Abstract
Many terpenoid compounds have been extracted from different tissues of Salvia guaranitica. However, the molecular genetic basis of terpene biosynthesis pathways is virtually unknown. In this study, approximately 4 Gb of raw data were generated from the transcriptome of S. guaranitica leaves using Illumina HiSeq 2000 sequencing. After filtering and removing the adapter sequences from the raw data, the number of reads reached 32 million, comprising 186 million of high-quality nucleotide bases. A total of 61,400 unigenes were assembled de novo and annotated for establishing a valid database for studying terpenoid biosynthesis. We identified 267 unigenes that are putatively involved in terpenoid metabolism (including, 198 mevalonate and methyl-erythritol phosphate (MEP) pathways, terpenoid backbone biosynthesis genes and 69 terpene synthases genes). Moreover, three terpene synthase genes were studied for their functions in terpenoid biosynthesis by using transgenic Arabidopsis; most transgenic Arabidopsis plants expressing these terpene synthetic genes produced increased amounts of terpenoids compared with wild-type control. The combined data analyses from the transcriptome and metabolome provide new insights into our understanding of the complex metabolic genes in terpenoid-rich blue anise sage, and our study paves the way for the future metabolic engineering of the biosynthesis of useful terpene compounds in S. guaranitica.
Keywords: Salvia guaranitica, transcriptome, terpene synthase genes, transgenic Arabidopsis, functional characterization
1. Introduction
Blue Anise Sage (Salvia guaranitica L.), belongs to the genus Salvia, which is one of the economically best-known genera due to its vast medicinal properties and rich aromatic oils. The genus Salvia (tribe Mentheae) is the largest of the Lamiaceae family, which comprises nearly 1,000 species. Salvia plants are widely distributed in three regions around the world but mainly exist in Central and South America (~500 species), West Asia (~200 species) and East Asia (~100 species), while the other Salvia species are spread throughout the world.1 Most of these plants contain various medicinally active components used throughout history in folk medicine, e.g. S. officinalis, S. japonica, S. santolinifolia, S. hydrangea, S. tomentosa, S. tuxtlensis, S. miltiorrhiza, S. chloroleuca, S. nipponica, S. fruticosa, S. aureus, S. przewalskii, S. epidermindis, S. isensis, S. lavandulifolia, S. glabrescens, S. allagospadonopsis, S. macrochlamys and S. recognita. Recently, Salvia species have become a valuable source for pharmaceutical research for identifying and discovering biologically active compounds.2 Essential oils of Salvia species exhibit significant bioactivities, including antimicrobial, antimutagenic, anticancer, antioxidant, anti-inflammatory, choleretic and antimicrobial activities. Salvia essential oils contain more than 100 active compounds with pharmacological effects, and they can be categorized into monoterpenes, sesquiterpenes, diterpenes and triterpenes.2 During their biosynthesis, these terpenoids are sequentially built up from the isoprene units (C5) building block, isopentyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). These components are condensed in a sequential manner by prenyltransferases, resulting in the formation of prenyl diphosphates, such as geranyl diphosphate (GPP), farnesyl pyrophosphate (FPP) and geranylgeranyl pyrophosphate (GGPP).3 These prenyl diphosphates are the immediate precursors for the biosynthesis of mono-, sesqui-, di- and tetraterpenes. Despite the scientific and medicinal interests in these terpenoids of S. guaranitica, the genes that are related to the biosynthesis of these compounds have not yet been fully identified or understood. Plant secondary metabolites have significant use in the food and pharmaceutical industries, such as in fine chemicals, and cosmetics. The biosynthesis, regulation and metabolic engineering of useful secondary metabolites have been extensively studied.4 In recent years, next-generation sequencing (NGS)-based RNA sequencing (RNA-Seq) has become a powerful tool for discovering genes that are involved in the biosynthesis of various secondary metabolite pathways in medicinal plants.5 For example, the volatile terpenoid biosynthesis in Salvia officinalis,6 the phenylpropanoid and terpenoid biosynthesis pathways in Ocimum sanctum and Ocimum basilicum,7 the biosynthesis of active ingredients in Salvia miltiorrhiza,8 the essential oil biosynthesis in aromatic Cymbopogon flexuosus,9 the biosynthesis of carotenoids in Momordica cochinchinensis,10 the biosynthesis of cellulose and lignin in Cunninghamia lanceolata11 and the biosynthesis of tea-specific compounds, i.e. catechins, caffeine and theanine pathways in Camellia sinens,12 have been explored by using NGS. Characterization of plant terpene synthases is typically carried out by the production of the recombinant enzymes in Escherichia coli. This is often difficult due to enzyme solubility and codon usage issues. Furthermore, plant terpene synthases that are localized to the plastids, such as diterpene synthases, must be abridged in a more or less experimental approach to ameliorate expression.13,14 Transgenic Arabidopsis (A. thaliana) is very efficient and has been successfully used for the characterization of one sesquiterpene synthase (PmSTS) genes from Polygonum minus: β-sesquiphellandrene, and also has been successfully used for the characterization the strawberry linalool/nerolidol synthase (monoterpene) and taxadiene synthase.15,16 Here, we characterized genes that are involved in terpenoid biosynthesis in S. guaranitica and determined their biological significance in S. guaranitica for terpenoid production in various tissues. In this study, a transcriptome database was established for S. guaranitica leaves using NGS technology to identify and to characterize genes that are related to the terpenoid biosynthesis pathway. The criteria used to achieve these objectives and to elucidate the complex metabolic pathways and genes for the understanding of terpenoid production in S. guaranitica included the following: (i) transcriptome analysis of leaves using Illumina HiSeq 2000 sequencing; (ii) GC-MS analysis for six fresh plant parts (old leaves, young leaves, stems, flowers, bud flowers and roots); (iii) characterization of three terpene genes in transgenic A. thaliana; (iv) qRT-PCR of highly expressed genes that are involved in the biosynthesis of terpenoids; (v) and the combination of data from the transcriptome, qRT-PCR and metabolome with GC-MS for revealing the functions of metabolic genes that are involved in the biosynthesis of valuable terpenoids.
2. Materials and methods
2.1. Plant materials and tissue collection
Seedlings of Salvia guaranitica L. were collected from the Wuhan Botanical Garden, China, and grown at National Key Laboratory of Crop Genetic Improvement farm of Huazhong Agricultural University, Wuhan, China. Different tissues were sampled from one-year-old S. guaranitica plants. For RNA-Seq, three biological replicates from leaves were sampled and handled. Each replicate consisted of two young and two old leaves from the same plant. For qRT-PCR, three biological replicates were collected from the following six parts (old leaves, young leaves, stems, flowers, bud flowers and roots). All samples were directly frozen in liquid nitrogen and then stored at −80 °C until RNA extraction. Furthermore, another three biological replicates from the individual six fresh parts were collected for isolation of the essential oil.
2.2. Isolation of chemical compounds
The correct method to reduce technical variability throughout a sampling procedure is essential to stop cell metabolism and to avoid leaking of metabolites during the various preparation steps before the actual metabolite extraction. Therefore, three biological replicates from each of the six fresh parts were immediately frozen on dry ice. In the laboratory, the frozen three biological replicates from each of the three fresh part samples were homogenized with a mortar and pestle in liquid nitrogen, after which the plant material (ca. 10 g) was directly soaked in n-hexane as a solvent in Amber storage bottles, 60 ml screw-top vials with silicone/PTFE septum lids (http://www.sigmaaldrich.com) were used to reduce loss of volatiles to the headspace then incubated with shaking at 37 °C and 200 rpm for 72 h. Afterward, the solvent was transferred using a glass pipette to a 10-ml glass centrifuge tube with screw-top vials with silicone/PTFE septum lids and centrifuged at 5,000 rpm for 10 min at 4 °C to remove plant debris. The supernatant was pipetted into glass vials with a screw cap, and oil was concentrated until remaining 1.5 ml of concentrated oils under a stream of nitrogen gas in a nitrogen evaporator (Organomation) with a water bath at room temperature (Toption-China-WD-12). The concentrated oils transferred to a fresh crimp vial amber glass, 1.5 ml screw-top vials with silicone/PTFE septum lids were used to reduce a loss of volatiles to the headspace. For absolute oil recovery, the remaining film crude oil in the internal surface of concentrated glass vials was dissolved in the minimum volume of n-hexane, thoroughly mixed and transferred to the same fresh crimp vial amber glass, 1.5 ml. And the crimp vial was placed on the auto-sampler of the gas chromatography-mass spectrometer (GC-MS) system for GC-MS analysis, or each tube was covered with parafilm after closed with screw-top vials with silicone/PTFE septum lids and stored at −20 °C until GC-MS analysis.6
2.3. GC-MS analysis of essential oil components
GC analysis was performed using a Shimadzu model GCMS-QP2010 Ultra (Tokyo, Japan) system. An approximately 1 µl aliquot of each sample was injected (split ratios of 15: 1) into a GC-MS equipped with an HP-5 fused silica capillary column (30 m × 0.25 mm ID, 0.25 µm film thicknesses). And we used Helium as a carrier gas at a constant flow of 1.0 ml min−1. The mass spectra were monitored between 50 and 450 m/z. Temperature was initially under isothermal conditions at 60 °C for 10 min. Temperature was then increased at a rate of 4 °C min−1 to 220 °C, held isothermal at 220 °C for 10 min, increased by 1 °C min−1 to 240 °C, held isothermal at 240 °C for 2 min and finally held isothermal for 10 min at 350 °C. The identification of the volatile constituents were determined by parallel comparison of their recorded mass spectra with the data stored in the Wiley GC/MS Library (10th edition) (Wiley, New York, NY, USA), the volatile organic compounds (VOC) analysis S/W software and the NIST Library (2014 edition). The relative % amount of each component was calculated by comparing its average peak area to the total areas, as well as retention time index. All of the experiments were performed simultaneously three times under the same conditions for each isolation technique with total GC running time was 80 min.6
2.4. RNA extraction
Total RNAs from the three biological leaf replicates were extracted for RNA-Seq. Moreover, total RNAs from three biological replicates from each of the plant parts (old leaves, young leaves, stems, flowers, bud flowers and roots) were extracted for qRT-PCR. Additionally, total RNAs from three biological replicates of A. thaliana were extracted for semiquantitative RT-PCR using the TRIzol Reagent (Invitrogen, USA) and treated with DNase I (Takara). RNA quality was examined on 1% agarose gels, and the purity was analysed using a Nano-Photometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was determined using a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA pools were prepared for cDNA libraries by mixing equal volumes from the three RNAs replications in one tube.
