Abstract
Purpose:
To demonstrate the framework of a novel Hadamard-encoded spectral editing approach for simultaneously detecting multiple low-concentration brain metabolites in vivo at 3T.
Methods:
HERCULES (Hadamard Editing Resolves Chemicals Using Linear-combination Estimation of Spectra) is a four-step Hadamard-encoded editing scheme. 20-ms editing pulses are applied at: (A) 4.58 and 1.9 ppm; (B) 4.18 and 1.9 ppm; (C) 4.58 ppm; and (D) 4.18 ppm. Edited signals from γ-aminobutyric acid (GABA), glutathione (GSH), ascorbate (Asc), N- acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), aspartate (Asp), lactate (Lac), and likely 2-hydroxyglutarate (2-HG) are separated with reduced signal overlap into distinct Hadamard combinations: (A+B+C+D); (A+B-C-D); and (A-B+C-D). HERCULES uses a novel multiplexed linear-combination modeling approach, fitting all three Hadamard combinations at the same time, maximizing the amount of information used for model parameter estimation, in order to quantify the levels of these compounds. Fitting also allows estimation of the levels of total choline (tCho), myo-inositol (Ins), glutamate (Glu), and glutamine (Gln). Quantitative HERCULES results were compared between two grey- and white-matter-rich brain regions (11 min acquisition time each) in 10 healthy volunteers. Coefficients of variation (CV) of quantified measurements from the HERCULES fitting approach were compared against those from a single-spectrum fitting approach, and against estimates from short-TE PRESS data.
Results:
HERCULES successfully segregates overlapping resonances into separate Hadamard combinations, allowing for the estimation of levels of seven coupled metabolites that would usually require a single 11-min editing experiment each. Metabolite levels and CVs agree well with published values. CVs of quantified measurements from the multiplexed HERCULES fitting approach outperform single-spectrum fitting and short-TE PRESS for most of the edited metabolites, performing only slightly to moderately worse than the fitting method that gives the lowest CVs for tCho, NAA, NAAG, and Asp.
Conclusion:
HERCULES is a new experimental approach with the potential for simultaneous editing and multiplexed fitting of up to seven coupled low-concentration and six high- concentration metabolites within a single 11-min acquisition at 3T.
Keywords: Magnetic resonance spectroscopy, spectral editing, Hadamard encoding, GABA, GSH, NAAG
1. Introduction
Magnetic resonance spectroscopy (MRS) is the only methodology that can determine the levels of neurochemicals in living tissue non-invasively, which provides a unique window on the neurobiology of the human brain in health and pathology. At field strengths commonly used for clinical MR imaging, i.e. 3 Tesla and below, spectral overlap and insufficient spectral resolution hinder the reliable separation of low-concentration metabolites with coupled resonances from overlapping high-concentration metabolites.
J-difference editing techniques improve the selectivity of MRS experiments by removing unwanted overlying signals. The most common J-difference editing implementation, Mescher- Garwood Point Resolved Spectroscopy (MEGA-PRESS), consists of two sub-experiments that differ in the treatment of the spin system of interest (Edden et al., 2012; Mescher et al., 1998; Rothman et al., 1993). In the ‘ON’ experiment, a selective editing pulse refocuses the evolution of the targeted scalar coupling, whereas the same coupling is allowed to evolve freely in the ‘OFF’ experiment. Subtraction of the OFF from the ON spectrum yields the edited difference (‘DIFF’) spectrum, which only contains signals affected by the editing pulse. Overlapping signals unaffected by the editing experiments are removed upon subtraction.
MEGA-PRESS was initially introduced for detection of the primary inhibitory neurotransmitter γ-aminobutyric acid (GABA), and has been adapted to selectively isolate a number of other compounds implicated in several important physiological mechanisms: the antioxidants glutathione (GSH) and ascorbate (Asc) (Chan et al., 2016b; Emir et al., 2011a, 2011b; Terpstra et al., 2003, 2006, 2011); the neuronal marker N-acetylaspartate (NAA) (Chan et al., 2017; Edden et al., 2007); the glutamatergic modulators N-acetylaspartylglutamate (NAAG) and aspartate (Asp) (Chan et al., 2016a; Chan et al., 2017; Edden et al., 2007; Murdoch et al., 2013); the anaerobic product lactate (Lac) (Chan et al., 2017b; Star-Lack et al., 1998); and the oncometabolite 2-hydroxyglutarate (2-HG) (Andronesi et al., 2012; Choi et al., 2012; Pope et al., 2012).
J-difference editing prioritizes the resolution of a single low-concentration metabolite of interest over the assessment of a broad neurochemical profile. Historically, editing therefore proceeds at a rate of only one edited metabolite per ~10-min experiment, precluding the interrogation of a large number of editable metabolites within the scan time limits of typical MR protocols. To overcome this limitation, our group has recently pioneered multiplexed editing approaches using Hadamard-encoded editing schemes. Hadamard Encoding and Reconstruction of MEGA-Edited Spectroscopy (HERMES) simultaneously resolves signals from two or three coupled spin systems in a single 11 -min experiment without affecting the signal-to-noise-ratio, and has been implemented for GABA/GSH (Saleh et al., 2018, 2016), macromolecule-suppressed GABA/GSH (Oeltzschner et al., 2017), and NAA/NAAG/Asp (Chan et al., 2016a; Chan et al., 2017).
Here, we introduce HERCULES (Hadamard Editing Resolves Chemicals Using Linear- combination Estimation of Spectra), the combination of an advanced Hadamard-encoded editing technique with a novel multiplexed linear-modeling approach using simulated basis functions. We demonstrate HERCULES in the phantom and in grey- and white-matter-rich regions in healthy subjects, and compare the quantitative outcome to results from short-TE PRESS acquisitions. The HERCULES scheme segregates J-difference-edited signals of GABA, GSH, Asc, NAA, NAAG, Asp, and Lac into distinct Hadamard combinations, and additionally quantifies the major signals from total creatine (tCr), total choline (tCho), myo-inositol (Ins), glutamate (Glu), and glutamine (Gln) that are retained in the sum spectrum. In tumor tissue, signal from 2-HG may also be edited. In summary, HERCULES allows simultaneous detection of up to 13 metabolites in a single 11-min experiment at 3 T.
2. Material and Methods
2.1. Theory
2.1.1. HERCULES editing scheme
As shown in Fig. 1a, HERCULES consists of four sub-experiments with different editing pulse profiles that are designed to differentiate the evolution of up to eight editable metabolites: (A) dual-band editing at 4.58 and 1.9 ppm; (B) dual-band editing at 4.18 and 1.9 ppm; (C) singleband editing at 4.58 ppm; (D) single-band editing at 4.18 ppm. The resulting sub-spectra (Fig. 1b) are combined to give three orthogonal Hadamard combinations of interest: the sum spectrum (A+B+C+D); one difference-edited spectrum (A+B-C-D) of signals impacted by the 1.9 ppm lobe; and a second difference-edited spectrum (A-B+C-D) of signals impacted by the 4.58 and 4.18 ppm lobes.
Figure 1.
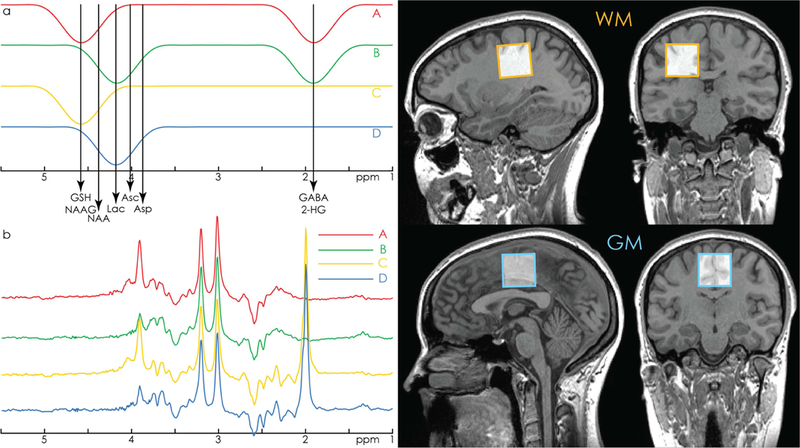
a. HERCULES editing scheme with editing lobes at 1.9 ppm (targeting GABA and 2- HG), 4.18 ppm (targeting Asp, Asc, Lac), and 4.58 ppm (targeting GSH and NAAG). The 4.58 and 4.18 ppm editing lobes are arranged symmetrically around the 4.38 ppm resonance of NAA. This design minimizes the contribution of highly concentrated NAA to the A-B+C-D difference spectrum, improving the segregation of the aspartyl signals of NAA and NAAG into orthogonal difference spectra. b. Example in vivo HERCULES sub-spectra A, B, C, and D, as produced by the editing scheme in 1a. Residual water signal has been filtered out. c. Example voxel placement in white-matter- and grey-matter-rich regions.
