Abstract
TRIM25 is a multi-domain, RING-type E3 ubiquitin ligase of the tripartite motif family that has important roles in multiple RNA-dependent processes. In particular, TRIM25 functions as an effector of RIG-I and ZAP, which are innate immune sensors that recognize viral RNA and induce ubiquitin-dependent anti-viral response mechanisms. TRIM25 is reported to also bind RNA, but the molecular details of this interaction or its relevance to anti-viral defense have not been elucidated. Here, we characterize the RNA-binding activity of TRIM25 and find that the protein binds both single-stranded and double-stranded RNA. Multiple regions of TRIM25 contribute to this functionality, including the C-terminal SPRY domain and a lysine-rich motif in the linker segment connecting the SPRY and coiled-coil domains. RNA binding modulates TRIM25’s ubiquitination activity in vitro, its localization in cells, and its anti-viral activity. Taken together with other studies, our results indicate that RNA binding by TRIM25 has at least three important functional consequences: by enhancing ubiquitination activity, either through allosteric effects or through clustering of multiple TRIM25 molecules; by modulating the multi-domain structure of the TRIM25 dimer, and thereby structural coupling of the SPRY and RBCC elements during the ubiquitination reaction; and by facilitating subcellular localization of the E3 ligase during virus infection.
Introduction
TRIM25 is a member of the tripartite motif family of E3 ubiquitin ligases that functions in multiple RNA-dependent pathways. It has been well established that TRIM25 plays a crucial role in the RIG-I anti-viral pathway, where TRIM25 is reported to bind viral RNA-activated RIG-I, leading to K63-linked polyubiquitination of the sensor to promote transcriptional upregulation of type I interferons (IFNs) [1, 2]. TRIM25 was also shown to be important for micro RNA processing, which also requires TRIM25’s E3 ligase activity [3, 4]. Most recently, TRIM25 was found to be an essential co-factor for messenger RNA (mRNA) degradation mediated by the ZAP protein [5, 6], which recognizes viral mRNAs with high CG-dinucleotide content [7]. TRIM25 is therefore an attractive model system for understanding the molecular mechanisms of how a single ubiquitin E3 enzyme is directed to specific cellular pathways to produce distinct biological outcomes.
The biophysical mechanism of TRIM25’s E3 ubiquitin ligase activity has now been elucidated. Like a typical TRIM protein, TRIM25 contains an N-terminal tripartite motif or RBCC motif, consisting of RING, B-box 1, B-box 2, and coiled-coil domains. Additionally, TRIM25 has a C-terminal SPRY domain that is required for recruitment to the RIG-I, ZAP, and RNA processing pathways [1-3, 5, 6]. The RBCC and SPRY domains are connected by a linker region (linker 2 or L2), part of which is integrated with the coiled-coil and likely facilitates functional coupling of these structural elements [8]. The basal oligomeric state of TRIM25 is a stable dimer mediated by the coiled-coil/L2 regions. This dimerization domain makes an elongated, anti-parallel scaffold, with the RINGs and B-boxes on opposite ends and separated by around 170 A, and the SPRY domains packed against the coiled-coil domain [8, 9]. The RING domain harbors the E3 ubiquitin ligase activity of TRIM25, and it is catalytically competent in vitro as an independent protein. The RING domain engages E2 conjugating enzymes and catalyzes polyubiquitination reactions as a dimer [10, 11]. In context of the full-length TRIM25 dimer, it was proposed that the two RING domains cannot interact with each other and that RING dimerization requires higher-order oligomerization or self-association of at least two coiled-coil mediated TRIM25 dimers [8]. Alternatively, the tandem B-boxes of TRIM25 may provide sufficient reach to allow the two RING domains within a coiled-coil-mediated TRIM25 dimer to interact with each other. Regardless, how TRIM25 self-association and RING activation occurs in a pathway-specific manner in cells is currently unknown.
One simple model is that TRIM25’s E2 ligase activity is indirectly stimulated by RNA signals; that is, TRIM25 is recruited as a downstream effector by the primary cellular RNA sensors RIG-I or ZAP. However, this simple model has been challenged by the finding that TRIM25 independently binds RNA, raising the possibility that RNA itself can induce TRIM25-mediated polyubiquitination [12, 13]. Here, we use biochemical, biophysical, and cell biological approaches to characterize this interaction and its functional significance for TRIM25’s anti-viral activity. We found that TRIM25 has a composite RNA binding site with energetically significant contributions from multiple regions of the protein. We further found that TRIM25’s RNA-binding activity is required for its for its subcellular localization, potentiation of RIG-I signaling, and its overall anti-viral activity in cells. Taken together with other studies, our results support a model wherein pathway-specific TRIM25 recruitment and activation results from a complex interplay of both protein-protein and protein-RNA interactions.
Results
RNA binding enhances TRIM25 E3 ligase activity in vitro
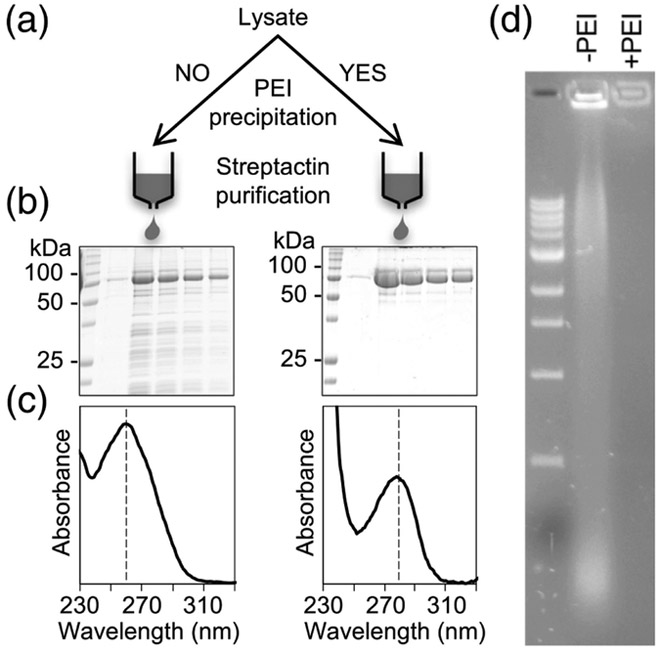
TRIM25 was first reported to harbor RNA-binding activity in pulldown studies [12]. We confirmed this in the course of purifying the protein for biochemical studies. Specifically, we found that recombinant Strep/FLAG-tagged TRIM25 overexpressed in insect cells co-eluted from affinity columns with large quantities of nucleic acid (Fig. 1). The protein samples were not very pure (Fig. 1b, left) and had a UV absorbance profile with a peak at 260 nm, with an A260/A280 ratio of ~1.9 (Fig. 1c, left). Use of polyethyleneimine (PEI) precipitation prior to affinity purification significantly reduced both the protein (Fig. 1b, right) and nucleic acid contaminations, giving an A260/A280 ratio of ~0.7 (Fig. 1c, right). Agarose gel electrophoresis confirmed the presence of significant amounts of nucleic acid in the non-PEI-treated but not the PEI-treated samples (Fig. 1d).
Fig. 1.
Recombinant TRIM25 co-purifies with nucleic acid. (a) Affinity purification scheme for Strep/FLAG-tagged TRIM25 expressed in insect cells. (b) Coomassie-stained SDS-PAGE gels of TRIM25 fractions after initial affinity chromatography step. (c) UV absorbance spectra of pooled protein fractions. Vertical dashed lines indicate the peak wavelength. (d) Visualization by agarose gel electrophoresis and SYBR-green staining of the co-purifying nucleic acid (−PEI) and its removal by polyethyleneimine treatment (+PEI).
