Abstract
OBJECTIVE:
To determine whether metabolic imaging using fluorescence lifetime imaging microscopy (FLIM) identifies metabolic differences between normal oocytes and those with metabolic dysfunction.
DESIGN:
Experimental study.
SETTING:
Academic research laboratories.
PATIENT(S):
None.
MAIN INTERVENTIONS:
Oocytes from mice with global knockout of Clpp (caseinolytic peptidase P; n=52) were compared to wild type (WT) oocytes (n=55) as a model of severe oocyte dysfunction. Oocytes from old mice (1-year old; n=29) were compared to oocytes from young mice (12-week-old; n=35) as a model of mild oocyte dysfunction.
MAIN OUTCOME MEASURE(S):
FLIM was used to measure the naturally occurring NADH and FAD autofluorescence in individual oocytes. Eight metabolic parameters were obtained from each measurement (4 per fluorophore): short (τ1) and long (τ2) fluorescence lifetime, fluorescence intensity (I), and fraction of the molecule engaged with enzyme (F). ROS levels and blastocyst development rates were measured to assess illumination safety.
RESULTS:
In Clpp-knockout oocytes compared to WT, FAD τ1 and τ2 were longer (p<0.0001) and I was higher (p<0.01), NADH τ2 was longer (p<0.0001), and F was lower (p<0.0001). In older oocytes compared to young ones, FAD τ1 was longer (p<0.001) and I was lower (p<0.01), while NADH τ1and τ2were shorter (p<0.0001 for both), I and F were lower (p<0.0001 and p<0.05, respectively). FLIM did not affect ROS levels or blastocyst development rates.
CONCLUSIONS:
FLIM parameters exhibit strong differentiation between Clpp-knockout vs WT, and old vs young oocytes. FLIM could potentially be used as a non-invasive tool to assess mitochondrial function in oocytes.
Keywords: mitochondria, mitochondrial unfolded protein response, CLPP, oocyte, aging, Fluorescence Lifetime Imaging Microscopy, FLIM
INTRODUCTION
Developing a precise and reliable method of assessing oocyte and embryo quality has long been a critical goal for assisted reproductive technologies (ART) (reviewed in (1)). Soon after the report of the first successful pregnancy with IVF (2) and the introduction of controlled ovarian stimulation (3), embryo grading systems based on embryo morphology and cleavage rate have been introduced (4–9). These approaches resulted in significant improvements in implantation and pregnancy rates (10); however, their accuracy remained limited, as 60% of fresh embryos transferred to women younger than 35 years old fail to implant (11). Implantation failure rate rises to 75% for women 41–42 years old, and to more than 80% for those who are 43–44 (11). Even in women undergoing IVF treatment using fresh embryos generated with donor eggs, the implantation rate remains at 50% (11). These data are consistent with the observation that in many cases, embryos with acceptable morphology will fail due to underlying metabolic defects or chromosomal anomalies; conversely, many morphologically or morphokinetically irregular embryos are capable of producing healthy babies (12).
Aneuploidy is the most common type of chromosome abnormality and is the leading cause of implantation failure, miscarriage, and congenital abnormalities in humans (13–16). To improve the accuracy of embryo viability assessment, a number of investigators employed pre-implantation genetic screening, (now called pre-implantation genetic testing for aneuploidy [PGT-A]) to diagnose embryo aneuploidy. Initial methods that utilized fluorescent in situ hybridization (FISH) were later found not to be clinically useful (17). More recently, with the introduction of whole genome amplification (WGA), combined with 24-chromosome polymerase chain reaction (PCR) or next generation sequencing (NGS), and efficient extended culture and embryo cryopreservation, a number of centers reached 65% sustained implantation rates and prospective randomized clinical trials demonstrated clinical benefits from PGT-A (18, 19). Nevertheless, one third of euploid embryos still fail to achieve sustained implantation, and it is likely that embryonic factors other than aneuploidy impact IVF success.
Mitochondrial function is essential for oocyte and embryo viability (20). More recently, methods have emerged to quantify mitochondrial DNA (mtDNA) copy number in trophoectoderm biopsies, taken for PGT-A (21). This approach has aimed to use mtDNA number as a reflection of overall mitochondrial function for embryos. While some reports have indicated that especially high mtDNA counts are associated with poor embryo prognosis (21, 22), contradictory reports have reported no effect (23, 24). Some have also proposed that the utility of this technique is intrinsically limited by high level of natural variation in mtDNA levels (25).
Nicotinamide adenine dinucleotide (NAD+/NADH) and flavin adenine dinucleotide (FAD/FADH2) are two cofactors that are involved in cellular respiration (reviewed in (26)). They accept high-energy electrons that they transport to electron transport chain (ETC), where the energy is used to generate adenosine triphosphate (ATP) molecules. When NAD+ and FAD accept electrons they are reduced to NADH (by receiving 1 proton [H+] and 2 electrons), and FADH2 (by receiving 2 H+ and 2 electrons), respectively. NADH molecules are formed during glycolysis (which does not require oxygen) in the cytoplasm, and during pyruvate decarboxylation into Acetyl Coenzyme A, and the Kreb’s cycle, both of which occur in the mitochondrial matrix. FADH2 molecules are formed in the Kreb’s cycle. At perfect efficiency under aerobic conditions, a single glucose atom results in the generation of 10 NADH and 2 FADH2 molecules. NADH and FADH2 are then oxidized at the ETC, located in the inner mitochondrial matrix, generating 3 and 2 ATPs per molecule, respectively (26).
In this study, we evaluate the potential of fluorescence lifetime imaging microscopy (FLIM)-based metabolic imaging (27) of NADH and FAD as a tool for measuring mitochondrial metabolic state in oocytes for potential application in IVF. Because NADH and FAD are central to cellular respiration, differences in metabolic state directly affect measurable fluorescence properties of these molecules (28). Additionally, these molecules are naturally fluorescent, avoiding the need for foreign probes or specialized reagents. We performed FLIM measurements using time-correlated single photon counting (TCSPC) (29). FLIM via TCSPC uses fast electronics to record the sub-nanosecond arrival time of every photon in an image, relative to the laser pulse that generated that photon. The analysis of these arrival times allows detailed information to be extracted about the fluorophores under study. Such approaches have been extensively used to characterize the metabolic state of cancer cells (30), other cell lines (31), stem cells (32), mammalian tissues (33), and during germ cell differentiation in C. elegans (32). However, we are unaware of any studies using these advance methods on mammalian eggs and embryos.