Each difference-edited Hadamard combination can be thought of as a separate MEGA-edited spectrum. The (A+B-C-D) combination is a typical GABA+-edited spectrum containing contributions from GABA+macromolecules (3.0 ppm, ‘GABA+’), NAA/NAAG methyl (2.0 ppm), Glu/Gln/GSH (2.25/3.75 ppm), and 2-HG (4.0 ppm, only present in tumor tissue). The (A-B+C-D) combination is a DEW (Double Editing With) -like (Emir et al., 2011a, 2011b; Terpstra et al., 2006) spectrum, where editing pulses are applied to a different set of metabolites in each half of the experiment. As such, it contains clearly separated edited signals from Lac (1.33 ppm) and GSH (2.95 ppm), slightly overlapping edited signals from the aspartyl protons of NAA, NAAG and Asp (between 2.5 and 2.7 ppm), and an edited signal from Asc (3.73 ppm) that is close to the 3.75 ppm signal from GSH.
One key feature of this editing scheme is that the editing lobes at 4.58 and 4.18 ppm are symmetrically placed around the 4.38 ppm resonance of NAA, analogous to the symmetrical suppression of macromolecular resonances in the Henry editing scheme for the detection of GABA (Henry et al., 2001). This arrangement retains the NAA signal in the sum spectrum while suppressing its contribution to the (A-B+C-D) spectrum, providing a mechanism to differentiate the (usually very strong) NAA-aspartyl signal from the (usually weaker) partially overlapping aspartyl signals of Asp and NAAG.
2.1.2. HERCULES spectral fitting /simulations
In Hadamard-encoded editing, more than one difference spectrum is extracted from a single acquired dataset. This multiplexed editing approach motivates a multiplexed analysis pipeline. Given that a substantial subset of compounds contributes signal to more than one Hadamard combination spectrum, it is expected that the quantification model will be more reliably constrained by considering all acquired information at the same time. Therefore, instead of fitting each Hadamard combination separately (as is usual for the fitting of single-edited difference spectra or un-edited spectra), HERCULES models the concatenation of all three Hadamard combinations together with HERCULES basis functions that are simulated for 21 metabolite spin systems. This simultaneous fit incorporates all the available spectral information to constrain the model, exploiting the separation of overlapping resonances into orthogonal Hadamard combinations.
2.2. Phantom and in vivo data
2.2.1. Acquisition
All data were acquired using a 32-channel head matrix coil on a 3T Siemens Prisma scanner (Siemens Healthineers, Erlangen, Germany).
Phantom data were acquired from a 3 × 3 × 3 cm3 voxel in a 1-liter phosphate-buffered saline phantom at room temperature (pH = 7.2) containing 1.5 mM Asc, 1.5 mM Asp, 2.5 mM ChoCl, 10 mM Cr, 2 mM GABA, 4 mM Gln, 10 mM Glu, 2 mM GSH, 6 mM Ins, 10 mM NAA, and 2 mM Lac (NAAG was not included due to its extremely high cost). In vivo data were acquired from 10 healthy adults (5 male, mean age 29.3 ± 2.9 y) in 3 × 3 × 3 cm3 voxels: one white- matter-rich (WM) voxel in the centrum semiovale, and one grey-matter-rich (GM) voxel over the cingulate cortex (Fig. 1c). From each voxel, HERCULES data were acquired with the following parameters: TR/TE = 2000/80 ms; 20-ms editing pulses with a single-lobe full-width half-maximum (FWHM) bandwidth of 61.9 Hz; 2048 data points; 2 kHz spectral width; CHESS water suppression; 320 averages (i.e. 80 repetitions of the A-B-C-D cycle shown in Fig. 1a). HERCULES reference data were acquired without water suppression with the same parameters (8 averages).
For comparison, short-TE PRESS data were acquired with the vendor-native sequence using the same parameters as HERCULES, except: TE = 30 ms; 64 averages. Short-TE water-reference PRESS data were acquired with the same parameters (8 averages).
For validation purposes, GABA/GSH HERMES (Saleh et al., 2016) data were acquired in a white-matter voxel in one subject. The acquisition parameters were the same as for HERCULES, except for the editing pattern: (A) dual-band editing at 4.56 and 1.9 ppm; (B) single-band editing at 4.56; (C) single-band editing at 1.9 ppm; (D) no editing pulse. HERMES water-reference data were acquired with the same parameters (8 averages).
2.2.2. Data processing and spectral fitting
HERCULES (and HERMES) data were processed in MATLAB (The MathWorks, Natick, MA) with a pipeline based on functions from the FID-A toolkit (Simpson et al., 2017). The optimal coil combination was calculated from the water-unsuppressed short-TE PRESS data, weighting the individual channels with the ratio of the signal to the square of the noise (Hall et al., 2014). Eddy-current correction was performed using the water-unsuppressed HERCULES/HERMES data. Within each sub-experiment, all averages were aligned to each other using frequency-and- phase correction in the time domain using spectral registration (Near et al., 2014), and then averaged and linebroadened with a 1-Hz exponential filter. Residual water signals were filtered from each sub-spectrum with a Hankel singular value decomposition (HSVD) filter (Barkhuijsen et al., 1987), and each sub-spectrum was subsequently baseline-corrected. To minimize subtraction artefacts in the Hadamard combination spectra, a set of frequency and phase shifts applied to sub-spectra B, C, and D was determined. This optimal set was found by determining the global minimum of the standard deviation of the trivial HERCULES combination (A-B- C+D), which should not contain any edited signals. After applying the calculated frequency corrections, the matrix containing the A, B, C and D sub-spectra was left-multiplied by the 4 × 4 Hadamard matrix to give the four resulting Hadamard combinations (including the unused combination A-B-C+D).
Short-TE PRESS data underwent the same pre-processing steps, including: coil combination and eddy-current correction based on the water-unsuppressed short-TE PRESS data, spectral registration, averaging, 1-Hz filtering, and removal of residual water.
HERCULES and short-TE PRESS basis functions were calculated in MATLAB using the FID-A toolkit. Full density-matrix simulations of the four HERCULES sub-experiments A, B, C, D, and the vendor-native PRESS sequence were computed for 21 metabolite spin systems: Asc, Asp, creatine (Cr), GABA, glycerophosphorylcholine (GPC), GSH, Gln, Glu, glycine (Gly), Ins, Lac, NAAG, NAA, phosphorylcholine (PCh), phosphocreatine (PCr), phosphorylethanolamine (PE), scyllo-inositol (Scy), serine (Ser), taurine (Tau), 2-HG, and residual water. Chemical shifts and coupling constants were taken from literature (Govind et al., 2015; Govindaraju et al., 2000; Near et al., 2013). To account for the spatially varying editing efficiency within the voxel due to chemical shift displacement effects (Edden and Barker, 2007; Kaiser et al., 2008; Near et al.,2013), a fast computation algorithm for spatially localized MRS was employed (Zhang et al., 2017). All simulations were performed on a three-dimensional grid of 21 × 21 × 21 spatial points within a 3 × 3 × 3 cm3 voxel, using ideal excitation and shaped refocusing pulses. For each metabolite, the vector containing the A, B, C, and D sub-spectra was left-multiplied by the 4 × 4 Hadamard matrix. In a final step, the resulting four Hadamard combinations were concatenated to produce a single basis function per metabolite for the entire HERCULES experiment. To mimic the macromolecular contamination of the 3 ppm GABA signal (due to co-editing of the coupled 1.7 ppm macromolecular resonance), a single Gaussian peak (10 Hz linewidth) with a total area of 40% of the 3 ppm GABA integral was added to the (A+B-C-D) basis function of GABA for the in vivo analysis. Since detectable levels of 2-HG are only present in tumor tissue, the 2-HG basis function was removed from the HERCULES basis set for the analysis of the in vivo data from healthy subjects.
