Abstract
Mitophagy, a selective autophagy of mitochondria, clears up damaged mitochondria to maintain cell homeostasis. We performed high-content analysis (HCA) to detect the increase of PINK1, an essential protein controlling mitophagy, in hepatic cells treated with several nanoparticles (NPs). PINK1 immunofluorescence-based HCA was more sensitive than assays and detections for cell viability and mitochondrial functions. Of which, superparamagnetic iron oxide (SPIO)-NPs or graphene oxide-quantum dots (GO-QDs) was selected as representatives for positive or negative inducer of mitophagy. SPIO-NPs, but not GO-QDs, activated PINK1-dependent mitophagy as demonstrated by recruitment of PARKIN to mitochondria and degradation of injured mitochondria. SPIO-NPs caused the loss of mitochondrial membrane potential, decrease in ATP, and increase in mitochondrial reactive oxide species and Ca2+. Blocking mitophagy with PARKIN siRNA aggravated the cytotoxicity of SPIO-NPs. Taken together, PINK1 immunofluorescence-based HCA is considered to be an early, sensitive, and reliable approach to evaluate the bioimpacts of NPs.
Keywords: Mitophagy, Mitochondrial quality, High-content analysis, Nanosafety assessment, Superparamagnetic iron oxide nanoparticles, Graphene oxide-quantum dots
There is a great potential that nanotechnology will supply some solutions to urgent problems such as environmental protection, energy, water cleanup, and medical development.1–3 However, along with the rapid development of nanomaterials (NMs) in medicine and manufacturing industries, the likelihood of the harm of NMs to the susceptible population is greatly increased when potential detrimental NMs cannot be identified accurately and rapidly.3,4 Understanding of the adverse effects and underlying mechanisms of NMs is helpful for the development of novel NMs with optimal benefit-to-risk ratio and to ensure the safety of NMs for application.
Up to now, there is no mature evaluation system for nanosafety, although several useful models have been proposed. Of which, the three tiered model seems to be well-developed and accepted in determining oxidative stress. In this model, antioxidant defensive response is recommended to be tested (Tier 1), followed by inflammatory response (Tier 2) and cytotoxicity (Tier 3).4 While most toxicological endpoints are used for examining higher tiers of oxidative stress, multi-parametric assays with more endpoints are suggested to be used for toxicity testing at Tiers 1 and 2.4,5 Besides antioxidant defensive response, other protective mechanisms also play important roles in anti-exogenous stress, such as DNA damage repair,6 endoplasmic reticulum stress,7 and autophagy.8 Developing suitable biomarkers or toxicological endpoints will contribute to establishing a systematic screening toolbox for comprehensive and precise assessment of NMs for potential adverse effects.
Autophagy degrades the harmful cellular components such as damaged organelles and aggregated proteins, and recycles them to maintain cell survival and homeostasis.9 Mitochondria control the energy metabolism and regulate calcium homeostasis, lipid synthesis, and cell death.10 When mitochondria are damaged by internal or external stresses, mitophagy (a selective autophagy of mitochondria) is triggered to remove excessive or damaged mitochondria to maintain homeostasis.10 In mammalian cells, while mitochondria are depolarized, PARKIN (encoded by PARK2 gene) is recruited to the injured mitochondria through PTEN-induced putative kinase 1 (PINK1), and then PARKIN conjugates ubiquitins onto proteins on the mitochondrial outer membrane (MOM) to subsequently induce mitophagy.11 Mutants of PINK1 or PARKIN, failing to mediate mitophagy, are associated with Parkinson disease, which can be a result of damaged mitochondria accumulation.12 Besides, mitophagy has been shown to play essential roles in other diseases, such as cardiac hypertrophy and dysfunction,13 neuromuscular disorders,14 tumors,15 and liver diseases.16 However, there is no report on the effects and mechanisms of NMs-induced mitophagy in hepatic cells.
Generally, mitophagy is triggered to clear up injured mitochondria to overcome stress at the early stage. With the increase of stress, mitophagy fails to maintain the cellular homeostasis, and cells will go through various forms of death.17 Thus, we hypothesized that mitophagy would act as a new sensitive biomarker for nanosafety assessment. As a novel imaging and statistics technology, high-content analysis (HCA) provides multi-parametric and quantitative analysis of each cell using multicolored fluorescence dyes or fluorescence-labeled antibodies.18 A multi-parametric cellular HCA system has been shown to be more in concordance with human toxicity than conventional assays for cytotoxicity.19,20
In the present study, using HCA, we explored whether mitophagy could be induced by some nanoparticles (NPs), including cadmium telluride (CdTe)-quantum dots (QDs), and gold (Au)-QDs, graphene oxide (GO)-QDs, cerium dioxide (CeO2)-NPs, silicon dioxide (SiO2)-NPs, superparamagnetic iron oxide (SPIO)-NPs, and zinc oxide (ZnO)-NPs. Subsequently, SPIO-NPs or GO-QDs was selected as positive or negative inducer of mitophagy for detailed studies. SPIO-NPs are usually used as contrast agent for magnetic resonance imaging (MRI).21 GO is also widely applied in biomedical field, such as drug delivery, gene carriers, photothermal and photodynamic therapies, and biosensors.22 Furthermore, using traditional methods and assays, we confirmed that mitophagy could be triggered by SPIO-NPs, rather than GO-QDs. Finally, we demonstrated that mitophagy is an early, sensitive, and suitable parameter in evaluating biocompatibility of NPs. The PINK1-dependent mitophagy-based HCA is considered to be a rapid, reliable, and high-throughput approach to incorporate into present toolbox for nanosafety assessment.
Methods
Additional methods are described in the Supporting Information.
Preparation and characterization
The SPIO-NPs were synthesized according to previous methods.23 CdTe-QDs and Au-QDs were gifts from Dr. Xiangyang Liu (Fujian Provincial Key Laboratory for Soft Functional Materials Research, Xiamen University). GO-QDs were obtained from JCNANO (Nanjing, China). CeO2-NPs, ZnO-NPs, and SiO2-NPs were purchased from Aladdin (Shanghai, China). The morphology of these NPs was observed using transmission electron microscopy (TEM) (FEI Tecnai, Blackwood, NJ, USA). Au-QDs and CdTe-QDs were captured with high-resolution TEM (JEOL, Peabody, MA, USA). The high-resolution TEM image of GO-QDs was provided by manufacturer. Malvern Zetasizer instrument (Worcestershire, UK) was used to measure the size and zeta potential of these NPs.