2.5. cDNA library preparation and sequencing
Three micrograms of RNA per sample were used for generating a sequencing library. cDNA was synthesized using an RNA Library Prep Kit for Illumina® (NEB, USA) for generated sequencing libraries according to the manufacturer’s instructions. The first strand of cDNA was synthesized in the presence of random hexamer primers and M-MuLV Reverse Transcriptase (RNase H), and the second strand of cDNA was synthesized in the presence of DNA Polymerase I and RNase H. The remaining cDNA was converted into blunt ends in the presence of exonuclease/polymerase activities. After the adenylation of three ends of DNA fragments, NEB Next, an adaptor with a hairpin loop structure, was ligated to prepare for hybridization. To select cDNA fragments of preferentially 150∼200 bp in length, the library fragments were purified using an AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μl of USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 95 °C for 5 min. Afterward, PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers and Index (X) Primer. Finally, PCR products were purified (AMPure XP system), and the library quality was assessed using an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Clustering of the index-coded samples was performed on a cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions (Novogene Experimental Department). After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2000 platform, and paired-end (PE) reads were generated.
2.6. Quality control
Raw data (raw reads) in the fastq format were first processed through in-house Perl scripts. During this step, clean data (clean reads) were obtained by removing reads containing adapters, reads containing ploy-N and low-quality reads from the raw data. At the same time, Q20, Q30, GC content and sequence duplication level of the clean data were calculated. All of the downstream analyses were based on high-quality clean data.
2.7. De novo transcriptome assembly
De novo assembly of the processed reads was carried out using Trinity program (Version: trinityaseq_r 2012-10-05)17,18 with the min_kmer_cov set to 2 by default and all other parameters set to default. The Trinity method consists of three software modules, (1) Inchworm, (2) Chrysalis and (3) Butterfly, applied sequentially to process large volumes of RNA-Seq reads. In the first step, read datasets were assembled into linear contigs by the first module (Inchworm program). The minimally overlapping contigs were then clustered into sets of connected components (build graph components) by the second module (Chrysalis program), and the transcripts were then constructed from each de Bruijn graph by the third software module (Butterfly program). Finally, the transcripts were clustered by a similarity of correct match length beyond 80% for longer transcripts or 90% for shorter transcripts using the multiple sequence alignment tool. The transcriptome data from S. guaranitica was submitted to the NCBI under submission ID (1955911). And the accession number from BankIt1954130 (KX869088) to BankIt1954278 (KX869125), and from BankIt1955703 (KX893913) to BankIt1955935 (KX894017) see Tables 1 and 2. And any inquiries about my submission should be sent to gb-admin@ncbi.nlm.nih.gov or sent to info@ncbi.nlm.nih.gov.
Table 1.
Transcript abundance of MEP, MVA and other terpenoid backbone biosynthesis pathway genes as per the S. guaranitica transcriptome data annotation
| Pathway | Gene name | Kegg entry | Gene bank accession ID | EC.No. | Read in leaf | FPKM |
|---|---|---|---|---|---|---|
| MEP | SgDXS 1 | K01662 | KX869088 | 2.2.1.7 | 8968.47 | 223.94 |
| SgDXS2 | K01662 | KX869089 | 2.2.1.7 | 40 | 41.89 | |
| SgDXS3 | K01662 | KX869090 | 2.2.1.7 | 169 | 3.68 | |
| SgDXS4 | K01662 | KX869091 | 2.2.1.7 | 3634.82 | 85.89 | |
| SgDXS5 | K01662 | KX869092 | 2.2.1.7 | 697.93 | 14.94 | |
| SgDXR 1 | K00099 | KX869093 | 1.1.1.267 | 4080.05 | 175.32 | |
| SgDXR 2 | K00099 | KX869094 | 1.1.1.267 | 158.7 | 37.97 | |
| SgMCT | K00991 | KX869095 | 2.7.7.60 | 563.54 | 25.17 | |
| SgCMK | K00919 | KX869096 | 2.7.1.148 | 1588.67 | 53.65 | |
| SgHDS 1 | K03526 | KX869097 | 1.17.7.1 | 350 | 13.57 | |
| SgHDS2 | K03526 | KX869098 | 1.17.7.1 | 123.02 | 8.35 | |
| SgHDS3 | K03526 | KX869099 | 1.17.7.1 | 11 | 4.08 | |
| SgHDS4 | K03526 | KX869100 | 1.17.7.1 | 17316.85 | 304.95 | |
| SgHDR 1 | K03527 | KX869101 | 1.17.1.2 | 4 | 2.72 | |
| SgHDR2 | K03527 | KX869102 | 1.17.1.2 | 850 | 175.65 | |
| SgHDR3 | K03527 | KX869103 | 1.17.1.2 | 5 | 2.7 | |
| SgHDR4 | K03527 | KX869104 | 1.17.1.2 | 1034 | 209.85 | |
| SgHDR5 | K03527 | KX869105 | 1.17.1.2 | 296 | 16.78 | |
| SgHDR6 | K03527 | KX869106 | 1.17.1.2 | 858.92 | 106.05 | |
| SgHDR7 | K03527 | KX869107 | 1.17.1.2 | 67 | 17.91 | |
| SgHDR8 | K03527 | KX869108 | 1.17.1.2 | 9 | 4.12 | |
| SgHDR9 | K03527 | KX869109 | 1.17.1.2 | 42686.56 | 1228.23 | |
| SgHDR10 | K03527 | KX869110 | 1.17.1.2 | 43 | 2.94 | |
| SgIDI1 | K01823 | KX869111 | 5.3.3.2 | 2 | 1.27 | |
| SgIDI2 | K01823 | KX869112 | 5.3.3.2 | 2344.77 | 98.18 | |
| SgIDI3 | K01823 | KX869113 | 5.3.3.2 | 1 | 1.44 | |
| MVA | SgAACT 1 | K00626 | KX869114 | 2.3.1.9 | 624.59 | 22.69 |
| SgAACT 2 | K00626 | KX869115 | 2.3.1.9 | 2001.02 | 67.38 | |
| SgHMGS | K01641 | KX869116 | 2.3.3.10 | 1897.92 | 61.34 | |
| SgHMGR1 | K00021 | KX869117 | 1.1.1.34 | 40 | 20.67 | |
| SgHMGR2 | K00021 | KX869118 | 1.1.1.34 | 2300.82 | 56.02 | |
| SgHMGR3 | K00021 | KX869119 | 1.1.1.34 | 23 | 4.09 | |
| SgHMGR4 | K00021 | KX869120 | 1.1.1.34 | 70 | 11.16 | |
| SgHMGR5 | K00021 | KX869121 | 1.1.1.34 | 144 | 25.27 | |
| SgHMGR6 | K00021 | KX869122 | 1.1.1.34 | 1691.49 | 68.13 | |
| SgHMGR7 | K00021 | KX869123 | 1.1.1.34 | 14 | 2.17 | |
| SgHMGR8 | K00021 | KX869124 | 1.1.1.34 | 39 | 20.82 | |
| SgHMGR9 | K00021 | KX869125 | 1.1.1.34 | 14 | 1.25 | |
| SgMVK1 | K00869 | KX893913 | 2.7.1.36 | 441.39 | 15.27 | |
| SgMVK2 | K00869 | KX893914 | 2.7.1.36 | 4 | 1.83 | |
| SgPMK | K00938 | KX893915 | 2.7.4.2 | 754 | 19.93 | |
| SgMDC | K01597 | KX893916 | 4.1.1.33 | 786.17 | 56.1 | |
| Monoterpene | SgGPPS | K14066 | KX893917 | 2.5.1.1 | 1292.8 | 47.81 |
| Sesqui and triterpene | SgFPPS | K00787 | KX893918 | 2.5.1.10 | 1909.92 | 80.21 |
| Diterpene | SgGGPSΙΙ1 | K13789 | KX893925 | 2.5.1.29 | 1393 | 60.95 |
| SgGGPSΙΙ2 | K13789 | KX893926 | 2.5.1.29 | 2153.95 | 74.07 | |
| SgGGPSΙΙ3 | K13789 | KX893919 | 2.5.1.29 | 2 | 0.67 | |
| SgGGPSΙΙ4 | K13789 | KX893920 | 2.5.1.29 | 125 | 9.99 | |
| SgGGPSΙΙ5 | K13789 | KX893921 | 2.5.1.29 | 54.01 | 25.45 | |
| SgGGPSΙΙ6 | K13789 | KX893927 | 2.5.1.29 | 180.01 | 12.7 | |
| SgGGPSΙΙ7 | K13789 | KX893922 | 2.5.1.29 | 25 | 5.21 | |
| SgGGPSΙΙ8 | K13789 | KX893923 | 2.5.1.29 | 44 | 6.97 | |
| SgGGPSΙΙ9 | K13789 | KX893924 | 2.5.1.29 | 16 | 7.69 | |
| SgGGPSΙΙ10 | K13789 | KX893928 | 2.5.1.29 | 673.01 | 28.66 | |
| Other terpenoid backbone biosynthesis | SgFOHSDR | K15890 | KX893929 | 1.1.1.216 | 241.13 | 9.68 |
| SgFOLK1 | K15892 | KX893930 | 2.7.1. | 998.85 | 85.9 | |
| SgFOLK2 | K15892 | KX893931 | 2.7.1. | 892.1 | 43.74 | |
| SgPCYOX1 | K05906 | KX893932 | 1.8.3.5 1.8.3.6 | 523.75 | 14.1 | |
| SgSTE24 | K06013 | KX893933 | 3.4.24.84 | 1467.53 | 48.98 | |
| SgCHLP1 | K10960 | KX893934 | 1.3.1.83 | 14506.79 | 419.11 | |
| SgCHLP2 | K10960 | KX893935 | 1.3.1.83 | 83 | 10.71 | |
| SgCHLP3 | K10960 | KX893936 | 1.3.1.83 | 675.74 | 26.22 | |
| SgFACE2 | K08658 | KX893937 | 3.4.22.- | 297 | 36.52 | |
| SgPCME1 | K15889 | KX893938 | 3.1.1.- | 232.82 | 11.54 | |
| SgPCME2 | K15889 | KX893939 | 3.1.1.- | 36 | 1.81 | |
| SgFNTB | K05954 | KX893940 | 2.5.1.58 | 626.09 | 21.79 | |
| SgSPS | K05356 | KX893941 | 2.5.1.84 2.5.1.85 | 6500.62 | 224.55 | |
| SgDHDDS1 | K11778 | KX893942 | 2.5.1.87 | 129.01 | 7.82 | |
| SgDHDDS2 | K11778 | KX893943 | 2.5.1.87 | 4818.35 | 227.35 | |
| SgDHDDS3 | K11778 | KX893944 | 2.5.1.87 | 778.03 | 31.45 | |
| SgDHDDS4 | K11778 | KX893945 | 2.5.1.87 | 3175 | 179.52 | |
| SgDHDDS5 | K11778 | KX893946 | 2.5.1.87 | 219 | 9.45 | |
| SgICMT1 | K00587 | KX893947 | 2.1.1.100 | 187 | 15.99 | |
| SgICMT2 | K00587 | KX893948 | 2.1.1.100 | 119 | 12.97 |
FPKM, fragments per kilobase of transcripts per million mapped fragments; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; IDI, isopentenyl-diphosphate delta-isomerase, AACT, acetyl-CoA C-acetyltransferase; HMGS, hydroxylmethylglutaryl-CoA synthase; HMGR, hydroxymethylglutaryl-CoA reductase (NADPH); MVK, mevalonate kinase; PMK, 5-phosphomevalonate kinase, MDC, mevalonate diphosphate decarboxylase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl pyrophosphate synthase; GGPS, geranylgeranyldiphosphate synthase, type II; FOHSDR, farnesol dehydrogenase; FOLK, farnesol kinase; PCYOX1, prenylcysteine oxidases/farnesylcysteine lyase; STE24, STE24 endopeptidases; CHLP, geranylgeranyl diphosphate reductase; FACE2, farnesylated protein-converting enzyme 2; PCME, prenylcysteine alpha-carboxyl methylesterase; FNTB, protein farnesyltransferase subunit beta; SPS, all-trans-nonaprenyl-diphosphate synthase; DHDDS, ditrans, polycis-polyprenyl diphosphate synthase; ICMT, protein-S-isoprenylcysteine O-methyltransferase.
