Abstract
Genetic variation contributes significantly to brain function and dysfunction, and studying the genetic factors responsible for neurological phenotypes is tremendously valuable for understanding brain development, physiology, and pathophysiology, as well as for advancements in disease diagnostics and therapeutics. Many genetic determinants of neurobiology are inherited from parents through the germline and are present in all cells of an individual, but others, known as somatic or mosaic mutations, may be acquired post-conception and are therefore present in only a subset of an individual’s cells. While the relationship between somatic mutation and cancer is clear, recent studies have also established a role for somatic mutations in several non-malignant neurological diseases of childhood, including cerebral cortical malformations and epilepsy disorders, autism spectrum disorder, and other neuropsychiatric diseases.
Cerebral cortical malformations
Cerebral cortical malformations represent a diverse set of disorders in which normal cortical development is perturbed leading to a physical malformation of the brain, often resulting in intractable epilepsy and developmental delay. Somatic mutations play a critical role in inherited forms of brain malformation such as tuberous sclerosis, in which one copy of TSC1 or TSC2 is mutated in every cell of the patient’s body, and somatic second hits lead to complete loss of gene function and formation of cortical tubers. In the cases of LIS1 and DCX, germline mutation results in lissencephaly while somatic loss-of-function mutations can result in subcortical band heterotopia, or a “double cortex” phenotype, due to the failure of cells marked by these somatic variants to migrate to their proper laminar position. Pathogenic somatic mutations have also been found in blood-derived DNA from cases of other neuronal migration defects associated with epilepsy and developmental delay, including periventricular nodular heterotopia, pachygyria, and perisylvian polymicrogyria.1,2
Causative somatic mutations have been identified in other types of cortical malformations, most notably the overgrowth syndromes megalencephaly and hemimegalencephaly and the related malformation focal cortical dysplasia, which together comprise some of the most common causes of pediatric intractable epilepsy. Megalencephaly, in which the entire head is abnormally large, hemimegalencephaly, in which half of the head and brain are asymmetrically enlarged, and focal cortical dysplasia, in which a localized area is found to have cortical disorganization, can all be caused by mosaic gain-of-function mutations in the PI3K-AKT3-mTOR pathway. In these diseases affected cells have a growth advantage and a balloon-shaped morphology, but do not have malignant potential. These distinct disorders are now thought to exist on a spectrum in which the same activating mutations can cause widespread overgrowth when occurring in earlier stages of development or focal disorganization when occurring later in more restricted progenitors.3,4 As such, initial studies of somatic mosaicism in these diseases identified causal somatic mTOR pathway mutations in blood and saliva samples,5 suggesting these mutations occurred early in development, before cells committed to the brain lineage. Later, additional studies demonstrated that some causal somatic variants could only be identified in brain-derived DNA.4,6 Indeed, in most cases of hemimegalencephaly and focal cortical dysplasia the causative somatic mutations are found only in DNA of a fraction of the cells in the brain lesion, and not in peripheral DNA.
Autism spectrum disorder
Somatic mutations can produce syndromic forms of autism. For example, cases of Rett syndrome in males have been documented in which the MECP2 mutation occurs in a mosaic state and is therefore compatible with life. Mounting evidence now suggests that somatic mutations can also impact risk and heritability of non-syndromic forms of autism. Importantly, somatic mutations could represent a genetic cause in a fraction of otherwise unsolved autism cases, since causative mutations might be restricted to the brain and not detectable in commonly studied sources of DNA such as blood and saliva. Localized somatic mutations that affect both brain function and structure might underlie physical “patches” of cortical disorganization that have been observed in autism,7 and might also contribute to the heterogeneity of the disorder, where certain cognitive functions are well preserved in some highfunctioning patients.
The role of de novo mutation in autism has been extensively explored over many years. More recently, multiple studies of whole-exome sequencing data from blood-derived DNA of thousands of autism families have shown that 5–22% of mutations that were previously thought to be de novo are actually likely to be somatic mutations.8,9 These studies have pointed toward an important role for somatic mutation in autism risk, with an excess of potentially diseasecausing somatic mutations occurring in autism cases compared to unaffected controls. Furthermore, nearly all studies on somatic mosaicism in autism conducted to date have used peripheral DNA rather than brain-derived DNA due to the scarcity of postmortem autism brain samples available for research, and as a result these studies have likely underestimated the degree to which brain somatic mutations impact autism risk. One study examined a set of known autism candidate genes on brain-derived DNA from 55 autism cases and 50 controls, identifying two autism cases and one Fragile X premutation case with deleterious likely causative somatic brain mutations, in addition to the discovery of deleterious somatic mutations in one case with unconfirmed autism and one case with social anxiety disorder.10 Further genotyping across multiple brain regions showed that some deleterious mosaic mutations are restricted to certain areas of the brain.