High-content analysis (HCA)
All cells were seeded on a 96-well imaging plate (PerkinElmer, Waltham, MA, USA) with glass bottom, at a density of 8000 cells per well. As for the concentration-effect ofNPs on mitophagy, cells were treated with NPs at 7.5, 15, 30, and 60 μg/mL for 12 h in complete medium containing 10% fetal bovine serum (FBS). As for the time-effect, cells were treated with 15 μg/mL SPIO-NPs or GO-QDs for 3, 6, 12, and 24 h. For mitophagy assay, cells were labeled with Mitotracker (red, Thermo Scientific, Hudson, NH, USA) and stained with anti-PINK1 primary antibody (anti-rabbit, Novus, Miami, FL, USA) with corresponding Alexa Fluor 488-labeled secondary antibody (green, Beyotime, Shanghai, China). After washing, cells were fixed in ice-cold methanol for 15 min at −20 °C and co-stained with 4’,6-diamidino-2-phenylindole (DAPI) for cell counts by nuclei staining. For mitochondrial reactive oxide species (mitoROS) measurement, cells were washed with HBSS/Ca2+/Mg2+ (Beyotime), and incubated with MitoSOX Red probe (Thermo Scientific) diluted in HBSS/Ca2+/Mg2+ for 15 min at 37 °C in the dark. This probe was excited at 510 nm and emitted at 580 nm. For mitochondrial membrane potential (MMP) measurement, cells were incubated with JC-1 probe (Beyotime) for 20 min at 37 °C and then washed with JC-1 staining buffer. JC-1 probe for MMP was excited at 488 nm and the monomer of JC-1 was detected at 520 nm (green fluorescence) and the aggregated JC-1 was detected at 590 nm (red fluorescence). Mitochondrial depolarization was indicated by an increase in the green/red fluorescence ratio. As for mitochondrial Ca2+ measurement, cells were incubated with Rhod-2 AM probe (Abcam, Cambridge, MA, USA) for 45 min at 37 °C, protected from light. Rhod-2 AM probe was excited at 561 nm and emitted at 593 nm.
All the fluorescence intensity and images were obtained using Opera Phenix HCA confocal microscopy (PerkinElmer). In each well, more than 24 imaging fields were captured at the same exposure time. Each treatment had at least three replicates. For each measured cell, the following calculations were performed automatically by Columbus™ analysis system (PerkinElmer): (a) The cell nuclei were assigned at the DAPI channel; (b) The average fluorescence intensity was calculated by dividing the brightness of the entire field of cells to the according area.
Results
Physical-chemical characterization
In order to investigate whether PINK1-dependent mitophagy could be triggered by NPs, we first characterized a group of NPs that were employed in this study, including CeO2-NPs, ZnO-NPs, SPIO-NPs, Au-QDs, CdTe-QDs, GO-QDs, and SiO2-NPs. The shapes of these NPs were characterized with TEM or high- resolution TEM. Au-QDs, CdTe-QDs, and SPIO-NPs were spherical and relatively uniform; GO-QDs were single layered and had narrow-size distribution; CeO2-NPs, ZnO-NPs, and SiO2- NPs were amorphous (Figure S1). The hydrodynamic size and zeta potential were measured when these NPs were dispersed in water and cell culture medium containing 10% FBS, respectively; these data are presented in Supplement Table S1. Of note, the hydrodynamic size of SPIO-NPs was 31.3±1.4 nm and 142.8±2.3 nm; the zeta potential was close to +48.9 mV and −8.3 mV, respectively. The hydrodynamic size of GO-QDs was 5.2±0.1 nm and 9.4±0.2 nm; the zeta potential was −13.7 mV and −8.9 mV in water and cell culture medium containing 10% FBS, respectively.
PINK1 immunofluorescence-based HCA for several NPs
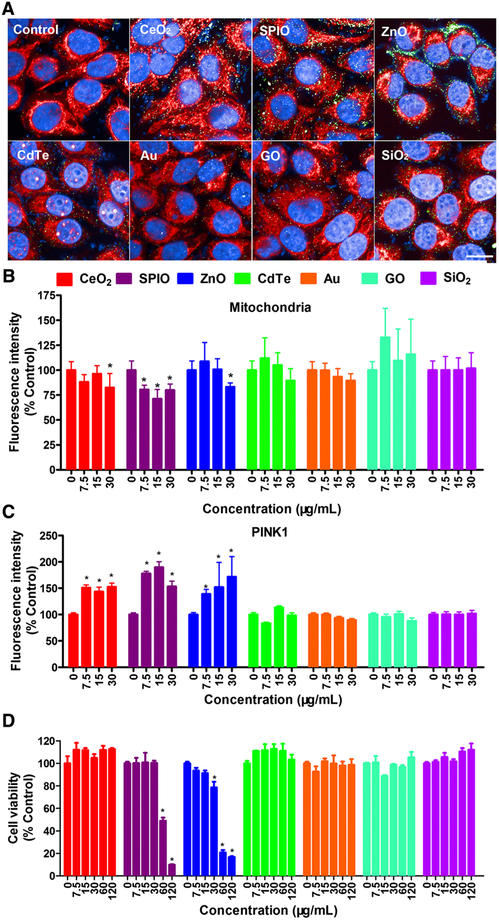
As a kinase regulating mitochondrial quality, PINK1 plays a protective role in structure and functions of mitochondria.24 HCA, a cell-based multi-parametric image analysis technique, can be applied to detect cell viability, oxidative stress, and mitochondrial Ca2+ to generate large data sets that can be used to create predictive models for nanosafety evaluation.18 To monitor whether the PINK1-dependent mitophagy can be triggered by different NPs, human normal hepatic L02 cells were treated with 7.5, 15, and 30 μg/mL CeO2-NPs, ZnO-NPs, SPIO-NPs, Au-QDs, CdTe-QDs, GO-QDs, and SiO2-NPs for 12 h, followed by PINK1 immunofluorescence-based HCA. As shown in Figure 1, A-C, groups of CeO2-NPs, ZnO-NPs, and SPIO-NPs decreased the fluorescence signals of mitochondria (Figure 1, A and B), and increased the fluorescence signals of PINK1 protein (Figure 1, A and C). Meanwhile, other NPs had no significant effects on the fluorescence of mitochondria and PINK1 protein. These results suggested that PINK1 immunofluorescence-based HCA may be a suitable method for NPs-induced PINK1-dependent mitophagy and thus hold the potential in assessing NPs’ bioeffects.
Figure 1.
Screening of PINK1-related mitophagy using HCA in L02 cells treated with different NMs (CeO2-NPs, ZnO-NPs, SPIO-NPs, Au-QDs, CdTe-QDs, GO-QDs, and SiO2-NPs). (A-C) L02 cells were treated with these NPs at 7.5, 15, and 30 μg/mL for 12 h and then labeled with Mitotracker (red) and stained with anti-PINK1 primary antibody with corresponding Alexa Fluor 488-labeled secondary antibody (green). The nuclei were stained with DAPI, and then imaged and analyzed using Opera Phenix HCA confocal microscopy. Representative HCA images (A) and the average fluorescence intensity of mitochondria (B) and PINK1 protein (C) are shown. Scale bar is 20 μm. Data are expressed as mean ± SD. Each treatment had at least three replicates. * P<0.05, compared to control. (D) Cell viability of L02 cells when treated with different concentrations of CeO2-NPs, ZnO-NPs, SPIO-NPs, Au-QDs, CdTe-QDs, GO-QDs, and SiO2-NPs at 7.5, 15, 30, 60, and 120 μg/mL for 12 h. The results are expressed as means ± SD (n=4). * P<0.05, compared to control.