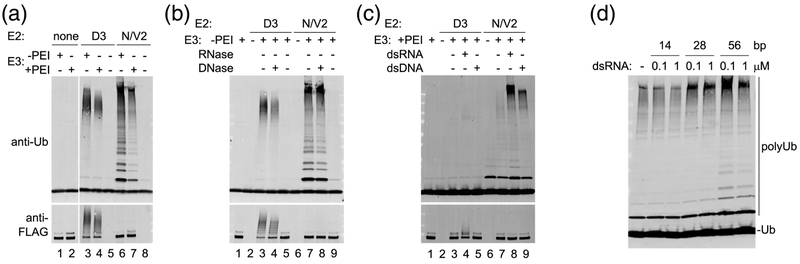
Interestingly, ubiquitination assays revealed that the presence of nucleic acid significantly enhanced the enzymatic activity of TRIM25 in vitro (Fig 2a, compare lane 2 to lane 3 and lane 5 to lane 6). This effect was observed with both Ube2D3 (also known as Ubc5c, which promotes in vitro auto-ubiquitination of TRIM25; Fig. 2a, lanes 2-4) and Ube2N/Ube2V2 (Ubc13/Mms2, which promotes synthesis of free polyubiquitin chains; Fig. 2a, lanes 5-7). Incubation of the non-PEI-treated sample with RNase A abrogated ubiquitination in vitro (Fig. 2b, lanes 5 and 9), whereas incubation with DNase I had little or no effect (Fig. 2b, lanes 4 and 8). This result confirmed that the co-purifying nucleic acids were indeed RNA. Conversely, incubation of PEI-treated samples with dsRNA significantly enhanced in vitro ubiquitination (Fig. 3c, lanes 4 and 8). Incubation with dsDNA also enhanced activity, but to a lesser extent (Fig. 3c, lanes 5 and 9); this result likely reflects the commonly observed non-specific interaction of RNA-binding proteins with DNA in vitro. The above data also indicate that RNA enhancement of TRIM25’s E3 ligase activity is independent of the E2 enzyme, although it is more pronounced with Ube2N/Ube2V2 than Ube2D3.
Fig. 2.
RNA enhances TRIM25’s catalytic activity in vitro. (a) Ubiquitination activities of −200 nM TRIM25 purified with (lanes 1, 3, and 6) or without (lanes 2, 4, and 7) PEI precipitation. TRIM25 synthesizes anchored (self-attached) polyubiquitin (polyUb) chains with 1 μM Ube2D3 and unanchored chains with 0.280 μM Ube2N/Ube2V2 [11]. Reactions contained 100 nM E1, 40 μM Ub, and 5 mM Mg-ATP. (b) TRIM25 purified in the absence of PEI treatment was pre-incubated with RNase A (lanes 5 and 9), DNase I (lanes 4 and 8), or buffer control (lanes 3 and 7) prior to setting up ubiquitination assays. (c) TRIM25 purified with PEI treatment was pre-incubated with 500 ng of dsRNA (lanes 4 and 8), 500 ng of dsDNA (lanes 5 and 9), or buffer control (lanes 3 and 7) prior to ubiquitination assays. (d) TRIM25 purified with PEI treatment was pre-incubated with the indicated concentrations of 14, 28, or 56-bp dsRNA prior to ubiquitination with Ube2N/Ube2V2 as E2.
Fig. 3.
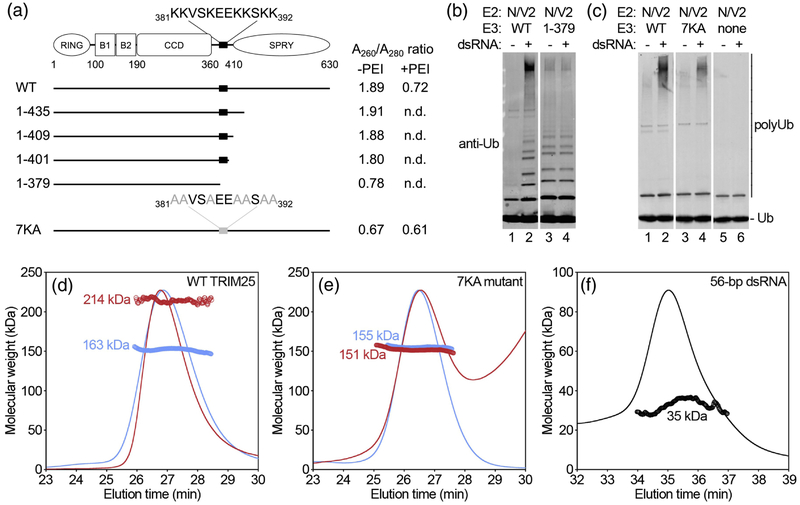
Identification of the 7K motif that mediates TRIM25 binding to RNA. (a) Schematic of TRIM25’s domain organization and summary of constructs used to map the RNA binding site. Observed A260/A280 UV absorbance ratios with and without PEI treatment are indicated. The lysine-rich sequence spanning residues 381-392 is highlighted. Abbreviated domain names: B1 = B-box 1; B2 = B-box 2; CCD = coiled-coil dimerization domain. (b-c) Ubiquitination activities of the indicated constructs (lanes 3 and 4) in the presence or absence of 1 μM 28-bp dsRNA, along with corresponding WT control assayed in parallel (lanes 1 and 2). (d-e) SEC-MALS analysis of the indicated purified TRIM25 constructs prior to (blue) and after incubation with 56-bp dsRNA (red). Curves show the refractive index of the eluting species normalized to the peak intensity. Open circles indicate calculated molecular weights in kDa. (f) SEC-MALS profile of the RNA alone, shown for reference.
The enzymatic activity of TRIM25 is thought to require higher-order oligomerization of the coiled-coil-mediated dimers to allow for RING domain dimerization [10, 11], and so a simple model for RNA-induced ubiquitination in vitro is through clustering of TRIM25 molecules. Typical dsRNA-binding proteins have a footprint of around 12-16 bp, and indeed, 14-bp dsRNA did not enhance the ubiquitination activity of PEI-treated TRIM25, whereas 28-bp and 56-bp dsRNA did (Fig. 3d). Thus, our data support a model wherein dsRNA promotes in vitro self-association of TRIM25 and RING domain activation, provided that the RNA is of sufficient length to bind more than one coiled-coil-mediated TRIM25 dimer.
RNA binding requires a lysine-rich motif in the L2 linker
The RNA-binding activity of TRIM25 was shown to require the C-terminal SPRY domain [13]. Our results confirm this because a TRIM25 construct spanning only the RBCC domains (amino-acid residues 1-379) did not co-purify with nucleic acid, as evidenced by a low A260/A280 ratio after affinity purification without PEI treatment (Fig. 3a). Correspondingly, the in vitro ubiquitination activity of this RBCC construct was insensitive to added dsRNA (Fig. 3b, compare lanes 3 and 4; WT control, compare lanes 1 and 2). (Note that the ubiquitination experiment was performed with Ube2N/Ube2V2, which generates free or unattached polyubiquitin chains, in order to preclude having to account for differences in the number of potential lysine acceptors). Interestingly, systematic extension of the RBCC construct (residues 1-401, 1-409, and 1-435) identified a lysine-rich sequence in the L2 linker that connects the coiled-coil and SPRY domains (381KKVSKEEKKSKK392, hereafter termed ‘7K motif’), the presence of which resulted in elevated A260/A280 ratios in the affinity-purified fractions even in the absence of the SPRY domain (Fig. 3a). This 7K motif resembles a segment in ribosomal protein S30 (18KVAKQEKKKKK28), in which the lysine sidechains mediate ionic contacts with the phosphodiester backbone of 18S ribosomal RNA [14]. We therefore surmised that the TRIM25 7K motif might make equivalent contacts with bound RNA, and indeed alanine substitution of all 7 lysines in context of full-length TRIM25 (TRIM25 7KA) resulted in a low A260/A280 ratio even in the absence of PEI precipitation (Fig. 3a). Consistent with this result, the ubiquitination activity of the purified 7KA mutant protein did not respond to dsRNA to the same extent as wildtype (WT) TRIM25 (Fig. 3c, compare lanes 3 and 4; WT control, compare lanes 1 and 2). However, dsRNA-dependent ubiquitination was still observed for the 7KA protein, indicating that this mutant still retained some ability to bind RNA, likely because it still retained the SPRY domain (see also below).