There is evidence for somatic structural variants playing a role in autism pathogenesis as well. Fluorescence in-situ hybridization has allowed for the assessment of large-scale chromosomal mosaicism, and case reports have linked the autistic phenotype to mosaic chromosome 3q tetrasomy11 and mosaic ring chromosome 17.12 Additionally, autistic individuals may have an increased burden of low-level mosaic aneuploidy.13
Other neuropsychiatric disorders of childhood
Studies on brain DNA from schizophrenia patients have suggested an increase in mosaic aneuploidy of chromosomes 1, 18, and X.14 Small but significant mosaic changes in the copy numbers of many genes associated with psychiatric disease have also been found in schizophrenia postmortem brains when compared to controls. In general, children with developmental disorders have a much higher burden of large mosaic structural variants in peripheral DNA than healthy children.15 However, most specific neurodevelopmental and psychiatric disorders have not yet been assessed for somatic mosaicism, and new deep sequencing technologies that can identify smaller scale and lower allele fraction mutations have not yet been applied to most neurological diseases. Additional studies of both peripheral and brain-derived DNA are needed to determine the contribution of somatic mosaicism to other diseases of brain development and function, such as schizophrenia, intellectual disability, movement disorders.
Relationship between mosaic fraction and disease
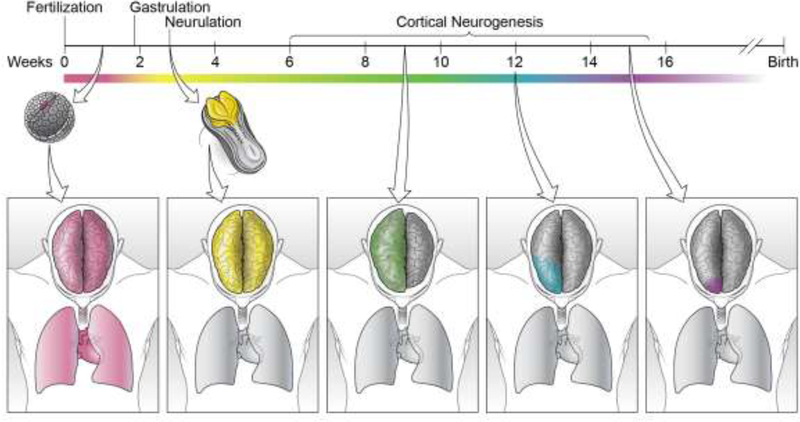
Somatic mutations that occur in the earliest stages of embryonic development can be present in nearly every cell of the body, while mutations that occur in a neural progenitor cell during development might be shared by only a small group of cells in the brain, and mutations that occur in postmitotic neurons would be present in only a single cell. The vast majority of somatic mutations are neutral, while others may be lethal to the mutated cell and therefore never expand clonally, and still others may lead to a growth advantage. Depending on the particular cell that acquires a somatic mutation during development, and depending on the nature of the mutation, a wide range of distributions for a single somatic mutation are possible (Figure 1).
Figure 1.
Developmental timing of somatic mutation acquisition determines distribution in the adult body. A somatic mutation that occurs in a developing embryo prior to gastrulation (far left, magenta) will have a broad distribution throughout the adult body and will be present in tissues derived from all three developmental germ layers. A somatic mutation that occurs during neurulation (center left, yellow) will be present throughout the central nervous system of the adult but absent in non-neural tissues. This mutation will only be detectable through studying CNS-derived DNA. Somatic mutations that occur early during cortical neurogenesis may be present in large regions of the adult brain (center, green), and may cause disease phenotypes, such as in hemimegalencephaly. The later a mutation occurs within cortical development, the more restricted its distribution will be in the adult brain. Mutations occurring during the middle of cortical neurogenesis may have varied distributions throughout portions of the cortex (center right, blue), and mutations occurring late in cortical development may have very restricted distributions within very small regions of cortex (right, purple), as in focal cortical dysplasias. These mutations will only be detected by studying DNA derived from the affected brain region.
