Abstract
Tools that allow inducible and reversible depletion of target proteins are critical for biological studies. The plant-derived auxin-inducible degradation system (AID) enables the degradation of target proteins tagged with the AID motif. This system has been recently employed in mammalian cells as well as in C. elegans and Drosophila. To test the utility of the AID approach in the nervous system, we used circadian locomotor rhythms as a model and applied the AID method to temporally and spatially degrade PERIOD (PER), a critical pacemaker protein in Drosophila. We found that the period locus can be efficiently tagged with the AID motif by CRISPR/Cas9-based genome editing without disrupting PER function. Moreover, we demonstrated that the AID system could be used to induce rapid and efficient protein degradation in the nervous system as shown by effects on circadian and sleep behaviors. Furthermore, the protein degradation by AID was rapidly reversible after auxin removal. Together, our results show that the AID system provides a powerful tool for behavior studies in Drosophila.
Keywords: auxin-inducible degradation, CRISPR/Cas9, Drosophila melanogaster, PERIOD, circadian rhythm
Graphical Abstract
Tools that allow rapid temporal and spatial depletion of target proteins are critical for biological studies. The auxin-inducible degradation system (AID) has been recently developed to enable the degradation of target proteins tagged with the AID motif. Here we used the Drosophila circadian locomotor rhythms as a proof of principle to test the potential use of the AID system in the nervous system. We report that the AID system can be used to elicit rapid and reversible degradation of PERIOD protein in the Drosophila nervous system.
Introduction
Various experimental tools for precise spatial and temporal control of gene expression have been developed that allow interrogation of gene function in living organisms. Temporally restricted genetic manipulations are critical for biological studies, and several strategies for temporally controlling gene expression have been developed. Some that depend on temperature, chemical ligand, or light triggers have been adapted for use in Drosophila [1–6]. Often, these methods cause side effects. For example, elevated temperatures may result in unwanted physiological changes [7, 8]. In addition, the most frequently used techniques in Drosophila to deplete gene products in specific stages and tissues have been achieved through RNA interference (RNAi) techniques, FLP-mediated excision of FRT flanked exon techniques, or CRISPR/Cas9-mediated deletion [9–12], approaches that manipulate protein expression at the level of genome or the transcriptome, meaning that they are indirect and sometimes irreversible. Additionally, there are often longer time frames for protein depletion than the process under investigation [13]. Therefore, techniques that directly enable rapid protein depletion in a controllable (inducible and reversible) manner are particularly desirable as this would allow the study of protein function at organismal level.
Recently, a variety of tools have been successfully applied to induce direct and specific protein degradation. Among these methods, the plant-derived auxin-inducible degradation (AID) system [14–17] is a promising tool for the rapid and conditional control of protein clearance. The AID method has been successfully applied to mammalian cells as well as C. elegans and Drosophila [18–22]. Using the AID system, proteins are tagged with the AID degron peptide. Upon addition of auxin, the AID-tagged protein is recruited to an engineered E3 ubiquitin ligase SCF complex, containing the TIR1 F-box protein from plants, which binds target proteins only in the presence of the plant hormone auxin [14]. Expression of TIR1 in non-plant cells is sufficient to result in the formation of the SCF complex, which then binds AID-tagged proteins in an auxin-dependent manner, leading to polyubiquitination and degradation via the proteasome [14]. In Drosophila, the AID system has been successfully employed in somatic and germline tissues at various stages of development [20, 22]. Given that the central nervous system (CNS) is generally protected by a blood brain barrier, the functionality of the AID system within the CNS cannot be deduced automatically from the previously published AID system tests. Here we used circadian locomotor rhythms as a model to evaluate utility of the AID method for behavioral studies in Drosophila.
The circadian rhythm is a roughly 24-hour cycle in the physiological processes of organisms from fungi to mammals. Endogenous molecular oscillators, built into the circadian neuronal network, generate these circadian rhythms. In Drosophila, a network of approximately 150 clock neurons comprises the circadian clock, and robust rhythmic expression of clock genes is observed in these cells [23–25]. The clock neuron network in the brain consists of three groups of dorsal neurons (DN1s, DN2s, and DN3s), three groups of lateral neurons (large LNvs, small LNvs, and LNds), and lateral posterior neurons (LPNs) [23–25]. The neuropeptide PIGMENT DISPERSING FACTOR (PDF), the main neuromodulator of the circadian neuronal network, is expressed in the LNvs [23]. The core clock mechanism consists of negative transcription translation feedback loops. The CLOCK/CYCLE (CLK/CYC) heterodimer acts as a transcription factor to initiate rhythmic transcription of hundreds of clock-controlled genes, such as period and timeless [23]. At a certain concentration, PERIOD (PER) dimerizes with TIMELESS (TIM) and inhibits the transcription activity of the CLK/CYC. The PER protein, encoded by the first gene known to control behavior of any kind, is an essential component of the circadian clock and is well characterized in Drosophila [23, 26–28].
Here we used the circadian locomotor behavior as a model to test the potential applicability of the AID method in the nervous system. By tagging the period locus with the AID motif and feeding flies a low concentration of auxin, we were able to temporally and spatially deplete PER and thus modulate circadian behavior rhythms. These results demonstrate that the AID system will be a useful functional tool for behavior and neuroscience studies.
Results
Design strategy for the AID system in the nervous system of Drosophila
To examine whether the plant-derived AID system is functional in the nervous system of Drosophila, we chose tim-Gal4, a pan-clock tissue driver [29], and the period locus to target in the circadian rhythm system (Fig. 1A). We first applied CRISPR/Cas9-mediated homologous recombination to fuse the AID motif together with an EGFP tag into the period locus after the region encoding the C terminus of the protein. Two highly specific genomic target sites (period-gRNA1 and period-gRNA2) flanking the stop codon of the period gene were selected using the online flyCRISPR Optimal Target Finder Tool [30] (Fig. 1B). The targeting sequences were cloned into a plasmid under the transcriptional control of the U6 promoter to generate targeting chiRNAs. For homology-directed repair we used a double-stranded DNA plasmid donor with two homology arms of approximately 1 Kb (Fig. 1B). The two protospacer adjacent motifs (PAM) of targeting sites on homology arms were mutated to prevent cutting by Cas9 after homology-directed repair events (Fig. 1C). chiRNAs and donor plasmids were mixed and injected into nos-Cas9 fly embryos. PCR and sequencing results demonstrated a high percentage of positive KI integration in P0 flies (13 out of 23; 56.5%) (Fig. 1C). Thus, the endogenous period locus was site-specifically and efficiently edited with CRISPR/Cas9.
Figure 1. Generation of the period-AID-EGFP allele by CRISPR/Cas9-induced homologous recombination.