To confirm that TRIM25 binds RNA directly, we performed SEC-MALS (size exclusion chromatography coupled with multi-angle light scattering) experiments, which allow direct measurements of particle mass, complex formation, and sample homogeneity. In control experiments, purified WT TRIM25 eluted in the absence of added RNA as a single major peak with a measured mass consistent with a dimer (Fig. 3d, blue; calculated mass from sequence = 2 × 75 kDa =150 kDa). In the presence of an equimolar ratio of 56-bp dsRNA, the major peak did not significantly change in elution position (indicating that the particle shape did not change considerably from the elongated dimer of the protein alone), but now had a mass increase of ~70 kDa (Fig. 3d, red), which is equivalent to two 56-bp dsRNA molecules (Fig. 3f). Thus, TRIM25 bound to dsRNA at a 1:1 molar ratio in this experiment. In contrast, the 7KA mutant had the same population-averaged mass in the absence (Fig. 3e, blue) and presence (Fig. 3e, red) of dsRNA, confirming that this protein was significantly deficient in RNA binding in vitro. Since larger oligomers of WT TRIM25 in the presence of 56-bp RNA were not observed by SEC-MALS, the enhancement of ubiquitination activity may be due to allosteric effects than clustering. Importantly, our results also indicated that the 7KA mutations had no effect on the structure of the TRIM25 dimer, or on the fundamental biophysical mechanism of RING E3 ligase activation.
TRIM25 binds RNA through multiple structural elements
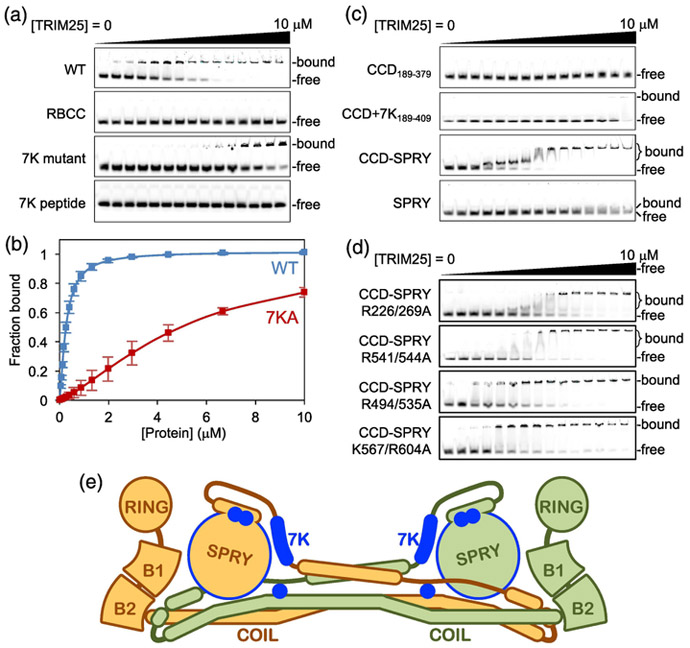
The above experiments revealed that the 7KA mutation did not completely abolish the ability of TRIM25 to interact with RNA and confirmed that the SPRY domain also contributed to RNA binding. To explore this further, we used an electrophoretic mobility shift assay (EMSA) to quantify TRIM25’s RNA binding activity in vitro. In this experiment, fluorescently-tagged 28-bp dsRNA (250 nM) was incubated with different concentrations of purified TRIM25 proteins (0-10 μM), and then the free and TRIM25-bound RNA fractions were separated on a native polyacrylamide gel and quantified by densitometry. In control experiments with full-length WT TRIM25, the gel scans gave the appearance of an all-or-none or highly cooperative binding mechanism (Fig. 4a, WT). However, this conclusion was not fully supported by the data because the TRIM25/RNA complexes did not enter the gel and the homogeneity of the bound fractions cannot be rigorously established. We therefore estimated the amounts of bound RNA through densitometric quantification of the unbound RNA bands, and performed curve fitting to a simple binding isotherm (see Materials and Methods for details). This revealed that TRIM25 bound to 28-bp dsRNA with an apparent dissociation constant (Kd) of 0.26 μM (Fig. 4b, blue and Table 1). Consistent with our purification data and with previous studies [13], the RBCC domain alone (residues 1-379) that lacks the SPRY domain did not bind RNA (Fig. 4a, RBCC). The 7KA mutant retained some binding, but with about 20-fold loss in affinity (Kd = 4.9 μM) (Fig. 4a, 7KA mutant; Fig. 4b, red; and Table 1). Interestingly, the lysine-rich peptide alone (Fig. 4a, 7K peptide) also did not bind. This result demonstrated that the TRIM25 7K motif does not constitute an independent RNA-binding element. Our interpretation of these data is that the presence of the SPRY domain is required for the 7K motif to adopt the appropriate structural configuration for binding RNA.
Fig. 4.
Mapping of TRIM25 RNA binding elements. (a) Representative EMSA results with 0.25 μM of 28-bp fluorescent dsRNA oligo incubated with 0, 0.05, 0.08, 0.12, 0.17, 0.26, 0.39, 0.59, 0.88, 1.32, 2.0, 3.0, 4.4, 7.0, and 10 μM of purified TRIM25 WT, ΔSPRY, 7KA mutant, or 7K peptide, which is an 18-mer peptide containing the lysine-rich sequence. Migration positions are indicated for unbound oligo and bound complexes, which did not enter the gel. (b) Unbound fractions were quantified by densitometry and used to estimate the bound fractions, which were plotted as a function of total TRIM25 concentration. Curves indicate best fits to a standard binding isotherm. Error bars indicate the standard deviation of three independent experiments. (c-d) Representative EMSA results with the indicated TRIM25 constructs and mutants. (e) Diagram of the TRIM25 dimer, with locations of elements that contribute significantly to RNA binding indicated in blue.
Table 1.
RNA binding affinities of TRIM25 constructs.
| Protein construct | Residues | Kd (μM) | Relative affinitya | Hill coefficient |
|---|---|---|---|---|
| WT | 1-630 | 0.26 ± 0.03b | 1.00 | 1.44 ± 0.10 |
| 7KA mutant | 1-630 | 4.89 ± 0.03 | 0.05 | 1.73 ±0.24 |
| ΔSPRY | 1-379 | no binding | ||
| 7K peptide | 379-394 | no binding | ||
| CCD | 189-379 | no binding | ||
| CCD+7K | 189-409 | no binding | ||
| CCD-SPRY | 189-630 | 0.11 ±0.02 | 2.36 | 2.24 ±0.04 |
| SPRY | 410-630 | 3.47 ± 1.17 | 0.07 | 1.73 ± 0.92 |
| CCD-SPRY R226/269A | 189-630 | 0.45 ±0.18 | 0.58 | 1.31 ± 0.10 |
| CCD-SPRY R541/544A | 189-630 | 0.27 ±0.02 | 0.96 | 2.50 ±0.55 |
| CCD-SPRY R494/53 5A | 189-630 | 0.22 ± 0.03 | 1.18 | 1.28 ±0.07 |
| CCD-SPRY R567/604A | 189-630 | 0.47 ± 0.17 | 0.55 | 1.54 ± 0.20 |
Compared to WT.
Average values and standard deviations were calculated from at least three independent trials.