Table 2.
Transcript abundance of TPS genes as per the S. guaranitica transcriptome
| Terpene synthase | Kegg entry | Gene bank accession ID | Annotation | Length (bp) | E.C. no. | Read in leaf | FPKM |
|---|---|---|---|---|---|---|---|
| Monoterpene | K12467 | KX893949 | Myrcene/ocimene synthase | 371 | 4.2.3.15 | 7 | 1.92 |
| K12467 | KX893950 | Myrcene/ocimene synthase | 223 | 4.2.3.15 | 3 | 4.41 | |
| K12467 | KX893951 | Myrcene/ocimene synthase | 217 | 4.2.3.15 | 0 | 0 | |
| K12467 | KX893952 | Myrcene/ocimene synthase | 208 | 4.2.3.15 | 3 | 6.39 | |
| K12467 | KX893953 | Myrcene/ocimene synthase | 449 | 4.2.3.15 | 11 | 2.05 | |
| K12467 | KX893954 | Myrcene/ocimene synthase | 366 | 4.2.3.15 | 5 | 1.41 | |
| K15095 | KX893955 | (+)-neomenthol dehydrogenase1 | 2831 | 1.1.1.208 | 2112.05 | 97.37 | |
| K15095 | KX893956 | (+)-neomenthol dehydrogenase2 | 1065 | 1.1.1.208 | 122.03 | 9.02 | |
| K15095 | KX893957 | (+)-neomenthol dehydrogenase3 | 1660 | 1.1.1.208 | 3417.92 | 106.62 | |
| K15095 | KX893958 | (+)-neomenthol dehydrogenase | 352 | 1.1.1.208 | 4 | 1.24 | |
| K15095 | KX893959 | (+)-neomenthol dehydrogenase | 216 | 1.1.1.208 | 21.01 | 36.39 | |
| K15095 | KX893960 | (+)-neomenthol dehydrogenase | 232 | 1.1.1.208 | 7 | 8.53 | |
| K07385 | KX893961 | 1, 8-cineole synthase | 303 | 4.2.3.108 | 3 | 1.37 | |
| K07385 | KX893962 | 1, 8-cineole synthase | 230 | 4.2.3.108 | 3 | 3.8 | |
| K07385 | KX893963 | 1, 8-cineole synthase | 268 | 4.2.3.108 | 7 | 4.76 | |
| K07385 | KX893964 | 1, 8-cineole synthase | 2338 | 4.2.3.108 | 896.71 | 29.15 | |
| K15086 | KX893965 | (3S)-linalool synthase1 | 2099 | 4.2.3.25 | 2191.73 | 64.89 | |
| K15086 | KX893966 | (3S)-linalool synthase2 | 1251 | 4.2.3.25 | 521 | 13.14 | |
| K17982 | KX893967 | (E, E)-geranyl linalool synthase | 2541 | 4.2.3.144 | 1277.71 | 22.13 | |
| K15099 | KX893968 | Geraniol isomerase synthase | 823 | 1.14.13.152 | 33 | 2.36 | |
| Sesquiterpene | K15891 | KX893969 | Farnesol dehydrogenase | 1385 | 1.1.1.216 | 241.13 | 9.68 |
| K14184 | KX893970 | α-humulene/β-caryophyllene synthase | 497 | 4.2.3.57 | 11 | 1.71 | |
| K14184 | KX893971 | α-humulene/β-caryophyllene synthase | 212 | 4.2.3.57 | 2 | 3.83 | |
| K14184 | KX893972 | α-humulene/β-caryophyllene synthase | 299 | 4.2.3.57 | 2 | 0.95 | |
| K14184 | KX893973 | α-humulene/β-caryophyllene synthase | 1425 | 4.2.3.57 | 374 | 14.89 | |
| K14181 | KX893974 | Valencene synthase (TPS-V) | 1653 | 4.2.3.73 4.2.3.86 | 1901 | 51.85 | |
| K14179 | KX893975 | Germacrene- A synthase (TPS-1) | 858 | 4.2.3.23 | 192 | 13.53 | |
| K15809 | KX893976 | Cis-muuroladiene synthase | 1695 | 4.2.3.67 | 619.04 | 19.21 | |
| K15803 | KX893977 | Germacrene-d synthase (TPS-6) | 1764 | 4.2.3.22 4.2.3.75 | 116 | 3.35 | |
| K15806 | KX893978 | Selinene synthase (TPS-3) | 1680 | 4.2.3.76 | 2141.91 | 59.35 | |
| K14183 | KX893979 | Gamma-cadinene synthase | 1533 | 4.2.3.13 | 67 | 2.07 | |
| k01000 | KX893980 | Bicyclogermacrene synthase (TPS-4) | 1326 | 4.2.3.100 | 2020.06 | 64.65 | |
| Diterpene | K13070 | KX893981 | Momilactone-A synthase | 374 | 1.1.1.295 | 8 | 2.15 |
| K13070 | KX893982 | Momilactone-A synthase | 347 | 1.1.1.295 | 5 | 1.6 | |
| K04124 | KX893983 | Gibberellin 3-beta-dioxygenase | 815 | 1.14.11.15 | 19 | 1.41 | |
| K04124 | KX893984 | Gibberellin 3-beta-dioxygenase | 319 | 1.14.11.15 | 8 | 3.17 | |
| K04125 | KX893985 | Gibberellin 2-oxidase | 1307 | 1.14.11.13 | 480.99 | 31.01 | |
| K04125 | KX893986 | Gibberellin 2-oxidase | 214 | 1.14.11.13 | 1 | 1.82 | |
| K04120 | KX893987 | Ent-copalyl diphosphate synthase | 1298 | 5.5.1.13 | 69 | 2.87 | |
| K04120 | KX893988 | Ent-copalyl diphosphate synthase | 2497 | 5.5.1.13 | 2616.28 | 51.8 | |
| K04120 | KX893989 | Ent-copalyl diphosphate synthase | 642 | 5.5.1.13 | 16 | 1.66 | |
| K04120 | KX893990 | Ent-copalyl diphosphate synthase | 213 | 5.5.1.13 | 1 | 1.87 | |
| K04120 | KX893991 | Ent-copalyl diphosphate synthase | 255 | 5.5.1.13 | 2 | 1.63 | |
| K04120 | KX893992 | Ent-copalyl diphosphate synthase | 345 | 5.5.1.13 | 6 | 1.94 | |
| K04120 | KX893993 | Ent-copalyl diphosphate synthase | 691 | 5.5.1.13 | 21 | 3.06 | |
| K04120 | KX893994 | Ent-copalyl diphosphate synthase | 300 | 5.5.1.13 | 4 | 1.88 | |
| K04121 | KX893995 | Ent-kaurene synthase-1 | 326 | 4.2.3.19 | 11 | 4.11 | |
| K04121 | KX893996 | Ent-kaurene synthase-5 | 252 | 4.2.3.19 | 6 | 5.14 | |
| K04121 | KX893997 | Ent-kaurene synthase-3 | 227 | 4.2.3.19 | 2 | 2.7 | |
| K04121 | KX893998 | Ent-kaurene synthase-4 | 1541 | 4.2.3.19 | 126 | 4.46 | |
| K04121 | KX893999 | Ent-kaurene synthase-2 | 1151 | 4.2.3.19 | 65 | 3.21 | |
| K04121 | KX894000 | Ent-kaurene synthase-6 | 1646 | 4.2.3.19 | 164 | 7.12 | |
| K04121 | KX894001 | Ent-kaurene synthase-7 | 1743 | 4.2.3.19 | 372 | 15.7 | |
| K04123 | KX894002 | Ent-kaurenoic acid hydroxylase | 2463 | 1.14.13.79 | 361.97 | 8.26 | |
| K04123 | KX894003 | Ent-kaurenoic acid hydroxylase | 1766 | 1.14.13.79 | 253 | 7.36 | |
| K16085 | KX894004 | 9beta-pimara-7, 15-diene oxidase | 434 | 1.14.13.144 | 133.16 | 26.44 | |
| K16083 | KX894005 | Ent-isokaurene C2-hydroxylase | 415 | 1.14.13.143 | 19 | 4.11 | |
| K05282 | KX894006 | Gibberellin 20-oxidase-1 | 354 | 1.14.11.12 | 2 | 0.61 | |
| K05282 | KX894007 | Gibberellin 20-oxidase-5 | 213 | 1.14.11.12 | 1 | 1.87 | |
| K05282 | KX894008 | Gibberellin 20-oxidase-3 | 256 | 1.14.11.12 | 2 | 1.61 | |
| K05282 | KX894009 | Gibberellin 20-oxidase-4 | 433 | 1.14.11.12 | 7 | 1.4 | |
| K05282 | KX894010 | Gibberellin 20-oxidase-2 | 1367 | 1.14.11.12 | 566.34 | 22.38 | |
| Triterpene | K15813 | KX894011 | Beta-amyrin synthase | 2745 | 5.4.99.39 | 2784.23 | 92.71 |