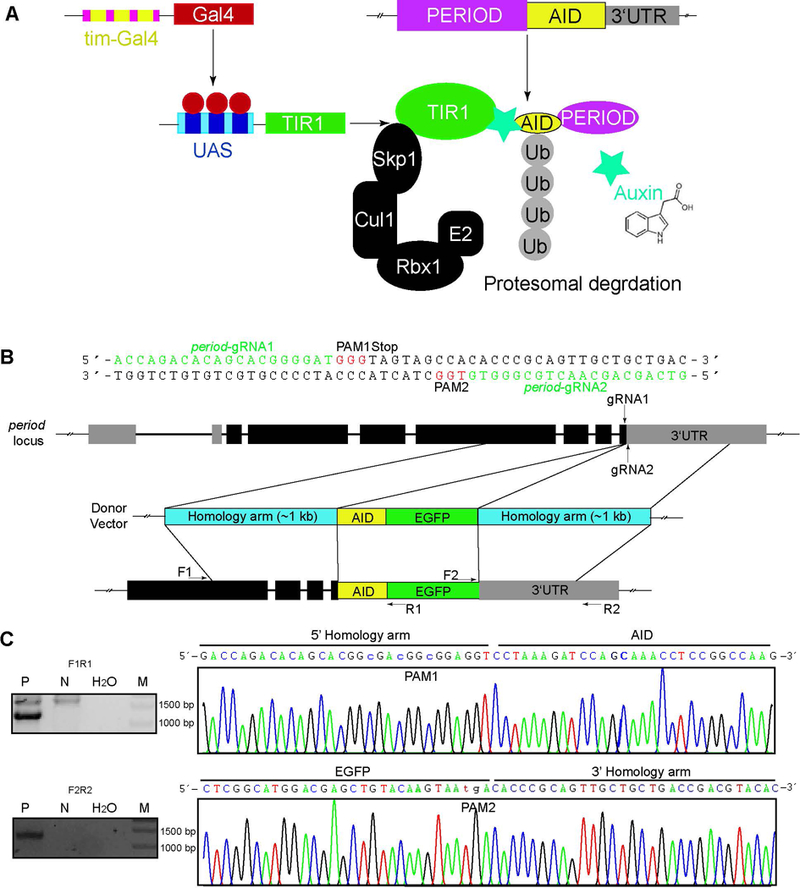
(A) A schematic of AID in the circadian rhythm system. The Gal4-UAS system is used to introduce TIR1 from rice (OsTIR1), which includes an F box that allows integration into an SCF ubiquitin ligase complex together with the endogenous Drosophila proteins. The endogenous PER protein is fused with the AID (PER-AID). In the presence of auxin, PER-AID is recruited to OsTIR1, resulting in its polyubiquitinylation and proteasomal degradation. (B) Strategy for tagging the period gene with AID-EGFP. The two target sites located in the last exon and downstream of the 3’UTR are shown. The circle donor vector containing two homology arms was designed to insert the AID-EGFP tag before the stop codon of period. (C) PCR analysis of 5’ and 3’ integration of AID-EGFP and representative sequencing results of the period-AID-EGFP allele. P, positive (y,sc,v,period-AID-EGFP), N, negative (y,sc,v), M, maker.
Expression and subcellular localization of the AID- and EGFP-tagged PER protein in adult brains
To determine the expression pattern of the AID-EGFP-fused PER protein, we used anti-GFP and anti-PER antibodies to detect EGFP and the PER protein in period-AID-EGFP homozygous flies. We entrained flies under regular light:dark (LD) cycles and dissected fly brains at circadian time 1 (CT1) under the first day of constant darkness, a time of high PER abundance. The expression pattern of EGFP was very similar to that of endogenous PER in the fly brain (Fig. 2A, B). The subcellular localization of the tagged PER protein (as revealed by GFP staining) in the circadian neurons was the same as that of the wild-type PER (Fig. 2C, D). Thus, the AID-EGFP-tagged PER is expressed with the same pattern as the endogenous PER.
Figure 2. Expression pattern of the AID-EGFP-tagged PER protein in period-AID-EGFP and control fly brains.
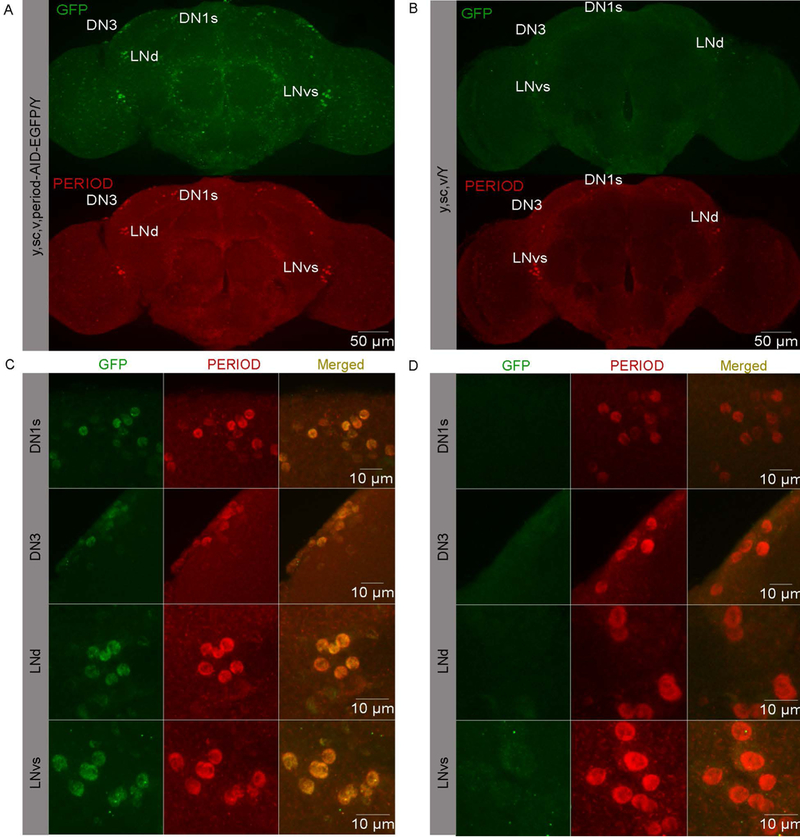
(A-B) Whole adult brains of A) period-AID-EGFP and B) control flies stained with anti-GFP and anti-PER at CT1. (C-D) Specific circadian neurons stained with anti-GFP and anti-PER in C) period-AID-EGFP and D) control flies. LNd, dorsal lateral neuron; lLNv, large ventral LN; sLNv, small ventral LN. DN, dorsal neuron.
PER fused with AID-EGFP at the C terminal is fully functional
We next evaluated whether the fusion of AID-EGFP alters PER function. Under 12:12 h LD conditions, Drosophila gradually increase their circadian locomotor activity in advance of the lights-on and lights-off signals and exhibit peaks of activity during dawn and dusk, a phenomenon termed anticipation [23, 24, 31, 32]. The per01 mutant flies lose morning and evening anticipation [33–35](Fig. 3A). Both morning and evening anticipation were normal in period-AID-EGFP flies (Fig. 3B–C). Moreover, the period-AID-EGFP allele was able to rescue the anticipatory behavior in female per01 flies (Fig. 3D–H). Drosophila sleep is also under circadian regulation, and per01 mutants have disrupted rhythms of sleep [24]. The circadian rhythm and sleep patterns of wild-type and period-AID-EGFP flies were comparable (Table 1 and Fig. 3I). Thus, these circadian and sleep behavior results indicate that the modification of the period allele by fusion to the sequence encoding AID-EGFP had no obvious effects on PER function.
Figure 3. Locomotor and sleep behaviors of period-AID-EGFP flies are normal.
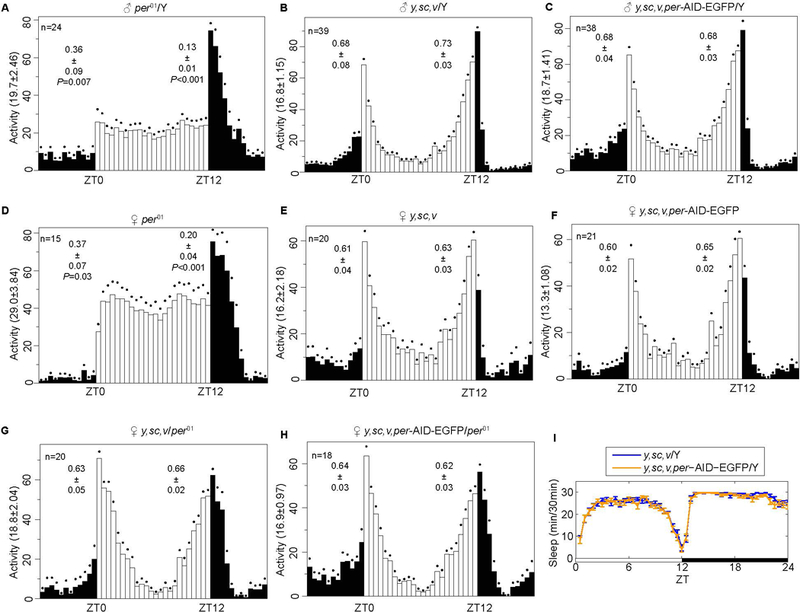
(A-H) Average activities for A) male per01 flies, B) male controls, C) male y,sc,v,period-AID-EGFP flies, D) female per01 flies, E) female controls, F) female y,sc,v,period-AID-EGFP flies, G) female y,sc,v/per01 flies and H) female y,sc,v,period-AID-EGFP/per01 flies during 12 h of light and 12 h of dark at 25 ℃. (I) Sleep patterns of male y,sc,v,period-AID-EGFP /Y and control flies. White bars represent day, black bars represent night, Zeitgeber time (ZT) is indicated on the x axes. The dots above the bars indicate SEM. The total numbers of each genotype are shown in the figure. The morning and evening anticipation signals are shown as means ± SEM. A Student’s two-tailed t-test was employed to determine significance compared to controls (y,sc,v/Y or y,sc,v). P values are shown in the figures.