Here, we show that this technique is able to detect and quantitatively characterize known defects in mitochondrial function in oocytes, using two mouse models of mitochondrial dysfunction. To investigate severe metabolic dysfunction, we used oocytes from mice with global germline deletion of the mitochondrial protease, Clpp. The deletion of this gene results in female infertility associated with severe mitochondrial dysfunction and metabolic abnormalities in oocytes (34). To investigate a milder form of metabolic dysfunction, we compared metabolic imaging measurements of oocytes from old and young mice. For each of these model systems, we compared the performance of FLIM with measurements of mtDNA amount. Finally, we performed an initial characterization of the safety of FLIM illumination. Given the intensity of light used for FLIM, the primary concern is the generation of reactive oxygen species (ROS) (35). We found that FLIM illumination of embryos results in no measurable increase in ROS, and that it has no measurable impact on blastocyst development rates.
METHODS
FLIM-based metabolic imaging of NADH and FAD
FLIM measurements were performed on a Nikon microscope using two-photon excitation from a Ti:Sapphire pulsed laser (M-squared lasers) with an 80-MHz repetition rate and ~150-fs pulse width, a galvanometer scanner, TCSPC module (SPC-150, Becker & Hickl) and a hybrid single-photon counting detector (HPM-100–40, Becker & Hickl). The wavelength of excitation was set to 750 nm for NADH and 845 nm for FAD, with powers of 45mW and 75mW, respectively (measured after the objective). Optical bandpass filters were positioned in front of the detector—460/50nm for NADH and 550/88nm for FAD (Chroma Technology). Imaging was performed with a 20X Nikon objective with 0.75 numerical aperture (NA). The size of the equipment was similar to a typical microscope (with approximately 3×5 feet base area).
At each time point, NADH and FAD images were acquired at three different focal planes within the oocytes, separate by a distance of 7µm, with an integration time of 60 seconds for (11–12 integrated scans of the field of view) each plane. Pixel size was 0.8µm/pixel, and scanner dwell time was 5µs/pixel. NADH/FAD complete acquisitions were orchestrated with a combination of custom software written in Labview and Becker Hickl acquisition software. In total, each complete metabolic measurement took approximately 7 minutes to acquire.
Mouse breeding and genotyping
We maintained the mice according to the Yale University animal research requirements. Food and water were available ad libitum and animals were housed under a 12-hour light-dark cycle. We obtained Institutional Animal Care and Use Committee approval prior to the initiation of the study (protocol number 2011–11207).
To obtain oocytes from old reproductive age mice, we purchased 7-month-old C57BL/6J retired breeder female mice from Jackson Laboratories (Bar Harbor, ME, USA). For the “old” group, we sacrificed mice at 12 months of age to retrieve oocytes.”Young” oocytes came from 12-week-old mice.
We acquired Clpp+/− male and female mice from the founder line IST13563G11 (line G) in the inbred C57BL/6J genetic background from Georg Auburger, PhD (Goethe University Medical School, Frankfort am Main, Germany) (36) and we bred them to obtain Clpp−/− (knockout) mice. The mouse genotyping was carried out as previously described (34).
Oocyte collection
Mouse oocytes were collected using standard protocols (37). Briefly, we superovulated female mice by intra-peritoneal injection of 5 IU of pregnant mare serum gonadotropin (PMSG; Sigma, St. Louis, MO) to stimulate follicle development. We then euthanized the mice 44h later by CO2 inhalation, removed the ovaries, and isolated cumulus oophorus complexes (COCs) by puncturing the ovaries with a 26–1/2 G needle under the dissecting microscope (Olympus SZH-ILLK). Cumulus cell were stripped by 75µm pipettes and GV oocytes (arrested at the prophase of the first meiotic division) were collected.
Sample preparation and FLIM acquisition
Mouse oocytes were imaged in Vitrolife Primo Vision 9-well dishes with 330µm diameter microwells, with four oocytes per well. Each well contained one 50µL droplet of α-minimum essential medium (MEMα; Life Technologies, Grand Island, NY) supplemented with 20mM Hepes, 75µg/ml penicillin G (Sigma), 50µg/ml streptomycin sulfate (Sigma), 0.1% polyvinyl alcohol (PVA; Sigma), and 10µM milrinone (Sigma), covered with Vitrolife Ovoil paraffin oil. An on-stage incubator system (Ibidi GmbH) was used to keep oocytes at 37±0.5ºC during imaging. Oocytes were retrieved with a micropipette after metabolic imaging and frozen in separate tubes for subsequent mtDNA copy number measurements. During this process, the correspondence was maintained so that metabolic measurements could be compared to mtDNA measurements in the same oocyte.
Quantification of mtDNA copy number in oocytes
To quantify mtDNA levels in GV oocytes, the Cox3 fragment was amplified and subcloned into pCR™2.1-TOPO® - cloning vector (Invitrogen) as previously described (30). One Shot TOP10 Chemically Competent E. coli were transformed and grown overnight at 37ºC. Recombinant plasmids were purified using the Qiagen plasmid isolation kit and the inserted mtDNA fragment was confirmed by DNA sequence analysis. Plasmid DNA was quantified using the NanoDrop 2000 spectrophotometer (Thermo Scientific). A standard curve from 108 to 101 plasmid molecules was generated by serial 10-fold dilutions.
Individual GV oocytes were lysed in 10μL lysis solution containing 125μg/mL Proteinase K and 17μM SDS in sterile water by incubating at 55ºC for 2 hours. Then Proteinase K was inactivated by heating the lysis mix at 95ºC for 10 min and the mix was used directly for downstream PCR. Reactions were performed in triplicates. Each 10µL reaction contained 5µL of SYBR Green supermix (Bio-Rad Laboratories, Hercules, CA), approximately 0.3µM of each primer, and 1/3 of total oocyte DNA. Oocyte mtDNA copy numbers were extrapolated from the standard curve.
