDNA binding sites on A3H are different.
To examine further if the RNA and DNA could both remain bound to A3H, we tested if A3H could bind the short dsRNA mediating dimerization and a DNA oligonucleotide at the same time. To visualize whether this occurred, we incubated A3H with increasing amounts of a 118 nt ssDNA and then treated the A3H-DNA complex with RNase A. If the DNA binding displaced the RNA, then the RNA would be sensitive to degradation by RNase A. However, we found that increasing amounts of DNA had no effect on the amount of the RNA bound to the A3H WT or the A3H Y112A/Y113A mutant, although the mutant bound less RNA overall (Figure 5G). These results further support the interpretation that A3H has different binding sites for RNA and DNA.
Discussion
A3H is unique in the APOBEC3 family of enzymes. Human A3H has the highest number of SNPs compared to other human A3s [20, 32]. Recently this was shown to be the case in pigtailed macaques as well [30]. A3H is also the only single-domain A3 that is able to strongly restrict HIV-1 [69]. Due to these different SNPs and resulting haplotypes, A3H is also the only A3 enzyme that has a positive role in suppressing HIV and negative role in contributing to cancer mutagenesis [1, 13, 20]. Other A3s involved in cancer mutagenesis, A3B and possibly A3A, do not become encapsidated in HIV-1 virions from CD4+ T cells and restrict HIV-1 [1]. The A3H haplotype I, which is not active against HIV-1 can contribute to mutagenesis in breast and lung cancer [13]. A3H haplotype I is also localized to the nucleus, in contrast to other HIV-1 restrictive haplotypes that are in the cytoplasm (haplotypes II, V, and VII) [26, 70].
Recently, four publications have identified another unique feature of A3H. A3H dimerizes through interactions with a dsRNA molecule and in the absence of protein-protein contacts [21, 22, 30, 31]. Despite this important finding, the data resulting from these publications were not in agreement on several issues regarding A3H activity. Namely, some reports found that the loop 7 W115 residue was most important for mediating dimerization [21, 30, 31], whereas another report found loop 7 W115 and helix 6 R175/R176 were involved [22]. There was also disagreement on whether the bound RNA was an allosteric inhibitor of deamination [22], direct inhibitor of deamination [21], or did not inhibit deamination of DNA [30] and whether monomers of A3H were active [21, 22]. Finally, these data based on crystallographic analyses were in disagreement with a previous publication from our lab that proposed that loop 7 residues Y112 and Y113 mediated tetramerization of A3H [49]. To reconcile the differences, we undertook a biochemical analysis of A3H haplotype II produced from Sf9 insect cells.
RNA-mediated dimerization of A3H involves loop 7 and helix 6 and is required for catalytic activity
In collective agreement with crystallographic studies, we found that both loop 7 W115 and helix 6 R175/R176 were involved in the RNA-mediated dimerization of A3H. However, that an RNA molecule is required for A3H stability and activity was not a consistent observation in recent studies. One study using A3H in 293T cell lysates found that monomeric helix 6 mutants were more active than A3H WT, despite lower cellular expression [22]. This is probable since the monomer would enable more free units of A3H to interact and deaminate ssDNA, despite the decreased stability. In agreement with our study, Matsuoka et al. found that mutation of A3H amino acids that interacted directly with the dsRNA resulted in loss of protein stability and proteasomal degradation in cells [31]. Another study also found that a stabilized A3H mutant (m1) with a W115A mutation was active [21]. We observed no activity from the W115A or R175E/R176E monomer forms of A3H, although they were able to bind ssDNA (Figure 3C-D). With the A3H from 293T cell lysates, the mutants may be stable enough in lysates to demonstrate activity [21, 22], but perhaps not stable enough to maintain activity throughout our purification (Figure 2). We additionally found that mutation of loop 7 Y112/Y113 residues resulted in partial disruption of cellular RNA binding (Figure 1), but A3H stability was retained, thereby enabling the Y112A/Y113A mutant to be used as a tool to study the effects of RNAmediated dimerization on A3H activity and nucleic acid binding.