Spectral fitting was then performed with in-house software developed in MATLAB, using a multiplexed least-squares linear-combination modeling approach. The linear-combination model function contained 49 parameters: twenty amplitude weighting coefficients (one for each metabolite in the basis set); two coefficients describing the Lorentzian and Gaussian contributions to the Voigt lineshape (added to all basis functions); six coefficients describing linear baseline terms for the three Hadamard combinations; twenty coefficients allowing a small frequency shift for each basis function (Provencher, 1993); and one coefficient allowing for a global frequency shift between the basis functions and the in vivo spectra. The fitting problem was solved using a nonlinear least-squares minimization algorithm (‘trust region reflective’, implemented in the lsqcurvefit function of MATLAB) over a fitting interval of 1.02 ppm to 4.50 ppm. Non-linear baseline functions were estimated separately for each Hadamard combination through five iterations of the following cycle: fitting the signal with the HERCULES basis functions; modeling a smooth spline through the fit residual; subtracting the spline from the initial signal; and feeding the spline-corrected spectrum back into the fit routine. The sum of the five spline iterations was then subtracted from the original spectrum, and the resulting baseline- free spectrum was fitted to yield the final linear-combination model parameters used for further analysis. Short-TE PRESS data were fit with the simulated single-spectra basis sets in the same way.
All metabolite levels were expressed as ratios to the total creatine signal (tCr = Cr+PCr). Coefficients of variation (CV) of the metabolite ratios across all subjects were calculated for each metabolite.
2.2.3. Comparison of simultaneous and single-spectrum fitting
In order to investigate the advantage of simultaneous fitting of multiplexed HERCULES data, the edited sum spectrum and each of the two difference spectra were also separately fitted using a single-spectrum fitting approach. Basis sets for this separate modeling were generated by treating the sub-spectra of the previously created HERCULES basis functions as separate functions with independent amplitude parameters. The respective linear-combination model function for the sum and the two difference spectra contained the same parameters as the HERCULES model except for the baseline parameters for the ‘other’ spectra; that is, all metabolite basis functions were included in each basis set, regardless of whether they showed any signal in the difference spectra. Since the two difference spectra contain no common reference signal, the tCr signal from the sum spectrum was used as the reference to quantify the signals in the difference spectra.
For each metabolite, the fitting approach that produced the lowest CV (across all subjects and regions) was determined.
3. Results
Simulated equimolar spectra for the eight edited metabolites are shown in Fig. 2, demonstrating that the HERCULES scheme successfully differentiates overlapping edited signals. Since the columns of a Hadamard matrix are orthogonal, signals that are affected by different editing pulses are segregated into separate difference spectra without cross-contamination. As an example, the edited signal of GABA at 3.01 ppm appears in the (A+B-C-D) spectrum, while the edited signal of GSH at 2.95 ppm appears in the (A-B+C-D) spectrum, but not vice versa. The symmetrical arrangement of the 4.58 and 4.18 ppm editing pulses provides an additional mechanism of distinction: it directs the (relatively strong) 2.6-ppm aspartyl signal of NAA into the sum spectrum, while the overlapping (lower-concentration) aspartyl signal of NAAG appears in the (A-B+C-D) spectrum, where it can be more reliably quantified.
Figure 2.
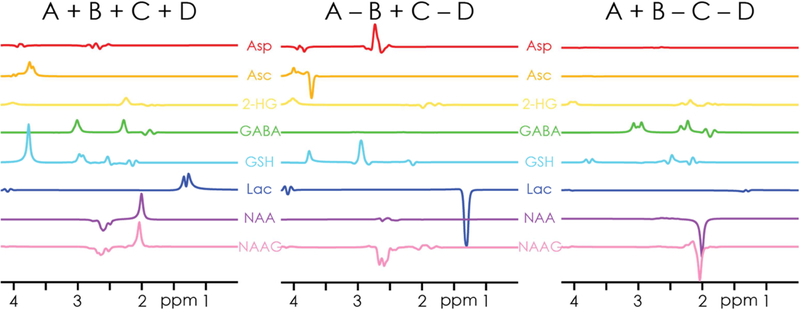
Simulated basis functions of the Hadamard combinations for the eight coupled metabolites edited in the HERCULES experiment. Note the segregation of overlapping signals (e.g., GSH and GABA around 3 ppm) into orthogonal difference spectra (GSH in A-B+C-D, GABA in A+B-C-D), and the differentiation of the NAA- and NAAG-aspartyl singles at 2.6 ppm in the A-B+C-D spectrum. The signal intensities represent equimolar concentrations.
The phantom HERCULES data and model are shown in Fig. 3, indicating successful implementation of the intended editing scheme and clearly edited target resonances. A small offset from the ideal NAA-aspartyl suppression (potentially from minimal mis-adjustment of the editing pulses to account for the water shift in phantom) is indicated by the slightly different lineshape of the aspartyl multiplet, and the underfit 3.9 ppm Cr resonance. With respect to 10 mM tCr, phantom metabolite levels were (not accounting for differences in T2 relaxation) calculated to be 1.4 mM for Asc (nominally 1.5 mM), 1.4 mM for Asp (1.5 mM), 2.4 mM for tCho (2.5 mM), 2.6 mM for GABA (2 mM), 6.7 mM for Gln (4 mM), 12.1 mM for Glu (10 mM), 3.5 mM for GSH (2 mM), 7.1 mM for Ins (6 mM), 18 mM for NAA (10 mM), and 2.4 mM for Lac (2 mM).
Figure 3.
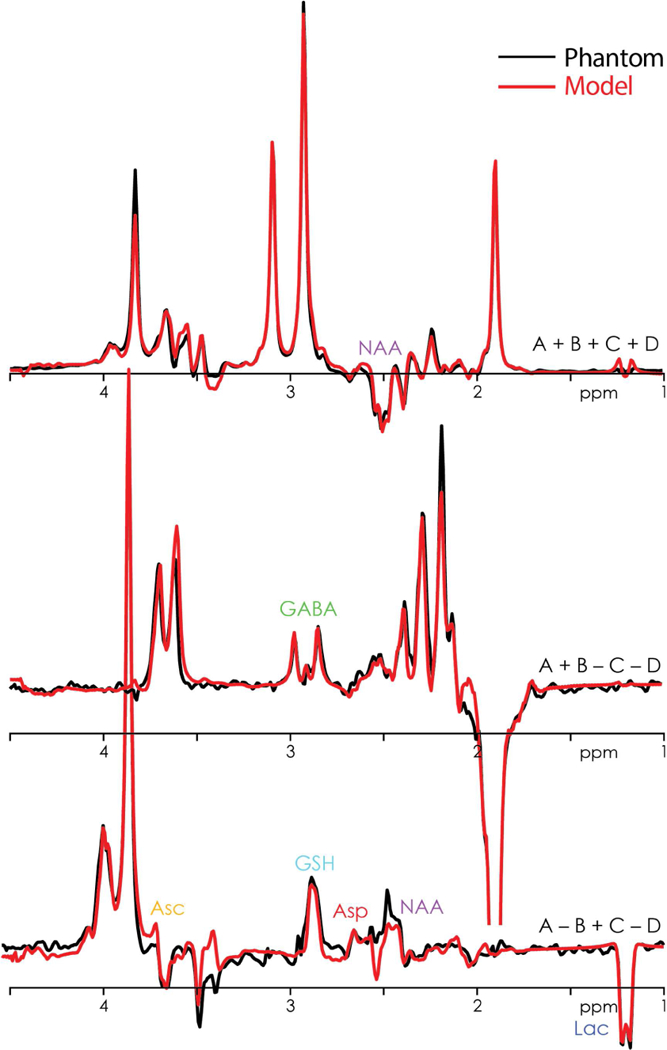
Phantom HERCULES data (black) showing the three Hadamard combinations overlaid by the multiplexed linear-combination model (red). The key spectral features of the eight edited metabolites are clearly visible. b. Overlay of the respective GABA- and GSH-edited difference spectra from HERCULES (orange) and HERMES (blue) from a white-matter voxel in one subject.
In vivo data from two white-matter HERCULES datasets and one grey-matter short-TE PRESS dataset were excluded from quantitative analysis due to severe lipid contamination and/or subject movement. The three Hadamard combinations and fits are shown for one subject (grey-matter voxel) in Fig. 4a. The overlay of the GABA- and GSH edited difference spectra of HERCULES and HERMES from a white-matter voxel in one subject is shown in Fig. 4b. The good agreement between the different methods further indicates that GABA- and GSH-editing in HERCULES is equivalent to HERMES.
Figure 4.