In order to compare the sensitivity of the PINK1 immunofluorescence-based HCA with MTS assay for cell viability, we treated L02 cells with NPs at 7.5, 15, 30, 60, and 120 μg/mL for 12 h. As shown in Figure 1, D, the cell viability was significantly decreased only by treatments with SPIO-NPs at 60 and 120 μg/mL and ZnO-NPs at 30, 60, and 120 μg/mL. Therefore, PINK1 immunofluorescence-based HCA was proved to be more sensitive than cell viability assay to detect the bioeffects of NPs as the mitophagy responses were detected at earlier time points and lower concentrations by HCA.
Mitophagy triggered by SPIO-NPs, rather than GO-QDs, in a time-dependent manner
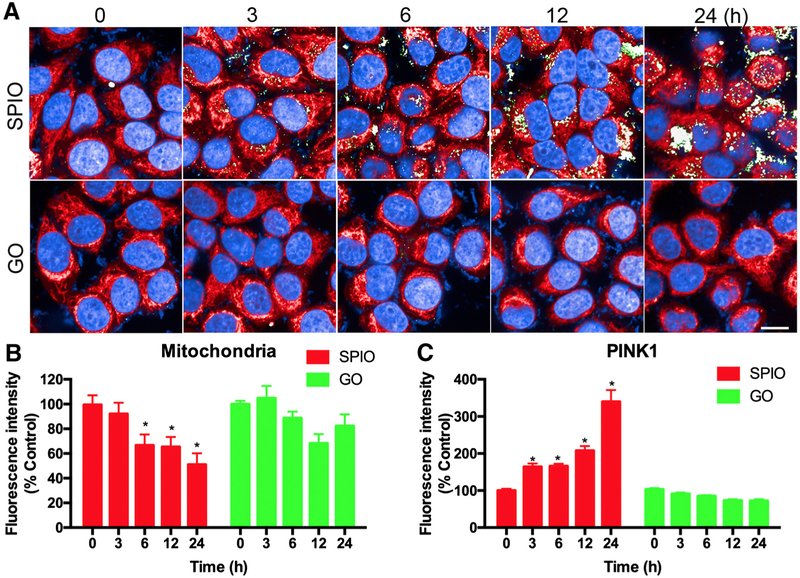
To further validate the PINK1 immunofluorescence-based HCA application for nanosafety assessment, we chose SPIO-NPs or GO-QDs as positive or negative inducer for PINK1-dependent mitophagy, respectively. With treatment at 15 μg/mL concentration, SPIO-NPs induced mitophagy but did not decrease cell viability, while GO-QDs had no significant effect on both (Figure 1, B), thus this concentration was selected for further analysis. HCA was performed to detect the dynamic change of mitochondria and PINK1 protein after L02 cells were treated with the two NPs from 3 to 24 h (Figure 2, A). Each cell was identified through DAPI stained nuclei (blue). Quantitative analysis of mitochondria (red) or PINK1 protein (green) was performed using the mean value of fluorescence intensity per cell. The results of mitochondrial staining indicated that the quantities of mitochondria were decreased by SPIO-NPs, but not by GO-QDs, from 6 to 24 h (Figure 2, B). On the other hand, SPIO-NPs treatment caused a significant time-dependent increase in PINK1 protein as early as at 3 h, while GO-QDs did not alter the signals of PINK1 protein at any time point (Figure 2, C). The PINK1 immunofluorescence-based HCA might be used as an effective, early, and sensitive approach for nanosafety assessment.25 In addition, we also examined whether PINK1-dependent mitophagy can be induced in different cells. Mouse fibroblast NIH3T3 cells and human epithelial HEK cells were treated with SPIO-NPs (15 μg/mL) and subjected to HCA analysis. As shown in Figure S2, SPIO-NPs increased the expression of PINK1 and decreased the mitochondrial mass in both cells, indicating that SPIO-NPs-induced mitophagy can occur in different types of cells, such as fibroblast/epithelial cells.
Figure 2.
HCA for mitochondria and PINK1 immunofluorescence under SPIO-NPs or GO-QDs treatment. L02 cells were treated with 15 μg/mL SPIO-NPs or GO-QDs for 3, 6, 12, and 24 h and then labeled with Mitotracker (red) and anti-PINK1 primary antibody with its corresponding Alexa Fluor 488-labeled secondary antibody (green). The nuclei were stained with DAPI, and then imaged and analyzed using Opera Phenix HCA confocal microscopy. Representative HCA images (A) and the average fluorescence intensity of mitochondria (B) and PINK1 protein (C) are shown. Scale bar is 20 μm. Data are expressed as mean ± SD. Each treatment had at least three replicates. * P<0.05, compared to control.
SPIO-NPs-induced mitophagy confirmed by conventional methods
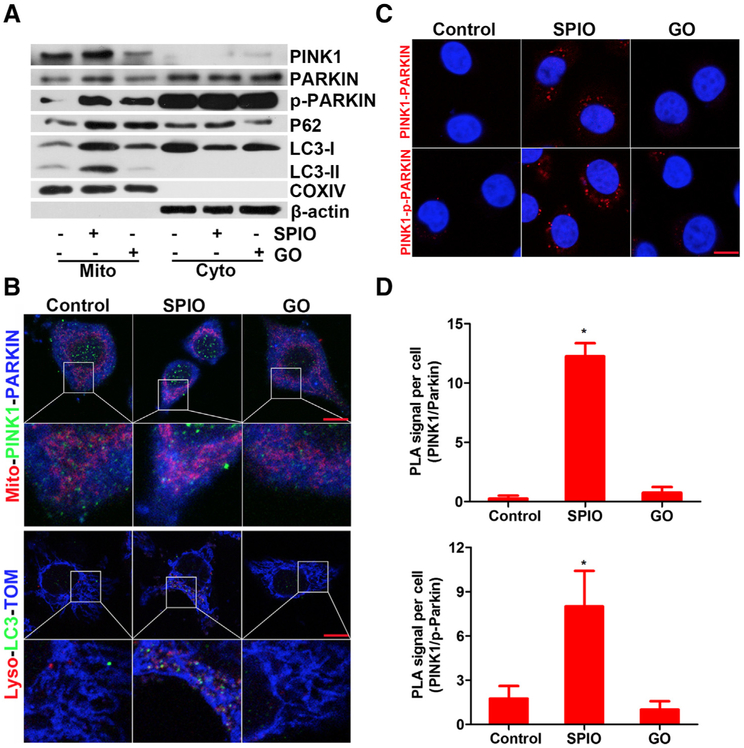
To verify PINK1-dependent mitophagy induced by SPIO-NPs, a number of conventional molecular and biochemical approaches were applied. First, we separated the mitochondrial fraction from the total cell lysate to investigate the expression and translocation of the proteins related to PINK1-dependent mitophagy.26,27 Using Western blot, we found that the levels of mitochondrial PINK1 and PARKIN were increased upon 15 μg/mL SPIO-NPs treatment for 12 h (Figures 3, A and S3). In addition, the phosphorylated form of mitochondrial PARKIN (p-PARKIN) at Ser65 was elevated by SPIO-NPs treatment, compared to control (Figures 3, A and S3). GO-QDs treatment also increased the p-PARKIN in mitochondrial fraction, but to a lesser extent. Additionally, treatment with SPIO-NPs or GO-QDs increased the level of P62 in mitochondria, compared to control. SPIO-NPs also caused an increase in the level of LC3-II in mitochondria, accompanied by a decreased level of it in the cytoplasm; GO-QDs, however, did not elevate the level of LC3-II in mitochondrial fraction (Figures 3, A and S3). Taken together, treatment of SPIO-NPs, but not GO-QDs, significantly increased the mitochondrial proportions of PINK1, PARKIN, and LC3-II, indicating that the mitochondria fused with phagophore to form mitophagosome and went through mitophagy in hepatic cells.