Importantly, although the A3H Y112A/Y113A, W115A, and R175E/R176E mutants still maintained an interaction with DNA, they were not catalytically active (Figure 2). The A3H W115A and Y112A/Y113A mutants even bound the ssDNA with a 3-fold higher affinity than A3H WT (Figure 3A, C). These data support that interaction of ssDNA with loop 7, when a dsRNA is not bound, enables higher affinity interaction with ssDNA. The A3H R175E/R176E mutant bound ssDNA ~2-fold less than A3H WT suggesting that helix 6 is required for high affinity ssDNA binding in the presence or absence of a dsRNA (Figure 3D). Overall, the RNAmediated dimerization appeared to be not only essential for stability, but also for enzyme activity. The A3H WT was active when bound to the cellular RNA and in the absence of any RNase A treatment. Degradation of the RNA molecules to the approximately 12 nt protected RNA only increased enzyme activity 4-fold, consistent with data from Bohn et al. [30] (Figure 2A). Altogether, the data demonstrate that for A3H WT, the dsRNA binding site is different from the DNA binding site, consistent with A3H WT being able to bind RNA and DNA at the same time (Figure 5G). For A3H WT there is still an outstanding question of how substrate recognition is achieved since loop 7 is also required for motif recognition on substrate ssDNA [50, 51]. Due to the close association of both RNA and DNA with loop 7, there may be some shifting of the RNA to bind helix 6 in the presence of DNA, but this remains to be shown experimentally.
A3H interacts differently with ssDNA and Alu RNA
In comparison to the Y112A/Y113A A3H, the RNA-mediated dimerization of A3H WT resulted in a lower affinity interaction with ssDNA, RNA, and incomplete saturation of bound ssDNA (Figure 3, Figure 4). Despite these results, AFM analysis of A3H WT on ssDNA showed extensive oligomerization, in contrast to A3H Y112A/Y113A that formed primarily monomers and dimers (Figure 5). The data with A3H WT and Y112A/Y113A are similar to data obtained with A3G WT and loop 7 mutant F126A/W127A where oligomerization of A3G WT prevented saturated binding to ssDNA [66]. For A3G, the oligomers had a decreased mobility once bound to ssDNA, resulting in less ability to accommodate additional A3G molecules, in comparison to monomeric A3G F126A/W127A [66]. This also resulted in a higher affinity interaction of the monomeric mutant with ssDNA than A3G WT [66, 71]. With A3G however, the oligomerization was due to protein-protein contacts [47]. For A3H, not only is the dimer mediated by dsRNA [21, 22, 30, 31], but A3H can undergo intersegmental transfer on ssDNA, which means that it can move from one ssDNA segment to another through a doubly bound state [49]. As a result, the AFM data showing A3H forms large oligomers on ssDNA could be due to inter-strand ssDNA binding, rather than intra-strand ssDNA binding mediated by protein-protein interactions. Although this may be occurring for some molecules, the data with A3H Y112A/Y113A support an additional model where loop 7 Y112/Y113 residues mediated tetramerization of A3H since the Y112A/Y113A A3H bound to ssDNA forms a discrete peak with primarily monomers and dimers (Figure 5). These data are consistent with a previous study showing that loop 7 Y112/Y113 residues mediated A3H tetramerization [49]. The dynamic analysis of these oligomerization states using single molecule optical tweezers was in agreement with the AFM and rotational anisotropy measurements and showed that the A3H WT was less able to alter the ssDNA persistence and contour lengths than the A3H Y112A/Y113A (Figure 4). Although both could form dimers or oligomers at high local concentration, the A3H Y112A/Y113A was more able to alter the ssDNA structure and dynamics, even stabilizing DNA hairpins (Figure 4). Collectively these data support the model that the enhanced number of individual molecules of A3H Y112A/Y113A that can interact with ssDNA enables it to have more of an effect on ssDNA kinetics than A3H WT (Figure 4 and Figure 5).
Despite A3H WT also having a lower affinity binding with Alu RNA in comparison with the A3H Y112A/Y113A, we did not find that WT and A3H Y112A/Y113A had the same properties when bound to Alu RNA and ssDNA (Figure 5). Rather, the RNA-mediated dimerization of A3H seemed to have an effect on binding the Alu RNA, perhaps due to competitive interactions for the RNA binding site. The A3H WT was not able to form large oligomers on RNA, suggesting that there are fewer protein-protein interactions or less ability to form inter-strand associations, in support of the RNA and DNA binding sites on A3H being different (Figure 5). In contrast, the Y112A/Y113A formed large oligomers on the Alu RNA, suggesting that in the absence of a high affinity interaction with the dsRNA mediating dimerization that nonspecific interactions with RNA were able to occur that enabled dimerization and oligomerization (Figure 5).