Table 1.
Locomotor activity of flies.
| Auxin (mM) | Genotypes | Tau | Power | Rhythmic flies | Total flies | % Rhythmic |
|---|---|---|---|---|---|---|
| y,sc,v/Y | 24.43±0.21 | 95.40±9.94 | 12 | 12 | 100 | |
| y,sc,v,per-AID-EGFP/Y | 24.73±0.21 | 107.20±9.67 | 18 | 18 | 100 | |
| 0 | AID-TG4-TIR1 | 24.26±0.07 | 88.07 ±5.92 | 35 | 43 | 81 |
| 25 | AID-TG4-TIR | 23.53±0.80 | 98.33 ±28.00 | 4 | 45 | 9 |
| 100 | AID-TG4-TIR | 23.77±0.67 | 38.27 ±12.21 | 3 | 47 | 6 |
| 400 | AID-TG4-TIR | 24.1 | 115.8 | 1 | 41 | 2.5 |
| 0 | AID-TIR2 | 24.14±0.08 | 114.90 ±6.07 | 34 | 38 | 89 |
| 25 | AID-TIR | 24.03±0.06 | 80.86 ±6.39 | 35 | 37 | 95 |
| 100 | AID-TIR | 24.28±0.06 | 88.00 ±4.89 | 43 | 44 | 98 |
| 400 | AID-TIR | 24.42±0.13 | 65.80±7.05 | 26 | 18 | 76 |
| 0 | AID-TG43 | 24.81±0.06 | 97.78 ±5.54 | 37 | 38 | 97 |
| 25 | AID-TG4 | 24.88±0.06 | 92.92 ±5.59 | 31 | 39 | 79 |
| 100 | AID-TG4 | 24.96±0.05 | 77.37 ±4.65 | 45 | 56 | 80 |
| 400 | AID-TG4 | 25.20±0.08 | 76.52±6.07 | 23 | 29 | 79 |
| 0 | TG4-TIR4 | 24.59±0.12 | 88.00 ±4.99 | 38 | 40 | 95 |
| 25 | TG4-TIR | 25.13±0.20 | 84.74 ±4.61 | 38 | 47 | 81 |
| 100 | TG4-TIR | 25.65±0.60 | 69.67 ±5.59 | 26 | 43 | 60 |
| 400 | TG4-TIR | 25.11±0.24 | 55.54±6.17 | 21 | 40 | 53 |
| 0 | TIR/+5 | 24.71±0.17 | 107.80 ±5.46 | 41 | 42 | 98 |
| 25 | TIR/+ | 25.08±0.20 | 83.13 ±5.85 | 36 | 40 | 90 |
| 100 | TIR/+ | 24.77±0.16 | 78.00 ±4.48 | 43 | 44 | 98 |
| 400 | TIR/+ | 24.87±0.20 | 80.29±5.09 | 29 | 36 | 81 |
| 0 | TG4/+6 | 24.97±0.07 | 93.12 ±6.01 | 24 | 26 | 92 |
| 25 | TG4/+ | 25.08±0.20 | 91.54 ±6.42 | 24 | 27 | 89 |
| 100 | TG4/+ | 25.11±0.09 | 80.82 ±6.74 | 28 | 38 | 74 |
| 400 | TG4/+ | 25.25±0.14 | 75.02±6.89 | 12 | 15 | 80 |
| 0 | AID-PG4-TIR7 | 24.13±0.06 | 124.30±7.73 | 36 | 38 | 95 |
| 25 | AID-PG4-TIR | 24.62±0.30 | 38.46±2.44 | 9 | 34 | 26 |
| 100 | AID-PG4-TIR | 24.10±0.29 | 36.78±9.95 | 4 | 40 | 10 |
| 400 | AID-PG4-TIR | 24.5 | 25.7 | 1 | 17 | 6 |
| 0 | AID-PG48 | 24.63±0.06 | 124.60±7.36 | 22 | 24 | 92 |
| 25 | AID-PG4 | 24.70±0.13 | 88.73±8.59 | 18 | 20 | 90 |
| 100 | AID-PG4 | 24.88±0.17 | 79.56±8.56 | 10 | 10 | 100 |
| 400 | AID-PG4 | 25.14±0.11 | 85.91±6.52 | 9 | 10 | 90 |
| 0 | PG4-TIR9 | 24.91±0.19 | 107.80±6.37 | 36 | 38 | 95 |
| 25 | PG4-TIR | 25.46±0.24 | 78.56±5.43 | 28 | 38 | 74 |
| 100 | PG4-TIR | 24.84±0.20 | 75.18±7.07 | 24 | 38 | 63 |
| 400 | PG4-TIR | 24.94±0.24 | 77.20±5.31 | 15 | 26 | 58 |
| 0 | PG4/+10 | 25.16±0.26 | 110.60±9.86 | 17 | 18 | 94 |
| 25 | PG4/+ | 25.34±0.17 | 101.40±7.13 | 18 | 19 | 95 |
| 100 | PG4/+ | 25.15±0.13 | 76.38±8.90 | 11 | 12 | 92 |
| 400 | PG4/+ | 25.44±0.67 | 86.01±8.17 | 12 | 12 | 100 |
AID-TG4-TIR: y,sc,v,per-AID-EGFP/Y; tim-Gal4/+; UAS-TIR1/+
AID-TIR: y,sc,v,per-AID-EGFP/Y;; UAS-TIR1/+
AID-TG4: y,sc,v,per-AID-EGFP/Y; tim-Gal4/+
TG4-TIR: y,sc,v /Y; tim-Gal4/+; UAS-TIR1/+
TIR: y,sc,v /Y;; UAS-TIR1/+
TG4: y,sc,v /Y; tim-Gal4/+
AID-PG4-TIR: y,sc,v,per-AID-EGFP/Y; pdf-Gal4/+; UAS-TIR1/+
AID-PG4: y,sc,v,per-AID-EGFP/Y; pdf-Gal4/+
PG4-TIR: y,sc,v /Y; pdf-Gal4/+; UAS-TIR1/+
PG4: y,sc,v /Y; pdf-Gal4/+
PER abundance oscillates in circadian neurons and peaks during the late night/early day [36, 37]. To determine whether period-AID-EGFP flies show normal PER oscillation, we co-stained the brains for GFP and with PER antiserum. We found that the oscillating expression of GFP was comparable to PER in the circadian neuronal groups that we examined (Fig. 4A, B). Indeed, the oscillation of tagged PER in circadian neurons was similar with previous studies [38–40]. Furthermore, circadian clocks have been shown to operate in many peripheral tissues, such as the Malpighian tubules [41]. We observed AID-EGFP-labeled PER-positive cells in the Malpighian tubules, which also exhibit oscillations of both GFP and PER signals (Fig. 4C, D). Taken together, these data indicate that the period-AID-EGFP allele encodes a tagged-PER protein that functions normally.
Figure 4. period-AID-EGFP flies functional normally on the molecular level.