Given the apparent requirement for both L2 and SPRY regions, we performed EMSA experiments on a series of TRIM25 constructs and full-length mutants to more fully delineate the elements that contribute to TRIM25’s RNA binding activity in vitro (Fig. 4c-d; results are summarized in Table 1). These experiments revealed that: (1) the RING and B-box domains were not required for RNA binding, and indeed may even act inhibitory because deletion of these domains (CCD-SPRY) enhanced the apparent RNA-binding affinity by 2-fold; (2) the coiled-coil/L2 dimerization domain (CCD+7K) did not independently bind to RNA in the absence of the 7K motif, but alanine substitutions for exposed arginine residues (Arg226 and Arg229) in this domain in context of full-length protein resulted in 2-fold loss of affinity; (3) the isolated SPRY domain bound RNA, with affinity similar to the full-length 7KA mutant (Kd = 3.5 μM); and (4) alanine substitution of SPRY residues Lys567 and Arg604 in context of the CCD-SPRY fragment also resulted in 2-fold loss of RNA binding. Thus, our aggregate data indicated that both the SPRY domain and the 7K motif in the L2 linker are the major determinants of RNA binding and likely provide direct contacts to the bound RNA.
TRIM25 can bind both single-stranded and double-stranded RNA
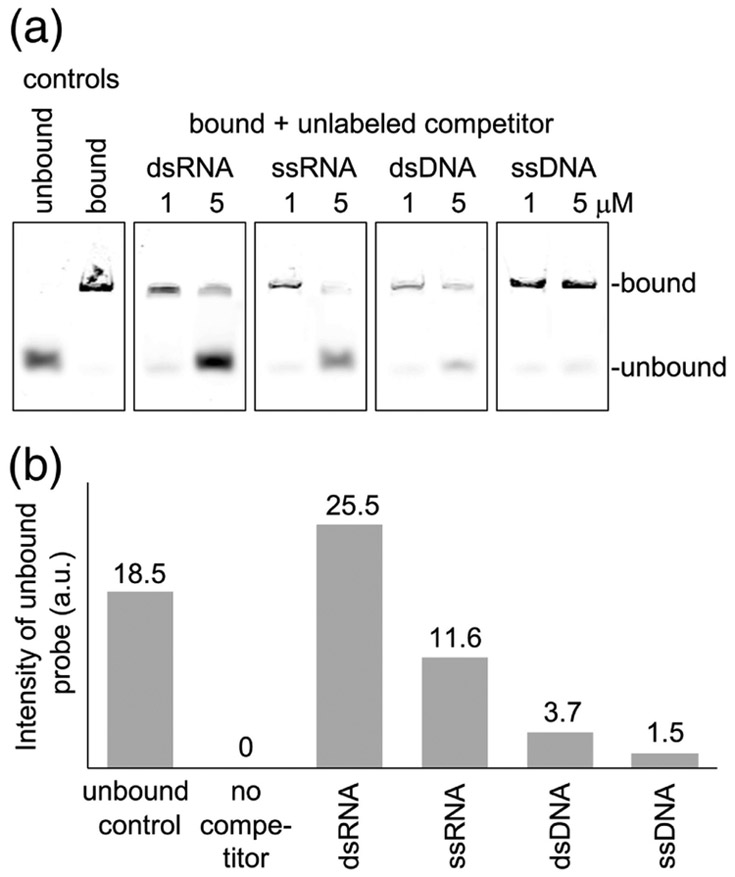
We have not found evidence for sequence-specific binding by TRIM25 as yet, but did observe a clear preference for RNA over DNA in EMSA experiments. Pre-formed complexes of TRIM25 and fluorescently-labeled dsRNA were incubated with unlabeled dsRNA, ssRNA, dsDNA, and ssDNA (all of the same lengths and sequence), and displacement of the labeled probe was quantified by polyacrylamide gel electrophoresis and densitometry (Fig. 5). Results showed that unlabeled ssRNA displaced about half the amount of labeled probe displaced by dsRNA, when compared on a molar basis. Since dsRNA has twice the number of strands as ssRNA, this result indicated that ssRNA was as competent as dsRNA in binding TRIM25 on a per-strand basis. In contrast, dsDNA displaced about 10-fold less probe compared to dsRNA, and ssDNA displaced also about half less than dsDNA. Thus, TRIM25 can bind both dsRNA and ssRNA in vitro and preferentially binds both forms of RNA over DNA.
Fig. 5.
Competition binding experiments. (a) The indicated concentrations of unlabeled competitor 28-bp oligos were incubated with 0.25 μM of TRIM25 in complex with fluorescent 28-bp RNA and electrophoretic migration of the fluorescent probe was analyzed. (b) The amounts of fluorescent probe displaced by 5 μM competitor were quantified and plotted. Experiments were repeated three times independently, and representative results are shown.
RNA binding is required for TRIM25’s overall anti-viral activity
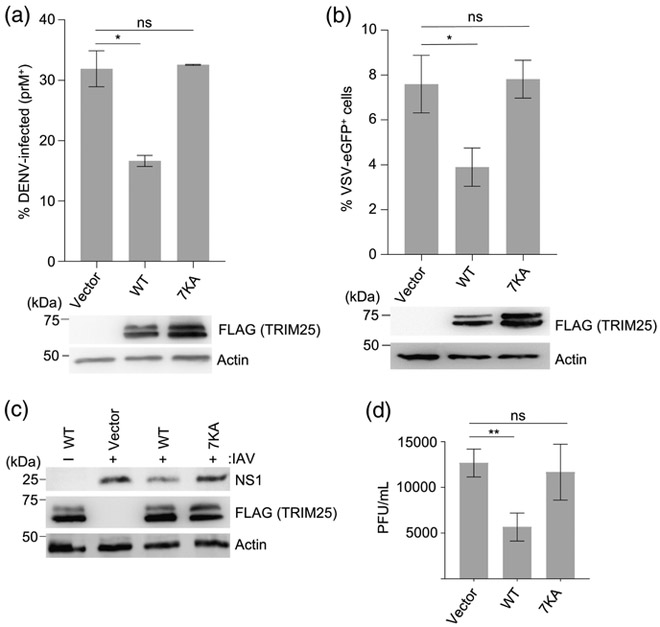
We next tested whether TRIM25’s RNA-binding ability is required for its anti-viral function. To this end, we reconstituted TRIM25-knockout (KO) HEK 293T cells [11] by transfection with either empty vector, FLAG-tagged WT TRIM25, or the 7KA mutant. Results showed that WT TRIM25, but not the 7KA mutant, effectively suppressed replication of dengue virus (DENV), a member of the Flaviviridae family, as monitored by expression levels of the viral prM protein in the infected cells (Fig. 6a). Specifically, DENV prM was expressed in only 15% of TRIM25-KO cells that were reconstituted with WT TRIM25, but in more than 30% of cells reconstituted with the 7KA mutant or vector only control. Furthermore, cells that expressed FLAG-tagged WT TRIM25, but not cells expressing the 7KA mutant, suppressed the replication of eGFP-expressing vesicular stomatitis virus (VSV-eGFP, Rhabdoviridae), as indicated by a decrease in VSV-eGFP-positive cells (Fig. 6b). We also tested replication of influenza A virus (IAV, Orthomyxoviridae) in reconstituted TRIM25-KO cells by measuring the abundance of the IAV non-structural protein 1 (NS1) in the whole cell lysates (WCLs) of infected cells through immunoblotting (Fig. 6c). This showed reduced NS1 protein abundance in TRIM25-KO cells complemented with WT TRIM25 as compared to vector-transfected control cells. In contrast, cells reconstituted with the 7KA mutant had comparable NS1 protein expression levels as vector-transfected control. Consistent with these results, IAV titers in the supernatants of WT TRIM25-expressing cells were significantly reduced, whereas cells reconstituted with the 7KA mutant had similar titers as cells reconstituted with empty vector (Fig. 6d). We conclude that the RNA-binding ability of TRIM25 is, indeed, important for its overall anti-viral activity.
Fig. 6.