FLIM data analyses
Data was analyzed using custom Matlab (Natick, MA) code. For each of the three focal planes acquired at every time point, a machine-learning algorithm (based on the ‘Weka Segmentation’ ImageJ plugin) was used on the intensity image to segment out the cytoplasmic region of each oocyte. Photons collected from the oocyte at the three focal planes were combined together into a single photon arrival histogram. The resulting histogram was fit to a sum of two exponentials:
Here, I is the intensity of the decay (photons detected within oocyte, per pixel area, per field of view scan), F is the fraction of photons coming from fluorophores with lifetime τ1, the remaining fluorophores (1-F) having a lifetime of τ2, and B is a background term (not coming from the fluorophore of interest). Fits were performed for both NADH and FAD, providing a total of eight quantitative parameters for characterizing the metabolic state of each oocyte: I, F, τ1, and τ2, for both NADH and FAD.
Repeated measurements on the same oocytes over a 70 min period revealed small decreases in measured metabolic parameters over time (fractions engaged < 1%, τ2’s < 2%, intensities < 8%, τ1’s < 22%). Linear regression of this data was used to calibrate and correct for this time variation in subsequent measurements. Performing this correction produced quantitative changes in the metabolic parameters, but did not qualitatively influence the conclusions of the manuscript.
Kolmogorov-Smirnov tests were performed on group distributions for each individual parameter to determine distribution normality. T-tests were performed between groups for each individual parameter, taking p< 0.05 to be statistically significant. In the Clpp experiment, we also derived a test condition by using a support machine vector algorithm. This method uses the data sets to calculate a hyperplane that optimally separates the two data sets, 55 Clpp+/+ and 52 Clpp−/− oocytes. Data points were deemed positive or negative based on whether they lie above or below the hyperplane.
Reactive oxygen species, blastocyst development, and calculating light absorption
Groups of embryos were incubated on the microscope and subjected to the following carrying degrees of FLIM illumination: 1 metabolic measurement, 3 measurements (1 per day), 24 measurements (1 every 2 hours), and 44 measurements (1 every 2 hours). For groups receiving 1, 3, and 24 measurements, we used CD1 embryos frozen at the 2-cell stage. The 44-measurement group entailed a longer incubation (88 hours); hence, we used a different strain of embryos, (B6C3F1 Female + B6D2F1 Male), that were frozen at the 1-cell stage. All embryos for these safety experiments were purchased from EmbryoTech. Reactive oxygen species measurements at the blastocyst stage and blastocysts development rates were subsequently measured. For each experiment, measurements of illuminated embryos were compared to control groups of embryos incubated in the same dish, but receiving no illumination.
For ROS studies, embryos were stained at blastocyst with an ROS reporter dye, HC-DCFDA (Sigma 7722-84-1). Staining was performed according to the Sigma’s provided protocol, incubating embryos in MEMα containing 25µM HC-DCFDA for 15 min. Embryos were then transferred to MEMα media, and HC-DCFDA fluorescence was imaged using 900nm illumination and a 550/88 emission filter. One ROS measurement was generated for each blastocyst by calculating the total photons within the intracellular region, and dividing by the intracellular area to get an average intensity value. Autofluorescence from FAD was measured before staining with the same imaging conditions, and the average autofluorescence was subtracted from ROS values for each group.
Blastocyst development rates were recorded for all embryo groups. Illuminated embryo rates were compared with non-illuminated embryos grown on the microscope, and also with embryos cultured in a table-top incubator (Panasonic MCO5MPA).
We also performed physical calculations to estimate the amount of light energy absorbed by FAD in an oocyte during an FAD FLIM acquisition. We first derived an estimate from first principles, considering one and two-photon absorption cross-sections, absorption due to pulsed illumination, confocal volume, and other factors (38). We then performed a corroborating empirical estimate, deriving absorbed energy from our measured photon counts. These values were then compared to similar estimates of energy absorbed during a standard morphological assessment with brightfield.
RESULTS
Clpp−/− vs Clpp+/+ oocytes
We first tested the hypothesis that FLIM-based metabolic imaging can accurately detect severe metabolic dysfunction in oocytes, and began by studying Clpp+/+ (wildtype) and Clpp−/− (knockout) oocytes. We imaged a total of 55 Clpp+/+ oocytes and 52 Clpp−/− oocytes, four at a time in microwells. For each microwell, we obtained FLIM data on NADH and FAD, segmented out the individual oocytes using the intensity images, fit the oocytes histogram of photon arrival times with a model, and obtained a total of eight metabolic parameters: fluorescence intensity (I), short lifetime (τ1), long lifetime (τ2), and fraction engaged with enzyme (F), for both NADH and FAD (Figure 1). Thus, FLIM of NADH and FAD provided both subcellular morphological information and a quantitative characterization of mitochondrial metabolism.
Figure 1:
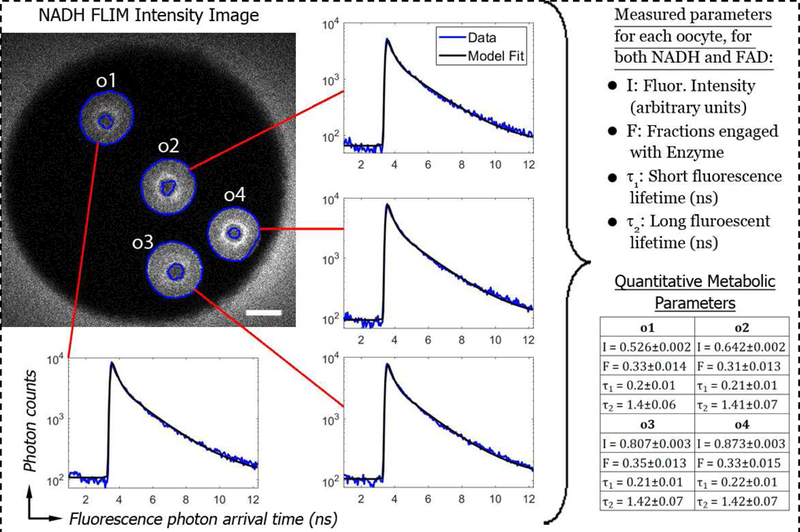
(Upper Left) NADH intensity image of four Clpp+/+ (wildtype) oocytes in one microwell of a 9-well dish. The cytoplasm of the oocytes was segmented (blue line boundaries), and oocytes were individually identified (here labelled o1, o2, o3, and o4). 50um scale bar. (Inserted Histograms) Histograms of photon counts vs fluorescence photon arrival time were constructed for each oocyte (blue curves) and fit with a model containing two fluorescent species (black lines). (Right) These fits allow four parameters to be measured. Similar fits were also performed for FAD fluorescence, allowing an additional four parameters to be measured.