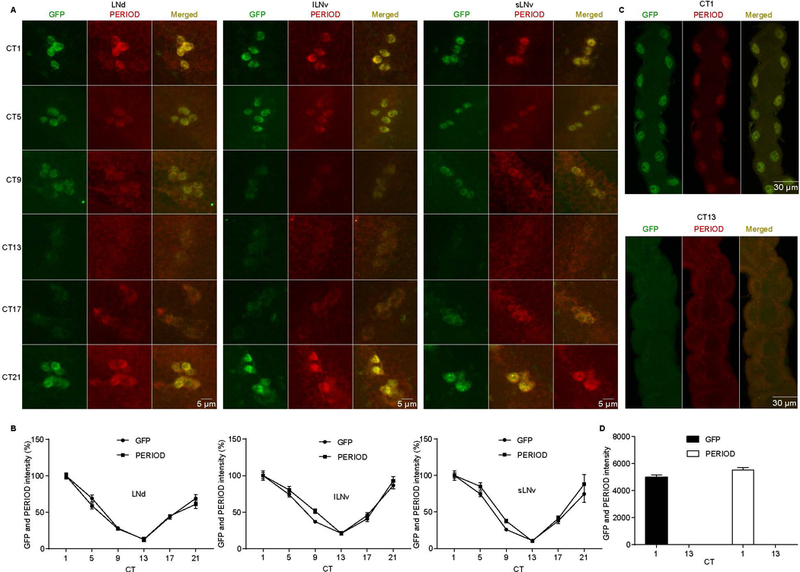
(A) Oscillation of GFP and PER in period-AID-EGFP flies. Adult brains were dissected at CT 1, 5, 9, 13, 17, and 21 at DD1 and circadian neurons were immunostained with anti-GFP and anti-PER. (B) Quantification of GFP and PER average intensity in each clock cell group (n=10). Signal was normalized to the value at CT1 which was set as 100%. CT is indicated on the x axes. (C) Malpighian tubules from period-AID-EGFP flies collected at the indicated circadian time on DD1 were immunostained with anti-GFP and anti-PER. (D) Quantification of GFP and PER intensity in Malpighian tubules (n=8). CT is indicated on the x axes. LNd, dorsal lateral neuron; lLNv, large ventral LN; sLNv, small ventral LN.
Endogenous PER protein can be conditionally depleted by the AID system
To test whether the AID system can be used to induce PER degradation, we used the GAL4-UAS system to express the TIR1 protein in all circadian tissues from the tim-Gal4 driver. When fed flies 3-indole acetic acid sodium salt (IAA) from two different suppliers to induce PER degradation, most of the flies died within two days (data not shown). We then tried the auxin analog 1-naphthaleneacetic acid (1-NAA, hereafter referred to as auxin), and observed no toxicity. We then fed flies with different concentrations of auxin (0, 25, 100, 400 mM). The period-AID-EGFP flies with TIR overexpression fed auxin lost morning and evening anticipation under LD condition and became arrhythmic under DD condition (Fig. 5A, B and Table 1). These behavior defects were observed at both concentrations of auxin for 25 and 100 mM, except the flies fed with 400 mM auxin led to decrease of rhythmicity even in control flies (Table 1). These circadian phenotypes upon auxin treatment in the TIR-expressing flies were identical to those of per01 mutant flies [26, 33–35]. Moreover, we found that auxin-treated y,sc,v,per-AID-EGFP/Y;tim-Gal4/+;UAS-TIR1/+ (AID-TG4-TIR, experimental) flies also had sleep defects similar to per01 mutants [24] (Fig. 5C). These behavior defects indicated that the molecular clock is disrupted in the period-AID-EGFP flies with TIR overexpression fed auxin, presumably due to PER degradation.
Figure 5. The AID system enables behavioral studies in nervous system of adult Drosophila.
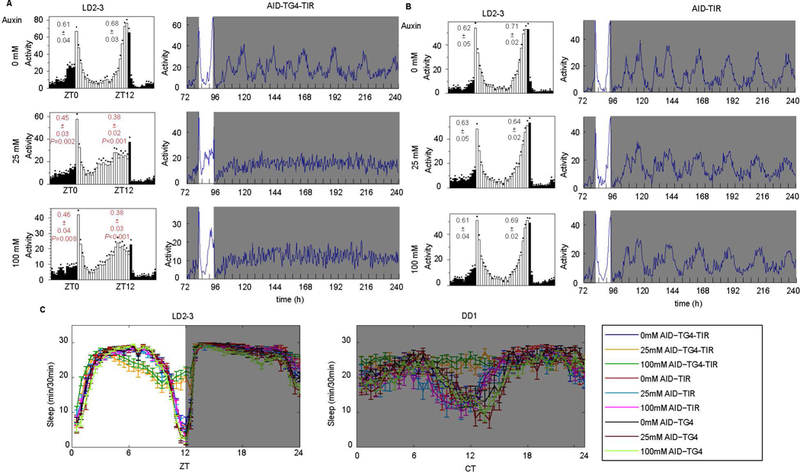
(A-B) The averaged activity profiles during LD and actograms throughout the behavioral analyses of A) AID-TG4-TIR flies and B) controls fed with indicated concentrations of auxin. The morning and evening anticipation signals are shown as means ± SEM. A Student’s two-tailed t-test was employed to determine significance of the difference between treatment and control groups. P values are shown in the figures. (C) Sleep patterns of the AID-TG4-TIR and control flies fed with auxin.
To confirm PER is degraded by the AID system, we dissected fly brains at the CT0 and stained with both GFP and PER antibodies. As expected, the level of the PER protein was drastically decreased in the circadian neurons in which the TIR1 protein was expressed, even at the low concentration of auxin (Fig. 6A). In fact, the PER signal was too low to be detected (Fig. 6B). The staining with antibodies against another pacemaker protein VRILLE as a marker (anti-VRI) confirmed that the absence of PER signal is not caused by the loss of circadian neurons (Fig. 6C). To further understand the kinetics of PER degradation upon auxin feeding, we fed flies 25mM auxin at ZT12 and examine PER abundance after 4 hours, 8 hours and 12 hours (Fig.7A). PER abundance started to increase after light off in the control flies and reached peak around late night. Interestingly, we found that AID-mediated PER degradation occurred rapidly within 8 hours after auxin feeding, indicating that the function of auxin in the CNS is highly efficient (Fig. 7A–B).
Figure 6. The AID system enables degradation of proteins in nervous system of adult Drosophila.
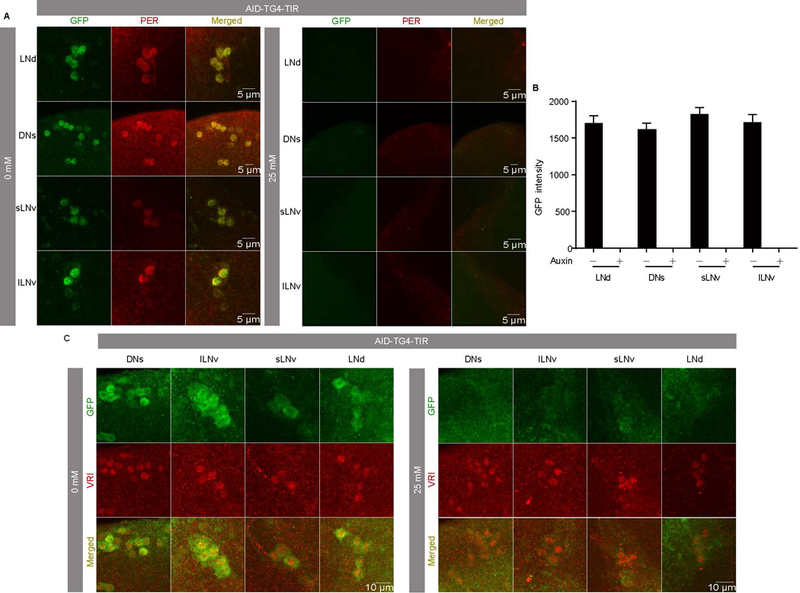
(A) Representative LNd, DN1, and LNvs stained with GFP and PER at CT1 to confirm degradation of PER in AID-TG4-TIR flies after 0 mM and 25 mM auxin treatments. (B) GFP intensities from analyses of AID-TG4-TIR flies after 0 mM (n=8) and 25 mM (n=10) auxin treatments. Flies were fed with auxin for 3 days and then transferred to constant dark (DD) condition. Samples were collected for staining on the first day of DD at CT1. (C) Representative LNd, DNs, and LNvs stained with GFP and VRI at ZT18 after the third day with or without auxin treatment to confirm the normality of circadian neurons (n=8).