| K15813 | KX894012 | Beta-amyrin synthase | 2739 | 5.4.99.39 | 2606.94 | 46.69 | |
| K00511 | KX894013 | Squalene monooxygenase | 1972 | 1.14.13.132 | 1324.67 | 35.44 | |
| K00511 | KX894014 | Squalene monooxygenase | 1909 | 1.14.13.132 | 1101.83 | 43.23 | |
| K00801 | KX894015 | Farnesyl-diphosphate farnesyltransferase | 1664 | 2.5.1.21 | 2268.89 | 108.66 | |
| K00801 | KX894016 | Farnesyl-diphosphate farnesyltransferase | 728 | 2.5.1.21 | 327.94 | 29.06 | |
| K15822 | KX894017 | Camelliol C synthase | 312 | 5.4.99.38 | 12 | 5.66 |
2.8. Annotation of unigenes
Unigenes were used as query sequences to search the annotation databases, including the NCBI non-redundant protein sequences database (NR) (http://www.ncbi.nlm.nih.gov/) and Swiss-Prot (a manually annotated and reviewed protein sequence database) (http://www.ebi.ac.uk/uniprot/), based on sequence homology to entries in the Gene Ontology (GO) database (http://www.geneontology.org/). Unigene sequences from S. guaranitica were categorized into three general sections: biological process (BP), cellular component (CC) and molecular function (MF). Additionally, the unigenes were used as query sequences for searching the Kyoto Encyclopedia of Genes and Genome (KEGG) pathways database (http://www.genome.jp/kegg/) and the Pfam (Protein family) database (http://pfam.sanger.ac.uk/).
2.9. Differential expression analysis
Expression levels of unigenes were normalized and calculated as the values of fragments per kilobase of transcripts per million mapped fragments (FPKM) during the assembly and clustering process. Differential expression analysis of unigenes was performed using the DESeq R package (1.10.1). DESeq provides statistical routines for assessing the differential genes expression in leaf tissues and assigns genes as differential expressed when the P-value < 0.05. P-value results were corrected using the Benjamini and Hochberg approach for controlling the false discovery rate (FDR).19
2.10. Quantitative real-time PCR (qRT-PCR) analysis
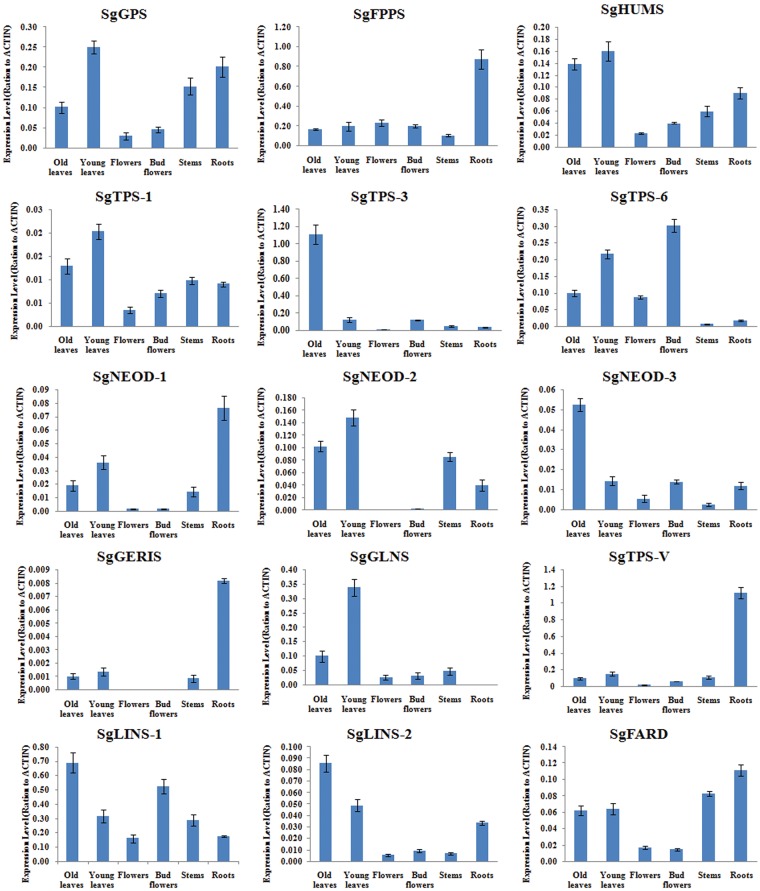
Quantitative real-time PCR was performed using an IQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad, USA) as described previously60 with SYBR Green Master (ROX) (Newbio Industry, China) following the manufacturer’s instructions at a total reaction volume of 20 µl. Gene-specific primers for SgActin as a reference gene and for the other 15 gene (SgGPPS, SgFPPS, SgHUMS, SgNEOD-1, SgNEOD-2, SgNEOD-3, SgTPS-1, SgTPS-3, SgTPS-6, SgLINS-1, SgLINS -2, SgGLNS, SgGERIS, SgTPS-V and SgFARD) involved in the biosynthesis of terpenes were designed using the primer designing tools of IDTdna (http://www.idtdna.com), as listed in Supplementary Table S1. The quantitative RT-PCR conditions were set as standard conditions: 95 °C for 3 min, 40 cycles of amplification (95 °C for 10 s, 60 or 58 °C for 30 s and 72 °C for 20 s), and a final extension at 65 °C for 1 min. The values are means ± SE of three replicates was normalized using SgActin as a reference gene. The relative expression levels were calculated by comparing the cycle thresholds (CTs) of the target genes with that of the reference gene SgActin using the 2-ΔΔCt method.6,20,21 The sizes of amplification products were 140–160 bp. The quantified data were analysed using the Bio-Rad IQTM 5 Multicolor Real-Time Manager software. Finally, the relative expression levels of SgGPPS, SgFPPS, SgHUMS, SgNEOD-1, SgNEOD-2, SgNEOD-3, SgTPS-1, SgTPS-3, SgTPS-6, SgLINS-1, SgLINS-2, SgGLNS, SgGERIS, SgTPS-V and SgFARD were detected.
2.11. Identification of simple sequence repeats (SSRs)
All of the transcripts of S. guaranitica were analysed with the MISA program version 1.0 (http://pgrc.ipk-gatersleben.de/misa/misa.html) for the detection of simple sequence repeat (SSR) motifs that have mono- to hexanucleotide repeats. In addition, primers for each SSR were designed using Primer3 version 2.3.5 (http://primer3.sourceforge.net/releases.php). The minimum number of SSR repeat units during analysis was ≥24 for mono- and dinucleotides and was 8, 7, 7 and 9 for tri-, tetra-, penta- and hexanucleotide repeats, respectively. The default parameters corresponding to each unit size of the minimum number of repetitions were 1-10, 2-6, 3, 5, 4, 5, 5, 5 and 6-5 for Unigene SSR detection.
2.12. Full-length terpene synthase cDNA clones and vectors
Full-length cDNAs for SgFPPS, SgGPPS and SgLINS were obtained by PCR amplification using short and long gene-specific primers (Supplementary Table S2, Fig. S1) based on RNA-Seq sequence information from the transcriptome sequencing of S. guaranitica leaves. Leaf cDNA was used as a template for the initial PCR amplification and performed using short primers with the KOD-Plus DNA polymerase (Novagen) under the following PCR conditions: 3 min at 94 °C followed by 10 s at 98 °C; 30 s at 60, 60 and 59 °C (different annealing temperatures), 1.30 min at 68 °C, and then 10 min at 68 °C. This process was repeated for 35 cycles. The first PCR products was used as a template for PCR cloning using long primers with the KOD-Plus DNA polymerase for the Gateway pDONR221 vector. The amplified PCR products were purified and cloned into the Gateway entry vector pDONR221 using BP Clonase (Invitrogen, USA). The resulting pDONR221 constructs harbouring target genes were sequenced, and Gateway LR Clonase (Invitrogen, USA) was used for recombination into the destination vector pB2GW7 for A. thaliana transformation. All final constructs containing SgFPPS, SgGPPS and SgLINS were confirmed by sequencing.