In cells, the RNA bound to A3H has been shown to be advantageous because it contributes to A3H cytoplasmic localization and ability to be virion encapsidated in an HIV-1 infected cell [21, 22, 30]. Since nuclear forms of A3H (haplotype I) can induce DNA damage through formation of uracils the cytoplasmic localization provides a protective function for genomic stability [13, 21, 22]. However, further studies need to be done on how A3H selects RNAs for binding. For instance, we found that although the RNA mediating dimerization does not inhibit A3H deamination activity, A3H WT is less able to bind and oligomerize on Alu RNA than the A3H Y112A/Y113A mutant, suggesting that there is competitive binding of RNA to A3H WT (Figure 3 and Figure 5). This is despite oligomerization on RNA being favorable for HIV-1 encapsidation and restriction [26].
Conclusions
Overall, the biochemical analysis of A3H WT and mutants demonstrates that both loop 7 and helix 6 mutants can independently disrupt the dsRNA-dependent dimerization of A3H. Importantly, A3H requires this dsRNA-mediated dimer for enzymatic activity on, but not binding to, ssDNA. Nonetheless, the RNA-mediated dimer comes at an expense to A3H nucleic acid binding affinity and ability to alter the structure of ssDNA, the latter of which may be beneficial when deaminating HIV-1 (−) DNA that has significant secondary structure. However, the lesser ability of A3H to stably bind ssDNA may be compensated for in the HIV virion where the high concentration of components and small volume in the capsid may facilitate ssDNA interactions that result in deamination-dependent and -independent restriction of HIV-1 [22, 52]. For A3H haplotypes that have “off-target” deaminase activity that results in genomic mutagenesis the lesser ability of A3H to stably bind and alter the structure of ssDNA and may be crucial to genomic integrity. Altogether, these data increase our understanding of how the dsRNA-mediated dimerization of A3H effects deamination of and interaction with ssDNA.
MATERIALS AND METHODS
Cloning and site-directed mutagenesis
A3H haplotype II was made by site-directed mutagenesis (QuickChange site-directed mutagenesis protocol, Agilent Technologies) using A3H haplotype I (NCBI accession number BC069023) as a template and is referred to as the WT. The GST-A3H WT was cloned into pFAST-bac1 vector (Life Technologies) using EcoRI and NotI restriction sites. A3H mutants (Y112A/Y113A, W115A, R175E/R176E) were made using the WT construct as a template. Specifically, A3H Y112A/Y113A was made using the forward primer 5′ATC TTC GCC TCC CGC CTG GCC GCT CAC TGG TGC AAG CCC CAG and the reverse primer 5′ CTG GGG CTT GCA CCA GTG AGC GGC CAG GCG GGA GGC GAA GAT, A3H W115A was made using the forward primer 5′ GCC TCC CGC CTG TAC TAC CAC GCT TGC AAG CCC CAG CAG and the reverse primer 5′ CTG CTG GGG CTT GCA AGC GTG GTA GTA CAG GCG GGA GGC, and R175E/R176E was made using the forward primer 5′ CAG TCG AGC CAT AAA GGA AGA GCT TGA CAG GAT AAA G and the reverse primer 5′ CTT TAT CCT GTC AAG CTC TTC CTT TAT GGC TCG ACT G. For producing RNA in vitro, the 91 nt Alu sequence derived from 7SL RNA [72] was cloned into pSP72 vector (Promega) using XhoI and HindIII sites and the RNA was produced by SP6 RNA polymerase-directed in vitro transcription (Ambion) with a fluorescein RNA labeling NTP mix containing Fluorescein-12-UTP (Merck). All constructed plasmids were verified by DNA sequencing. Primers were obtained from Integrated DNA Technologies.