Figure 7. Rapid AID mediated degradation of PER indicated by a kinetic assay.
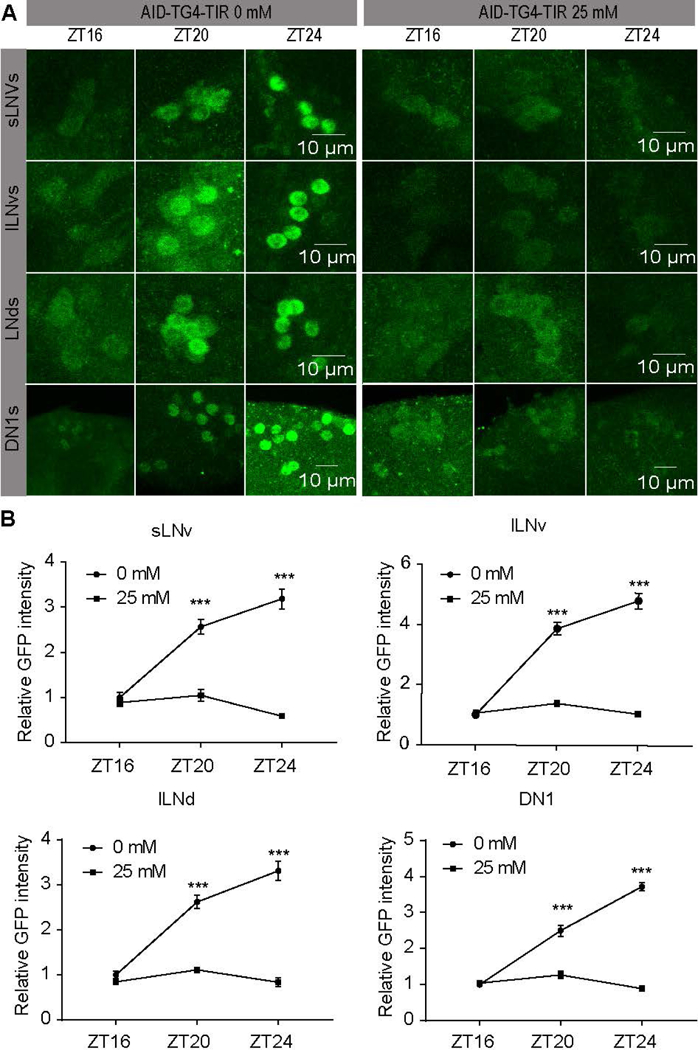
(A) AID-TG4-TIR flies were feed with 0 mM or 25 mM auxin from ZT12, and then dissection for staining with anti-GFP at ZT16 (n=8), ZT20 (n=10), ZT24 (n=10). Representative LNd, DN1, and LNvs were shown. (B) GFP intensities from analyses of (A). Each time point was normalized to the ZT16 of flies without auxin feeding. The GFP intensity was shown as mean ± SEM. A Student’s two-tailed t-test was employed to determine significance compared to controls and treatments at the same time point. *** P<0.001
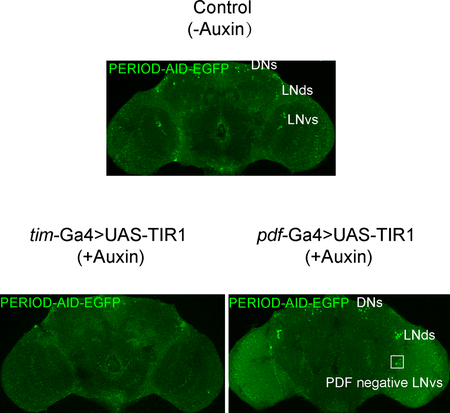
PER expression in the PDF-positive LNvs specifically controls the morning activity, whereas expression of PER in other circadian neurons, including LNds, controls the evening activity [33, 34]. Therefore, to further confirm the specificity of the AID system, we used pdf-GAL4 to express TIR and depleted PER only in PDF-positive LNvs. Upon auxin treatment, arrhythmic behavior under DD was observed in these flies (Fig. 8A and Table 1). Interestingly, under LD, the auxin treatment only disrupted the morning anticipation but not the evening anticipation in these flies, indicating that PER was not affected in the evening neurons. Imaging confirmed that PER protein was only absent in PDF-positive LNvs of the y,sc,v,per-AID-EGFP/Y;pdf-Gal4/+;UAS-TIR1/+ (AID-PG4-TIR) flies treated with auxin (Fig. 8B–F).
Figure 8. The AID system does not cause non-specific degradation of proteins in nervous system of adult Drosophila.
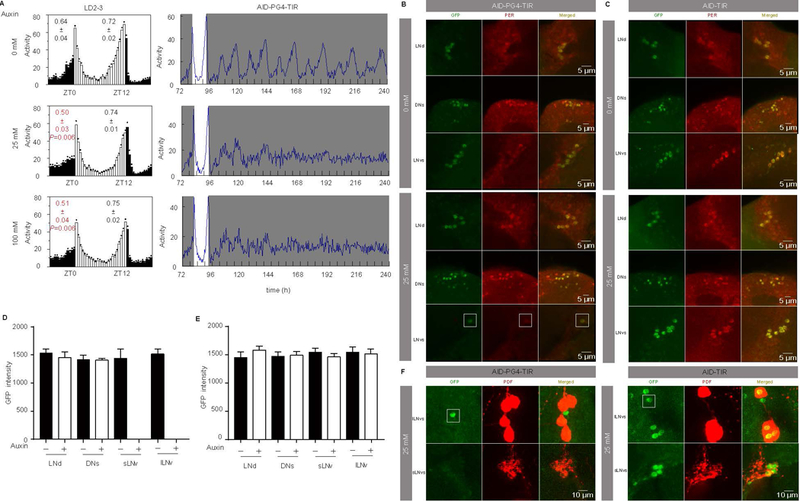
(A) Averaged activity profiles during LD and actograms throughout the behavioral analyses of AID-PG4-TIR flies treated with 0, 25, or 100 mM auxin. (B-C) Representative LNd, DN1, and LNvs stained with GFP and PER at CT1 to confirm degradation of PER in AID-PG4-TIR flies and control flies after 0 mM and 25 mM auxin treatments. (D-E) GFP intensities from analyses of AID-PG4-TIR flies and control flies after 0 mM (n=8) and 25 mM (n=10) auxin treatments. Flies were fed with auxin for 3 days and then transferred to constant dark (DD) condition. Samples were collected and stained on the first day of DD at CT1. Note that in AID-PG4-TIR flies treated with 25 mM auxin, there was only one GFP-positive neuron in the LNvs, which is a PDF-negative neuron, indicated with a box. The morning and evening anticipation signals are shown as means ± SEM. A Student’s two-tailed t-test was employed to determine significance of the difference between treatment and control groups. P values are shown in the figures. (F) AID-PG4-TIR or control flies treated with 25 mM auxin for three days and then stained with GFP and PDF at CT1 to confirm that the remain neuron is exactly the PDF negative LNv (n=8).
In a control experiment, we analyzed period-AID-EGFP/Y;;UAS-TIR1/+ flies, which do not express GAL4. These flies should not express TIR1. In these flies, auxin treatment caused no substantial locomotor activity or sleep behavior defects (Fig. 8B–E). This result indicates that the observed behavior defect is a direct consequence of the TIR1-mediated PER depletion and is not due to a side effect of the auxin treatment. Therefore, the AID system can be specially applied to induce protein depletion in the nervous system enabling behavior or other physiology studies in the absence of a single protein.
The AID-mediated protein degradation in the nervous system is reversible
Finally, to determine how quickly AID-mediated PER degradation causes a phenotype, flies fed normal food were transferred to food supplemented with auxin, while flies were first treated with auxin and then switched to normal food. In the control flies, the behavior was normal throughout the experiment as were control flies continuously fed with normal food or 25 mM auxin-supplemented food (Fig. 9). By contrast, we found that AID-mediated PER degradation caused a phenotype in the treatment groups within 12 hours even in flies fed a low concentration of auxin (Fig. 10A).