RNA binding is important for TRIM25’s ability to inhibit the replication of representative members of three different RNA virus families. (a) TRIM25-KO HEK 293T cells were transiently transfected with either empty vector, FLAG-tagged TRIM25 (WT), or the 7KA mutant (7KA). Twenty-four hours later, cells were infected with DENV (MOI 4) for 24 h, and then stained for the viral prM protein. The percentage of prM-positive cells were determined by flow cytometry. Error bars represent the standard deviation of three biological replicates. (b) HEK 293T cells, transfected as in (A), were infected with VSV-eGFP (MOI 0.01). Seventeen hours later, the percentage of eGFP-positive cells were determined by flow cytometry. Error bars represent the standard deviation of three biological replicates. (c) TRIM25-KO HEK 293T cells were transfected as in (a). Twenty-four hours later, cells were infected with IAV (MOI 0.5) for 72 h, and then immunoblotted with anti-NS1 to determine expression levels of the viral NS1 protein. For all viral assays, whole cell lysates (WCLs) were further immunoblotted with anti-FLAG antibody to confirm expression of TRIM25 WT and 7KA proteins. Actin was used as loading control. (d) IAV replication in TRIM25-KO HEK 293T cells that were transfected as in (a), and infected with IAV (MOI of 0.1) for 24 h. Error bars represent the standard deviation of three biological replicates. *P <0.05; **P <0.01; ns, statistically not significant.
TRIM25 RNA binding is required to potentiate RNA-independent RIG-I signaling
Mechanistically, one interpretation of our data is that TRIM25 does not have inherent RNA specificity, but then gains this by interaction with RNA receptors, such as RIG-I or ZAP. The RIG-I receptor has a well-characterized affinity for 5’-triphosphorylated blunt-ended, double-stranded RNA (reviewed in detail in [15]), whereas ZAP was recently shown to bind CG dinucleotide motifs in mRNA [7]. Therefore, an attractive model is that TRIM25 is recruited by RIG-I or ZAP in order to bind to the same RNA molecule and generate a supercomplex for RNA-dependent ubiquitination, i.e., TRIM25 is a co-receptor of RNA. We tested this model in context of RIG-I using EMSA but found that TRIM25 did not show a clear preference in binding RIG-I/RNA complexes over RNA alone in vitro.
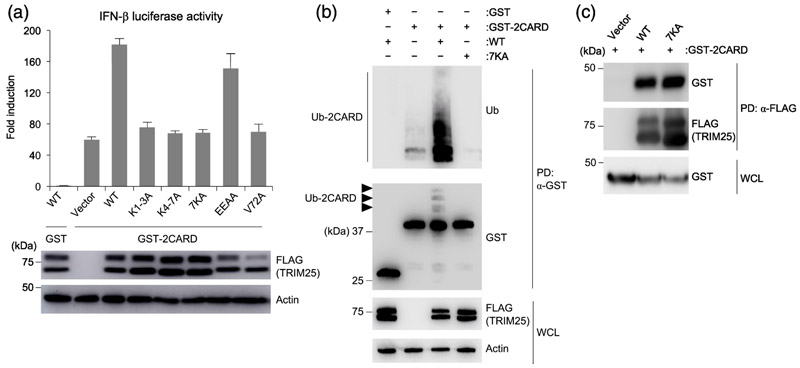
Overexpression of GST-fused RIG-I 2CARD (GST-2CARD) robustly induces IFN without the need for a viral RNA input, and this activity is established to be promoted by TRIM25 [1, 16, 17]. We therefore tested whether the TRIM25 7KA mutant can still potentiate the IFN-inducing activity of GST-2CARD (Fig. 7a). GST-2CARD overexpression in the presence of WT TRIM25 induced IFN-β promoter activity by ~180-fold above background, as opposed to ~60-fold induction in the absence of TRIM25 (co-expression with empty vector). Somewhat surprisingly, IFN-β promoter activation by GST-2CARD in the presence of 7KA was similar to co-expression of empty vector or the negative control V72A TRIM25 mutant that we previously showed to be deficient in RING dimerization [11]. This result suggested that the RNA-binding activity of TRIM25 was still important for its ability to potentiate the GST-2CARD-mediated signaling activity in cells, even though GST-2CARD does not bind RNA and its IFN-inducing activity is independent of a viral RNA signal. Since GST-2CARD is thought to mimic the viral RNA-activated form of full-length RIG-I, this result implies that TRIM25 and RIG-I do not necessarily need to bind the same RNA molecule to induce an IFN response. This raises the possibility that two independent RNA inputs may allow for full TRIM25/RIG-I activation in cells.
Fig. 7.
RNA-binding activity of TRIM25 is required for GST-2CARD ubiquitination and induction of interferon. (a) TRIM25-KO HEK 293T cells were transfected with plasmids encoding IFN-β-luciferase, β-galactosidase, and GST or GST-fused RIG-I 2CARD (GST-2CARD) together with empty vector or the indicated FLAG-tagged TRIM25 constructs (WT; K1-3A, K4-7A, 7KA that respectively replaced the first 3, second 4, and all 7 lysines in the 7K motif with alanine; EEAA that replaced two Glu residues in the 7K motif with alanine; and previously described V72A mutant that is impaired in ubiquitination activity [11]). Twenty-four hours later, IFN-β promoter activity was measured by a luciferase assay, and values were normalized to β-galactosidase levels. Whole cell lysates (WCL) were analyzed by immunoblotting with anti-FLAG to determine ectopic TRIM25 expression, with actin as loading control. (b) HEK 293T cells were transfected with GST or GST-2CARD together with empty vector or the indicated FLAG-tagged TRIM25 constructs (WT or 7KA mutant). Forty-eight hours later, WCL were subjected to pulldown with anti-GST followed by immunoblotting with anti-Ub and anti-GST antibodies. WCL were further immunoblotted with anti-FLAG to determine expression levels of the TRIM25 WT and 7KA proteins. Actin was used as a loading control. (c) HEK 293T cells were transfected as in (B). Forty-eight hours later, WCL were subjected to pulldown with anti-FLAG followed by immunoblotting with anti-GST and anti-FLAG antibodies. WCLs were further immunoblotted with anti-GST to determine expression levels of GST-2CARD.
TRIM25 potentiates the IFN-inducing activity of GST-2CARD by modifying the 2CARD with K63-linked polyubiquitin [1]. To determine if the 7KA mutation disrupts this activity, we overexpressed GST-2CARD and examined its ubiquitination status in the presence of WT or mutant TRIM25. As shown in Fig. 7b, GST-2CARD was robustly ubiquitinated when co-expressed with WT TRIM25 but not with the 7KA mutant or vector control (compare lane 3 to lanes 2 and 4). Thus, loss of RNA binding caused by 7KA correlates well with loss in TRIM25’s ability to ubiquitinate RIG-I 2CARD and potentiate IFN induction. Importantly, we confirmed that the impaired ability of TRIM25 7KA in ubiquitinating the 2CARD was not due to a defect in 2CARD-binding of this mutant because both WT and 7KA mutant TRIM25 co-immunoprecipitated GST-2CARD with about the same efficiency (Fig. 7c). Thus, RNA binding by TRIM25 appears to directly regulate its ability to ubiquitinate substrates, consistent with our in vitro ubiquitination assays and recent studies [13]. Interestingly, our observation that interaction between TRIM25 and GST-2CARD is readily detectable by co-immunoprecipitation from cell lysates but not by biochemical reconstitution from purified components suggests that the cellular environment promotes stable association of these two proteins.