We first compared the morphology of Clpp+/+ and Clpp−/− oocytes. While brightfield images of Clpp+/+ and Clpp−/− oocytes appear quite similar (compare left images of Figure 2A and2B), the NADH intensity images are markedly different, with many Clpp−/− oocytes displaying a strong NADH signal around their periphery (compare right images of Figure 2A and2B). Because NADH is highly enriched in mitochondria (39), the brighter regions of the NADH images reflect the concentration of mitochondria, indicating that the subcellular localization of mitochondria is often perturbed in Clpp−/− oocytes. Conversely, Clpp+/+ oocytes did not exhibit such mitochondrial anomalies, and consistently showed perinuclear localization of the mitochondria, which has been previously observed to occur during oocyte maturation (40, 41). We therefore conclude that NADH imaging of oocytes provides novel morphological information that is inaccessible to conventional brightfeld microscopy.
Figure 2:
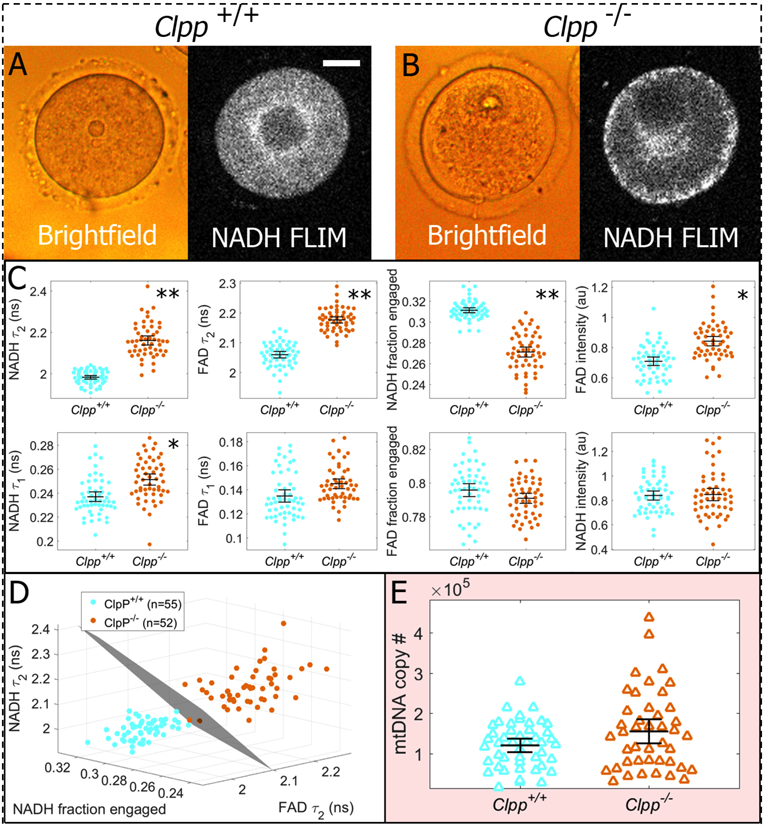
Comparative assessments of Clpp+/+ and Clpp−/− oocytes. A) Brightfield images show gross morphological features of oocytes, while NADH FLIM intensity images reflect the mitochondrial distribution. B) Obvious defects in Clpp−/− oocytes were not observed with brightfield, but fluorescence imaging revealed aberrant mitochondrial distributions in many oocytes. C) Metabolic imaging of Clpp+/+ (n=55) and Clpp−/− oocytes (n=52) detected highly significant differences in five of the eight parameters measured. Parameters are plotted in order of decreasing separation, with asterisks indicating the following p-values: (*: p<10−11) and (**: p<10−26) D) If the three most sensitive metabolic parameters (NADH long lifetime, NADH fraction engaged, and FAD long lifetime) are represented in a 3D plot, we can fit a plane that perfectly separates the two data sets. E) Clpp+/+ oocytes (n=46) and Clpp−/− oocytes (n=43) were lysed to obtain individual mtDNA copy number measurements. T-tests on mtDNA measurements showed only a marginally significant difference with a p-value of 0.042. Error bars represent standard errors.
We next investigated biochemical differences between mitochondria in Clpp−/− and Clpp+/+ oocytes. Five of the eight measured FLIM parameters revealed extremely significant differences between Clpp+/+ and Clpp−/− oocytes (Figure 2C): NADH long lifetime (p = 2e-32), FAD long lifetime (p = 9e-31), NADH fraction engaged (p = 2e-26), FAD intensity (p = 3e-15), and FAD short lifetime (p = 4e-11). The support vector machine calculation performed on the three most differentiating parameters (NADH long lifetime, FAD long lifetime, and NADH fraction engaged) produced a plane that perfectly separated the data, producing a test sensitivity and specificity of 1 (Figure 2D), demonstrating that the differences in their metabolism can be easily resolved with FLIM.
In contrast, comparing mtDNA copy number, measured by qRT-PCR, revealed only a marginally significant difference between Clpp−/− and Clpp+/+ oocytes (p = 0.042) (Figure 2E). As mtDNA measurements were taken on the same oocytes as the FLIM measurements, we were able to investigate the correlation between them. However, we did not observe strong correlations between FLIM parameters and mtDNA. NADH τ1 showed the highest correlation, with a Pearson coefficient of only 0.34.