Protein expression and purification
The pFAST-bac1vectors were used to produce recombinant baculovirus according to the protocol for the Bac-to-Bac system (Life Technologies). Recombinant GST-A3H WT baculovirus was used to infect Sf9 cells at a multiplicity of infection of 10, and cells were harvested after 72 h. GST-tagged Y112A/Y113A, W115A, and R175E/R176E were produced by using the recombinant GST-A3H baculovirus to infect Sf9 cells at a multiplicity of infection of 5, 1, and 10, respectively, and cells were harvested after 72 h, except for GST-R175E/R176E, which was harvested after 48 h. Cells lysates were treated with 100 μg/mL of RNase A (Qiagen), unless indicated otherwise. Lysates were cleared by centrifugation and then incubated with Glutathione-Sepharose 4B resin (GE Healthcare) at 4 °C overnight and subjected to a series of salt washes (0.25M to 1M NaCl) as described previously [47]. For A3H WT and Y112A/Y113A, on-column cleavage from the GST tag with Thrombin (GE Healthcare) was performed at 21 °C for 18 h in thrombin digestion buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, and 2 mM DTT). Due to low expression of A3H W115A and R175E/R176E, enzymes were eluted with the GST tag in elution buffer (100 mM Tris, pH 8.8, 150 mM NaCl, 10% glycerol, and 50 mM reduced glutathione). GST-A3H WT and GST-A3H Y112A/Y113A were also produced in parallel with GST-A3H W115A and GST-A3H R175E/R176E for comparative analysis. Eluted GST-A3H was further purified using a UNO Q1 cation exchange chromatography column (Bio-Rad). The enzymes were loaded onto the column in a low-salt running buffer (50 mM Tris, pH 8.0, 50 mM NaCl, 10% glycerol, and 2 mM DTT) and eluted using a salt gradient. GST-A3H proteins were eluted from the column at ~300 mM NaCl. Proteins were assessed to be 90% pure by SDS-PAGE (Supplementary Figure S2).
Size exclusion chromatography
The oligomerization states of GST-A3H (WT, Y112A/Y113A, W115A, and R175E/R176E) were determined by SEC using 10 μg of purified protein. The column (0.5 cm diameter and 16 cm height) was prepared by pouring a 10 mL Superdex 200 (GE Healthcare) resin bed. The running buffer contained 20 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, and 5 mM DTT. A gel filtration standard (Bio-Rad) was used to generate a calibration curve from which the apparent molecular weights of eluted proteins were determined. The column was run using a Bio-Rad BioLogic DuoFlow, however, due to the low amount of protein applied, the chromatograms from the 10 ml Superdex 200 column were constructed by analyzing the integrated gel band intensities of the protein in each fraction after resolution by SDS-PAGE. The gels for each panel were resolved, stained with Oriole fluorescent gel stain, and scanned in parallel.
Nuclease treatment assay
To examine the nature of the nucleic acids present in purified A3H, the same amount of A3H (3.5 μg for WT, GST-WT, Y112A/Y113A, GST-Y112A/Y113A, GST-W115A, GST-R165E/R176E) was or was not treated with Proteinase K (New England Biolabs) for 15 min at room temperature followed by heat inactivation for 10 min at 95°C. Then the sample was or was not treated with RNase A (Roche Applied Science) or DNase I (New England Biolabs) for 15 min at room temperature. For RNase A treatments in the presence of a 118 nt ssDNA oligonucleotide, 3.5 µg (10 μM) A3H was incubated with 10, 50, or 100 μM DNA for 3 min at room temperature before RNase A treatment. The 118 nt ssDNA oligonucleotide has been previously described [49]. At the end of the reactions, samples were mixed with an equal volume of formamide containing 5 mM EDTA and resolved on a 20% (v/v) denaturing polyacrylamide gel. The resolved nucleic acids were stained with SYBR-GOLD (Invitrogen) and gel images were obtained using a Chemidoc-MP imaging system (Bio-Rad).