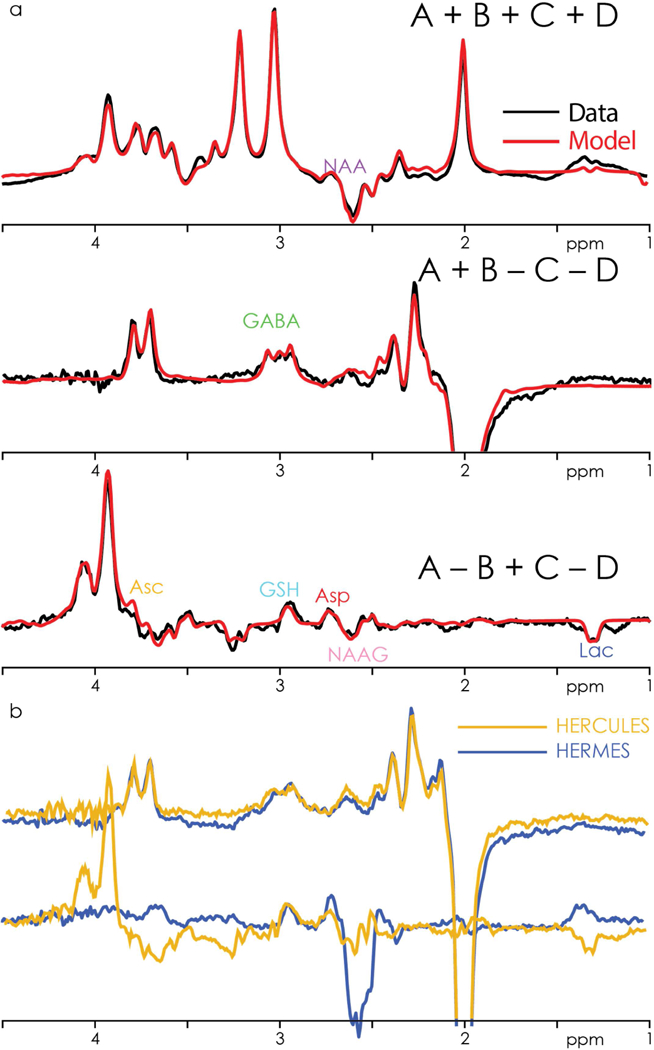
a. In vivo HERCULES data (black) from one subject (grey-matter voxel), showing the three Hadamard combinations overlaid by the multiplexed linear-combination model (red). The key spectral features of the eight edited metabolites are clearly visible. b. Overlay of the respective GABA- and GSH-edited difference spectra from HERCULES (orange) and HERMES (blue) from a white-matter voxel in one subject.
Individual model components determined by the HERCULES fitting approach are shown in Fig. 5. In addition to the seven edited metabolites (excluding 2-HG), signals from Gln, Glu, Ins, and Cr- and Cho-containing compounds can also be resolved. Quantitative results from the displayed WM and GM regions and all subjects are summarized for eleven metabolites in Fig. 6, demonstrating several significant between-region differences: higher levels of Asp, GABA+, and Glu in GM; higher levels of NAAG, Gln, and tCho in WM.
Figure 5.
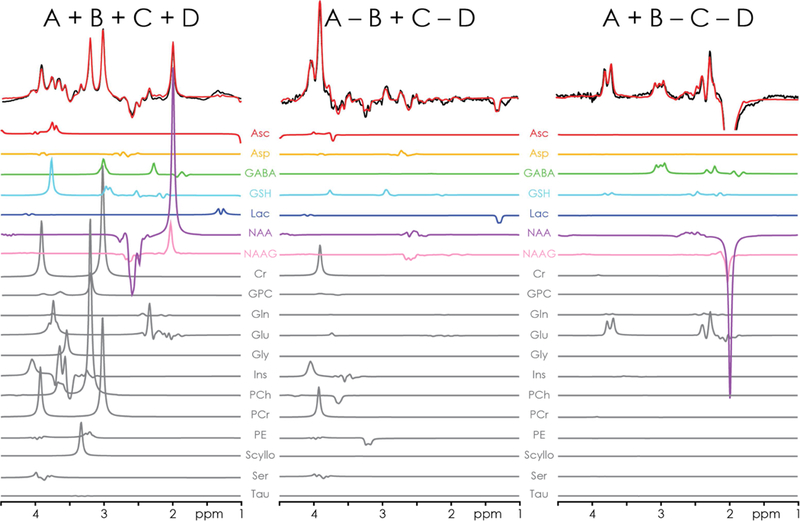
Multiplexed linear modeling of the in vivo HERCULES spectra from Fig. 4, using concatenated HERCULES basis functions. The basis set includes the seven edited metabolites after removing 2-HG from the basis set (Asc, Asp, GABA, GSH, Lac, NAA, NAAG). Together with five ‘conventionally accessible’ metabolites (Gln, Glu, Ins, tCr, tCho), a total of 12 metabolites can be quantified within a single HERCULES experiment (13 when including 2-HG in tumor).
Figure 6.
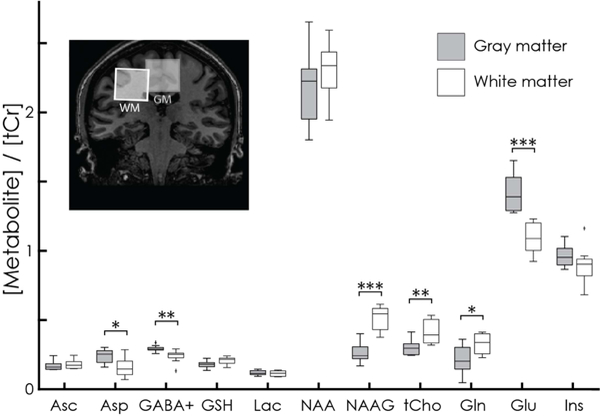
Quantitative analysis of HERCULES data with respect to total creatine (tCr = Cr+PCr) for eleven metabolites. The central horizontal line of each boxplot indicates the median; the top and bottom edges represent the 25th and 75th percentile, and the whiskers indicate the most extreme values. Significant differences between the GM and WM voxels appear for Asp (P = 0.019), GABA+ (P = 0.007), NAAG (P < 0.001), tCho (P = 0.007), Gln (P = 0.027), and Glu (P < 0.001).
All fitting approaches - simultaneous; sum only; (A+B-C-D) only; (A-B+C-D) only - yielded reasonable model fits, as shown for one dataset in Fig. 7. Metabolite levels are listed for each metabolite for the HERCULES fitting approach in Table 1, along with the CVs for each metabolite for all fitting approaches, as well as the fitting approach that yielded the lowest mean CV for the respective metabolite. The HERCULES simultaneous modeling approach produced the lowest mean CV for the majority of metabolites, and was only slightly to moderately worse than fitting the PRESS spectrum for NAAG and NAA, and the sum spectrum for tCho.
Figure 7.
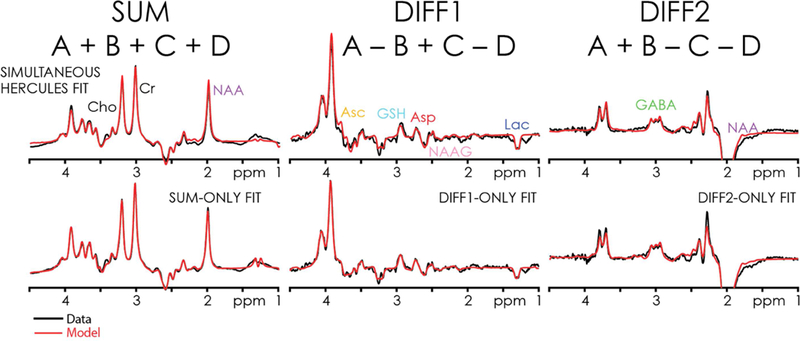
HERCULES data modeling of the dataset from Figs. 4 and 5, for all four modeling approaches: HERCULES simultaneous modeling; and separate modeling of the sum and difference spectra. Red lines represent the best-fit model. Black lines represent in vivo data.
Table 1.