2.13. Arabidopsis plant growth conditions and preparation of Agrobacterium cultures for floral-dip transformation
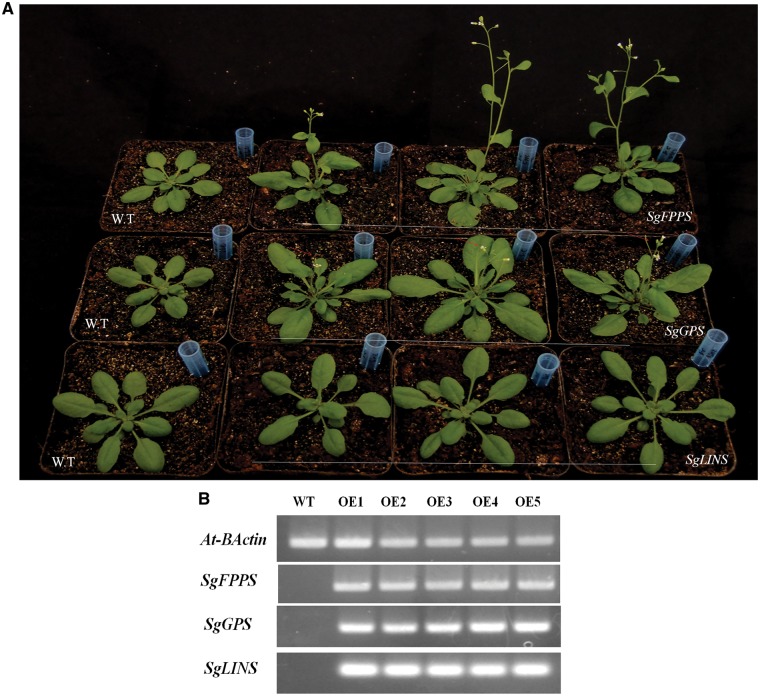
Ecotype of A. thaliana plant seeds Columbia-0 (Col-0) were pre-germinated by adding 1 ml sterilized water on some seed at 1.5 ml Eppendorf tube, then incubated at 4 °C for three days at the refrigerator. After that A. thaliana seeds had been growing in our Key Lab growth chamber at a temperature of 22 °C day/20 °C night with humidity of 50–70%, and photoperiod at 16 h day/8 h night, with a light density of 100–150 μmoles m −2 s −1 using fluorescent bulbs. After 2 months plants were ready for floral-dip transformation, and one week after the primary inflorescences were clipped. Plant watering was stopped 3 days prior to transformation for improved and increase the transformation efficiency. In addition, the constructs of pB2GW7 vectors with all inserted genes were introduced into Agrobacterium tumefaciens strain EHA105 by direct electroporation. Recombinant A. tumefaciens was grown for 2 days at 28 °C in solid LB media supplemented with 50 μg/ml each of rifampicin and spectinomycin. An individual colony of each sample was inoculated into 1.0 ml of liquid medium and grown at 28 °C under 200 rpm agitation overnight with the same media composition. After 24 h, 1.0 ml of each sample of liquid medium was transferred to a 250-ml conical flask containing 50 ml of LB media supplemented with the same compositions; the samples were grown at 28 °C in a shaker overnight until an optical density of 0.6–8.0 (OD 600) was reached. Overnight cell cultures were harvested by centrifugation at 5,000 rpm for 10 min at 4 °C, and the pellet was resuspended in the floral-dip inoculation medium contained 5% sucrose and 0.05% Silwet. A. thaliana was transformed by soaked the secondary inflorescences in the inoculation medium and stirred gently to allow the intake of Agrobacterium harbouring the pB2GW7 vector into the flower gynoecium. The transformed plants were kept in the dark and wrapped with plastic cover overnight to maintain humidity. The next day, the plants were returned back to their normal growth conditions. The transformation was repeated after 1 week to increase the transformation efficiency. Plants were grown for additional 4–5 weeks until all of the siliques became brown and dry. The seeds were harvested and stored at 4 °C under desiccation.15,16 BASTA was used for selection of transformant seedlings which were also confirmed with PCR for positive transgenic lines, more than 10 positive plant lines from each gene were analysed for terpenoid profiling and target gene expression.
2.14. Semiquantitative RT-PCR analysis
Semiquantitative real-time PCR was performed on an Eppendorf PCR (Eppendorf Mastercycler-Nexus GSX1, POCD Scientific, Australia) system with a total reaction volume of 25 µl. A gene-specific primer for AT-B-actin was used as a reference gene, and the other three gene-specific primers for SgFPPS, SgGPPS and SoLINS which are involved in the biosynthesis of terpenes, were designed using the primer designing tools of IDTdna (http://www.idtdna.com/scitools/ Applications/RealTimePCR/); the primer sequences are listed in (Supplementary Table S1). The semiquantitative RT-PCR conditions were as follows: predenaturation step at 95 °C for 4 min, 35 cycles of amplification (95 °C for 30 s, 60or 59 °C for 30 s and 72 °C for 1 min), and a final extension step at 72 °C for 10 min. The PCR products were resolved on 1% agarose gel, and the expression levels of AT-BActin, SgFPPS, SgGPPS and SgLINS genes were detected.
2.15. Metabolite extraction from transgenic A. thaliana leaves
Terpenoid compounds from non-transgenic A. thaliana leaves (control) and transgenic A. thaliana leaves containing either SgFPPS, SgGPPS and SgLINS expression constructs were extracted and isolated. For this, 15 leaves from each transgenic A. thaliana line (three leaf from each plant) were homogenized in liquid nitrogen with a mortar and pestle, after which the plant material powder was directly soaked in n-hexane as a solvent in Amber storage bottles, 30 ml screw-top vials with silicone/PTFE septum lids (http://www.sigmaaldrich.com) were used to reduce loss of volatiles to the headspace then incubated with shaking at 37 °C and 200 rpm for 72 h. Afterward, the solvent was transferred using a glass pipette to a 10-ml glass centrifuge tube with screw-top vials with silicone/PTFE septum lids and centrifuged at 5,000 rpm for 10 min at 4 °C to remove plant debris. The supernatant was pipetted into glass vials with a screw cap and oil was concentrated until remaining 1.5 ml of concentrated oils under a stream of nitrogen gas with a nitrogen evaporator (Organomation) and water bath at room temperature (Toption-China-WD-12). The concentrated oils transferred to a fresh crimp vial amber glass, 1.5 ml screw-top vials with silicone/PTFE septum lids were used to reduce a loss of volatiles to the headspace. For absolute oil recovery, the remaining film crude oil in the internal surface of concentrated glass vials was dissolved in the minimum volume of n-hexane, thoroughly mixed and transferred to the same fresh crimp vial amber glass, 1.5 ml. And the crimp vial was placed on the auto-sampler of the GC-MS system for GC-MS analysis, or each tube was covered with parafilm after closed with screw-top vials with silicone/PTFE septum lids and stored at −20 °C until GC-MS analysis. The same programme and standard conditions that were used for GC-MS analysis with S. officinalis essential oil components were applied.6
3. Results and discussion
3.1. Identification of essential oil components
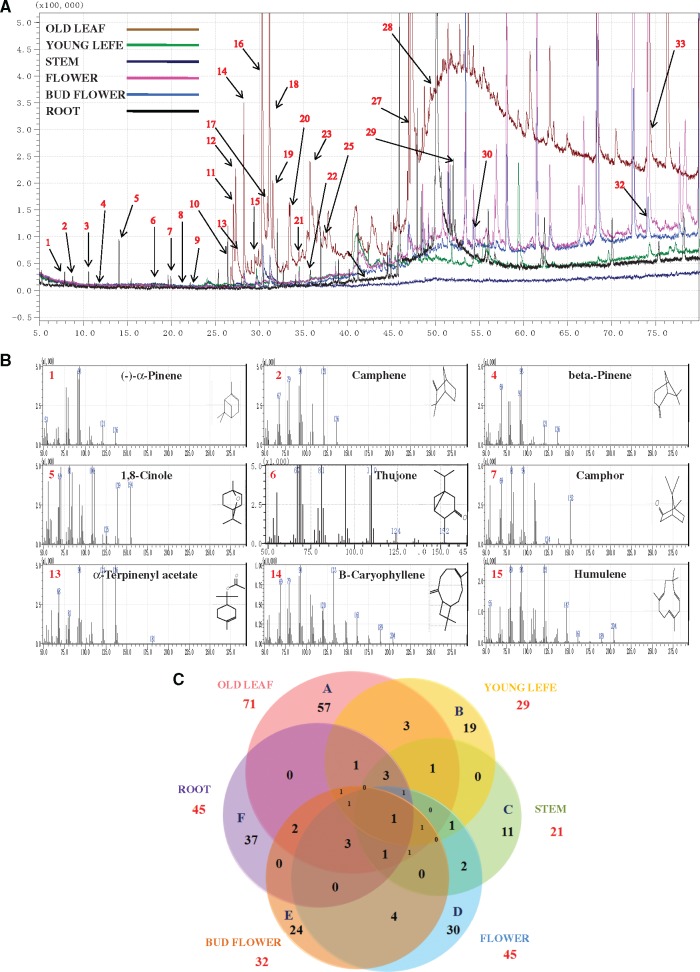
For GC-MS analysis, 204 compounds were identified using n-hexane extracts from six fresh parts of S. guaranitica. The numbers of obtained compounds from old leaves, young leaves, stems, flowers, bud flowers and roots were 71 (98.73%), 29 (74.58%), 21 (83.87%), 45 (93.51%), 32 (80.06%) and 45 (96.79%), respectively. The results of the qualitative and quantitative analyses of all compounds from the essential oils are reported in (Table 3 and Supplementary Table S3). The identified compounds are listed based on the retention time, compounds mass and percentage of peak area (Fig. 1A and B). In old leaves, one triterpene was shown as the main compound (32.31%), followed by the group of diterpene (21.16%) and sesquiterpenes group (16.7%). In young leaves, the sesquiterpenes compounds were observed to be the main group (25.78%), followed by one diterpene and one monoterpene compounds represented (11.45 and 0.19%), respectively. The main compound in the stem was one triterpene (0.15%). Furthermore, in flowers the sesquiterpenes compounds were observed to be the main group (0.42%), followed by one diterpene compound (0.01%).
Table 3.