In vitro deamination assay
The 85 nt ssDNA substrate had the following sequence: 5′AAA GTG AAA GTG ATA CTC AAA TTT AAA AGT dT-Fluorescein AGA TAG AAG GTG ATA CTC AAA TAT GAA AGT TAG TAA GAT GTG TAA GTA TGT TAA. The ssDNA substrate (100 nM), which contained two A3H deamination motifs (5′CTC, with the underlined C being deaminated), was incubated with 800 nM of A3H (WT, GST-WT, Y112A/Y113A, GST-Y112A/Y113A, GST-W115A, and GST-R175E/R176E) for 40 min at 37 °C in buffer that contained 50 mM Tris pH 7.5, 40 mM KCl, 0–10 mM MgCl2, and 2 mM DTT. A3H catalyzed deaminations were detected by treating the ssDNA with Uracil DNA Glycosylase (New England Biolabs) and heating under alkaline conditions before resolving the Fluorescein dT-labeled ssDNA on a 16% (v/v) denaturing polyacrylamide gel. For reactions under single-hit conditions (<15% substrate usage), a processivity factor was determined by dividing the quantified total amount of deaminations occurring at both sites on the same ssDNA by a calculated theoretical value of deaminations that would occur at both sites if the deamination events were uncorrelated (not processive) [24]. A processivity factor of 1 means that the enzyme is not processive and that the deamination events were uncorrelated. Gel images were obtained using a Chemidoc-MP imaging system (Bio-Rad) and integrated gel band intensities were analyzed using ImageQuant (GE Healthcare).
Steady-state fluorescence polarization
The apparent Kd values of A3H, Y112A/Y113A, GST-W115A, and GST-R175E/R176E for fluorescein labeled ssDNA (118 nt) or Alu RNA were determined using steady state fluorescence polarization to measure rotational anisotropy. Reactions (50 μL) were conducted in deamination buffer (50 mM Tris, pH 7.5, 40 mM KCl, 10 mM MgCl2, and 2 mM DTT) and contained 50 nM fluorescein labeled DNA or RNA and increasing amounts of A3H. A QuantaMaster QM-4 spectrofluorometer (Photon Technology International) with a dual emission channel was used to collect data and calculate anisotropy. Measurements were performed at 21°C. Samples were excited with vertically polarized light at 495 nm (6 nm band pass) and vertical and horizontal emissions were measured at 520 nm (6 nm band pass). The Kd was obtained by fitting to a rectangular hyperbola or sigmoidal curve equation using SigmaPlot 11.2 software.
Optical tweezers
An 8.1 kbp dsDNA with biotin and digoxigenin labeled ends was constructed as previously described [73]. The DNA is tethered between an anti-digoxigenin coated bead attached to a micropipette tip moved by a piezo electric stage and a streptavidin coated bead held in a stationary dual beam optical trap. By moving the piezo electric stage to extend the DNA while measuring the deflection of the trap laser, which measures the force exerted on the DNA by the trap, the force extension curve of the DNA was obtained. The DNA was incubated with T7 DNA polymerase and held at a tension of 50 pN to trigger exonuclease activity, removing one stand from the free 3′ end. The remaining ssDNA was incubated with 1 μM A3H in buffer (10 mM HEPES, 50 mM Na, pH 7.5) while held at a constant force of 30 pN for 5 min. The protein was then removed from the sample by clean buffer to allow bound protein to dissociate for 5 min. The change in ssDNA extension was measured during both the incubation and dissociation process, and force extension curves were measured for the ssDNA before incubation, after incubation, and after dissociation.
Atomic force microscopy
AFM measurements were carried out at ambient conditions on a Model 4500 AFM instrument (Agilent Technologies, Chandler, AZ) operating in intermittent contact mode. A conical silicon cantilever (Applied NanoScience, Tempe, AZ) with a force constant of approximately 50 N/m, resonant frequency of approximately 170 kHz, and a with a curvature radius of <10 nm was used for AFM measurements. All measurements were taken with the ratio of the set-point oscillation amplitude to free air oscillation amplitude of 0.85–0.90, and resonant amplitude in the range of 510 V. In addition, all measurements were performed at ambient conditions with the instrument mounted in a vibration isolation system. The scan rate was 0.5–1.0 Hz (512 pixels per line) for all images. Protein solutions (25 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM DTT) were prepared as described previously [47]. Briefly, 10 µL of protein or protein/nucleotide was dropped onto freshly cleaved mica, incubated 20–60 s, rinsed 3–5 times with ddH20, and gently blown dry with nitrogen. The net Z-axis volume (nm3) for the protein and protein/nucleotide complexes were determined using the Particle and Pore Analysis module in Scanning Probe Image Processor (SPIP) softwareV5.1.6 (Image Metrology A/S, Horsholm, Denmark).