RNA binding mediates TRIM25 localization to RNA stress granules
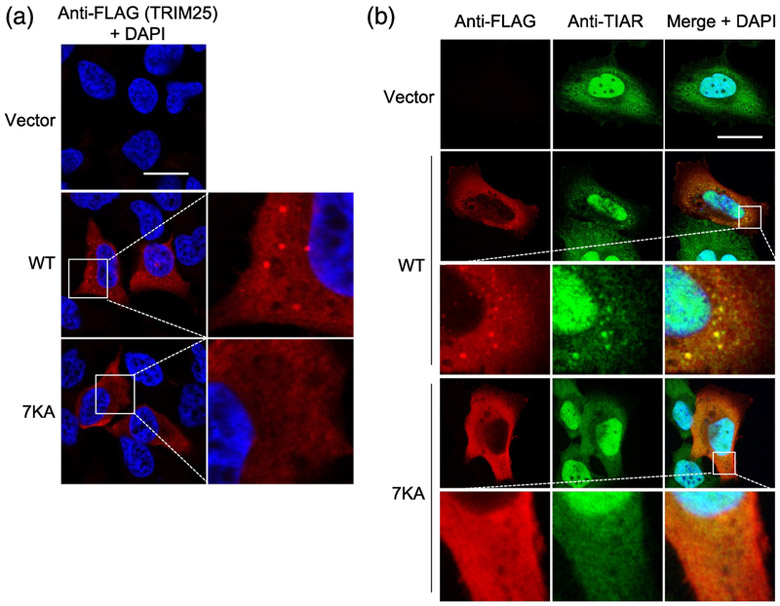
Previous studies have shown that TRIM25 assembles into cytoplasmic puncta, and evidence suggests that these puncta have the characteristics of RNA stress granules and comprise co-localization sites with RIG-I upon pathway activation [1, 18, 19]. We therefore also considered whether RNA binding allows TRIM25 to localize to these punctate cellular compartments. Overexpression of WT TRIM25 resulted in prominent punctate staining, whereas the 7KA mutant had a diffuse distribution (Fig. 8a). Co-staining the cells for endogenous TIAR1, a stress granule marker protein, showed co-localization with WT TRIM25 in the punctate structures, but not with 7KA, which remained diffusely distributed (Fig. 8b). Comparing vector-transfected and WT TRIM25-expressing cells, it appeared that overexpression of WT TRIM25 induced TIAR-stained puncta formation, which is indicative of stress granule induction, although this requires further investigation. Overall, these results indicated that the RNA-binding activity of TRIM25 is important for its localization to RNA stress granules. Since RNA granules were recently shown to be sites of interaction with activated RIG-I during viral infection, we propose that the RNA-mediated co-localization facilitates functional interaction between RIG-I and TRIM25, as previously suggested [19].
Fig. 8.
RNA binding is important for TRIM25’s subcellular localization. (a) Localization of TRIM25 WT and 7KA mutant proteins in HeLa cells that were transiently transfected with empty vector, or the indicated FLAG-tagged TRIM25 constructs and, 48 h later, stained with anti-FLAG antibody (red). Scale bar, 20 μm. DAPI-stained nuclei are seen in blue. (b) HeLa cells, transfected as in (a), were co-stained with anti-FLAG (red) and anti-TIAR (green) antibodies. DAPI-stained nuclei are in blue. Scale bar, 20 μm.
Discussion
TRIM25 functions in multiple RNA-dependent ubiquitination mechanisms, and was previously identified in an unbiased screen as an RNA-binding protein [12]. Our studies provide biochemical proof, using purified components, that TRIM25 indeed directly binds to RNA with around 250 nM affinity. Interestingly, our results further show that TRIM25’s RNA-binding activity not only requires the SPRY domain as reported [13], but also a lysine-rich ‘7K motif’ in the L2 region that links the SPRY domain to the RBCC scaffold. Alanine substitution of all seven lysines in this motif reduces RNA-binding affinity of full-length TRIM25 to that of the isolated SPRY domain, but the isolated 7K motif does not have independent RNA-binding activity. Our interpretation of these results is that the segment of the L2 linker that contains the 7K motif adopts a binding-competent configuration only in context of the full-length TRIM25 dimer. In other TRIM proteins, the L2 linker has been shown to facilitate structural and mechanistic coupling of the SPRY and RBCC regions [20, 21], and we therefore propose that RNA modulates this functional coupling in TRIM25.
Our proposed model is consistent with a recent crystal structure of the coiled-coil/SPRY fragment of TRIM25. In that structure, the two SPRY domains in the TRIM25 dimer were observed to pack against the coiled-coil scaffold, and this packing configuration is important for TRIM25’s ability to polyubiquitinate RIG-I in vitro [9]. Importantly, the 7K motif is not observed in this structure, because it is within a disordered polypeptide segment that is 75 amino-acid residues long. We therefore speculate that RNA binding to both the SPRY domain and the 7K motif would have an important impact on the structure of the TRIM25 dimer and thereby regulate substrate recognition by the SPRY domain. Since the SPRY binds near the ends of the coiled-coil where the RING and B-boxes are located [9], it is plausible that RNA binding may allosterically affect RING domain dimerization and activation as well. Furthermore, the architecture of the TRIM25 dimer – bivalent, with multiple domains connected by long, flexible linkers – also suggest that long RNA molecules may induce clustering of multiple dimers and formation of an extended ubiquitination network. Such a scenario would be facilitated by cellular compartments that are highly enriched in RNA, such as the cytoplasmic stress granules where TRIM25 is localized upon overexpression (this study) or during virus infection [18, 19].
RNA binding by TRIM25 could promote dual recognition of viral RNA signals. In the simplest model, TRIM25 could function as a co-receptor that binds the same RNA molecule as the primary receptor, for example by co-binding to 5’-triphosphorylated viral RNA in the RIG-I pathway. However, our experiments indicate that – at least in context of RIG-I activation – TRIM25 need not bind to the same RNA molecule as RIG-I, because TRIM25’s RNA-binding activity is still required for IFN-β induction by GST-2CARD, which does not require input from viral RNA. It therefore appears that TRIM25 binds to some physiological RNA species to facilitate RIG-I signaling. Our data also suggest that functional interactions between TRIM25 and GST-2CARD occur much more readily in cells, and further studies are now required to determine why this is so and the role of RNA binding (if any) in this process. Importantly, the 7KA mutant cannot ubiquitinate GST-2CARD despite retaining the ability to bind, showing a direct correlation with TRIM25’s RNA binding and ubiquitination activities in cells. With regards to full-length RIG-I, recent studies showed that its activation by viral RNA induces RIG-I co-localization with TRIM25 in RNA stress granules [19]. Our results also support this model and further reveal that the RNA-binding activity of TRIM25 is important for this localization.
Whether the co-receptor model holds true for other TRIM25-mediated pathways remains to be established. In this regard, TRIM25 was also recently implicated in the ZAP pathway, which induces degradation of CG-rich viral mRNAs [5-7]. Notably, TRIM25’s cellular RNA binding activity was initially reported in a study to identify mRNA binding proteins in the absence of viral infection [12]. It is therefore possible that TRIM25 is promiscuously but benignly associated with cellular mRNA, and that subsequent ZAP binding somehow triggers ubiquitin-dependent signaling to degrade the CG-rich subpopulation. However, it is not yet clear whether or how TRIM25’s ubiquitination activity regulates ZAP, with some studies showing an absolute requirement for the RING domain and others not [5, 6]. Interestingly, ZAP is also reported to localize to RNA granules where it recruits deadenylase and exonuclease complexes, as well as mRNA decapping enzymes [22, 23]. In human cells, a variant of ZAP is reported to functionally associate with RIG-I [24], suggesting the possibility that TRIM25’s overall anti-viral activity may in fact reflect an intersection of the RIG-I and ZAP pathways.