Old vs young oocytes
We next compared oocytes from old and young mothers to determine whether FLIM-based metabolic imaging can detect mild metabolic dysfunction. We imaged a total of 29 oocytes from old (1-year old) mice and 35 oocytes from young (12-wek-old) mice. Neither bright field images nor NADH intensity images revealed obvious differences between the morphology of young and old oocytes (Figure 3A, 3B). In contrast, four of the eight measured FLIM parameters were highly significantly different between young and old oocytes (Figure 3C): NADH short lifetime (p = 4e-7), NADH long lifetime (p = 2e-4), NADH intensity (p = 2e-3), and FAD long lifetime (p = 0.02). Simultaneously plotting the NADH short lifetime, NADH long lifetime, NADH intensity, for each oocyte on one three-dimensional graph revealed a clear separation between young and old oocytes (Figure 3D). Sensitivity or specificity was not calculated however, as age is not a disease condition, but rather only correlates with oocyte failure.
Figure 3:
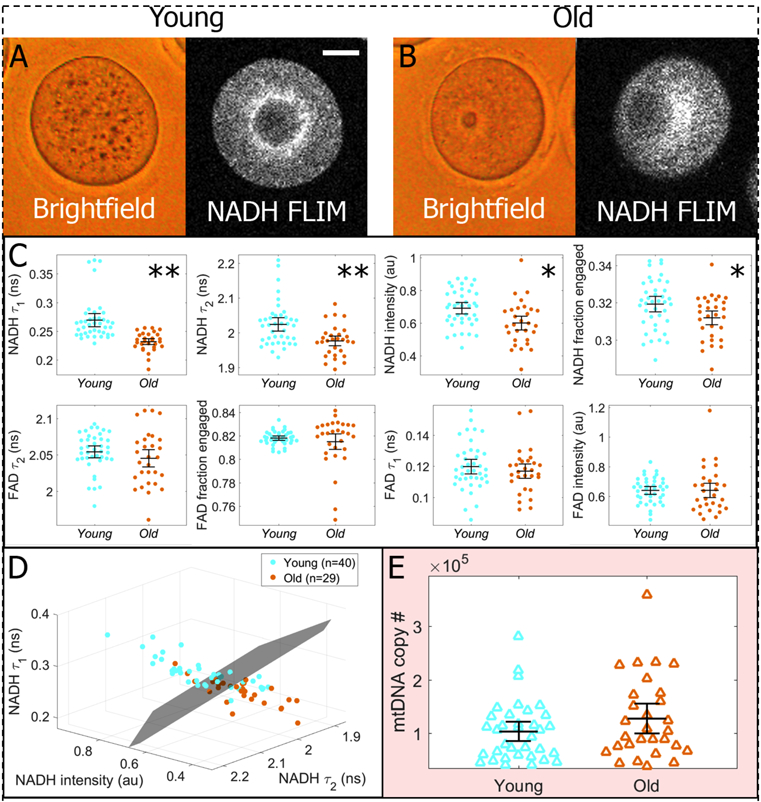
Comparative assessments of oocytes from old (12-month) and young (3-month) mice. A,B) Neither brightfield images nor FLIM intensity images revealed obvious differences in morphology or mitochondrial distribution. 20um bar. C) Conversely, metabolic imaging measurements on the same oocytes effectively differentiated young (n=35) and old (n=29) groups, with four of the eight parameters showing significant differences. Parameters are plotted in order of decreasing separation, with asterisks indicating the following p-values: (*:p<0.02) and (**: p<10−3). P-values were not as low as with Clpp, the more severe case of metabolic dysfunction. D) If the three most sensitive metabolic parameters (NADH intensity NADH short lifetime, and NADH long lifetime) are represented in a 3D plot, we can draw a plane that effectively separates the two data sets; however, there is more overlap between the distributions than in the more extreme case of Clpp. E) mtDNA copy number measurements were taken on young (n=35) and old (n=29) oocytes, and no significant difference between the two groups was observed (p=0.14). Error bars represent standard errors.
In contrast to our findings using FLIM, mtDNA copy number was not significantly different between young and old oocytes (p = 0.14) (Figure 3E). We also found no significant correlation between mtDNA and any of the FLIM parameters (highest Pearson correlation coefficient measured was 0.1 for FAD fraction engaged).
Safety of FLIM illumination
We observed no significant difference in ROS levels between illuminated and non-illuminated embryos for any of the administered photodoses (Figure 4A). Embryo numbers for the photodoses were: for 1 measurement, n=33 illuminated and n=33 non-illuminated embryos; for 3 measurements, n=33 illuminated and n=33 non-illuminated embryos; for 24 measurements, n=18 illuminated and n=18 non-illuminated embryos; and for 44 measurements, n=37 illuminated and n=39 non-illuminated embryos.