Quantitative results of HERCULES and PRESS (with respect to total creatine, tCr) for the GM-rich and WM-rich voxels; statistical assessment of GM-WM differences (two-tailed t- test, no correction for multiple comparison); coefficients of variation (CV) for HERCULES, the HERCULES single-spectrum fitting methods, and PRESS; and the fitting method that gave the lowest overall CV. Values in brackets denote standard deviations. All CV values are given in %.
| Asc | Asp | GABA+ | GSH | Lac | NAA | NAAG | tCho | Gln | Glu | Ins | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HERCULES | GM Met/tCr (SD) | 0.171 (0.034) | 0.237 (0.049) | 0.293 (0.022) | 0. 182 (0.028) | 0.119 (0.017) | 2.182 (0.261) | 0.265 (0.077) | 0.3G4 (0.057) | 0.219 (0.1G2) | 1.416 (0.125) | 0.965 (0.076) |
| CV HERCULES | 20 | 21 | 8 | 16 | 14 | 12 | 29 | 19 | 46 | 9 | 8 | |
| CV A-B+C-D | 30 | 77 | 134 | 22 | 45 | 89 | 61 | 85 | 75 | 212 | 23 | |
| CV A+B-C-D | 177 | 131 | 14 | 31 | 165 | 14 | 28 | 57 | 35 | 55 | 66 | |
| CV SUM | 75 | 146 | 50 | 32 | 118 | 31 | 32 | 11 | 37 | 9 | 12 | |
| WM Met/tCr (SD) | 0.181 (0.034) | 0.160 (0.071) | 0.239 (0.049) | 0.208 (0.029) | 0.115 (0.021) | 2.305 (0.211) | 0.520 (0.095) | 0.416 (0.088) | 0.330 (0.076) | 1.093 (0.117) | 0.897 (0.138) | |
| CV HERCULES | 19 | 45 | 20 | 14 | 18 | 9 | 18 | 21 | 23 | 11 | 15 | |
| CV A-B+C-D | 43 | 142 | 52 | 21 | 59 | 90 | 44 | 74 | 130 | 243 | 24 | |
| CV A+B-C-D | 143 | 83 | 20 | 36 | 149 | 27 | 58 | 74 | 40 | 63 | 141 | |
| CV SUM | 159 | 33 | 53 | 31 | 92 | 57 | 33 | 14 | 79 | 20 | 11 | |
| p (GM vs. WM) | 0.531 | 0.019* | 0.007** | 0.071 | 0.664 | 0.295 | <0.001*** | 0.007** | 0.027* | <0.001*** | 0.202 | |
| PRESS | GM Met/tCr (SD) | 0.168 (0.068) | 0.399 (0.112) | 0.212 (0.075) | 0.305 (0.056) | 0.264 (0.068) | 1.215 (0.106) | 0.362 (0.048) | 0.250 (0.016) | 0.022 (0.048) | 1.285 (0.140) | 0.595 (0.170) |
| CV | 41 | 28 | 35 | 18 | 26 | 9 | 13 | 6 | 223 | 11 | 29 | |
| WM Met/tCr (SD) | 0.261 (0.100) | 0.391 (0.127) | 0.121 (0.107) | 0.277 (0.056) | 0.195 (0.075) | 1.291 (0.114) | 0.418 (0.084) | 0.282 (0.019) | 0.005 (0.014) | 1.001 (0.153) | 0.677 (0.105) | |
| CV | 38 | 33 | 88 | 20 | 38 | 9 | 20 | 7 | 316 | 15 | 16 | |
| p (GM vs. WM) | 0.030* | 0.893 | 0.050* | 0.275 | 0.051 | 0.149 | 0.093 | 0.001** | 0.295 | <0.001*** | 0.221 | |
| MEAN CV | HERCULES | 19 | 33 | 14 | 15 | 16 | 11 | 24 | 20 | 35 | 10 | 12 |
| A-B+C-D | 37 | 110 | 93 | 22 | 52 | 90 | 53 | 80 | 103 | 228 | 24 | |
| A+B-C-D | 160 | 107 | 17 | 34 | 157 | 21 | 43 | 66 | 38 | 59 | 104 | |
| SUM | 117 | 90 | 52 | 32 | 105 | 44 | 33 | 13 | 58 | 15 | 12 | |
| PRESS | 40 | 31 | 62 | 19 | 32 | 9 | 17 | 7 | 270 | 13 | 23 | |
| Lowest CV | HERC | PRESS | HERC | HERC | HERC | PRESS | PRESS | PRESS | HERC | HERC | HERC |
P < 0.05;
P < 0.01;
P < 0.001
4. Discussion
The HERCULES experiment combines several spectral editing concepts to maximally exploit the available information. While DEW-MEGA-PRESS (Terpstra et al., 2006) is capable of editing more than one compound simultaneously, it is restricted to spin systems with resolved editing-target and detected resonances, and fails if the detected resonances overlap. Hadamard- encoded techniques effectively address this problem by segregating overlapping edited resonances into separate difference spectra. HERCULES combines these advantages: it exploits Hadamard-encoding for separation of, for example, GABA and GSH, and DEW to edit more signals into one HERCULES difference spectrum. HERCULES further implements a symmetrical editing pulse arrangement for NAA-aspartyl suppression in the (A-B+C-D) spectrum, improving its DEW characteristics by mitigating overlap with NAAG and Asp. The phantom and in vivo HERCULES data demonstrate the successful implementation of the multiplexed editing scheme, segregating overlapping coupled resonances into the intended orthogonal Hadamard combinations. The HERCULES fitting approach yielded the lowest overall CVs for all but four metabolites, being only slightly to moderately worse than PRESS for NAAG and NAA, tCho, and Asp.
Overall, the results of the HERCULES quantification provide a preliminary validation of the quantitative potential of this approach, as several expected differences in metabolite levels between GM and WM regions were reproduced. Higher levels of GABA+ and Glu in GM, likely due to the higher density of inhibitory and excitatory cell bodies, have been shown previously (Bhattacharyya et al., 2011; Choi et al., 2006; Hnilicova et al., 2016; Jensen et al., 2005). In contrast to our findings, higher GSH levels in GM have been reported at 7T (Srinivasan et al., 2010). The ~10% higher levels of NAA in WM compared to GM are consistent with results obtained from spectroscopic imaging (parietal WM and cortical GM), as well as increased tCho (Horská et al., 2002; McLean et al., 2000; Pan et al., 1998). Higher concentrations of NAAG in WM have been demonstrated (Pouwels and Frahm, 1997), and the NAA:NAAG ratio measured in WM with HERCULES (about 4.4:1) is in excellent agreement with published values (Chan et al., 2016a; Chan et al., 2017a; Edden et al., 2007; Pouwels and Frahm, 1997). Tissue-specific levels of Asp have not been demonstrated yet, but given its roles as a neurotransmitter at the N- methyl-D-aspartate receptor and in metabolism, higher levels in GM are not unreasonable.
Mean tCr ratios for tCho, Glu, and Ins were within the expected ranges (Govindaraju et al., 2000), while the NAA/tCr ratio (~2.3) was measured to be consistently high, especially when compared to the results from the short-TE PRESS data (NAA/tCr ~ 1.3). This can be explained by the notably longer in-vivo T2 values of NAA compared to Cr-containing compounds (Wyss et al., 2018), which will cause the T2-uncorrected NAA/tCr ratio to assume larger values with increasing echo time. With the exception of tCho (20%), the major metabolites showed low CVs around 10%. Mean CVs (24%) for NAAG were slightly higher than the ~16% observed with previous MEGA-PRESS and HERMES experiments in mostly white-matter voxels (Chan et al., 2016a; Chan et al., 2017; Edden et al., 2007), however, HERCULES CVs in the white-matter region (18%) agreed well with these values. Lac CVs (16%) were lower than the ~20% observed in a recent DEW study (Chan et al., 2017). While the CVs for GABA+ and GSH (14% and 15%) obtained with HERCULES were comparable to MEGA-PRESS (Bogner et al., 2010; Geramita et al., 2011; Mikkelsen et al., 2017; Terpstra et al., 2003), HERCULES quantification yields higher mean tCr ratios for both compounds. Mean tCr ratios for Asc agreed with expectations from literature (Emir et al., 2011b; Terpstra et al., 2010; Terpstra and Gruetter, 2004), and mean CVs (19%) are in agreement with these reports. Finally, mean CVs for Asp (33%) were slightly higher than the ~20% obtained with a three-metabolite HERMES editing scheme (Chan et al., 2017a), and Asp/tCr ratios were in agreement with preliminary results from a single volunteer obtained with MEGA-PRESS editing (Murdoch et al., 2013). It should be noted that both the HERMES and the MEGA-PRESS experiments used in the Asc and Asp studies were optimized for editing of Asc (Asp), while HERCULES maximized the number of edited metabolites, achieving only about two thirds (one third) of the maximum Asc (Asp) editing efficiency and, hence, SNR for the respective metabolite (cf. Fig. 1a, see below). A summary of CVs with literature values is given in Table 2.
Table 2.