Supplementary Material
Highlights.
An RNA mediates dimerization of the cytidine deaminase, APOBEC3H
The RNA remains bound to APOBEC3H in the presence of substrate DNA
The APOBEC3H RNA-mediated dimer decreases the affinity for ssDNA and other RNAs
The core amino acids that disrupt dimerization also disrupt protein stability
The APOBEC3H RNA-mediated dimer prevents oligomerization on RNA, but not DNA
RNA is an integral part of APOBEC3H structure and function
ACKNOWLEDGEMENTS
We acknowledge Jason Maley from the Saskatchewan Structural Sciences Center (Saskatoon, Saskatchewan) for obtaining the AFM data and M. Nabuan Naufer for assistance with DNA construct preparation for the optical tweezers experiments.
FUNDING
This work was supported by the Natural Sciences and Engineering Council of Canada Discovery Grant [RGPIN-2016–04113 to L.C.]; the Canadian Institutes of Health Research Grant [MOP137090 to L.C.]; and National Institutes of Health Grant [GM072462 to M.C.W.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: single-stranded (ss), Activation Induced Cytidine Deaminase (AID), double-stranded (ds), APOBEC3H (A3H), size exclusion chromatography (SEC), atomic force microscopy (AFM), dissociation constant (Kd).
REFERENCES
- [1].Adolph MB, Love RP, Chelico L. Biochemical Basis of APOBEC3 Deoxycytidine Deaminase Activity on Diverse DNA Substrates. ACS Infect Dis 2018;4:224–38. [DOI] [PubMed] [Google Scholar]
- [2].Siriwardena SU, Chen K, Bhagwat AS. Functions and Malfunctions of Mammalian DNACytosine Deaminases. Chemical reviews 2016;116:12688–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med 2010;2:594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, et al. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 2015;6:6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 2010;17:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem 2012;287:40875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Arias JF, Koyama T, Kinomoto M, Tokunaga K. Retroelements versus APOBEC3 family members: No great escape from the magnificent seven. Front Microbiol 2012;3:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Suspène R, Mussil B, Laude H, Caval V, Berry N, Bouzidi MS, et al. Self-cytoplasmic DNA upregulates the mutator enzyme APOBEC3A leading to chromosomal DNA damage. Nucleic Acids Res 2017;45:3231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 2010;38:4274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol 2009;83:9474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol 2008;26:481–511. [DOI] [PubMed] [Google Scholar]
- [12].Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol 2008;26:261–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun 2016;7:12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov 2015;5:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013;45:970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013;494:366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013;45:977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2013;2:e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell cycle (Georgetown, Tex) 2013;12:762–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 2008;4:249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ito F, Yang H, Xiao X, Li SX, Wolfe A, Zirkle B, et al. Understanding the Structure, Multimerization, Subcellular Localization and mC Selectivity of a Genomic Mutator and AntiHIV Factor APOBEC3H. Sci Rep 2018;8:3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shaban NM, Shi K, Lauer KV, Carpenter MA, Richards CM, Salamango D, et al. The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNAMediated Dimerization Mechanism. Molecular cell 2018;69:75–86 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A 2003;100:4102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol 2006;13:392–9. [DOI] [PubMed] [Google Scholar]
- [25].Larijani M, Petrov AP, Kolenchenko O, Berru M, Krylov SN, Martin A. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequenceindependent manner. Mol Cell Biol 2007;27:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, et al. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol 2014;426:1296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, et al. Highmolecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A 2006;103:15588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, et al. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 2015;11:e1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].York A, Kutluay SB, Errando M, Bieniasz PD. The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog 2016;12:e1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bohn JA, Thummar K, York A, Raymond A, Brown WC, Bieniasz PD, et al. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat Commun 2017;8:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Matsuoka T, Nagae T, Ode H, Awazu H, Kurosawa T, Hamano A, et al. Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA. Nucleic Acids Res 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 2011;85:3142–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol 2009;83:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Monajemi M, Woodworth CF, Zipperlen K, Gallant M, Grant MD, Larijani M. Positioning of APOBEC3G/F mutational hotspots in the human immunodeficiency virus genome favors reduced recognition by CD8+ T cells. PLoS One 2014;9:e93428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Squires KD, Monajemi M, Woodworth CF, Grant MD, Larijani M. Impact of APOBEC Mutations on CD8+ T Cell Recognition of HIV Epitopes Varies Depending on the Restricting HLA. J Acquir Immune Defic Syndr 2015;70:172–8. [DOI] [PubMed] [Google Scholar]