Materials and Methods
Protein and RNA preparation
Full-length human TRIM25 used for ubiquitination and binding studies was expressed using a baculovirus system and purified to homogeneity as described previously [11]. In brief, cells were resuspended in lysis buffer (50 mM Tris, pH 8.0, 20 mM NaCl, 200 mM (NH4)2SO4, 10% (v/v) glycerol, 1.5% (v/v) Triton X-100, 1 mM tris(2-carboxyethyl)phosphine (TCEP)) containing 2 mM PMSF, 2-3 protease inhibitor cocktail tablets (Roche), and 25 U/mL of Benzonase (Sigma). Cells were lysed using a motorized Dounce homogenizer which was calibrated not to disrupt nuclei (and thereby prevent DNA release) at 30 strokes. Clarified cell lysates were treated with PEI to remove nucleic acid contamination, and the Strep/FLAG-tagged protein was purified on a StrepTactin resin (GE Healthcare) and then on a Superdex 200 gel filtration column (GE Heathcare) [11]. The following protein constructs were expressed and purified in a similar manner: RBCC constructs spanning residues 1-379, 1-401, and 1-435, and the full-length 7KA mutant. Nucleic acid-contaminated TRIM25 used for experiments in Fig. 2 was purified in a similar manner, but (NH4)2SO4 and Benzonase were omitted from the lysis buffer and the PEI precipitation step was not performed. CCD constructs were expressed in E. coli as SUMO-fusion proteins and purified to homogeneity as described previously [8]. CCD-SPRY and SPRY proteins were also expressed in as SUMO-fusion constructs. Cell pellets were resuspended in lysis buffer (20 mM Tris pH 8, 150 mM NaCl, 1 mM TCEP, 20 mM imidazole pH 8, 20 U/mL Benzonase, and 1 protease inhibitor cocktail tablet) and lysed by a microfluidizer. Proteins were purified to homogeneity by a combination Ni-NTA affinity, Source 15Q ion exchange, and size exclusion chromatography into the final buffer (20 mM sodium phosphate, pH 8, 100 mM NaCl, 1 mM TCEP).
RNA oligonucleotides were purchased from Integrated DNA Technologies either in single-stranded or double-stranded form. Sequences of the “sense” strands are (5’ to 3’): 14-mer, AUGGCUA GCUGGAG; 28-mer, AUGGCUA GCUGGAG CCACCCG CAGUUCG; 58-mer, AUGGCUA GCUGGAG CCACCCG CAGUUCG UACAAGG AUGACGA UGACAAG GGUACAA. DNA oligos had identical sequences, except that T was substituted for U.
In vitro ubiquitination assays
TRIM25 constructs were incubated at 37 °C with E1 (100 nM), E2 (1 μM Ube2D3 or 0.28 μM Ube2N/Ube2V2), and ubiquitin (40 μM) in reaction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM TCEP, 5 mM ATP, 10 mM MgCl2). Reactions were stopped by addition of SDS-PAGE sample buffer and boiling for 10 min. Immunoblots were performed with anti-Ub (1:2,000; P4D1, Santa Cruz Biotechnology) and anti-FLAG M2 (1:5,000; Sigma). Experiments in Fig. 2 used RNase A (Qiagen), DNase I (Sigma), dsRNA ladder (NEB), and 100-bp dsDNA ladder (NEB).
SEC-MALS
Size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) measurements were performed on a Dionex UltiMate 3000 HPLC system (ThermoFisher) with in-line miniDAWN TREOS three-angle light scattering detector (Wyatt Technology) and Optilab T-rEX differential refractometer (Wyatt Technology). Sample volumes of 100 μL at 20 μM concentration were applied to a Superdex 200 HR 10/300 GL column (GE Healthcare) and developed at a flow rate of 0.4 mL/min in 50 mM Tris, pH 9, 200 mM NaCl, 0.5 mM TCEP. Data were recorded and processed using ASTRA software (Wyatt Technology). A single dn/dc value of 0.185 mL/g was used for all samples, as the typical dn/dc values of proteins (0.16-0.20 mL/g) and RNA (0.17-0.19 mL/g) are quite similar [25].
EMSA
The fluorescent 28-bp dsRNA duplex used in EMSA and competition experiments contained a 5’-linked IRDye 800CW fluorophore attached to the sense strand (Integrated DNA Technologies). Note that all TRIM25 proteins used for binding assays were highly pure with minimal nucleic acid contamination, as assessed by SDS-PAGE, UV absorbance profiles, and A260/A280 ratios. For EMSAs used to evaluate RNA binding, individual TRIM25 constructs at 0 to 10 μM were incubated with 250 nM fluorescent RNA ligand for 30 min on ice in binding buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP, 5% glycerol, 0.3 mg/mL BSA). After incubation, 5 μL of each sample was resolved on a 6% polyacrylamide gel in 0.5× TBE for 30 min at 50 V. The RNA was imaged on an Odyssey Classic infrared scanner (Licor). The fractions unbound RNA bands were quantified with the Licor Image StudioLite software and plotted as fraction RNA bound vs. protein concentration. The data were fit to a simple binding isotherm model, using the equation: fraction bound = [TRIM25]n/(Kdn + [TRIM25]n), where Kd = dissociation constant and n = Hill coefficient. To evaluate TRIM25 binding specificity for RNA or DNA, fluorescent 28-mer dsRNA (250 nM) was saturated with TRIM25 (4 μM). Non-fluorescent 28-mer competitors at 1 μM or 5 μM, with the same nucleotide sequence, were then added to the reaction mix and allowed to incubate on ice for an additional 30 min. Samples were resolved by gel electrophoresis and displaced RNA was quantified as above.
Cell culture and viruses
HeLa (ATCC), HEK 293T (ATCC) and TRIM25-KO HEK 293T [11] cells were cultured in Dulbecco’s modified Eagle’s mediuam (DMEM, Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS, Hyclone), 1% (v/v) penicillin-streptomycin (Life Technologies) and 1% (v/v) L-glutamine (Life Technologies). All cell lines were regularly tested for mycoplasma contamination.
IAV (PR/8/1934(H1N1)) was purchased from Charles River Laboratories (cat #10100374). DENV serotype 2 (strain 16681) has been previously described [26]. VSV-eGFP was kindly provided by Sean Whelan (Harvard).
Viral infection assays
TRIM25-KO HEK 293T cells [11], seeded into 12-well plates (~5 × 105 cells/well), were transfected with 2 μg of pCMV empty vector or the indicated FLAG-tagged TRIM25 constructs using linear polyethylenimine (PEI) (1 mg/mL solution in 20 mM Tris pH 6.8; Polysciences), according to the manufacturer’s instructions. At 24 h post-transfection, cells were infected with the indicated viruses.
For DENV replication experiments, the reconstituted TRIM25-KO cells were infected with an MOI of 4 in serum-free medium (OPTI-MEM, Life Technologies). After 2 h, medium was replaced with supplemented DMEM. At 24 h post-infection, cells were harvested and stained for DENV prM protein via flow cytometry as previously described [26]. Briefly, cells were washed once in phosphate-buffered saline (PBS) and then fixed in 1% (w/v) paraformaldehyde (PFA, in PBS) for 30 min, followed by permeabilization in 0.1% saponin (in 2% FBS in PBS) for 30 min. Cells were incubated for 1 h with anti-prM (2H2, Merck Millipore) conjugated to DyLight 633 using a commercial kit (Thermo Scientific). Cells were washed twice with PBS, re-suspended in 1% PFA (w/v) (in PBS), and then analyzed on a BD LSRII flow cytometer. Analysis was performed using FlowJo software. For experiments with VSV-eGFP, cells were infected at MOI of 0.01 and analyzed for eGFP expression by using flow cytometry seventeen hours later. For experiments with IAV, cells were infected at MOI of 0.5. At the indicated times post-infection, cells were lysed in NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM HEPES, pH 7.4, supplemented with protease inhibitors). Cell debris was pelleted by centrifugation at 13,000 rpm for 20 minutes at 4 °C. Whole cell lysates were mixed with 6× SDS-PAGE loading buffer and heated at 95 °C for 5 min. Protein expression of IAV NS1 was determined by immunoblotting using anti-NS1. IAV titers were determined by plaque assay as previously described [27].