Figure 3.
SPIO-NPs, but not GO-QDs, trigger PINK1-dependent mitophagy in hepatic cells. L02 cells were exposed to SPIO-NPs or GO-QDs at 15 μg/mL for 12 h. (A) The expression level of mitophagy-related proteins including PINK1, PARKIN, p-PARKIN, P62, and LC3-II in mitochondria and cytoplasm was detected; COXIV or β-actin was used as loading control for mitochondrial or cytoplasmic fraction. (B) SPIO-NPs-induced PINK1-dependent mitophagy was imaged. Upper panel, the cells were incubated with the primary antibody for anti-PINK1 or anti-PARKIN followed by staining with Alexa Fluor 488-labeled (green) or 405-labeled (blue) secondary antibody, respectively. Mitochondria were labeled with Mitotracker (red). Lower panel, representative confocal microscopy images of LC3 (green, stained with primary antibody for anti-LC3 and Alexa Fluor 488-labeled secondary antibody); lysosomes were stained with Lysotracker (red); and mitochondria were stained with anti-TOM antibody and Alexa Fluor 405-labeled secondary antibody (blue). TOM protein is amarker of MOM. (C) PLA images of PINK1-PARKIN and PINK1-p-PARKIN proximity and interaction; the nuclei were stained with DAPI. Scale bars are 20 μm. (D) The average numbers of PINK1-PARKIN (upper) and PINK1-p-PARKIN (lower) PLA red foci were counted. Data are expressed as mean ± SD, n=4. * P<0.05, compared to control.
Next, confocal laser scanning microscopy was performed to study the distribution and co-localization of PINK1, PARKIN, and LC3. As indicated in Figures 3, B and S4, A, PINK1 was up-regulated and partially localized at mitochondria of SPIO-NPs-treated L02 cells; however, under this experimental condition, PINK1 was not augmented in GO-QDs-treated cells. SPIO-NPs treatment caused mitochondria quantity decrease and mitochondria translocation to the prinuclear space. The intensity of PARKIN was also enhanced upon the treatment of SPIO-NPs (Figure S4,) B. PINK1 and PARKIN co-located at the mitochondria, suggesting that PARKIN was recruited to mitochondria by PINK1 upon SPIO-NPs treatment (Figure 3, B, upper panel). Co-localization of LC3 protein with lysosome and mitochondria was also observed in SPIO-NPs-treated cells (Figures 3, B, lower panel and S4, C, D), suggesting that damaged mitochondria fused with phagophore to form mitophagosome. Moreover, the interaction of PINK1 and PARKIN was examined using PLA. As shown in Figure 3, C and D, compared to control or GO-QDs, SPIO-NPs-treated cells presented significantly more red loci, indicating stronger interaction between PINK1 and PARKIN, which further supported that SPIO-NPs triggered PINK1-dependent mitophagy in hepatic cells.
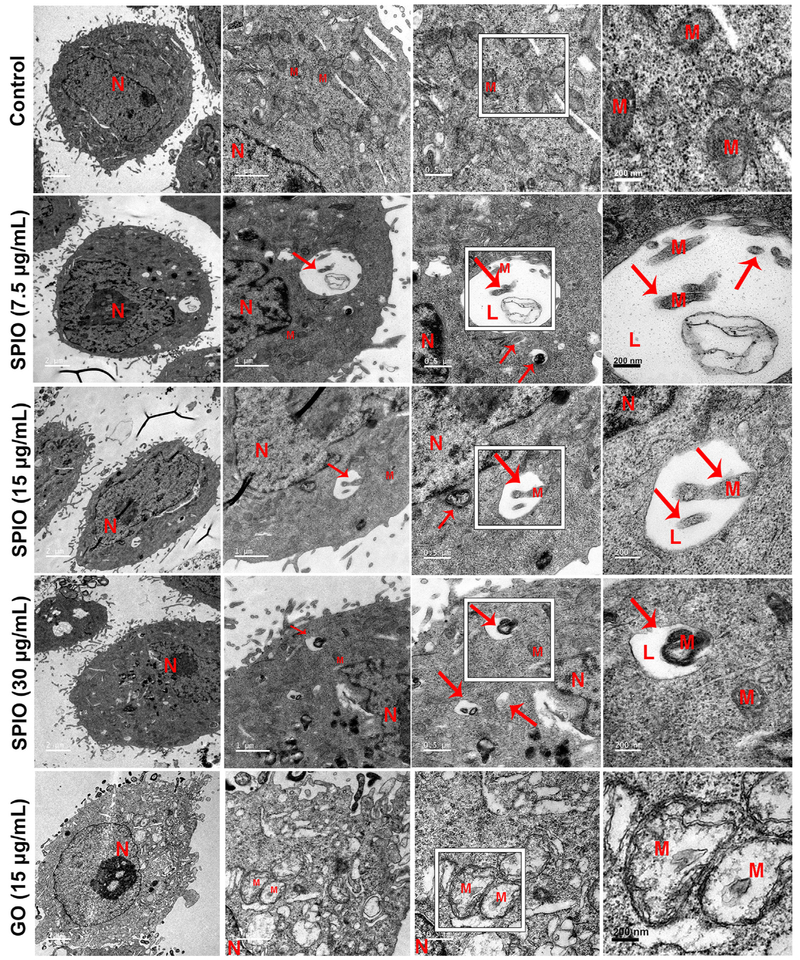
Finally, SPIO-NPs-induced mitophagy in L02 cells was examined by TEM. Cellular TEM directly detected the formation of mitolysosome, which contained the damaged mitochondria in cells treated with SPIO-NPs (7.5, 15, and 30 μg/mL) (Figure 4). SPIO-NPs also disrupted the cristae of mitochondria. In contrast, GO-QDs had little effect on the cellular ultrastructure (Figure 4). Altogether, the increase in PINK1, PARKIN, and LC3-II, the mitochondrial translocation of PINK1 and PARKIN, and the formation of autophagsome induced by SPIO-NPs treatment detected by conventional methods were consistent with the results obtained from PINK1 immunofluorescence-based HCA.