The major chemical compositions in the essential oils of S. guaranitica
| N | Compound name | Retention time (min.) | Retention time index | Formula | Molecular Mass (g mol-1) | Terpene type | Old leaf |
Young leaf |
Stem |
Flower |
Bud Flower |
Root |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Peak area | % Peak area | % Peak area | % Peak area | % Peak area | % Peak area | O.S.S | |||||||
| 1 | α-Pinene | 7.323 | 939 | C10H16 | 136.234 | Mon | – | – | – | – | – | 0.96 | SO, SL, SA, SF, SC |
| 2 | Camphene | 8.367 | 951 | C10H16 | 136.234 | Mon | – | – | – | – | – | 0.55 | SO, SL, SA, SF, SC |
| 3 | laevo-β-Pinene | 10.108 | 974 | C10H16 | 136.234 | Mon | – | – | – | – | – | 0.97 | SO, SL, SA, SF, SC |
| 4 | beta.-Pinene | 11.27 | 980 | C10H16 | 136.234 | Mon | – | – | – | – | – | 0.36 | SO, SL, SA, SF, SC |
| 5 | 1,8-cineol | 13.99 | 1030 | C10H18O | 154.2493 | Mon | – | – | – | – | – | 2.61 | SO, SL, SA, SF |
| 6 | Thujone | 18.35 | 1112 | C10H16O | 152.2334 | Mon | – | – | – | – | – | 0.35 | SO, SF |
| 7 | (-)-Camphor | 20.325 | 1141 | C10H16O | 152.2334 | Mono | – | – | – | – | – | 1.14 | SO, SL, SA, SF |
| 8 | (+)-borneol | 21.485 | 1152 | C10H18O | 154.2493 | Mono | – | – | – | – | – | 0.53 | SO, SL, SA, SF, SC |
| 9 | Cis-α-terpineol | 22.648 | 1209 | C10H18O | 154.2493 | Mono | – | – | – | – | – | 0.13 | SO |
| 10 | Farnesan | 26.943 | 1376 | C15H32 | 212.4146 | Sesqui | – | – | – | – | – | 0.22 | |
| 11 | (-)-.beta.-Bourbonene | 27.023 | 1386 | C15H24 | 204.351 | Sesqui | 0.72 | – | – | – | – | – | SO |
| 12 | (E)-β-Elemene | 27.29 | 1389 | C15H24 | 204.3511 | Sesqui | 0.95 | – | – | – | – | – | |
| 13 | α-Terpineol acetate | 27.638 | 1351 | C12H20O2 | 196.286 | Mono | – | – | – | – | – | 0.21 | |
| 14 | β-Caryophyllene | 28.225 | 1420 | C15H24 | 204.3511 | Sesqui | 1.33 | 1.98 | – | 0.2 | – | 0.31 | SO, SF |
| 15 | Humulene | 29.46 | 1454 | C15H24 | 204.357 | Sesqui | 0.07 | – | – | – | – | – | SO, SL SA |
| 16 | (-)-Germacrene D | 30.293 | 1481 | C15H24 | 204.3511 | Sesqui | 5.43 | 10.7 | – | – | – | – | SO |
| 17 | pi- α-Muurolene | 30.908 | 1496 | C15H24 | 204.3511 | Sesqui | 0.41 | – | – | – | – | – | SO, SL SA |
| 18 | 1-Ethenyl-1-methyl-2, 4-bis-(1methylethenyl) cyclohexane | 31.195 | 1449 | C15H24 | 204.351105 | Sesqui | – | – | – | 0.22 | – | – | SO |
| 19 | δ-Cadinene | 31.518 | 1507 | C15H24 | 204.3511 | Sesqui | 0.67 | – | – | – | – | – | |
| 20 | Germacrene-A | 33.407 | 1510 | C15H26O | 222.3663 | Sesqui | 0.49 | – | – | – | – | – | |
| 21 | Caryophyllene oxide | 33.462 | 1546 | C15H24 | 204.3511 | Sesqui | 0.43 | 4.16 | – | – | – | – | SO, SA, SL, SN |
| 22 | Ledol | 35.185 | 1600 | C15H26O | 222.3663 | Sesqui | – | – | – | – | – | 1.56 | SO, SL, SA |
| 23 | α-Cadinol | 35.727 | 1653 | C15H26O | 222.3663 | Sesqui | 0.54 | – | – | – | – | – | |
| 24 | Indene, 6-methyl-3a, 4, 7, 7a-tetrahydro | 35.85 | 1113 | C10H14 | 134.2182 | Mono | – | 0.19 | – | – | – | – | |
| 25 | Longipinocarveol, trans- | 37.823 | 1618 | C15H24O | 220.350494 | Sesqui | 0.78 | – | – | – | – | – | |
| 26 | Trans-bisabolene epoxide | 37.93 | 1529 | C15H24O | 220.350494 | Sesqui | 0.14 | – | – | – | – | SO | |
| 27 | trans-phytol, (E) Phytol | 47.172 | 2110 | C20H40O | 296.531 | Diter | 21.11 | 11.45 | – | – | 0.48 | – | |
| 28 | Phytan | 49.165 | 1811 | C20H42 | 282.5475 | Diter | 0.05 | – | – | – | – | – | |
| 29 | Caryophyllene oxide | 52.368 | 1582 | C15H24O | 220.3505 | Sesqui | – | – | – | – | – | 1.91 | SO, SA, SL, SN |
| 30 | δ-Decalactone | 54.81 | 1504 | C10H18O2 | 170.248703 | Mono | – | – | – | – | 0.28 | – | |
| 31 | Kauran-18-al, 17-(acetyloxy)-, (4.beta.)- | 68.623 | 2040 | C22H34O3 | 346.503601 | Diter | – | – | – | 0.01 | – | – | |
| 32 | Peppermint camphor | 74.07 | 1179 | C10H20O | 156.265198 | Mono | – | – | – | – | 0.83 | – | |
| 33 | Squalene | 74.235 | 2831 | C30H50 | 410.718 | Tri | 32.31 | – | 0.15 | – | – | – | SO |
| Total percentage ( % ) of Monoterpenes | 0.19 | 1.11 | 7.81 | ||||||||||
| Total percentage ( % ) of sesquiterpenes | 16.7 | 25.78 | 0.42 | 4.0 | |||||||||
| Total percentage ( % ) of diterpenes | 21.16 | 11.45 | 0.01 | 0.48 | |||||||||
| Total percentage ( % ) of triterpenes | 32.31 | 0.15 | |||||||||||
RT, retention time; OSS, other salvia species; SA, Salvia acetabulosa; SL, Salvia leriifolia; SF, Salvia fruticosa; SN, Salvia nemorosa; SC, Salvia compressa; SO, Salvia officinalis; Mono, monoterpene; Sesqui, sesquiterpene; Dit, diterpene; Tri, triterpene; –, terpene compounds not detected.
Figure 1.
Typical GC-MS mass spectragraphs for terpenoids from old leaf, young leaf, stem, bud flower, flower and root of S. guaranitica. (A) GC-MS Peak of the essential oil, (B) mass spectrum of GC peak with retention time for the major compound, (C) Six-way Venn diagram to show the number of unique and common compounds in the essential oil extracts from old leaf (A), young leaf (B), stem (C), flower (D), bud flower (E) and root (F) of S. guaranitica.
On the other hand, in bud flowers, the monoterpenes compounds were shown as the main group (1.11%), followed by one diterpene and one sesquiterpene compound represented (0.48 and 0.03%), respectively. Finally, Monoterpenes from the main group of compounds (7.81%) found in the roots, followed by sesquiterpene group represented 4.0%. Moreover, the six hexane extracts from the different tissue essential oils have unique, common and major compounds. For example, the extracts of old leaf essential oils (A) had 57 unique compounds, three common compounds shared with extracts from young leaf essential oils, one common compound shared with extracts from stem essential oils, three common compounds shared with extracts from flower essential oils and two common compounds shared with extracts from bud flower essential oils. Furthermore, the young leaf essential oils (B) contained 19 unique compounds. While the stem essential oils (C) contained 11 unique compounds and two common compounds shared with extracts from flower essential oils and three common compounds shared with extracts from root essential oils. Also, the extracts from flower essential oils (D) had 30 unique compounds, four common compounds shared with extracts from bud flower essential oils. Moreover, the extracts from bud flower essential oils (E) and the root essential oils (F) had 24 and 37 unique compounds, respectively (Fig. 1C). On the other hand, we found one common compound named (Cyclooctasiloxane, hexadecamethyl) shared with all six plant parts. Additionally, we found some other common compounds shared among all six plant parts, such as (trans-phytol, 2-methyloctacosane, β-caryophyllene, cyclohexasiloxane, dodecanethiol-, cycloheptasiloxane, tetradecane- methyl- and cyclononasiloxane, octa deca methyl-) (Supplementary Table S3 and Fig. 1C). Regarding the major compounds, squalene (32.31%) was the major compound in the essential oils extracts of old leaves, followed by trans-phytol (21.11%), (-)-germacrene D (5.43%), n-octadecanal (5.15%), 8-isopropenyl-1, 5-dimethyl-1, 5-cyclodecadiene (4.88%) and β-caryophyllene (1.33%), whereas the essential oil of young leaves was also characterized by trans-phytol (11.45%), followed by (-)-germacrene D (10.7%), 8-isopropenyl-1, 5-dimethyl-1, 5-cyclodecadiene (8.8%), caryophyllene oxide (4.16%), 3-methyl-cis-3a, 4, 7, 7a-tetrahydroindan (3.49%) and β-caryophyllene (1.98%). N-octadecanal was characterized as the major compound in stem extracts (38.78%), followed by undecane, 2-methyl (1.27%), and then squalene (0.15%). Furthermore, the essential oils of flowers was also characterized by 2-methyloctacosane (5.34%), followed by 10-methyleicosane (1.9%), and tetracosane, 2-methyl (1.7%). Moreover, the essential oil of bud flowers was also characterized by peppermint camphor (0.83%) as a major compound, followed by trans-phytol (0.48%), angelicoidenol (0.28%) and longi borneol (0.03%). Finally, the root essential oil was characterized by 1,8-cineol (2.61) as a major compound, followed by caryophyllene oxide (1.91), ledol (1.56), (-)-camphor (1.14), laevo-β-pinene (0.97) and α-pinene (0.96) (Table 3). When comparing the composition of the six essential oil extracts of S. guaranitica, we deduced that some common compounds exist at different levels within the parts of S. guaranitica (Fig. 1A). Additionally, some of the compounds that have been found in S. guaranitica were detected in the other Salvia plant species (Table 3 and Supplementary Table S3).6,13,14,22 Therefore, we suggest that plant parts can have a major effect on the metabolic composition of their essential oils.23,24 From the previous GC-MS data, an important question has been raised, why do the triterpenes, sesquiterpenes and monoterpene compounds of S. guaranitica mostly accumulated in old leaves, young leaves and roots, respectively? This question was difficult to answer before conducting the present work because there was a lack of information at the genetic level regarding the terpenoid biosynthetic pathway and how these compounds are synthesized in S. guaranitica.