Comparison of HERCULES CVs (mean value of grey-matter CVs and white-matter CVs) for low-concentration edited metabolites with values calculated from previous reports in literature. All CV values are given in %.
| Metabolite | HERCULES CV [%] | Literature CV [%] | References | Remarks |
|---|---|---|---|---|
| Asc | 19 | 22-24 | (Emir et al., 2011b) | DEW @ 4T |
| 13 | (Terpstra et al., 2006) | DEW @ 4T | ||
| 23 | (Terpstra and Gruetter, 2004) | MEGA @ 4T | ||
| Asp | 33 | 19 | (Chan et al., 2017) | HERMES @ 3T |
| GABA+ | 14 | 12 | (Mikkelsen et al., 2017) | MEGA @ 3T |
| 13 | (Bogner et al., 2010) | MEGA @ 3T | ||
| 9 | (Geramita et al., 2011) | MEGA @ 3T | ||
| GSH | 15 | 15 | (Terpstra et al., 2003) | DEW @ 4T |
| 16-40 | (Emir et al., 2011b) | DEW @ 4T | ||
| 16 | (Chan et al., 2017) | DEW @ 3T | ||
| NAAG | 24 | 14 (WM) | (Edden et al., 2007) | MEGA @ 3T |
| (GM 29; WM 18) | 18 (WM) | (Chan et al., 2016a) | HERMES @ 3T | |
| 16 (WM) | (Chan et al., 2017) | HERMES @ 3T | ||
| Lac | 16 | 15-29 | (Emir et al., 2011b) | DEW @ 4T |
| 23 | (Chan et al., 2017) | DEW @ 3T | ||
| Gln | 35 | 26-53 | (Goryawala et al., 2016) | MRSI @ 3T |
| 18 | (Gonenc et al., 2010) | Multi-TE @ 3T | ||
| 24 | (Jensen et al., 2009) | 2D-JPRESS @ 4T | ||
Quantitative comparison with the short-TE PRESS data yielded results in generally broad agreement with HERCULES. Interestingly, PRESS achieved the lowest mean CV for the estimation of Asp, NAAG and NAA, and surprisingly low mean CVs for GSH. PRESS, however, failed to detect a significant difference in NAAG levels between gray and white matter, and estimated levels of Asp, Lac, and GSH to be 1.5 to 3 times as high as the HERCULES estimates which agreed substantially better with literature values. This suggests that PRESS appears to estimate these metabolites consistently, but incorrectly. A potential reason for this overestimation is the fact that no macromolecular resonances were included in the PRESS analysis. More sophisticated fitting algorithms like LCModel may benefit short-TE quantification by including internally simulated macromolecular and lipid resonances, but also often make a priori assumptions by default about the levels of low-concentration metabolites. For the comparison with the newly developed, assumption-free HERCULES fitting approach, we therefore chose to perform the PRESS analysis without any constraints on the absolute or relative amplitudes of these metabolites. Future studies will aim to improve the HERCULES model by including macromolecular and lipid resonances to allow more complex studies on the relationship between unedited (short-TE) and edited metabolite estimates.
An interesting secondary finding of this study was the improved separation of Gln and Glu in HERCULES compared to short-TE PRESS. Higher Gln/tCr levels in WM compared to GM have been previously observed from whole-brain short-TE MRSI at 3T, with similar CVs of ~20% in WM and ~40-60% in GM (Goryawala et al., 2016). Studies using two-dimensional or multi-TE methods have reported CVs around 20% (Gonenc et al., 2010; Jensen et al., 2009). In combination with the information from the (A+B-C-D) difference spectrum, HERCULES is likely to gain additional power for distinguishing Gln and Glu from the sum spectrum. Since the Gln and Glu resonances are partially inverted in the experiments with 1.9 ppm editing pulses (A and B), and freely evolving in the other experiments (C and D), the sum spectrum can be thought of as an artificially created TE-averaged (over two ‘virtual’ echo times) spectrum. This mitigates the overlap between the multiplets compared to short-TE PRESS (see Supplementary Figure 1), and may benefit their separate estimation.
While editing of 2-HG was not demonstrated in vitro (due to extensive costs for a phantom) or in vivo (due to unavailability of eligible patients), it is demonstrated that the edited 4.02 ppm resonance in the (A+B-C-D) difference spectrum is only overlapped by distinctly lower- intensity multiplets from Lac and NAAG (see Supplementary Figure 2). This suggests that 2-HG levels commonly encountered in IDH-mutated glioma (5-35 mM, Andronesi et al., 2012) can be reliably detected. Compared to standard MEGA-PRESS, HERCULES offers the additional benefit of being able to separate potentially elevated contributions of Lac by taking the inverted 1.3 ppm Lac signal in the (A-B+C-D) difference spectrum into account.
Higher metabolite CVs (i.e. less reliable fitting), in particular for the weak coupled signals, can be reasonably explained by a current lack of adequate spectral alignment routines for HERCULES spectra, and the relatively basic fitting algorithm used in this proof-of-principle work. Edited quantification is highly susceptible to subtraction artefacts resulting from motion or scanner drift (Edden et al., 2016; Harris et al., 2014), and improved frequency-and-phase correction and fitting routines (see below) will improve the agreement with results from single-metabolite editing. Although the novel post-processing routine used in this study notably reduced Cho subtraction artefacts, underlying baseline modulations and remaining spectral imperfections persisted, potentially influencing the estimates of the coupled metabolites. The demand for adequate post-processing has long been recognized and addressed for MEGA-PRESS of GABA, but is less well-explored for editing of other metabolites. Four-step editing schemes produce spectra with fewer shared landmark features used for spectral alignment, which makes them substantially more prone to misalignment and subtraction artefacts than two-step MEGA editing. Further experiment-specific optimization for multiplexed editing schemes is therefore required, as has been recently proposed for HERMES of GABA and GSH (Mikkelsen et al., 2017).
A comprehensive editing scheme like HERCULES is best evaluated with a linear-combination modeling approach that uses all the available information across the different Hadamard combinations (and the same may be true for all difference-edited data). Since the individual metabolite basis functions for each Hadamard combination are coupled to each other in amplitude, the information from a well-resolved resonance in one spectrum is used as a form of prior knowledge in another spectrum. As an example, the NAA-aspartyl fit in the sum spectrum (A+B+C+D) and the NAAG-aspartyl fit in the (A-B+C-D) difference spectrum each constrain the smaller signal contribution to the other spectrum, and the fitting of the N-acetyl methyl in the sum and (A+B-C-D) spectra constrains the total signal, allowing for a more precise estimation of the levels of NAA and NAAG.
As noted above, more sophisticated modeling algorithms (such as LCModel) may include more prior knowledge about lineshape or baseline modeling, and may aid in, for example, the estimation of small phase corrections of the underlying baseline. As mentioned, lipid and macromolecular signals have - with the exception of the 3 ppm signal added to the GABA basis function - not been considered in this initial version of the HERCULES fitting algorithm (and are absorbed into the splined baseline). The inclusion of a simulated or measured lipid and macromolecular background into the basis set is very likely to improve the modeling of the HERCULES spectra.
In its current implementation, the HERCULES editing scheme uses 20-ms editing pulses in all sub-experiments. The corresponding bandwidth implies suboptimal editing efficiency for the Asc and Asp resonances (cf. Fig 1a). Using editing pulses with a broader 4.1-ppm lobe in subexperiments B and D would increase sensitivity for Asc and Asp. Future HERCULES implementations are likely to be flexibly adapted to prioritize their specificity for some metabolites over others.
Finally, it should be noted that successful HERCULES data acquisition requires a high degree of frequency fidelity and stability. While the top of Gaussian editing pulses has a relatively benign profile (>95% inversion is achieved within ±0.07 ppm of the maximum), the symmetric suppression of the strong NAA-aspartyl signal is very susceptible to editing pulse frequency offsets. In macromolecule-suppressed MEGA-PRESS of GABA, even very small frequency offsets (< 5 Hz) break the symmetry of the pulse arrangement (Edden et al., 2016; Harris et al., 2014; Oeltzschner et al., 2017), and lead to substantial unwanted signal in the (A-B+C-D) difference spectrum (see the simulated behavior of the NAA- and NAAG-aspartyl signals in Supplementary Figure 3). Substantial NAA contamination will hinder the successful quantification of NAAG and Asp. Active measures for tracking motion- or drift-related frequency shifts should be considered as a remedy for similar effects on NAA suppression, such as by the use of prospective frequency correction (Edden et al., 2016).