Figure 4.
SPIO-NPs, rather than GO-QDs, cause mitophagy of hepatic cells as observed by TEM. After SPIO-NPs (7.5, 15, and 30 μg/mL) or GO-QDs (15 μg/mL) treatment for 12 h, L02 cells were fixed, cut, and imaged with TEM. The red arrows indicate that the damaged mitochondrion fuses into lysosome to form mitolysosome in SPIO-NPs-treated cells. Scale bars are 2, 1, 0.5 μm, and 200 nm, respectively.
Mitochondrial functions impaired by SPIO-NPs, rather than by GO-QDs
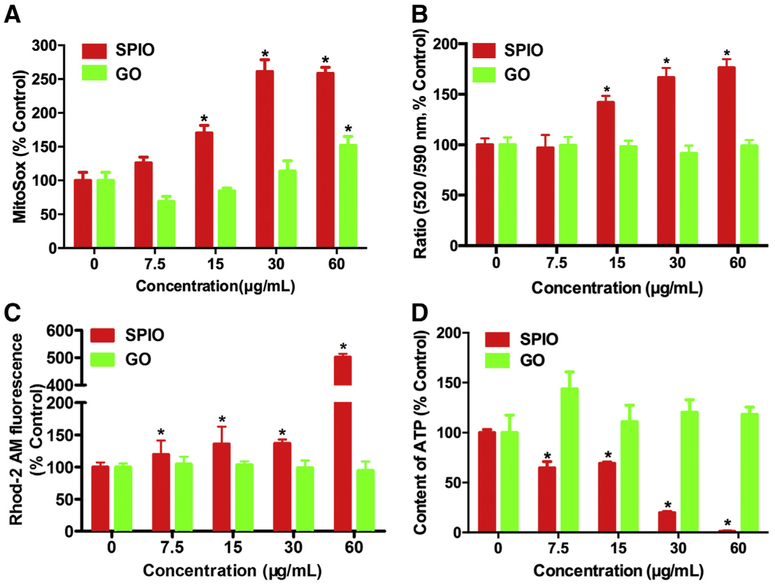
ROS, the byproduct of metabolism process, plays an important role in cellular homeostasis.28 It was of interest to investigate whether SPIO-NPs and GO-QDs provoke ROS, so we used MitoSOX fluorescent probe to measure the mitoROS level using HCA. As shown in Figure 5, A, treatment of SPIO-NPs for 12 h caused a significant and concentration-dependent increase in mitoROS, but such effect was only observed at the highest concentration for the treatment of GO-QDs. This result indicated that SPIO-NPs harbored greater capability to induce oxidative stress and mitochondrial damage, compared to GO-QDs.
Figure 5.
The levels of mitoROS, MMP, mitochondrial Ca2+, and intracellular ATP in hepatic cells treated with SPIO-NPs or GO-QDs. The L02 cells were treated with SPIO-NPs or GO-QDs at 7.5, 15, 30, and 60 μg/mL for 12 h. The cells were labeled with (A) MitoSOX probe for mitoROS, (B) JC-1 probe for MMP, (C) Rhod-2 AM probe for mitochondrial Ca2+, and then imaged and analyzed using Opera Phenix HCA confocal microscopy. The fluorescence intensity was used to indicate the level of each parameter within cells. MitoSOX probe was excited at 510 nm and emitted at 580 nm. JC-1 probe for MMP as excited at 488 nm and the monomer of JC-1 was detected at 520 nm (green fluorescence) and the aggregated JC-1 was detected at 590 nm (red fluorescence). Mitochondrial depolarization is indicated by an increase in the green/red fluorescence ratio. Rhod-2 AM probe was excited at 561 nm and emitted at 593 nm. In each well, more than 24 imaging fields were captured at the same exposure time. Data are expressed as mean ± SD. Each treatment had at least three replicates. * P<0.05, compared to control. (D) The ATP production was measured as luminescence value using a microplate luminometer upon SPIO-NPs or GO-QDs treatment. Data are expressed as mean ± SD of at least four replicates. * P<0.05, compared to control.
Evidence shows that excessive mitoROS disrupts the mitochondrial membranes, which is responsible for the loss of MMP.28 In the present study, MMP was detected with JC-1 probe using HCA. The green fluorescence, emitted from monomer of JC-1, indicates a lower MMP; while the red fluorescence, emitted from aggregation of JC-1, indicates a higher MMP. A significant loss of MMP was observed in cells treated with 15, 30, and 60 μg/mL SPIO-NPs for 12 h, as indicated by a 40.0%, 69.0%, and 77.4% increase in the ratio of JC-1 green/red fluorescence intensity, respectively. In contrast, GO-QDs treatment did not alter MMP of L02 cells (Figure 5, B).
Previous study has demonstrated that the increase of mitochondrial Ca2+ is associated with damage of mitochondria, and the disruption of Ca2+ homeostasis is linked to the induction of autophagy. 29 With Rhod-2 AM probe, we measured the mitochondrial Ca2+ level in the cells treated with SPIO-NPs or GO-QDs using HCA. Our results showed that SPIO-NPs significantly increased the level of mitochondrial Ca2+, compared to control. The concentration of mitochondrial Ca2+ stayed roughly the same in GO-QDs-treated cells as that in control cells (Figure 5, C).
As the power house of cell, mitochondria produce cellular energy in the form of ATP, so we measured the cellular ATP content in L02 cells. As shown in Figure 5, D, SPIO-NPs decreased the level of ATP in a concentration-dependent manner. The depletion of ATP suggested that the function of mitochondria was impaired in SPIO-NPs-treated cells. The ATP content was slightly increased by GO-QDs, but there was no significant change. All of these results suggested that mitochondrial functions of hepatic cells were adversely altered by SPIO-NPs, but not by GO-QDs. Moreover, compared with the fluorescence of mitochondria, MMP, and mitoROS, PINK1 immunofluorescence was significant increased by SPIO-NPs at the concentration as low as 7.5 μg/mL, which further supported that PINK1 immunofluorescence-based HCA can be used as an early and sensitive method for assessment of nanotoxicity.