Figure 4:
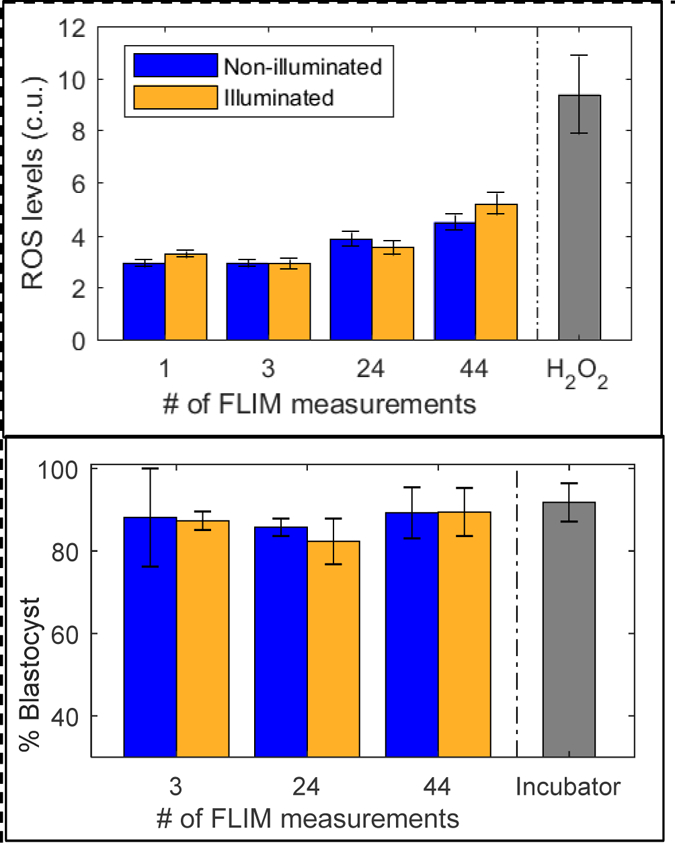
Evaluation of safety for varying photodoses of FLIM illumination. Top: Reactive oxygen species levels were measured via HC-DCFDA fluorescence (custom units). Significant differences between illuminated and non-illuminated embryos were not observed for any of the photodoses studied. Embryos exposed to 30mM H2O2 were measured as a positive control. Error standard error bars represent variation between individual embryo measurements. Bottom: Embryos were cultured on the microscope, and blastocyst development rates of illuminated embryos were compared to non-illuminated embryos in the same dish. Embryos cultured in a standard incubator were used as a control. FLIM illumination did not have any significant impact on blastocyst development rates. Standard error bars represent variation between experiment batches.
FLIM illumination resulted in no measurable impact on blastocyst development rates (Figure 4B). For each photodose, blastocyst rates were averaged between several replicates with the following numbers: for 3 measurements, n=39 illuminated and n= 37 non-illuminated batches; for 24 measurements, n=184 illuminated and n=205 non-illuminated batches; and for 44 measurements, n=52 illuminated and n=51 non-illuminated batches.
Theoretical estimates of light energy absorbed during an FAD FLIM acquisitions yielded values of 1.75 picojoules from first principles and 2.31 picojoules from our empirical estimate. By comparison, we estimate that oocytes absorb 2.62 picojoules of light energy during 10s of a standard morphological assessment.
DISCUSSION
Here we report the application of FLIM-based metabolic imaging of NADH and FAD as a diagnostic strategy for identifying metabolic dysfunction in oocytes. To adequately assess the diagnostic potential of FLIM, we utilized two models. First, we compared mice with a targeted deletion of Clpp to wild type mice. Clpp is a key factor in mitochondrial stress response, and its absence is associated with impaired mitochondrial function and loss of mitochondrial protein homeostasis (proteostasis), therefore generating a severe metabolic dysfunction (34). In addition, Clpp-knockout mice display elevated mtDNA copy number and infertility. The second model, compared oocytes from young (12-week-old) and old (1-year-old) mice provided an example of mild/moderate metabolic dysfunction, as previously published (42).
Metabolic imaging parameters exhibited clear separations between control and test groups for both old-young and Clpp knockout/wild type experiments. Aging resulted in decreased NADH intensity, which is in agreement with previous studies showing decreasing NADH levels in various cell types (43). Old oocytes also had lower NADH fraction engaged, NADH long and short lifetimes, and FAD long lifetime. Group separation was not as large as in the Clpp-deficient oocytes, consistent with the premise that aging is not as severe as a metabolic mutation, and there was significant overlap in the old and young parameter distributions. This observation is in agreement with what is known to be true in the clinical context—that young mothers have more viable eggs than old mothers on average, but young mothers have some non-viable eggs and old mothers have some viable ones.
Knockout of Clpp resulted in different parameter shifts: higher FAD intensity, higher FAD short and long lifetimes, and lower NADH fraction engaged. This difference in metabolic signature is not surprising, as these two types of metabolic dysfunction are likely due to very distinct mechanisms. Furthermore, the overall distributions of the two different strains were very different, indicating strain-dependence of metabolic states. This intriguing result suggests that ‘healthy’ metabolic function can be achieved via multiple metabolic configurations in different systems. For clinical diagnostic purposes, it is sufficient to show that within systems, this technique has high precision for differentiating metabolic states associated with good and bad prognosis. In human, we may expect that healthy metabolism would have consistent parameter ranges, and that various forms of metabolic defects would register as deviations from these ‘Goldilocks’ ranges (44).
At this early stage, we can make several first-order physiological interpretations of the data. Intensity changes primarily reflect changes in molecule concentrations, and fractions engaged provide information about enzyme activity. Changes in NADH τ2 and FAD τ1 (engaged lifetimes) can be caused by a change in the proportions of molecules engaged with different kinds of enzymes. To make more precise interpretations, experiments using controlled metabolic perturbations and biophysical modeling are underway.
We found that the differences observed between the groups using FLIM-parameters were much more significant compared to that achieved by mtDNA quantification. mtDNA copy number was significantly different between Clpp-knockout mouse oocytes compared to oocytes obtained from same age wild type mouse (with a p value of less than 0.05), while the mtDNA copy number of old and young mouse GV stage oocytes did not differ. Several factors could explain the limitations of mtDNA copy number as a diagnostic test. First, significant variation in mtDNA copy number exists between individual oocytes. Indeed, both in mouse and human, somewhere between 50,000 to 550,000 mtDNA copies is fund in oocytes, with considerable degree of variability between samples, and increasing variability with age in human (45–49). Our observations in the current study are consistent with previous reports on this subject. Second, mtDNA copy number measurement in a single cell (or 5 to 10 cells available in a trophoectoderm biopsy) is challenging due to low copy number. Investigators tackled this issue using different techniques. Some used qRT-PCR followed by normalization to Alu repeat sequences in nuclear DNA (21). Others measured mtDNA copy number by next generation sequencing (NGS) normalized to total nuclear DNA following whole genome amplification (WGA) (24), while we performed absolute quantification by cloning a segment of mtDNA, and using known copy number as standards (42). While each one of these approaches is justified scientifically, the level of intra- and inter-assay variability that they introduce remains to be established. Finally, mtDNA copy number is an indirect marker of cellular distress, as there are currently no experimental models showing a direct correlation (linear or otherwise) between cellular stress and mtDNA copy number. While targeted deletion of genes required for key mitochondrial functions have been associated with elevated mtDNA copy number (34), others reported low mtDNA copy number in animal models of mitochondrial distress (50). These challenges contribute to the current debate regarding the use of mtDNA copy number in embryo viability assessment. Initial studies from two independent groups strongly suggested that mtDNA copy number is higher in euploid embryos that fail to implant (22, 36). The findings of these two studies have been challenged by others who failed to find a similar association (23), and a recent report that showed that higher mtDNA copy number did not predict implantation failure in sibling embryos (in women undergoing double embryo transfer) (24).