4.1. Future developments
This study was designed to demonstrate the feasibility of the editing scheme and the multiplexed fitting approach of HERCULES. As such, several logical development steps can be taken to increase the specificity and accuracy of this method. Longer editing pulses (or higher B1 power) may help increase the pulse selectivity. This may improve editing efficiency for GSH and NAAG, as the 20-ms editing pulse at 4.18 ppm causes 15% of inversion at 4.58 ppm, with an attendant loss of edited signal. Simultaneous fitting of multiple difference spectra will benefit from further reduction of spectral imperfections and artefacts, brought about by tailored multiplexed frequency-and-phase correction schemes (Mikkelsen et al., 2017), real-time frequency adjustments during measurement (Edden et al., 2016), and/or the use of basis sets derived from the individual measurement-specific frequency-shift histogram (van der Veen et al., 2017). Modeling will also benefit from improved lineshape and decreased linewidth resulting from improved B0 shimming.
5. Conclusion
In summary, HERCULES is a novel acquisition and post-processing framework for the simultaneous editing and separation of seven low-concentration neurochemicals (including 2- HG) in addition to six high-concentration metabolites. Anticipated developments in the postprocessing and spectral fitting of this multiplexed acquisition and quantification approach may increase the quality of multiplexed HERCULES editing. Future studies should rigorously investigate the accuracy, reproducibility, and reliability of HERCULES in standard (or even smaller) voxels, and assess its sensitivity to motion to judge its potential to perform in pediatric, elderly and pathological cohorts. The combination of multiplexed editing and linear-combination modeling suggests that HERCULES could become a useful tool for probing several relevant biochemical systems in the human brain within clinically reasonable examination times.
Supplementary Material
6. Acknowledgements
The authors would like to express their gratitude to David Lythgoe and Diana Rotaru (King’s College, United Kingdom) for sharing their FID-A implementation of the fast spatial simulation algorithm. This work was supported by NIH grants R01 EB016089, R01 EB023963, U54 HD079123, and P41 EB015909. NAJP receives support from NIH grant K99/R00 MH107719.
Abbreviations:
- HERCULES
Hadamard Encoding Resolves Chemicals Using Linear- combination Estimation of Spectra
- GABA
γ-aminobutyric acid
- GSH
glutathione
- Asc
ascorbate
- NAA
N-acetylaspartate
- NAAG
N-acetylaspartylglutamate
- Asp
aspartate
- Lac
lactate
- 2-HG
2-hydroxyglutarate
- tCho
total choline
- Ins
myo-inositol
- Glu
glutamate
- Gln
glutamine
- tCr
total creatine
- DEW
Double Editing With
- HERMES
Hadamard Encoding and Reconstruction of MEGA-Edited Spectroscopy
- WM
white matter
- GM
grey matter
- IWR
interleaved water referencing
- GPC
glycerophosphorylcholine
- Gly
glycine
- PCh
phosphorylcholine
- PCr
phosphocreatine
- PE
phosphorylethanolamine
- Scy
scyllo- inositol
- Ser
serine
- Tau
taurine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG, 2012. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med 4, 116ra4. doi:10.1126/scitranslmed.3002693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhuijsen H, De Beer R, Van Ormondt D, 1987. Improved Algorithm for Noniterative Time-Domain Model Fitting to Exponentially Damped Magnetic Resonance Signals. J. Magn. Reson. 73, 553–557. [Google Scholar]
- Bhattacharyya PK, Phillips MD, Stone LA, Lowe MJ, 2011. In vivo magnetic resonance spectroscopy measurement of gray-matter and white-matter gamma-aminobutyric acid concentration in sensorimotor cortex using a motion-controlled MEGA point-resolved spectroscopy sequence. Magn. Reson. Imaging 29, 374–9. doi:10.1016/j.mri.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner W, Gruber S, Doelken M, Stadlbauer A, Ganslandt O, Boettcher U, Trattnig S, Doerfler A, Stefan H, Hammen T, 2010. In vivo quantification of intracerebral GABA by single-voxel 1H-MRS—How reproducible are the results? Eur. J. Radiol., PET/CT - An Imaging Challenge 73, 526–531. doi:10.1016/j.ejrad.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Chan KL, Puts NAJ, Schär M, Barker PB, Edden RAE, 2016a. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn. Reson. Med. 76, 11–19. doi :10.1002/mrm.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Puts NAJ, Snoussi K, Harris AD, Barker PB, Edden RAE, 2016b. Echo time optimization for J-difference editing of glutathione at 3T. Magn. Reson. Med. doi :10.1002/mrm.26122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Saleh MG, Oeltzschner G, Barker PB, Edden RAE, 2017. Simultaneous measurement of Aspartate, NAA, and NAAG using HERMES spectral editing at 3 Tesla. Neuroimage 155. doi:10.1016/j.neuroimage.2017.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Snoussi K, Edden RAE, Barker PB, 2017. Simultaneous detection of glutathione and lactate using spectral editing at 3 T. NMR Biomed. 30, e3800. doi :10.1002/nbm.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang X-L, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA, 2012. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 18, 624–629. doi:10.1038/nm.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I-Y, Lee S-P, Merkle H, Shen J, 2006. In vivo detection of gray and white matter differences in GABA concentration in the human brain. Neuroimage 33, 85–93. doi: 10.1016/j.neuroimage.2006.06.016 [DOI] [PubMed] [Google Scholar]
- Edden RA, Barker PB, 2007. Spatial effects in the detection of gamma-aminobutyric acid: improved sensitivity at high fields using inner volume saturation. Magn Reson Med 58, 1276–1282. doi :10.1002/mrm.21383 [DOI] [PubMed] [Google Scholar]
- Edden RA, Pomper MG, Barker PB, 2007. In vivo differentiation of N-acetyl aspartyl glutamate from N-acetyl aspartate at 3 Tesla. Magn Reson Med 57, 977–982. doi:10.1002/mrm.21234 [DOI] [PubMed] [Google Scholar]
- Edden RA, Puts NA, Barker PB, 2012. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med 68, 657–661. doi:10.1002/mrm.24391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Oeltzschner G, Harris AD, Puts NAJ, Chan KL, Boer VO, Schär M, Barker PB, 2016. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J. Magn. Reson. Imaging 44, 1474–1482. doi:10.1002/jmri.25304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emir UE, Deelchand D, Henry P-G, Terpstra M, 2011a. Noninvasive quantification of T2 and concentrations of ascorbate and glutathione in the human brain from the same double-edited spectra. NMR Biomed. 24, 263–269. doi:10.1002/nbm.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emir UE, Raatz S, McPherson S, Hodges JS, Torkelson C, Tawfik P, White T, Terpstra M, 2011b. Noninvasive quantification of ascorbate and glutathione concentration in the elderly human brain. NMR Biomed. 24, 888–894. doi:10.1002/nbm.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geramita M, van der Veen JW, Barnett AS, Savostyanova AA, Shen J, Weinberger DR, Marenco S, 2011. Reproducibility of prefrontal γ-aminobutyric acid measurements with J-edited spectroscopy. NMR Biomed. 24, 1089–1098. doi:10.1002/nbm.1662 [DOI] [PubMed] [Google Scholar]
- Gonenc A, Govind V, Sheriff S, Maudsley AA, 2010. Comparison of spectral fitting methods for overlapping J-coupled metabolite resonances. Magn. Reson. Med. 64, 623–8. doi: 10.1002/mrm.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryawala MZ, Sheriff S, Maudsley AA, 2016. Regional distributions of brain glutamate and glutamine in normal subjects. NMR Biomed. 29, 1108–1116. doi:10.1002/nbm.3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind V, Young K, Maudsley AA, 2015. Corrigendum: Proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153. NMR Biomed. 28, 923–924. doi:10.1002/nbm.3336 [DOI] [PubMed] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA, 2000. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed 13, 129–153. doi:10.1002/1099-1492(200005)13:3<129::AID-NBM619>3.0.CO;2-V[pii] [DOI] [PubMed] [Google Scholar]
- Hall EL, Stephenson MC, Price D, Morris PG, 2014. Methodology for improved detection of low concentration metabolites in MRS: Optimised combination of signals from multielement coil arrays. Neuroimage 86, 35–42. doi:10.1016/J.NEUROIMAGE.2013.04.077 [DOI] [PubMed] [Google Scholar]