Mitophagy plays a protective role in mitochondrial injury induced by SPIO-NPs
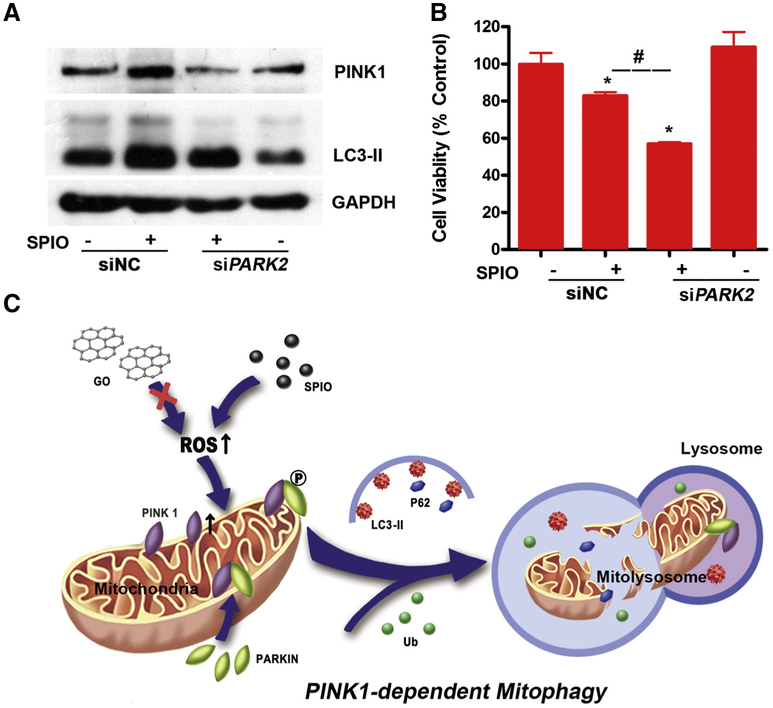
To investigate the role of mitophagy in SPIO-NPs-induced cytotoxicity, PARK2 siRNA (siPARK2) was employed to block the mitophagy. Knockdown of PARK2 inhibited the expression levels of PINK1 and LC3-II induced by SPIO-NPs, confirming the blockage of the mitophagy (Figures 6, A and S5). SPIO-NPs decreased 17% of the cell viability, while SPIO-NPs treatment with PARK2 siRNA decreased the cell viability further to 55% (Figure 6, B). Additionally, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), a well-known mitophagy inducer, was employed to treat L02 cells for 1, 3, and 6 h. Firstly, we confirmed that CCCP could induce PINK1-dependent mitophagy in L02 cells in a time-dependent manner, as evidenced by the increase of PINK1, PARKIN, and LC3-II using Western blot (Figure S6, A). After siPARK2 transfection, L02 cells were treated with CCCP for 12 h. Similar to SPIO-NPs, CCCP treatment inhibited the cell viability of L02 cells, which was also significantly enhanced by knockdown PARKIN (Figure S6, B). These findings indicated that PINK1-dependent mitophagy protected mitochondrial injury and cytotoxicity from SPIO-NPs.
Figure 6.
Knockdown of PARK2 aggregates SPIO-NPs-induced mitophagy and cytotoxicity of L02 cells. (A) Knockdown of PARK2, with transfection 50 nM siRNA for 12 h, decreased the up-regulation of the PINK1 and LC3-II induced by SPIO-NPs using Western blot. GAPDH was used as loading control. (B) Knockdown of PARK2 exacerbated the decrease of cell viability caused by SPIO-NPs using MTS assay. siNC stands for the siRNA negative control. All data are expressed as mean ± SD of four replicates. * P<0.05, compared to siNC; # P<0.05, compared to the SPIO-NPs-treated group. (C) The scheme of this study. SPIO-NPs, but not GO-QDs, trigger ROS overproduction and mitochondrial disrupt in hepatic cells, leading to the increase in PINK1 stability. PINK1 then recruits and phosphorylates PARKIN at Ser65. The p-PARKIN ubiquitinates the proteins at mitochondrial outer membrane and recruits the ubiquitins (Ub) to the damaged mitochondria that fuse into lysosomes with the help of LC3-II and P62, and finally making mitochondria go through mitophagy.
Discussion
In the present study, PINK1 immunofluorescence-based HCA was employed to detect mitophagy triggered by many NPs in various cells, including hepatic L02 cells, fibroblast NIH3T3 cells, and epithelial HEK cells. SPIO-NPs or GO-QDs were chosen as positive or negative inducer of mitophagy, respectively. We further validated PINK1-dependent mitophagy induced by SPIO-NPs, but not by GO-QDs, with traditional experiments, such as TEM, Western blot, PLA, and immunofluorescence confocal microscopy. Mechanically, we demonstrated that SPIO-NPs, rather than GO-QDs, activated phosphorylation and mitochondrial translocation of PARKIN, triggered the degradation of injured mitochondria, and resulted in PINK-dependent mitophagy. Consistently, SPIO-NPs treatment induced the decrease in MMP and ATP, as well as the increase in mitochondrial ROS and Ca2+. Moreover, the results of the present study showed that PINK1 immunofluorescence-based HCA was more sensitive than assays for cell viability and detections for mitochondrial functions. Herein, PINK1 immunofluorescence-based HCA is considered to be an early, sensitive, and reliable approach to evaluate the bioimpacts of NPs.
HCA is a multi-parametric imaging and quantitative statistics technology using multi-colored fluorescence dyes or fluorescence-labeled antibodies, which has been applied in many research fields, including biology, pharmacology, and nanotoxicology.18 Several probes for mitochondrial structure and functions are available to employ in HCA, such as Mitotracker for mitochondria, mitoSOX for mitoROS, Rhod-2 AM for mitochondrial Ca2+, and JC-1 for MMP.18 In this study, we developed a novel HCA method to monitor the effects of different types of NPs on mitochondria based on PINK1-dependent mitophagy. PINK1-dependent mitophagy was triggered by several NPs, including SPIO-NPs, CeO2-NPs, and ZnO-NPs as detected using HCA. This effect was further studied and confirmed by selected NPs using different conventional methods. Consistent with our finding, a recent study showed that ZnO-NPs also induced PINK1-dependent mitophagy in a mouse microglia BV-2 cell line.29 In our study, we found that mitochondrial dysfunction and mitochondrial ROS overproduction contributed to induction of mitophagy. In line with us, CeO2-NPs and ZnO-NPs were also proven to target mitochondrion to induce oxidative stress and mitochondrial disorder.30–32 All these results support that NPs-triggered mitophagy is closely linked to their targeting to mitochondrion and inducing mitochondrial dysfunction and ROS overproduction.
Our study demonstrated that several metal oxide NPs (such as SPIO-NPs) induced mitophagy, but non-metal NPs (such as GO-QDs) did not. Unlike SPIO-NPs, GO-QDs did not affect the cell viability and mitochondrial functions even at very high concentrations. Other studies also showed that GO-QDs and other stable and inert NMs are less toxic33–35 For example, no damaging effects were observed in mice treated with GO (60 mg/kg every 24 h for 5 days) via intra-abdominal injection, which presented very low or nearly no toxicity of GO for male reproduction.34 Chemically pure, few-layered GO and reduced GO did not induce significant cytotoxicity or genotoxicity in lung epithelial FE1 cells at relatively high doses (5-200 μg/mL).35 In contrast, there are many reports show that SPIO-NPs cause mitochondrial dysfunction, ROS production and mitochondria-related cell death.22,36,37 Although the detailed mechanisms warrant further investigations, one possible reason for the cellular dysfunctions caused by SPIO-NPs could be that as iron-containing NPs, SPIO-NPs release ions after they are swallowed by lysosomes. The newly formed ions may affect the intracellular oxide-reduction reactions and homeostasis of ROS inside cells mainly through the Fenton reaction.37 These results suggest that mitophagy would be easily induced by activated NPs that target and disrupt mitochondria, rather than by inert NPs.