- [36].Grant M, Larijani M. Evasion of adaptive immunity by HIV through the action of host APOBEC3G/F enzymes. AIDS Res Ther 2017;14:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 2017;21:494–506 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A 2008;105:5501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol 2010;84:10402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog 2014;10:e1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 2013;14:411–21. [DOI] [PubMed] [Google Scholar]
- [42].Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet 2014;10:e1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol 2008;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front Microbiol 2014;5:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].King JJ, Larijani M. A Novel Regulator of Activation-Induced Cytidine Deaminase/APOBECs in Immunity and Cancer: Schrödinger’s CATalytic Pocket. Front Immunol 2017;8:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aydin H, Taylor MW, Lee JE. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure (London, England : 1993) 2014;22:668–84. [DOI] [PubMed] [Google Scholar]
- [47].Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem 2010;285:16195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog 2009;5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Feng Y, Love RP, Ara A, Baig TT, Adolph MB, Chelico L. Natural Polymorphisms and Oligomerization of Human APOBEC3H Contribute to Single-stranded DNA Scanning Ability. J Biol Chem 2015;290:27188–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activationinduced cytidine deaminase. J Biol Chem 2009;284:22898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Logue EC, Bloch N, Dhuey E, Zhang R, Cao P, Herate C, et al. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS One 2014;9:e97062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mitra M, Singer D, Mano Y, Hritz J, Nam G, Gorelick RJ, et al. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology 2015;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 2008;456:121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maiti A, Myint W, Kanai T, Delviks-Frankenberry K, Sierra Rodriguez C, Pathak VK, et al. Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA. Nat Commun 2018;9:2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 2004;23:2451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yu Q, König R, Pillai S, Chiles K, Kearney M, Palmer S, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol 2004;11:435–42. [DOI] [PubMed] [Google Scholar]
- [57].Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 2004;14:1385–91. [DOI] [PubMed] [Google Scholar]
- [58].Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol 1995;262:232–56. [DOI] [PubMed] [Google Scholar]
- [59].Chelico L, Sacho EJ, Erie DA, Goodman MF. A model for oligomeric regulation of APOBEC3G cytosine deaminase-dependent restriction of HIV. J Biol Chem 2008;283:1378091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jameson DM, Ross JA. Fluorescence polarization/anisotropy in diagnostics and imaging. Chemical reviews 2010;110:2685–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One 2012;7:e38771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 2007;390:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Khatua AK, Taylor HE, Hildreth JE, Popik W. Inhibition of LINE-1 and Alu retrotransposition by exosomes encapsidating APOBEC3G and APOBEC3F. Virology 2010;400:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Koyama T, Arias JF, Iwabu Y, Yokoyama M, Fujita H, Sato H, et al. APOBEC3G oligomerization is associated with the inhibition of both Alu and LINE-1 retrotransposition. PLoS One 2013;8:e84228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996;271:795–9. [DOI] [PubMed] [Google Scholar]
- [66].Morse M, Huo R, Feng Y, Rouzina I, Chelico L, Williams MC. Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G. Nat Commun 2017;8:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ratcliff GC, Erie DA. A novel single-molecule study to determine protein--protein association constants. J Am Chem Soc 2001;123:5632–5. [DOI] [PubMed] [Google Scholar]
- [68].Shlyakhtenko LS, Lushnikov AY, Miyagi A, Li M, Harris RS, Lyubchenko YL. Atomic force microscopy studies of APOBEC3G oligomerization and dynamics. J Struct Biol 2013;184:217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 2011;85:11220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li MM, Emerman M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol 2011;85:8197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Feng Y, Chelico L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J Biol Chem 2011;286:11415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology 2008;5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Naufer MN, Murison DA, Rouzina I, Beuning PJ, Williams MC. Single-molecule mechanochemical characterization of E. coli pol III core catalytic activity. Protein Sci 2017;26:1413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.