Immunoblot analysis
Immunoblot analysis was performed as previously described [1]. The following antibodies were used: anti-NS1 (polyclonal rabbit, 1:1,000, provided by A. García-Sastre, Mount Sinai), anti-FLAG (1:2,000, M2, Sigma), and anti-β-actin (1:15,000, AC-15, Sigma).
IFN-β luciferase assay
The luciferase reporter assay was performed as described previously [11]. The pEBG vector expressing GST and GST-RIG-i 2CARD have been previously described [1].
In-cell ubiquitination assay
HEK 293T cells were seeded into 10 cm-dishes (~107 cells per dish) and transfected with 12 μg of empty vector, or FLAG-tagged TRIM25 WT or 7KA together with 7.5 μg of pEBG vector expressing GST or GST-2CARD. At the indicated times, cells were harvested and lysed in RIPA buffer (150 mM NaCl, 1% (v/v) NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8.0) supplemented with protease inhibitor cocktail (Sigma). Cell debris were pelleted by microcentrifugation at maximum speed for 20 min at 4 °C. GST pulldown was performed using Glutathione Sepharose 4B (GE Healthcare) as previously described [1].
Co-immunoprecipitation of TRIM25 and GST-2CARD
HEK 293T cells were seeded into 10 cm-dishes (~107 cells per dish) and transfected with 10 μg of empty vector, or FLAG-tagged TRIM25 WT or 7KA, together with 10 μg of pEBG expressing GST-2CARD. Cells were lysed in NP-40 buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% (v/v) NP-40) supplemented with protease inhibitor cocktail (Sigma). Co-immunoprecipitation was performed by incubating cell lysates with anti-FLAG M2 affinity gel (Sigma) for 4 h at 4 °C. After extensive washing of the beads with lysis buffer, bound proteins were eluted by heating samples in 2× Laemmli SDS sample buffer for 5 min at 95 °C, and then subjected to SDS-PAGE.
Confocal microscopy
HeLa cells grown on coverslips (Chemglass) in 24-well plates (~2 × 105 cells/well) were transfected with 1 μg of FLAG-tagged TRIM25 WT or 7KA mutant using Lipofectamine LTX and Plus reagent (Life Technologies) according to the manufacturer’s instruction. At 48 h post-transfection, cells were fixed with 4% (w/v) paraformaldehyde (PFA, Santa Cruz Biotechnology) for 20 min and permeabilized with 0.5% (v/v) Triton X-100 in phosphate-buffered saline (PBS). After blocking with 5% (v/v) fetal bovine serum (in PBS, Hyclone) for 1 h, cells were immunostained with anti-FLAG M2 (1:400, Sigma) and anti-TIAR (1:200, Cell Signaling), followed by incubation with secondary antibodies conjugated to AlexaFluor 488 or AlexaFluor 594 (both 1:400, Life Technologies). Cells were mounted in DAPI-containing Vectashield (Vector Labs) to stain nuclei. All laser scanning images were acquired on a Leica SP8 confocal microscope and analyzed as previously described [28].
Statistical analysis
An unpaired two-tailed Student’s t-test was used to compare differences between two unpaired experimental groups in all cases. A P value of <0.05 was considered statistically significant.
Highlights.
TRIM25 is a RING-type E3 ubiquitin ligase that binds RNA with high affinity
RNA binding involves the substrate-binding SPRY domain and a disordered lysine-rich motif
RNA modulates both activation of TRIM25’s E3 ligase activity and subcellular localization
TRIM25’s RNA binding activity facilitates cellular anti-viral defense
Acknowledgements
This study was supported by National Institutes of Health (NIH) grants R01-AI087846 and R01-AI27774 (to M.U.G.) and R01-GM112508 (to O.P.). J.G.S. was supported by NIH training grant T32-GM008136 and the Robert R. Wagner Fellowship Fund. K.M.J.S. was supported by a fellowship from the German Research Foundation (SP 1600/1-1). R.A.R. and M.A.Z. received support from NIH training grant T32-GM007183.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. , TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity, Nature 446 (2007) 916–920. [DOI] [PubMed] [Google Scholar]
- [2].Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, et al. , Roles of RIG-IN-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction, Proc. Natl. Acad. Sci. U.S.A 105 (2008) 16743–16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G, Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation, Cell Rep 9 (2014) 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Heikel G, Choudhury NR, Michlewski G, The role of Trim25 in development, disease and RNA metabolism, Biochem. Soc. Trans 44 (2016) 1045–1050. [DOI] [PubMed] [Google Scholar]
- [5].Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G, TRIM25 is required for the antiviral activity of zinc finger antiviral protein, J. Virol 91 (2017) e00088–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li MM, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, et al. , TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP), PLoS Pathog. 13 (2017)e1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takata MA, Goncalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, et al. , CG dinucleotide suppression enables antiviral defence targeting non-self RNA, Nature 550 (2017) 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O, The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer, Proc. Natl. Acad. Sci. U.S.A 111 (2014) 2494–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koliopoulos MG, Lethier M, van der Veen AG, Haubrich K, Hennig J, Kowalinski E, et al. , Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition, Nat. Commun 9 (2018) 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K, Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity, EMBO J. 35 (2016) 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sanchez JG, Chiang JJ, Sparrer KM, Alam SL, Chi M, Roganowicz MD, et al. , Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway, Cell Rep 16 (2016)1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, et al. , The RNA-binding protein repertoire of embryonic stem cells, Nat. Struct. Mol. Biol 20 (2013) 1122–1130. [DOI] [PubMed] [Google Scholar]
- [13].Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, et al. , RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination, BMC Biol 15 (2017) 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anger AM, Armache JP, Beminghausen O, Habeck M, Subklewe M, Wilson DN, et al. , Structures of the human and Drosophila 80S ribosome, Nature 497 (2013) 80–85. [DOI] [PubMed] [Google Scholar]
- [15].Schlee M, Master sensors of pathogenic RNA - RIG-I like receptors, Immunobiology 218 (2013)1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, et al. , Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity, Cell 141 (2010)315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Peisley A, Wu B, Xu H, Chen ZJ, Hur S, Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I, Nature 509 (2014) 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yoo JS, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, et al. , DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation, PLoS Pathog. 10 (2014) e1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sanchez-Aparicio MT, Ayllon J, Leo-Macias A, Wolff T, Garcia-Sastre A, Subcellular localizations of RIG-I, TRIM25, and MAVS complexes, J. Virol 91 (2017) e01155–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Roganowicz MD, Komurlu S, Mukherjee S, Plewka J, Alam SL, Skorupka KA, et al. , TRIM5α SPRY/coiled-coil interactions optimize avid retroviral capsid recognition, PLoS Pathog. 13 (2017) e1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Weinert C, Morger D, Djekic A, Grutter MG, Mittl PR. Crystal structure of TRIM20 C-terminal coiled-coil/B30.2 fragment: implications for the recognition of higher order oligomers, Sci. Rep 5 (2015) 10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, et al. , Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus, Proc. Natl. Acad. Sci. U.S.A 110 (2013) 12379–12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Turner M, Galloway A, Vigorito E, Noncoding RNA and its associated proteins as regulatory elements of the immune system, Nat. Immunol 15 (2014) 484–491. [DOI] [PubMed] [Google Scholar]
- [24].Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, et al. , ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses, Nat. Immunol 12 (2011) 37–44. [DOI] [PubMed] [Google Scholar]
- [25].Patel TR, Chodnowski G, Astha, Koul A, McKenna SA, Bujnicki JM, Structural studies of RNA-protein complexes: A hybrid approach involving hydrodynamics, scattering, and computational methods, Methods 118-119 (2017) 146–162. [DOI] [PubMed] [Google Scholar]
- [26].Chan YK, Gack MU, A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity, Nat. Immunol 17 (2016) 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, et al. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein, PLoS Pathog. 8 (2012) e1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol 2 (2017) 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]