Mitophagy is one type of selective autophagy.10 It maintains the function and quality of mitochondria by selectively swallowing and removing excessive and injured mitochondria.10 PINK1-dependent mitophagy is mostly investigated because of its important role in many diseases.13–16 Under normal status, PINK1 protein is located on the MOM and maintained at low level. PINK1 releases into cytoplasm, and is ultimately degraded by ubiquitin-proteasome pathway.26 Upon intracellular or extracellular stress, the expression of PINK1 is increased, and the excessive mitochondrial PINK1 recruits PARKIN from cytoplasm to mitochondria via phosphorylating Ser65 at PARKIN ubiquitin-like domain. As a mitochondrial E3 ligase, PARKIN is recruited and ubiquitinates proteins on the MOM and finally leads to mitochondrial elimination via mitophagy.27 In our study, PINK1, PARKIN, and p-PARKIN at Ser65 were up-regulated by SPIO-NPs in hepatic cells. Once MOM proteins are ubiquitinated by PARKIN, the ubiquitins are recruited to the damaged mitochondria that fuse to lysosomes with the help of LC3-II.27 Moreover, the increase of mitochondrial proportions of LC3-II and P62 was observed, indicating that the cells went through mitophagy caused by SPIO-NPs. Combined with PINK1 immunofluorescence-based HCA, our study confirmed that SPIO-NPs induced PINK1-dependent mitophagy in hepatic cells.
It has been reported that mitoROS triggers mitophagy to eliminate the damaged mitochondria and to alleviate the inflammatory response.38,39 One study has shown that mitoROS induced the expression PINK1, and the up-regulated PINK1 played a defensive role against oxidative stress by decreasing mitoROS production.40 The overproduction of mitoROS also can destroy the mitochondrial membrane, which is responsible for the loss of MMP.28 Our previous study has shown that SPIO-NPs induced mitoROS and impaired the mitochondrial electron transport chain in hepatocellular carcinoma cells.36 When MMP is lost, the ATP production of mitochondria will decline.28,36 In this study, SPIO-NPs caused the elevation of mitoROS, the loss of MMP, decrease of ATP production, and increase of mitochondrial Ca2+, all of which could potentially induce PINK1-dependent mitophagy triggered by SPIO-NPs.
It has been reported that PINK1-dependent mitophagy probably protects liver from alcohol-induced oxidative stress, injury, and steatosis.41 PARKIN knockout mice also display severe mitochondrial damage and dysfunction in hepatic cells.42 Previous studies have shown that PINK1 and PARKIN overexpression alleviated mitochondrial injury and restored the ATP production in fruit flies with mutant huntingtin protein.43 Consistently, our results showed that inhibition of PINK1-dependent mitophagy aggravated the cytotoxicity of SPIO-NPs as well as a well-known mitophagy inducer in hepatic cells. Taken together, PINK1-dependent mito-phagy might be a protective mechanism against SPIO-NPs-induced hepatotoxicity.
In conclusion, PINK1 immunofluorescence-based HCA was performed to detect mitophagy induced by some NPs, such as CeO2-NPs, ZnO-NPs and SPIO-NPs. SPIO-NPs or GO-QDs were chosen as positive or negative inducer of mitophagy, respectively, which was further validated by conventional methods. Specifically, SPIO-NPs, but not GO, caused the loss of MMP, decrease in ATP production, and increase in mitochondrial ROS and Ca2+, resulting in PINK1-dependent mitophagy (Figure 6, C). Moreover, through knockdown technique, PINK1-dependent mitophagy has been demonstrated to play a protective role in SPIO-NPs-induced hepatotoxicity. More importantly, we proved that NPs’ bioeffects could be evaluated with PINK1 immunofluorescence-based HCA, which was more sensitive than cell viability assay and mitochondrial function-related detections. Therefore, we propose mitophagy to be a sensitive biomarker for biocompatibility evaluation of NPs. Also, PINK1 immunofluorescence-based HCA has been proven to be an early, sensitive, and reliable screening method to incorporate into current nanosafety assessment system.
Supplementary Material
Acknowledgments
We appreciate Dr. Lei Guo (The National Center for Toxicological Research, USA) and Mr. Blake Xunbai Mei for English editing.
Funding: This study was supported by grants from the National Natural Science Foundation of China (81472997, 81573181, 81773465, and U1705281), the Major State Basic Research Development Program of China (2017YFA0205201), the Natural Science Foundation of Fujian Province of China (2015J01344), the Scientific Research Foundation (2016ZY003), the Fundamental Research Funds for the Central Universities (20720180045), the Open Research Fund (2016KF06) of State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, and the Intramural Research Program, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
Abbreviations:
- Au
gold
- [Ca2+]i
intracellular Ca2+
- CdTe
cadmium telluride
- CeO2
cerium dioxide
- DAPI
4’,6-diamidino-2-phenylindole
- DLS
dynamic light scattering
- GO
graphene oxide
- HCA
high-content analysis
- mitoROS
mitochondrial reactive oxide species
- MMP
mitochondrial membrane potential
- MOM
mitochondrial outer membrane
- MRI
magnetic resonance imaging
- NMs
nanomaterials
- NPs
nanoparticles
- PBS
phosphate buffered solution
- PINK1
PTEN-induced putative kinase 1
- PLA
proximity ligation assay
- QDs
quantum dots
- ROS
reactive oxide species
- SiO2
silicon dioxide
- siRNA
small interfering RNA
- SPIO
superparamagnetic iron oxide
- TEM
transmission electron microscopy
- ZnO
zinc oxide
Footnotes
Statement: There are no any commercial associations, current and within the past five years, that might pose a potential, perceived or real conflict of interest.
Conflict of Interests: None declared.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nano.2018.09.003.