3.2. Illumina sequencing and the de novo assembly of the S. guaranitica leaf transcriptome
In the past few years, the Illumina sequencing platform has become a powerful method for analysing and discovering the genomes of non-model plants.17,25In this context, to generate transcriptome sequences, complementary DNA (cDNA) libraries were prepared from leaf tissues of S. guaranitica, and cDNA was then sequenced using PE reads sequencing using an Illumina HiSeq 2000 platform. Previous reports involving Illumina sequencing reported that the use of PE sequencing showed significant improvement in the efficiency of de novo assembly and increased the depth of sequencing.10,26 The cDNA sequencing generated 4 Gb of raw data from S. guaranitica leaves. After filtering and removing the adapter sequences from the raw data, the number of reads was 32,862,861 (32.86 million), comprising of 186,299,510 high-quality nucleotide bases, with 96.32% Q20, 92.42% Q30 and 47.55% GC content. For further analysis, high-quality reads were selected, and the transcriptome was assembled using the Trinity program,18 which produced 179,369 transcripts with an N50 length of 1,603 bp, an N90 length of 462 bp and a mean length of 1,039 bp. Moreover, 61,400 unigenes could be detected with an N50 length of 1,334 bp, an N90 length of 277 bp and a mean length of 731 bp. The distribution of the assembled transcript length ranged from 200 to > 2,000 bases; the maximum number of transcripts (66,664 transcripts, 37.165%) ranged from 200 to 500 bp, followed by 48,716 transcripts (27.159%) ranging from 1,000 to 2,000 bp and then 40,323 transcripts (22.480%) ranging from 500 to 1,000 bp. On the contrary, the lowest number of transcripts (23,666 transcripts, 13.194%) was obtained for a size of more than 2,000 bp. In contrast, the assembled unigene lengths were distributed between 200 and > 2,000 bp. The maximum number of unigenes (37,659 unigenes, 37.165%) ranged from 200 to 500 bp, followed by 10,132 unigenes (16.501%) ranged from 500 to 1,000 bp, and then 8,777 unigenes (14.294%) ranging from 1,000 to 2,000 bp. Finally, the lowest number of unigenes (4,832 unigenes, 7.869%) was obtained for a size of >2,000 bp. The length distribution of the transcripts and unigenes are shown in (Supplementary Table S4 and Fig. S2). Our results are in agreement with those for Salvia officinalis, Boehmeria nivea, Curcuma longa, Medicago sativa, Centella asiatica and Apium graveolens in which the largest number of both transcript and unigene lengths were found to range between 75 and 500 bp.6,27,28
3.3. Functional annotation and classification of assembled S. guaranitica unigenes
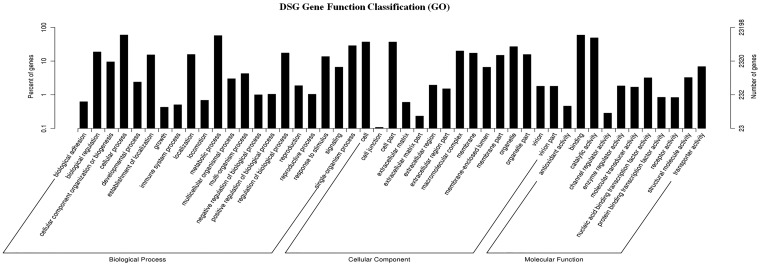
The total number of unigenes (61,400, 100% of all unigenes) was compared against the NR, NT, KO, Swiss-Prot, PFAM, GO and KEGG annotation database (Supplementary Table S5 and Fig. S3). The annotation percentage results in this research were higher than the annotation percentages in other non-model plant studies (58% in Carthamus tinctorius and 58. 01% in C. lanceolata).11,29,30The international standardized gene functional annotation system (GO Annotation) provides a powerful way to recognize the functions and properties of sequences that have not been characterized for an organism.31 The BLAST2 GO program was used to categorize the functions of these annotated unigenes, and a total of 23,198 unigenes (37.78% of all of the assembled unigenes) were mapped to at least one GO term. Based on sequence homology, the unigene sequences from S. guaranitica were categorized into 47 functional groups under three general sections: 60,139 were assigned to the BP, 42,494 were assigned to the CC and 29,574 were assigned to the MF sections. As a result, cellular process (13,830) and metabolic process (13,253) were the most enriched GO terms in the BP section. Regarding the CC section, the cell (8,590) and cell part (8,553) were the most enriched. Within the MF section, binding (13,723) and catalytic activities (11,368) were highly enriched (Fig. 2). These results revealed that the main GO classifications in the annotated unigenes were responsible for metabolism and fundamental biological regulation. These results were similar to previous results with the S. miltiorrhiza, S. officinalis transcriptome, and with the transcriptome of O. sanctum and O. basilicum (members of the same family), which have the highest percentages of metabolic process, cellular process, cell, cell part, binding and catalytic activity.6,32,33 Moreover, these results are in agreement way with previous studies on de novo transcriptome assembly in the tuberous root of sweet potato, transcriptome sequencing from S. officinalis, de novo transcriptome sequencing from Raphanus sativus and de novo characterization of roots from the Chinese medicinal plant Polygonum cuspidatum.6,30,34 The lowest percentage of unigenes categories included channel regulator activity (66), extracellular matrix parts (54) and cell junction (25). Therefore, the present work suggests that the enormous potential data that exist in the GO classifications can be used to identify the new genes.
Figure 2.
Functional annotation and classification of assembled unigenes in S. guaranitica. GO terms are summarized in three general sections of the BP, CC and MF.
3.4. KEGG analysis of S. guaranitica transcriptomes
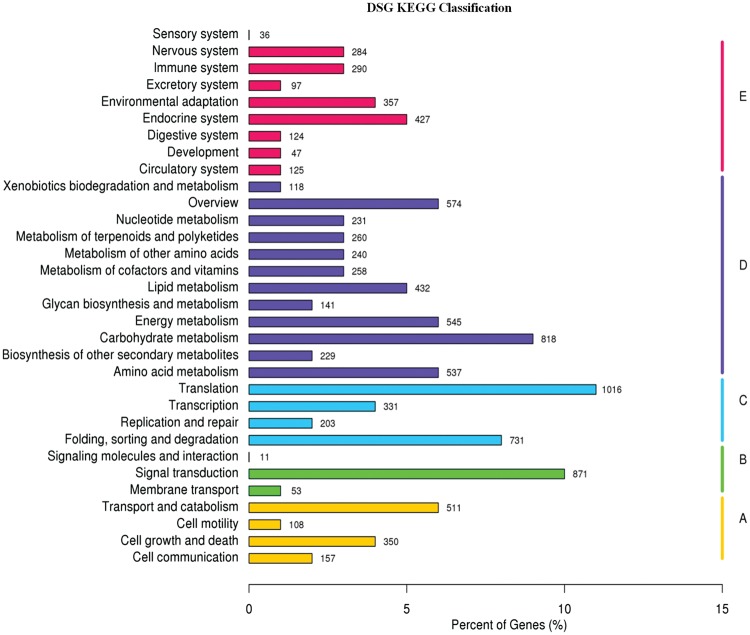
The KEGG pathway database can facilitate the understanding of the functional annotations of enzymes and the biological functions of genes regarding their networks.8,35 To identify active biological functional pathways in the leaf tissues of S. guaranitica, all 61,400 unigenes sequences were mapped in reference to the canonical pathways of KEGG, but 9,163 (14.92%) unigene sequence were assigned to 260 KEGG pathways. Furthermore, all transcripts were classified into five larger pathways categories, including cellular processes, environmental information processing, genetic information processing, metabolism and organismal systems (Fig. 3). The highest number of transcripts from S. guaranitica could be assigned to the metabolism category, followed by genetic information processing, organismal systems and cellular processes, whereas the lowest number of transcripts was related to the category environmental information processing. Interestingly, 570 transcripts of S. guaranitica were related to the biosynthesis of various secondary metabolite pathways, which were sorted into 26 subcategories, with phenylpropanoid biosynthesis (ko00940), terpenoid backbone biosynthesis (ko00900) and carotenoid biosynthesis (ko00906) representing the largest subcategories (Supplementary Table S6). These results were in agreement with previous results from the transcriptome of S. officinalis, O. sanctum and O. basilicum, which are members of the same family, and from de novo transcriptome sequencing from R. sativus, the transcriptome of which had the highest percentages of phenylpropanoid biosynthesis and terpenoid backbone biosynthesis.6,7,30
Figure 3.
KEGG classified into five largest categories pathways includes cellular processes (A), environmental information processing (B), genetic information processing (C), metabolism (D) and organismal systems (E).