While our findings regarding the use of FLIM for metabolic imaging of oocytes findings are quite promising, our study has a number of limitations. First, we used a mouse model, which allowed us to conduct our experiments in a genetically homogeneous system. While level of variations in humans remains to be determined through clinical studies, it is likely that mitochondrial states will not be much more heterogeneous in human, as mitochondrial state and function are under tight selective pressure. Second, we assessed FLIM parameters in GV stage oocytes, primarily because Clpp knockout mice do not generate mature oocytes or early embryos, and assessment at the GV stage allowed us to compare and contrast the two models. Whether metabolic distress in embryos results in similar deviations from normal remains to be investigated. Finally, and most importantly, our study only demonstrates a clear distinction between healthy and unhealthy cohorts (wild type vs. Clpp knockout and young vs. old). While these findings are promising, whether FLIM parameters will be able to differentiate embryos that are “more” or “less” likely to implant within the same cohort (i.e. embryos obtained from the same patient), will be important to investigate.
The discovery of biomarkers for human embryo viability is a central challenge for contemporary reproductive scientists. Our findings, combined with recently reported correlations between mtDNA copy number and human embryo viability, suggest that mitochondrial metabolic parameters may play a role in determining the viability of oocytes and potentially the implantation potential of euploid embryos. Further research is required to validate the accuracy and clinical potential of this test.
Acknowledgements
The authors would like to thank Becker Hickl for contributing a single-photon counting detector and TCSPC electronics to this research. This project was supported by the Blavatnik Biomedical Accelerator Grant at Harvard University. T.S. was supported by NSF Postdoctoral Research Fellowship in Biology grant (1308878). T.W. was supported by the National Natural Science Foundation of China (81501247). E.S. was supported by Award R01HD059909, from the National Institute of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Tim Sanchez and Dan Needleman are co-founders and stake holders at LuminOva, Inc.
Summary sentence: Mitochondrial dysfunction caused by reproductive aging (mild) and CLPP-deficiency (severe) is accurately detected by Fluorescence Lifetime Imaging Microscopy (FLIM).
REFERENCES
- 1.Bromer JG,Seli E. Assessment of embryo viability in assisted reproductive technologies: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol 20: 234–241, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Steptoe PC,Edwards RG. Birth after the reimplantation of a human embryo. Lancet 2: 366, 1978. [DOI] [PubMed] [Google Scholar]
- 3.Trounson A, Leeton J, Wood C, Webb J,Wood J. Pregnancies in humans by fertilization in vitro and embryo transfer in the controlled ovulatory cycle. Science 216: 681–2, 1981. [DOI] [PubMed] [Google Scholar]
- 4.Scott LA,Smith S. The successful use of pronuclear embryo transfer the day following oocyte retrieval. Hum Reprod 13: 1003– 1013, 1998. [DOI] [PubMed] [Google Scholar]
- 5.Tesarik J,Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear state morphology. Hum Reprod 14: 1318–1323, 1999. [DOI] [PubMed] [Google Scholar]
- 6.Veeck L, An Atlas of Human Gametes and Conceptuses: an illustrated reference for assisted reproductive technology 1999: Parthenon Publishing, New York, USA. [Google Scholar]
- 7.Gerris J, De Neubourg D, Mangelschots K,al. e. Prevention of twin pregnancy after in-vitro fertilization or intracytoplasmic sperm injection based on strict embryo criteria: a prospective randomized clinical trial. Hum Reprod 14: 2581–7, 1999. [DOI] [PubMed] [Google Scholar]
- 8.VanRoyan E, Mangelschots K, De Neubourg D, Valkenburg M, Van de Meerssche M, Ryckaert G, Eestermens W,Gerris J. Characterization of a top quality embryo, a step towards single-embryo transfer. Hum Reprod 14: 2345–2349, 1999. [DOI] [PubMed] [Google Scholar]
- 9.Gardner DK,Schoolcraft WB, In vitro culture of human blastocysts., in Towards Reprodcutive Certainty: Fertility and Genetics Beyond, M D. Jansen R, Editor. 1999, Carnforth: Parthenon publishing; p. 378–88. [Google Scholar]
- 10.Toner JP. Progress we can be proud of: U.S. trends in assisted reproduction over the first 20 years. Fertility & Sterility 78: 943–50, 2002. [DOI] [PubMed] [Google Scholar]
- 11.SART. Assisted reproductive technology success rates. National summary and fertility clinic reports Centers for disease control, USA., 2013. [Google Scholar]
- 12.Stecher A, Vanderzwalmen P, Zintz M, Wirleitner B, Schuff M, Spitzer D,Zech NH. Transfer of blastocysts with deviant morphological and morphokinetic parameters at early stages of in-vitro development: a case series.. Reprod Biomed Online 28 424–35, 2014. [DOI] [PubMed] [Google Scholar]
- 13.Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J,Zaragoza M. Human aneuploidy: incidence, origin, and etiology. Environmental & Molecular Mutagenesis 28: 167–75, 1996. [DOI] [PubMed] [Google Scholar]
- 14.Hassold T,Chiu D. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 70: 11–7, 1985. [DOI] [PubMed] [Google Scholar]
- 15.Hassold T, Hall H,Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 16: R203–8, 2007. [DOI] [PubMed] [Google Scholar]
- 16.Hassold T,Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Reviews Genetics 2: 280–291, 2001. [DOI] [PubMed] [Google Scholar]
- 17.Mastenbroek S, Twisk M, van der Veen F,Repping S. Preimplantation genetic screening: A systematic review and meta-analysis of RCTs. Hum Reprod Update 17 454–66, 2011. [DOI] [PubMed] [Google Scholar]
- 18.Scott RT, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, Tao X,Treff NR. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: A randomized controlled trial. Fertil Steril 100: 697–703, 2013. [DOI] [PubMed] [Google Scholar]