Although every gene in the genome is somatically mutated multiple times within an individual, most mutations are either benign or never reach a high enough mosaic fraction to cause disease. Even for a deleterious somatic mutation, the mosaic fraction, or fraction of cells possessing the mutation in a given tissue, might alter the degree to which the mutation is pathogenic in an individual. For example, somatic mutations that lead to a gain of function or growth advantage, such as those that cause cancer, might cause disease if they are present in only one cell. On the other hand, somatic mutations that lead to a loss of function might need to occur in a larger clonal fraction in order to cause disease. Therefore, for every deleterious somatic mutation there likely exists a threshold mosaic fraction above which the mutation will cause disease, but below which the mutation will have a clinically undetected phenotype. While this critical mosaic fraction for pathogenicity is largely unknown and likely varies depending on the specific disease, mutation, and genetic background of an individual, there is evidence that clinically significant cortical malformations can be caused by somatic mutations present in as few as 1% of cells.6 The threshold mosaic fraction to cause other diseases such as autism is likely higher, but is still an open area of investigation.
Acknowledgments
Figure 1 was illustrated by Ken Probst (Xavier Studio). R.E.R. is supported by National Institute of General Medical Sciences T32GM007753. C.A.W. is supported by the Manton Center for Orphan Disease Research, the Paul G. Allen Institute, and grants from the NINDS (R01 NS032457, R01 NS035129) and NIMH (U01 MH106883, UO1 MH106891). C.A.W. is a member of the National Academy of Sciences and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jamuar SS, Lam AT, Kircher M, et al. Somatic mutations in cerebral cortical malformations. N Engl J Med. 2014;371(8):733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirzaa GM, Conti V, Timms AE, et al. Characterisation of mutations of the phosphoinositide-3-kinase regulatory subunit, PIK3R2, in perisylvian polymicrogyria: a next-generation sequencing study. The Lancet Neurology. 2015;14(12):1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain : a journal of neurology. 2015;138(Pt 6):1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Gama AM, Woodworth MB, Hossain AA, et al. Somatic Mutations Activating the mTOR Pathway in Dorsal Telencephalic Progenitors Cause a Continuum of Cortical Dysplasias. Cell Rep. 2017;21(13):3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riviere JB, Mirzaa GM, O’Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nature genetics. 2012;44(8):934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima M, Saitsu H, Takei N, et al. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Annals of neurology. 2015;78(3):375–386. [DOI] [PubMed] [Google Scholar]
- 7.Stoner R, Chow ML, Boyle MP, et al. Patches of disorganization in the neocortex of children with autism. The New England journal of medicine. 2014;370(13):1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed D, Pevsner J. The Contribution of Mosaic Variants to Autism Spectrum Disorder. PLoS Genet. 2016;12(9):e1006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krupp DR, Barnard RA, Duffourd Y, et al. Exonic Mosaic Mutations Contribute Risk for Autism Spectrum Disorder. Am J Hum Genet. 2017;101(3):369–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Gama AM, Pochareddy S, Li M, et al. Targeted DNA Sequencing from Autism Spectrum Disorder Brains Implicates Multiple Genetic Mechanisms. Neuron. 2015;88(5):910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira G, Matoso E, Vicente A, et al. Partial tetrasomy of chromosome 3q and mosaicism in a child with autism. Journal of autism and developmental disorders. 2003;33(2):177–185. [DOI] [PubMed] [Google Scholar]
- 12.Havlovicova M, Novotna D, Kocarek E, et al. A girl with neurofibromatosis type 1,atypical autism and mosaic ring chromosome 17. American journal of medical genetics Part A. 2007;143A(1):76–81. [DOI] [PubMed] [Google Scholar]
- 13.Yurov YB, Iourov IY, Vorsanova SG, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2(6):e558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yurov YB, Vostrikov VM, Vorsanova SG, Monakhov VV, Iourov IY. Multicolor fluorescent in situ hybridization on post-mortem brain in schizophrenia as an approach for identification of low-level chromosomal aneuploidy in neuropsychiatric diseases. Brain & development. 2001;23 Suppl 1:S186–190. [DOI] [PubMed] [Google Scholar]
- 15.King DA, Jones WD, Crow YJ, et al. Mosaic structural variation in children with developmental disorders. Human molecular genetics. 2015;24(10):2733–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]