Figure 9. Control fly behavior is not altered by auxin.
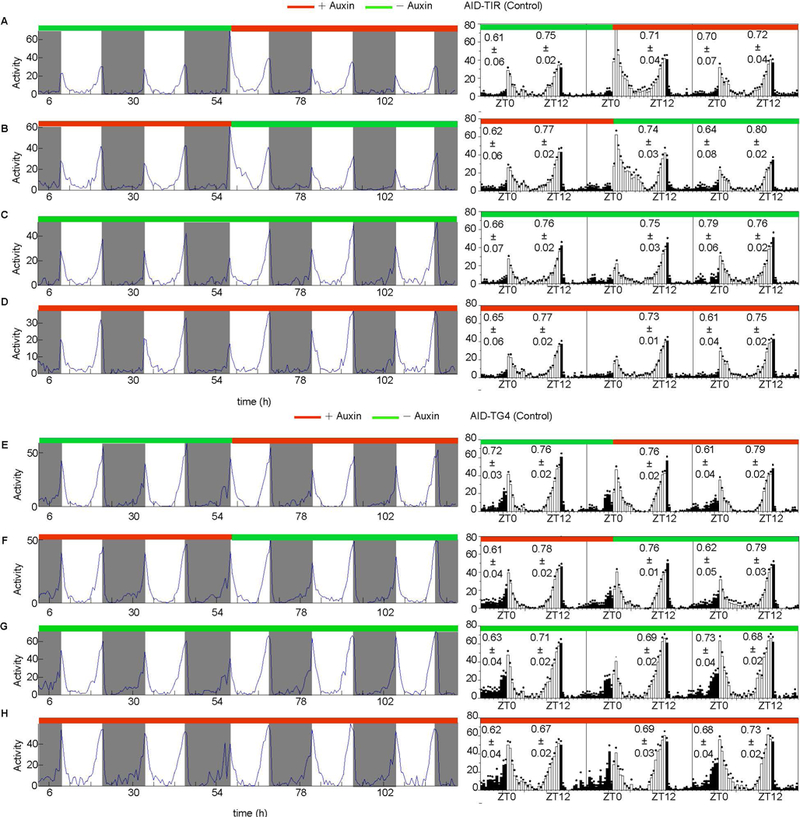
(A-D) Behaviors of y,sc,v,per-AID-EGFP/+; ;UAS-TIR1/+ control flies A) fed normal food for 3 days then transferred to 25 mM auxin-supplemented food for an additional 3 days (n=45), B) fed 25 mM auxin-supplemented food for 3 days then transferred to normal food for an additional 3 days (n=42), C) fed normal food for the study period (n=51), and D) fed 25 mM auxin-supplemented food for the study period (n=50). (E-H) Behaviors of y,sc,v,per-AID-EGFP/+; tim-Gal4/+ control flies E) fed normal food for 3 days then transferred to 25 mM auxin-supplemented food for an additional 3 days (n=32), F) fed 25 mM auxin-supplemented food for 3 days then transferred to normal food for an additional 3 days (n=48), G) fed normal food for the study period (n=32), and H) fed 25 mM auxin-supplemented food for the study period (n=40). Averaged activity profiles during the entire assay period are shown in the left panels. Average locomotor activity plots of a day before and a day after food transfer are shown in the right panels. The morning and evening anticipation signals are plotted as means ± SEM.
Figure 10. Reversible depletion of the AID-EGFP-tagged PER protein by the AID system.
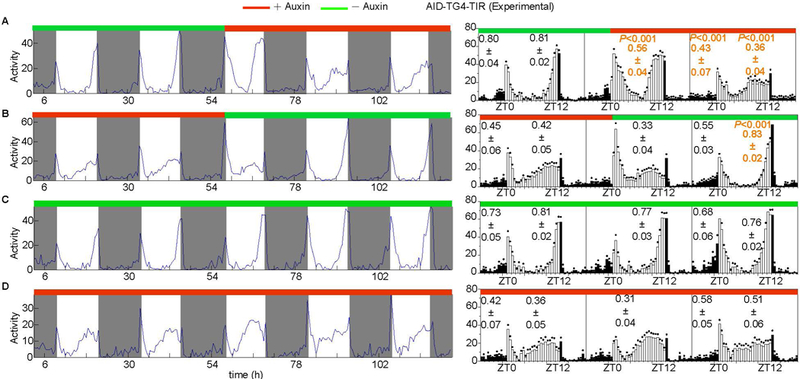
Behaviors of AID-TG4-TIR flies A) fed normal food for 3 days then transferred to 25 mM auxin-supplemented food for an additional 3 days (n=49), B) fed 25 mM auxin-supplemented food for 3 days then transferred to normal food for an additional 3 days (n=32), C) fed normal food for the study period (n=42), and D) fed 25 mM auxin-supplemented food for the study period (n=50). Averaged activity profiles during the entire assay period are shown in the left panels. Average locomotor activity plots of a day before and a day after food transfer are shown in the right panels. The morning and evening anticipation signals are plotted as means ± SEM. A Student’s two-tailed t-test was employed to compare the significance of the difference between the day before and after food transfer. P values are shown.
To determine the reversibility of the AID system, flies fed with 25 mM auxin for 3 days and then with normal food for 3 days were subjected to behavior analysis. In the AID-TG4-TIR flies, one day after auxin removal, the suppression decreased, and after 36 hours behavior was normal (Fig. 10B), demonstrating that the AID system acts in a reversible manner. AID-TG4-TIR flies continuously fed with normal food or 25 mM auxin food were used as positive controls (Fig. 10C, D).
Discussion
Recently, several different tools have been successfully applied to induce direct protein degradation. A common strategy utilizes the UBR1 E3 ligase pathway and N-end rule [42, 43]. Some strategies, such as tobacco virus protease (TEV) protease-mediated cleavage, the GFP nanobody coupled with a degradation signal (deGradFP), the photo-inactivation by FlAsH-FALI, the ligand-inducible destabilizing domain (DD) fused protein degradation, and the temperature-sensitive intein-mediated protein splicing, have been adapted to Drosophila [44–49]. Limitations are associated with these tools when applied for behavior studies. For example, temperature can cause unwanted physiological changes and have significant impacts on fly behavior that may prevent detection of the phenotype of interest [7, 8]. The reactive oxygen species generated by FlAsH-FALI might also have side effects. Although TEV and deGradFP act at the protein level, neither are particularly fast nor are they readily reversible. Most importantly, none of these tools have been employed in the nervous system. The AID system of plants is a small-molecule-inducible protein degradation system that has enabled rapid, conditional protein depletion in many organisms including Drosophila [18–22]. Here, we report for the first time that the AID system can induce rapid and reversible protein degradation in the nervous system enabling behavioral studies in Drosophila. Our analyses demonstrate that the AID method allows reversible conditional protein degradation with precise temporal control.
Here we targeted sites in the Drosophila endogenous period locus using the CRISPR/Cas9 system resulting in flies that express PER with AID-EGPF at the C terminus with very high efficiency. We found that the PER protein fused with the 287-amino acid AID-EGFP is fully functional at molecular and behavior levels. In addition, the fly line period-AID-EGFP we generated here will serve as an alternative in experiments that require detections of PER. The anti-PER antibody is not commercially available but GFP antibodies are.
Normal neuronal function requires well-balanced extracellular ions, nutrients, and metabolic homeostasis. Therefore, all organisms equipped with a complex nervous system have developed the so-called blood-brain barrier (BBB), an evolutionarily ancient structure that provides direct support and protection of the nervous system [50]. The auxin analog employed here must cross the BBB and readily penetrate the membranes of neurons in the fly brain. Synthetic auxin analogs 1-NAA and IAA activate the AID system with similar efficiencies and induce Vasa depletion in vasa-AID-EGFP flies [20]. Here, we observed toxicity with IAA. 1-NAA worked well in the adult flies at the relatively low concentration applied. Actually even at low concentration, 1-NAA rapidly triggers the degradation of PER in circadian neurons in less than 8 hours. As 1-NAA will likely cross the BBB in other organisms, the AID system is a promising tool for functional study of nervous systems in organisms in addition to the fly.