- Harris AD, Glaubitz B, Near J, John Evans C, Puts NA, Schmidt-Wilcke T, Tegenthoff M, Barker PB, Edden RA, 2014. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med 72, 941–948. doi:10.1002/mrm.25009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry PG, Dautry C, Hantraye P, Bloch G, 2001. Brain GABA editing without macromolecule contamination. Magn Reson Med 45, 517–520. doi:10.1002/1522-2594(200103)45:3<517::AID-MRM1068>3.0.CO;2-6[pii] [DOI] [PubMed] [Google Scholar]
- Hnilicová P, Považan M, Strasser B, Andronesi OC, Gajdošik M, Dydak U, Ukropec J, Dobrota D, Trattnig S, Bogner W, 2016. Spatial variability and reproducibility of GABA-edited MEGA-LASER 3D-MRSI in the brain at 3 T. NMR Biomed. 29, 1656–1665. doi:10.1002/nbm.3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horská A, Kaufmann WE, Brant LJ, Naidu S, Harris JC, Barker PB, 2002. In vivo quantitative proton MRSI study of brain development from childhood to adolescence. J. Magn. Reson. Imaging 15, 137–143. doi:10.1002/jmri.10057 [DOI] [PubMed] [Google Scholar]
- Jensen JE, deB. Frederick B, Renshaw PF, 2005. Grey and white matter GABA level differences in the human brain using two-dimensional,J-resolved spectroscopic imaging. NMR Biomed. 18, 570–576. doi:10.1002/nbm.994 [DOI] [PubMed] [Google Scholar]
- Jensen JE, Licata SC, Öngür D, Friedman SD, Prescot AP, Henry ME, Renshaw PF, 2009. Quantification of J -resolved proton spectra in two-dimensions with LCModel using GAMMA-simulated basis sets at 4 Tesla. NMR Biomed. 22, 762–769. doi: 10.1002/nbm.1390 [DOI] [PubMed] [Google Scholar]
- Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB, 2008. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 21, 22–32. doi:10.1002/nbm.1150 [DOI] [PubMed] [Google Scholar]
- McLean MA, Woermann FG, Barker GJ, Duncan JS, 2000. Quantitative analysis of short echo time1H-MRSI of cerebral gray and white matter. Magn. Reson. Med. 44, 401–411. doi:10.1002/1522-2594(200009)44:3<401::AID-MRM10>3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R, 1998. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 11, 266–272. doi:10.1002/(SICI)1099-1492(199810)11:6<266::AID-NBM530>3.0.C0;2-J [DOI] [PubMed] [Google Scholar]
- Mikkelsen M, Barker PB, Bhattacharyya PK, Brix MK, Buur PF, Cecil KM, Chan KL, Chen DY-T, Craven AR, Cuypers K, Dacko M, Duncan NW, Dydak U, Edmondson DA, Ende G, Ersland L, Gao F, Greenhouse I, Harris AD, He N, Heba S, Hoggard N, Hsu T-W, Jansen JFA, Kangarlu A, Lange T, Lebel RM, Li Y, Lin C-YE, Liou J-K, Lirng J-F, Liu F, Ma R, Maes C, Moreno-Ortega M, Murray SO, Noah S, Noeske R, Noseworthy MD, Oeltzschner G, Prisciandaro JJ, Puts NAJ, Roberts TPL, Sack M, Sailasuta N, Saleh MG, Schallmo M-P, Simard N, Swinnen SP, Tegenthoff M, Truong P, Wang G, Wilkinson ID, Wittsack H-J, Xu H, Yan F, Zhang C, Zipunnikov V, Zöllner HJ, Edden RAE, 2017Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage 159. doi: 10.1016/j.neuroimage.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M, Saleh MG, Near J, Chan KL, Gong T, Harris AD, Oeltzschner G, Puts NAJ, Cecil KM, Wilkinson ID, Edden RAE, 2017. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magn. Reson. Med. doi:10.1002/mrm.27027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JB, Wheaton AJ, Anderson R, 2013. MEGA-PRESSing Onward for More Metabolites: Aspartate, Lactate, and PE, in: Proceedings of the 21st Annual Meeting of ISMRM Salt Lake City, Utah, USA, p. Abstract 2026. [Google Scholar]
- Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P, 2014. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. doi:10.1002/mrm.25094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Evans CJ, Puts NAJ, Barker PB, Edden RAE, 2013. J-difference editing of gamma-aminobutyric acid (GABA): Simulated and experimental multiplet patterns. Magn. Reson. Med. 70, 1183–1191. doi:10.1002/mrm.24572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G, Chan KL, Saleh MG, Mikkelsen M, Puts NA, Edden RAE, 2017. Hadamard editing of glutathione and macromolecule-suppressed GABA. NMR Biomed. doi:10.1002/nbm.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeltzschner G, Snoussi K, Puts NA, Mikkelsen M, Harris AD, Pradhan S, Tsapkini K, Schär M, Barker PB, Edden RAE, 2017. Effects of eddy currents on selective spectral editing experiments at 3T. J. Magn. Reson. Imaging. doi:10.1002/jmri.25813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JW, Twieg DB, Hetherington HP, 1998. Quantitative spectroscopic imaging of the human brain. Magn. Reson. Med. 40, 363–369. doi:10.1002/mrm.1910400305 [DOI] [PubMed] [Google Scholar]
- Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, Wilson N, Driggers E, Jang HG, Su SM, Schenkein DP, Lai A, Cloughesy TF, Kornblum HI, Wu H, Fantin VR, Liau LM, 2012. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J. Neurooncol. 107, 197–205. doi: 10.1007/s11060-011-0737-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels PJ, Frahm J, 1997. Differential distribution of NAA and NAAG in human brain as determined by quantitative localized proton MRS. NMR Biomed 10, 73–78. [DOI] [PubMed] [Google Scholar]
- Provencher SW, 1993. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med. 30, 672–9. [DOI] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH, 1993. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc. Natl. Acad. Sci. U. S. A. 90, 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Mikkelsen M, Oeltzschner G, Chan KL, Berrington A, Barker PB, Edden RA, 2018. Simultaneous editing of GABA and glutathione at 7T using semi-LASER localization. Magn. Reson. Med. doi:10.1002/mrm.27044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Oeltzschner G, Chan KL, Puts NAJ, Mikkelsen M, Schär M, Harris AD, Edden RAE, 2016. Simultaneous edited MRS of GABA and glutathione. Neuroimage 142, 576–582. doi:10.1016/j.neuroimage.2016.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J, 2017. Advanced processing and simulation of MRS data using the FID appliance (FID-A)-An open source, MATLAB- based toolkit. Magn. Reson. Med. 77, 23–33. doi:10.1002/mrm.26091 [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Ratiney H, Hammond-Rosenbluth KE, Pelletier D, Nelson SJ, 2010. MR spectroscopic imaging of glutathione in the white and gray matter at 7 T with an application to multiple sclerosis. Magn. Reson. Imaging 28, 163–170. doi:10.1016/J.MRI.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB, 1998. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson 133, 243–254. doi:10.1006/jmre.1998.1458 [DOI] [PubMed] [Google Scholar]
- Terpstra M, Gruetter R, 2004. 1H NMR detection of vitamin C in human brain in vivo. Magn. Reson. Med. 51, 225–229. doi:10.1002/mrm.10715 [DOI] [PubMed] [Google Scholar]
- Terpstra M, Henry P-G, Gruetter R, 2003. Measurement of reduced glutathione (GSH) in human brain using LCModel analysis of difference-edited spectra. Magn. Reson. Med. 50, 19–23. doi: 10.1002/mrm.10499 [DOI] [PubMed] [Google Scholar]
- Terpstra M, Marjanska M, Henry P-G, Tkac I, Gruetter R, 2006. Detection of an antioxidant profile in the human brain in vivo via double editing with MEGA-PRESS. Magn. Reson. Med. 56, 1192–1199. doi:10.1002/mrm.21086 [DOI] [PubMed] [Google Scholar]
- Terpstra M, Torkelson C, Emir U, Hodges JS, Raatz S, 2011. Noninvasive quantification of human brain antioxidant concentrations after an intravenous bolus of vitamin C. NMR Biomed. 24, 521–528. doi:10.1002/nbm.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra M, Ugurbil K, Tkàc I, Tkac I, 2010. Noninvasive quantification of human brain ascorbate concentration using 1H NMR spectroscopy at 7 T. NMR Biomed. 23, 227–232. doi: 10.1002/nbm.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen JW, Marenco S, Berman KF, Shen J, 2017. Retrospective correction of frequency drift in spectral editing: The GABA editing example. NMR Biomed. e3725. doi: 10.1002/nbm.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss PO, Bianchini C, Scheidegger M, Giapitzakis IA, Hock A, Fuchs A, Henning A, 2018. In vivo estimation of transverse relaxation time constant (T2) of 17 human brain metabolites at 3T. Magn. Reson. Med. 80, 452–461. doi:10.1002/mrm.27067 [DOI] [PubMed] [Google Scholar]
- Zhang Y, An L, Shen J, 2017. Fast computation of full density matrix of multispin systems for spatially localized in vivo magnetic resonance spectroscopy. Med. Phys. 44, 4169–4178. doi :10.1002/mp.12375 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