References
- 1.Formoso P, Muzzalupo R, Tavano L, De Filpo G, Nicoletta FP. Nanotechnology for the environment and medicine. Mini-Rev Med Chem 2016;16(8):668–75. [PubMed] [Google Scholar]
- 2.Chen YX, Lavacchi A, Miller HA, Bevilacqua M, Filippi J, Innocenti M, et al. Nanotechnology makes biomass electrolysis more energy efficient than water electrolysis. Nat Commun 2014;54036. [DOI] [PubMed] [Google Scholar]
- 3.Chan WW, Chhowalla M, Glotzer S, Gogotsi Y, Hafner JH, Hammond PT, et al. Nanoscience and nanotechnology impacting diverse fields of science, engineering, and medicine. ACS Nano 2016;10(12):10615–7. [DOI] [PubMed] [Google Scholar]
- 4.Nel A, Xia T, Meng H, Wang X, Lin S, Ji Z, et al. Nanomaterial toxicity testing in the 21st century: use of a predictive toxicological approach and high-throughput screening. Acc Chem Res 2013;46(3):607–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nel AE, Nasser E, Godwin H, Avery D, Bahadori T, Bergeson L, et al. A multi-stakeholder perspective on the use of alternative test strategies for nanomaterial safety assessment. ACS Nano 2013;7(8):6422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong L, Gong H, Pan X, Chang C, Ou Z, Ye S, et al. P53 isoform delta113p53/delta133p53 promotes dna double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell Res 2015;25(3):351–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R, Huo L, Shi X, Bai R, Zhang Z, Zhao Y, et al. Endoplasmic reticulum stress induced by zinc oxide nanoparticles is an earlier biomarker for nanotoxicological evaluation. ACS Nano 2014;8(3):2562–74. [DOI] [PubMed] [Google Scholar]
- 8.Yang R, Ouyang Y, Li W, Wang P, Deng H, Song B, et al. Autophagy plays a protective role in tumor necrosis factor-alpha-induced apoptosis of bone marrow-derived mesenchymal stem cells. Stem Cells Dev 2016;25(10):788–97. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011;147(4):728–41. [DOI] [PubMed] [Google Scholar]
- 10.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 2005;8(1):3–5. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. Pink1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent parkin for mitophagy. J Cell Biol 2010;189(2):211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron 2010;68(2):201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao W, Li Y, Jia L, Pan L, Li H, Du J. Atg5 deficiency-mediated mitophagy aggravates cardiac inflammation and injury in response to Angiotensin Ii. Free Radio Biol Med 2014;69:108–15. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuhashi S, Nishino I. Phospholipid synthetic defect and mitophagy in muscle disease. Autophagy 2011;7(12):1559–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargini R, Garcia-Escudero V, Izquierdo M. Therapy mediated by mitophagy abrogates tumor progression. Autophagy 2011;7(5):466–76. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Liu XL, Nie J, Zhang J, Kimball SR, Zhang H, et al. Alcat1 controls mitochondrial etiology of fatty liver diseases, linking defective mitophagy to steatosis. Hepatology 2015;61(2):486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem 2012;393(7):547–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas CR, George S, Horst AM, Ji Z, Miller RJ, Peralta-Videa JR, et al. Nanomaterials in the environment: from materials to high-throughput screening to organisms. ACS Nano 2011;5(1):13–20. [DOI] [PubMed] [Google Scholar]
- 19.Brayden DJ, Cryan SA, Dawson KA, O’Brien PJ, Simpson JC. High-content analysis for drug delivery and nanoparticle applications. Drug Discov Today 2015;20(8):942–57. [DOI] [PubMed] [Google Scholar]
- 20.O’Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol 2006;80(9):580–604. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Wang L, Cao J, Huang Y, Lin Y, Wu X, et al. Functional investigations on embryonic stem cells labeled with clinically translatable iron oxide nanoparticles. Nanoscale 2014;6(15):9025–33. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Gao J, Ai H, Chen X. Applications and potential toxicity of magnetic iron oxide nanoparticles. Small 2013;9(9-10):1533–45. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Tan C, Zhang H, Wang L. Two-dimensional graphene analogues for biomedical applications. Chem Soc Rev 2015;44(9):2681–701. [DOI] [PubMed] [Google Scholar]
- 24.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila Pink1 mutants is complemented by Parkin. Nature 2006;441(7097):1157–61. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HT, Mi L, Wang T, Yuan L, Li XH, Dong LS, et al. Pink1/Parkin-mediated mitophagy play a protective role in manganese induced apoptosis in SH-SY5Y cells. Toxicol in Vitro 2016;34:212–9. [DOI] [PubMed] [Google Scholar]
- 26.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, et al. Mitochondrial processing peptidase regulates pink1 processing, import and Parkin recruitment. EMBO Rep 2012;13(4):378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. Pink1 phosphorylates ubiquitin to activate parkin e3 ubiquitin ligase activity. J Cell Biol 2014;205(2):143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ordureau A, Heo JM, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, et al. Defining roles of parkin and ubiquitin phosphorylation by Pink1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A 2015;112(21):6637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L, Wang J, Chen A, Liu J, Feng X, Shao L. Involvement of PINK1/parkin-mediated mitophagy in ZnO nanoparticle-induced toxicity in BV-2 cells. Int J Nanomedicine 2017;12:1891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma V, Anderson D, Dhawan A. Zinc oxide nanoparticles induce oxidative DNA damage and ROS-triggered mitochondria mediated apoptosis in human liver cells (HepG2). Apoptosis 2012;17(8):852–70. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Deng X, Zhang F, Chen D, Ding W. ZnO nanoparticle-induced oxidative stress triggers apoptosis by activating JNK signaling pathway in cultured primary astrocytes. Nanoscale Res Lett 2014;9(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torrano AA, Herrmann R, Strobel C, Rennhak M, Engelke H, Reller A, et al. Cell membrane penetration and mitochondrial targeting by platinum-decorated ceria nanoparticles. Nanoscale 2016;8(27):13352–67. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Peng C, Yang J, Lv M, Liu R, He D, et al. Uniform ultrasmall graphene oxide nanosheets with low cytotoxicity and high cellular uptake. ACS Appl Mater Interfaces 2013;5(5):1761–7. [DOI] [PubMed] [Google Scholar]
- 34.Liang S, Xu S, Zhang D, He J, Chu M. Reproductive toxicity of nanoscale graphene oxide in male mice. Nanotoxicology 2015;9(1):92–105. [DOI] [PubMed] [Google Scholar]
- 35.Bengtson S, Kling K, Madsen AM, Noergaard AW, Jacobsen NR, Clausen PA, et al. No cytotoxicity or genotoxicity of graphene and graphene oxide in murine lung epithelial FE1 cells in vitro. Environ Mol Mutagen 2016;57(6):469–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, Jiang S, Jin H, Chen S, Lin G, Yao H, et al. Mitochondrial electron transport chain identified as a novel molecular target of spio nanoparticles mediated cancer-specific cytotoxicity. Biomaterials 2016;83:102–14. [DOI] [PubMed] [Google Scholar]
- 37.Huang G, Chen H, Dong Y, Luo X, Yu H, Moore Z, et al. Superparamagnetic iron oxide nanoparticles: amplifying ROS stress to improve anticancer drug efficacy. Theranostics 2013;3(2):116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-β, and Bcl-2. Mol Cell 2007;25 (2):193–205. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Deng YZ, Naqvi NI. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe Oryzae. Autophagy 2013;9(11):1818–27. [DOI] [PubMed] [Google Scholar]
- 40.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in Nlrp3 inflammasome activation. Nature 2011;469(7329):221–5. [DOI] [PubMed] [Google Scholar]
- 41.Gautier CA, Kitada T, Shen J. Loss of Pink1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A 2008;105(32):11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams AJ, Ni H-M, Ding Y, Ding W-X. Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am J Physiol Gastrointest Liver Physiol 2015;309(5):G324–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalil B, Fissi NE, Aouane A, Cabirol-Pol M-J, Rival T, Liévens J-C. Pink1-induced mitophagy promotes neuroprotection in Huntington’s disease. Cell Death Dis 2015;6(1):e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.