Materials and methods
Drosophila strains, plasmid construction, and transgenesis
tim-Gal4 and pdf-Gal4 flies were used in previous study [31]. UAS-OsTIR1 (BL76123) and vasa-AID-EGFP (BL76126) flies were obtained from Bloomington Stock Center.
To generate the period-AID-EGFP fly line, the donor was constructed via the following overlap PCR steps. First, homology sequences were PCR amplified from genomic DNA of the yw fly strain using PrimeSTAR Max DNA polymerase (TaKaRa, R045, Kusatsu, Shiga, Japan). The AID-EGFP sequence was amplified from a vasa-AID-EGFP fly. To prevent cutting of the repair template, guide RNA recognition sites were eliminated on the plasmid. The following oligonucleotides were used: 5arm-f: 5’-ACATAAACCTCTGGCCACCGTTCT-3’ 5arm-r: 5’-GATCTTTAGGACCTCCGCCGTCGCCGTGCTGTGTCTGGTCCTC-3’ AID-EGFP-f: 5’-CGGCGGAGGTCCTAAAGATCCAGCCAAACCTCC-3’ AID-EGFP-r: 5’-TCAGCAGCAACTGCGGGTGTCATTACTTGTACAGCTCGTCCATGCC-3’ 3arm-f: 5’-ACACCCGCAGTTGCTGCTGACC-3’ 3arm-r: 5’-CAAGCTGCTCATCCTGCCCATTG-3’
Subsequently, 5arm, AID-EGFP, and 3arm fragments were mixed and amplified using 5arm-f and 3arm-r primer pairs. Finally, the PCR product of 5arm-AID-EGFP-3arm was cloned into the pCR-Blunt vector (ThermoFisher, k275020, Waltham, MA, USA) through blunt-end cloning. For chiRNAs construction, the two target site sequences were cloned into the pU6-BbsI-chiRNA plasmid (Addgene, #45946, Cambridge, MA, USA) as previously described [30] using the following oligonucleotides: PAM1-sense: 5’-CTTCGACCAGACACAGCACGGGGAT-3’ PAM1-antisense: 5’-AAACATCCCCGTGCTGTGTCTGGTC-3’ PAM2-sense: 5’-CTTCGTCAGCAGCAACTGCGGGTG-3’ PAM2-antisense: 5’-AAACCACCCGCAGTTGCTGCTGAC-3’
The donor and pU6-BbsI-chiRNA plasmids were mixed and injected into embryos of the nos-Cas9 attp2 fly line (Rainbow Transgenic Flies, Inc., Camarillo, CA, USA).
Screen for integration
After injection, a single F0 fly was collected and crossed with an FM7 balancer fly. In the F1 generations, 10–15 larval or pupa flies from the single cross were pooled together for genome extraction. The following primers for partial EGFP fragment amplification were used: EGFP-f: 5’-ctggtcgagctggacggcgacg-3’ EGFP-r: 5’-cacgaactccagcaggaccatg-3’
For the EGFP PCR positive single cross, 10–20 F1 flies were used to do single crossing with the FM7 balancer fly. When eggs were present in the food, the genome of a single F1 fly was extracted and amplified again using EGFP-f and EGFP-r primers. The following primers that flank the two homologous arms plus the primers in the AID-EGFP were used for amplification and sequencing: F1: 5’-CTCCTCCGTGGGCAGTTCCACG-3’ R1: 5’-aacagctcctcgcccttgctcac-3’ F2: 5’-ctgctgcccgacaaccactacct-3’ R2: 5’-GGATTCACCAAGGACCTCACCAGG-3’
Immunostaining
Adult flies (3–5 days old) were collected and fixed at room temperature for 1 h in 4% formaldehyde diluted in PBT (PBS with 0.1% Trition X-100). Then, the brains were dissected in PBT at indicated time points and fixed in 4% formaldehyde for another 20 min at room temperature. The brains were rinsed and washed with PBT three times (15 min each) and then were blocked in 10% normal goat serum in PBT for 2 h at room temperature. Then, the brains were incubated with primary antibody overnight at 4 ℃. Indicated dilutions of the following primary antibodies were made in PBT: rabbit anti-PER (1:1500, a gift from Michael Rosbash, Brandeis University) and mouse anti-GFP (1:400, from ThermoFish, #15379). After three 15-min washes, brains were incubated with secondary goat antibodies overnight at 4 ℃, followed by extensive washes. Finally, the brains were mounted in the Mounting Medium (Vector Laboratories, #H-1000, Burlingame, CA, USA). Imaging was performed on a Leica Confocal Microscope SP8 system. Confocal images were obtained at an optical section thickness of 1–2 μm and were analyzed with Image J.
Locomotor behavior assay
All the flies were raised on cornmeal/agar medium at 25 ℃ under a 12h:12 h LD cycle. Unless noted otherwise, male flies were monitored in a 12 h:12 h LD cycle at 25 ℃ for 3–4 days, followed by 6–7 days in DD using Trikinetics Drosophila Activity Monitors. Activity records were collected in 1-min bins and analyzed using Faas (Fly activity analysis suite) software developed by M. Boudinot and derived from the Brandeis Rhythm Package (http://hawk.bcm.tmc.edu). Circadian periods were determined by periodogram analysis [51]. A signal-processing toolbox was used to plot flyplot and sleep patterns [52]. The morning and evening anticipation were determined by assaying of the locomotor activity [53]. Statistical analyses were performed with two-tailed Student’s t-test.
Treatment with auxin and auxin analogs
The auxin 3-indole acetic acid sodium salt (IAA; Abcam, ab146403, Cambridge, MA, USA and Millipore Sigma, I5148, Burlington, MA, USA) stock solutions of 125 or 500 mM were prepared in water. The 1-naphthaleneacetic acid (1-NAA; Millipore Sigma, N0640, Burlington, MA, USA) stock solutions of 125, 500 mM or 2 M were prepared in absolute ethyl alcohol. Then the stock solution was mixed with sucrose agar food (5% sucrose and 2% agar) for behavior assay. Sucrose agar food has been used for circadian rhythms and sleep assay in Drosophila. It is easy to make and can last long (for weeks) during the behavior assay.
Acknowledgements
We thank Dr. Phillip Karpowicz for critical comments on the manuscript. We would like to thank Bloomington Drosophila Stock Center for providing fly stocks. The anti-PER antibody is a generous gift from Dr. Michael Rosbash. Wenfeng Chen’s work is supported by the National Natural Science Foundation of China (grant number 31601894), the Fujian Natural Science Foundation (grant. number 2017J0106) and the Chinese Government Scholarship (201706655001). Yong Zhang’s lab is supported by the National Institutes of Health COBRE Grant P20 GM103650.