- 19.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, Treff NR,Scott RJ. In vitro fertilization with single euploid blastocyst transfer: A randomized controlled trial. . Fertil Steril 100 100–107, 2013. [DOI] [PubMed] [Google Scholar]
- 20.Seli E Mitochondrial DNA as a biomarker for in-vitro fertilization outcome. Curr Opin Obstet Gynecol 28: 158–63, 2016. [DOI] [PubMed] [Google Scholar]
- 21.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S,Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet 11: e1005241, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, Díaz-Gimeno P, Valbuena D,Simón C. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril 104: 534–541, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL,Viotti M. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril 107: 34–42, 2017. [DOI] [PubMed] [Google Scholar]
- 24.Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, Morrison L, Morin SJ,Scott RTJ. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. . Hum Reprod 32: 954–62, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks KA, Howe DK, Leung A, Denver DR,Estes S. In vivo quantification reveals extensive natural variation in mitochondrial form and function in Caenorhabditis briggsae. PLoS One 7 e43837, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rr Scott, Zhang M,E S. Metabolism of the oocyte and the preimplantation embryo: implications for assisted reproduction. Curr Opin Obstet Gynecol 30: 163–170, 2018. [DOI] [PubMed] [Google Scholar]
- 27.Becker W Fluorescence lifetime imaging – techniques and applications. . J Microsc 247 119–36, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Heikal A Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. . Biomark Med 4 241–63. , 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker W, Advanced Time-Correlated Single Photon Counting Techniques 2005, Berlin Heidelberg: Springer-Verlag [Google Scholar]
- 30.Yu Q,Heikal A. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B 95: 46–57, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niesner R, Peker B, Schlüsche P, Gericke K-H. Noniterative biexponential fluorescence lifetime imaging in the investigation of cellular metabolism by means of NAD(P)H autofluorescence. Chemphyschem 5: 1141–9, 2004. [DOI] [PubMed] [Google Scholar]
- 32.Stringari C, Cinquin A, Cinquin O, Digman MA, Donovan PJ,Gratton E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. . Proc Natl Acad Sci U S A 108 13582–7, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skala M, Riching K, Bird D, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri K, Keely P,Ramanujam N. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt 12: 24014, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Babayev E, Jiang Z, Li G, Zhang M, Esencan E, Horvath T,Seli E. Mitochondrial unfolded protein response gene Clpp is required to maintain ovarian follicular reserve during aging, for oocyte competence, and development of pre-implantation embryos. Aging Cell In pess, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters BR,So P, Handbook of biomedical nonlinear optical microscopy 2008: Oxford University Press. [Google Scholar]
- 36.Gispert S, Parganlija D, Klinkenberg M, Dröse S, Wittig I, Mittelbronn M, Grzmil P, Koob S, Hamann A, Walter M, Büchel F, Adler T, Hrabé de Angelis M, Busch DH, Zell A, Reichert AS, Brandt U, Osiewacz HD, Jendrach M,Auburger G. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum Mol Genet 22: 4871–87, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N,Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci USA 102: 367–72, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denk W, Strickler JH,Webb WW. Two-photon laser scanning fluorescence microscopy. Science 248: 73– 6, 1990. [DOI] [PubMed] [Google Scholar]
- 39.Stein L,Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 23: 420–8, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Blerkom J,Runner M. Mitochondrial reorganization during resumption of arrested meiosis in the mouse oocyte. . Am J Anat 171: 335–55, 1984. [DOI] [PubMed] [Google Scholar]
- 41.Muggleton-Harris A,Brown J. Cytoplasmic factors influence mitochondrial reorganization and resumption of cleavage during culture of early mouse embryos. . Hum Reprod 3 1020–8, 1988. [DOI] [PubMed] [Google Scholar]
- 42.Babayev E, Wang T, Lowther K, Horvath T, Taylor HS,Seli E. Aging is associated with changes in mitochondrial dynamics, function and mtDNA quantity.. Maturitas Epub June 23, 2016. [DOI] [PMC free article] [PubMed]
- 43.Camacho-Pereira J, Tarragó MG, Chini CCS, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A,Chini EN. CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. . Cell Metab 23 1127–39. , 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leese HJ, Guerif F, Allgar V, Brison DR, Lundin K,Sturmey RG. Biological optimization, the Goldilocks principle, and how much is lagom in the preimplantation embryo. . Mol Reprod Dev 83 748–54, 2016. [DOI] [PubMed] [Google Scholar]
- 45.Steuerwald N, Barritt JA, Adler R, Malter H, Schimmel T, Cohen J,Brenner CA. Quantification of mtDNA in single oocytes, polar bodies and subcellular components by real-time rapid cycle fluorescence monitored PCR. Zygote 8: 209–215, 2000. [DOI] [PubMed] [Google Scholar]
- 46.Reynier P, May-Panloup P, Chrétien MF, Morgan CJ, Jean M, Savagner F, Barrière P,Malthièry Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod 7: 425–429, 2001. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Prosser R, Simonetti S, Sadlock J, Jagiello G,Schon EA. Rearranged mitochondrial genomes are present in human oocytes. Am J Hum Genet 57: 239–47, 1995. [PMC free article] [PubMed] [Google Scholar]
- 48.Pikó L,Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 123: 364–374, 1987. [DOI] [PubMed] [Google Scholar]
- 49.Chan CC, Liu VW, Lau EY, Yeung WS, Ng EH,Ho PC. Mitochondrial DNA content and 4977 bp deletion in unfertilized oocytes. Mol Hum Reprod 11: 843–846, 2005. [DOI] [PubMed] [Google Scholar]
- 50.Zhang M, Wang T, Esencen E, Jiang Z,Seli E. Disruption mitofusin 1 in oocyte results in defective ovarian follicle development and female infertility. SRI, 2018. [Google Scholar]