Abbreviations
- 1-NAA
1-naphthaleneacetic acid
- AID
auxin-inducible degradation
- BBB
blood-brain barrier
- chiRNA
chimeric single guide RNA
- CRISPR
clustered regularly interspaced short palindromic repeats
- gRNA
guide RNA
- CNS
central nervous system
- CT
circadian time
- EGFP
enhanced green fluorescent protein
- FlAsH-FALI
FlAsH-mediated fluorescein-assisted light inactivation
- FLP
flippase
- FRT
flippase recognition target
- IAA
3-indole acetic acid sodium salt
- LD
light:dark
- DD
dark:dark
- PG4
pdf-Gal4
- SCF
SKP1/CUL1/F-box
- TEV
tobacco etch virus
- TG4
tim-Gal4
- TIR1
transport inhibitor response 1
- UTR
untranslated region
- ZT
zeitgeber time
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Bello B, Resendez-Perez D & Gehring WJ (1998) Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system, Development. 125, 2193–202. [DOI] [PubMed] [Google Scholar]
- 2.Bieschke ET, Wheeler JC & Tower J (1998) Doxycycline-induced transgene expression during Drosophila development and aging, Molecular & general genetics : MGG. 258, 571–9. [DOI] [PubMed] [Google Scholar]
- 3.McGuire SE, Le PT, Osborn AJ, Matsumoto K & Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila, Science. 302, 1765–8. [DOI] [PubMed] [Google Scholar]
- 4.Han DD, Stein D & Stevens LM (2000) Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation, Development. 127, 573–83. [DOI] [PubMed] [Google Scholar]
- 5.Osterwalder T, Yoon KS, White BH & Keshishian H (2001) A conditional tissue-specific transgene expression system using inducible GAL4, Proceedings of the National Academy of Sciences of the United States of America. 98, 12596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roman G, Endo K, Zong L & Davis RL (2001) P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster, Proceedings of the National Academy of Sciences of the United States of America. 98, 12602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigrist SJ, Reiff DF, Thiel PR, Steinert JR & Schuster CM (2003) Experience-dependent strengthening of Drosophila neuromuscular junctions, The Journal of neuroscience : the official journal of the Society for Neuroscience. 23, 6546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S & Griffith LC (2016) Reorganization of Sleep by Temperature in Drosophila Requires Light, the Homeostat, and the Circadian Clock, Current biology : CB. 26, 882–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heigwer F, Port F & Boutros M (2018) RNA Interference (RNAi) Screening in Drosophila, Genetics. 208, 853–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn C & Handler AM (2005) Site-specific genomic targeting in Drosophila, Proceedings of the National Academy of Sciences of the United States of America. 102, 12483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo S & Ueda R (2013) Highly improved gene targeting by germline-specific Cas9 expression in Drosophila, Genetics. 195, 715–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Port F, Chen HM, Lee T & Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila, Proceedings of the National Academy of Sciences of the United States of America. 111, E2967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K & Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells, Nature. 411, 494–8. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T & Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells, Nature methods. 6, 917–22. [DOI] [PubMed] [Google Scholar]
- 15.Dharmasiri N, Dharmasiri S & Estelle M (2005) The F-box protein TIR1 is an auxin receptor, Nature. 435, 441–5. [DOI] [PubMed] [Google Scholar]
- 16.Kepinski S & Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor, Nature. 435, 446–51. [DOI] [PubMed] [Google Scholar]
- 17.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M & Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase, Nature. 446, 640–5. [DOI] [PubMed] [Google Scholar]
- 18.Natsume T, Kiyomitsu T, Saga Y & Kanemaki MT (2016) Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors, Cell reports. 15, 210–218. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Ward JD, Cheng Z & Dernburg AF (2015) The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans, Development. 142, 4374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bence M, Jankovics F, Lukacsovich T & Erdelyi M (2017) Combining the auxin-inducible degradation system with CRISPR/Cas9-based genome editing for the conditional depletion of endogenous Drosophila melanogaster proteins, The FEBS journal. 284, 1056–1069. [DOI] [PubMed] [Google Scholar]
- 21.Wood L, Booth DG, Vargiu G, Ohta S, deLima Alves F, Samejima K, Fukagawa T, Rappsilber J & Earnshaw WC (2016) Auxin/AID versus conventional knockouts: distinguishing the roles of CENP-T/W in mitotic kinetochore assembly and stability, Open biology. 6, 150230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trost M, Blattner AC & Lehner CF (2016) Regulated protein depletion by the auxin-inducible degradation system in Drosophila melanogaster, Fly. 10, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allada R & Chung BY (2010) Circadian organization of behavior and physiology in Drosophila, Annual review of physiology. 72, 605–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubowy C & Sehgal A (2017) Circadian Rhythms and Sleep in Drosophila melanogaster, Genetics. 205, 1373–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitabach MN & Taghert PH (2008) Organization of the Drosophila circadian control circuit, Current biology : CB. 18, R84–93. [DOI] [PubMed] [Google Scholar]
- 26.Konopka RJ & Benzer S (1971) Clock mutants of Drosophila melanogaster, Proceedings of the National Academy of Sciences of the United States of America. 68, 2112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bargiello TA, Jackson FR & Young MW (1984) Restoration of circadian behavioural rhythms by gene transfer in Drosophila, Nature. 312, 752–4. [DOI] [PubMed] [Google Scholar]
- 28.Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M & Hall JC (1984) P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster, Cell. 39, 369–76. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko M & Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections, The Journal of comparative neurology. 422, 66–94. [DOI] [PubMed] [Google Scholar]
- 30.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM & O’Connor-Giles KM (2014) Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila, Genetics. 196, 961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall JC (2003) Genetics and molecular biology of rhythms in Drosophila and other insects, Advances in genetics. 48, 1–280. [DOI] [PubMed] [Google Scholar]
- 32.Helfrich-Forster C (2000) Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster--sex-specific differences suggest a different quality of activity, Journal of biological rhythms. 15, 135–54. [DOI] [PubMed] [Google Scholar]
- 33.Grima B, Chelot E, Xia R & Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain, Nature. 431, 869–73. [DOI] [PubMed] [Google Scholar]
- 34.Stoleru D, Peng Y, Agosto J & Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila, Nature. 431, 862–8. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE & Emery P (2010) Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior, Current biology : CB. 20, 600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Ling J, Yuan C, Dubruille R & Emery P (2013) A role for Drosophila ATX2 in activation of PER translation and circadian behavior, Science. 340, 879–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim C & Allada R (2013) ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila, Science. 340, 875–9. [DOI] [PubMed] [Google Scholar]
- 38.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R & Choe J (2011) The novel gene twenty-four defines a critical translational step in the Drosophila clock, Nature. 470, 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grima B, Dognon A, Lamouroux A, Chelot E & Rouyer F (2012) CULLIN-3 controls TIMELESS oscillations in the Drosophila circadian clock, PLoS biology. 10, e1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Lamba P, Guo P & Emery P (2016) miR-124 Regulates the Phase of Drosophila Circadian Locomotor Behavior, The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 2007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunawardhana KL & Hardin PE (2017) VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila, Current biology : CB. 27, 3442–3453 e4. [DOI] [PubMed] [Google Scholar]
- 42.Kanemaki MT (2013) Frontiers of protein expression control with conditional degrons, Pflugers Archiv : European journal of physiology. 465, 419–25. [DOI] [PubMed] [Google Scholar]
- 43.Campbell AE & Bennett D (2016) Targeting protein function: the expanding toolkit for conditional disruption, The Biochemical journal. 473, 2573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ & Nasmyth K (2008) Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons, Developmental cell. 14, 239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caussinus E, Kanca O & Affolter M (2011) Fluorescent fusion protein knockout mediated by anti-GFP nanobody, Nature structural & molecular biology. 19, 117–21. [DOI] [PubMed] [Google Scholar]
- 46.Harder B, Schomburg A, Pflanz R, Kustner K, Gerlach N & Schuh R (2008) TEV protease-mediated cleavage in Drosophila as a tool to analyze protein functions in living organisms, BioTechniques. 44, 765–72. [DOI] [PubMed] [Google Scholar]
- 47.Zeidler MP, Tan C, Bellaiche Y, Cherry S, Hader S, Gayko U & Perrimon N (2004) Temperature-sensitive control of protein activity by conditionally splicing inteins, Nature biotechnology. 22, 871–6. [DOI] [PubMed] [Google Scholar]
- 48.Sethi S & Wang JW (2017) A versatile genetic tool for post-translational control of gene expression in Drosophila melanogaster, eLife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venken KJ, Kasprowicz J, Kuenen S, Yan J, Hassan BA & Verstreken P (2008) Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation, Nucleic acids research. 36, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ & Klambt C (2008) Organization and function of the blood-brain barrier in Drosophila, The Journal of neuroscience : the official journal of the Society for Neuroscience. 28, 587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klarsfeld A, Leloup JC & Rouyer F (2003) Circadian rhythms of locomotor activity in Drosophila, Behavioural processes. 64, 161–175. [DOI] [PubMed] [Google Scholar]
- 52.Levine JD, Funes P, Dowse HB & Hall JC (2002) Signal analysis of behavioral and molecular cycles, BMC neuroscience. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, Bai W, Zhou D & Zhao Z (2014) Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle, Nature communications. 5, 5549. [DOI] [PubMed] [Google Scholar]