3.5. Genes related to the biosynthesis of isoprenoids
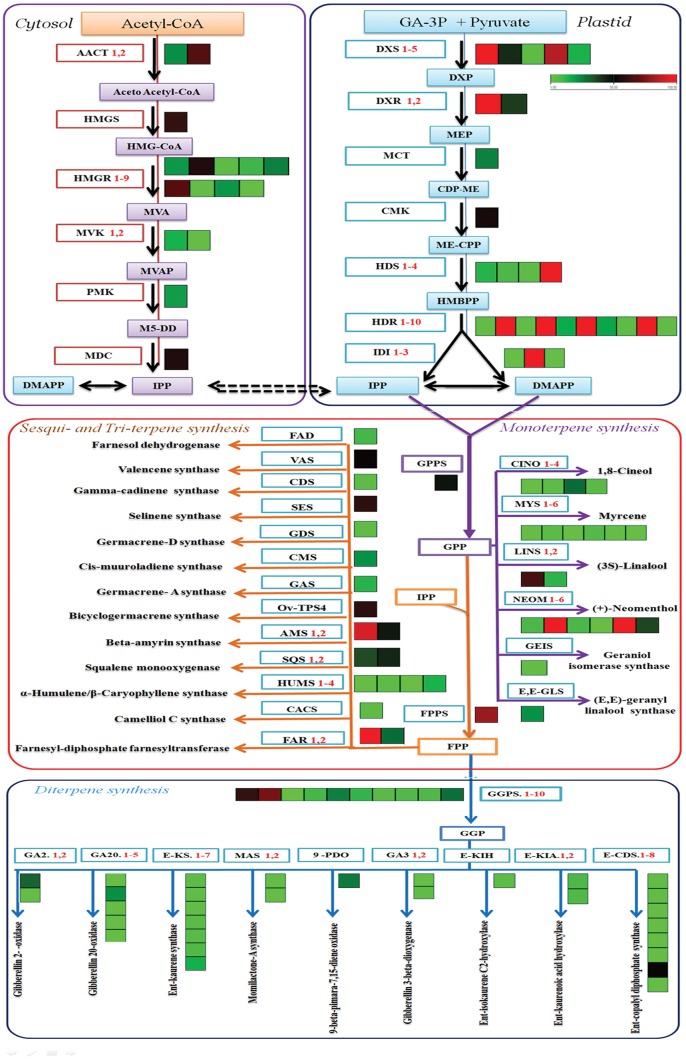
Various types of terpenoids were found in the essential oil extracts of S. guaranitica. The mixture contained mainly α-pinene, camphene, laevo-β-pinene, beta-pinene, 1,8-cineol, thujone, (-)-camphor, (+)-borneol, cis-α-terpineol, farnesan, (-)-beta-bourbonene, (E)-β-elemene, β-caryophyllene, humulene, (-)-germacrene D, pi-α-muurolene, δ-cadinene, germacrene-A, ledol, α-cadinol, trans-longipinocarveol, trans-phytol, phytan, kauran-18-al, 17-(acetyloxy)-, (4.beta.)- and squalene. Precursor molecules for terpenoid biosynthesis are derived from the cytosolic mevalonate (Ac-MVA) and plastidial MEP pathways. Therefore, queries against the Lamiaceae family transcriptome libraries were applied to identify and to determine the genes that encode the enzymes involved in the different steps of the terpenoid biosynthesis pathway, such as, IPPS (isopentyl diphosphate isomerase), DMAPPS (dimethylallyl diphosphate isomerase), GPPS (geranyl diphosphate synthases), FPPS (farnesyl pyrophosphate synthases) and GGPS (geranylgeranyl diphosphate synthases).36,37 Furthermore, we identified and estimated the expression levels of isoprenoid genes by using uniprot annotations against the transcriptome libraries (Table 1). From the annotation data analyses, we found many transcript genes related to isoprenoid biosynthesis from the MEP pathway with higher expression levels, including gene transcripts such as SgDXS1, 4 and 5 (1-deoxy-d-xylulose-5-phosphate synthase 1, 4 and 5), SgDXR1 (1-deoxy-d-xylulose-5-phosphate reductoisomerase 1), SgMCT (2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase), SgCMK (4-diphosphocytidyl-2-C-methyl-d-erythritol kinase), SgHDS 2 and 4 ((E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase 2 and 4) SgHDR 4, 6 and 9 (4-hydroxy-3-methylbut-2-enyl diphosphate reductase 4, 6 and 9), SgIDI 2 (isopentenyl-diphosphate delta-isomerase 2). Additionally, we obtained some gene transcripts that were related to isoprenoid biosynthesis from the MVA pathway with higher expression levels, such as, SgAACT 1 and 4 (acetyl-CoA C-acetyltransferase 1 and 4), SgHMGS (hydroxymethyl glutaryl-CoA synthase), SgHMGR 3 and 4 (hydroxymethyl glutaryl-CoA reductase (NADPH) 3 and 4), SgMVK (mevalonate kinase), SgPMK (phosphomevalonate kinase). Moreover, the transcriptome dataset of S. guaranitica presented other genes, such as SgGPPS, SgFPPS, and SgGGPSΙΙ2 that are the immediate precursor of the mono-, sesqui- and di-terpene biosynthesis pathway. The SgGPPS, SgFPPS and SgGGPSΙΙ2 genes were highly abundant in leaves and had higher values of fragments per kilobase of transcripts per million mapped fragments (FPKM), which were 47.81, 80.21 and 74.07, respectively (Fig. 4 and Table 1). Our results were similar to previously obtained results from the transcriptomes of S. officinalis, O. sanctum, O. basilicum and S. miltiorrhiza, which are members of the same family and have a higher number of transcripts for the isoprenoid biosynthesis genes related to the terpenoid biosynthesis pathway.6–8
Figure 4.
Representative terpenoid biosynthesis pathway with cognate heat maps for transcript levels of genes from S. guaranitica transcriptome data with substrates and products, coloured arrows connect substrates to their corresponding products. Green/red colour-coded heat maps represent relative transcript levels of different terpenoid genes determined by Illumina HiSeq 2000 sequencing; red, up-regulated; green, down-regulated. Transcript levels data represent by FPKM: fragments per kilobase of transcripts per million mapped fragments. MeV, multi-experiment Viewer software was used to depict transcript levels. DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; ISPF, 2-C-methyl-d-erythritol 2, 4-cyclodiphos-phate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductases; IDI, isopentenyl-diphosphate delta isomerase; AACT, acetyl-CoA C-acetyl transferase; HMGS, hydroxyl methyl glutaryl-CoA synthase; HMGR, hydroxymethyl glutaryl-CoA reductase (NADPH); MVK, mevalonate kinase; PMK, phospho-mevalonate kinase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl pyrophosphate synthase; GGPS, geranylgeranyl diphosphate synthase, type II; CINS, 1,8-cineole synthase; MYS, myrcene/ocimene synthase; LINS, (3S)-linalool synthase; NEOM, (+)-neomenthol dehydrogenase; SABI, (+)-sabinene synthase; TPS6, (-)-germacrene d synthase; AMS, beta-amyrin synthase; FARNESOL, farnesol dehydrogenase; SEQ, squalene monooxygenase; HUMS, α-humulene/β-caryophyllene synthase; FAR, farnesyl-diphosphate farnesyltransferase; GA2, gibberellin 2-oxidase; GA20, gibberellin 20-oxidase; E-KS, ent-kaurene synthase; MAS, momilactone-A synthase; GA3, gibberellin 3-beta-dioxygenase; E-KIA, ent-iso-kaurene C2-hydroxylase; E-KIH, ent-kaurenoic acid hydroxylase; E-CDS, ent-copalyl diphosphate synthase.
3.6. Genes related to terpene synthesis
Plants produce various terpenoid compounds with highly diverse structures. These compounds play an important role and functions in the interactions with environmental factors and in fundamental BPs.37,38 Multiple terpenoids are synthesized in plants by the expression of many terpene syntheses (TPSs) genes. Moreover, some TPS genes have the ability to catalyse the production of multiple products. Thus, the TPS genes family was classified according to phylogenetic relationships into eight subfamilies (TPS a, b, c, d, e/f, g and h), which comprises mono-, sesqui-, di- and triterpene synthases.39 Therefore, the annotation of transcriptome data from S. guaranitica against the Lamiaceae family and Arabidopsis revealed many terpene synthases involved in the terpenoid biosynthesis pathway, e.g. myrcene, (+)-neomenthol, 1, 8-cineole, (3S)-linalool, (E, E)-geranyl linalool, geraniol isomerase, farnesol, α-humulene, valencene, germacrene-A, cis-muuroladiene, selinene, gamma-cadinene, bicyclogermacrene, momilactone-A, gibberellin 3-beta-dioxygenase, gibberellin 2-oxidase, ent-copalyl diphosphate, ent-kaurene, ent-kaurenoic acid, 9 beta-pimara-7, 15-diene, ent-isokaurene C2-, gibberellin 20-, beta-amyrin, squalene, farnesyl- pyrophosphate and camelliol C. From the dataset, 69 TPS unigenes were identified and determined based on sequence similarities with a TPS sequence in the canonical annotation reference database. Twenty unigenes were annotated as being involved in monoterpene biosynthesis, including myrcene/ocimene synthase, (+)-neomenthol dehydrogenase, 1,8-cineole synthase, (3S)-linalool synthase, (E, E)-geranyl linalool synthase and geraniol isomerase synthase, and 12 other unigenes were annotated as being involved in sesquiterpene biosyntheses, including farnesol dehydrogenase, α-humulene/β-caryophyllene synthase, valencene synthase, germacrene-A synthase, cis-muuroladiene synthase, germacrene-D synthase, selinene synthase, gamma-cadinene synthase and bicyclo-germacrene synthase. Additionally, 30 unigenes were annotated as being involved in diterpene biosynthesis, including momilactone-A synthase, gibberellin 3-beta-dioxygenase, gibberellin 2-oxidase, ent-copalyl diphosphate synthase, ent-kaurene synthase, ent-kaurenoic acid hydroxylase, 9beta-pimara-7, 15-diene oxidase, ent-iso kaurene C2-hydroxylase and gibberellin 20-oxidase. Finally, seven unigenes were annotated as being involved in triterpene biosyntheses, including beta-amyrin synthase, camelliol C synthase, squalene monooxygenase and farnesyl-diphosphate farnesyl transferase, but some of these previous genes showed high abundance in leaves and higher FPKM values (Fig. 4 and Table 2). The previous compounds have significant pharmacological activities, such as anticancer, anti-HIV, antiviral, anti-inflammatory and antibacterial activities.40 Sesquiterpenoids are similar to triterpenoids as both share the same origin and originate from FPP. Triterpenoid compounds originate from the conversion of FDP into squalene by squalene synthase (SQS) and then to (S)-2, 3-epoxysqualene by squalene monooxygenase (SQE). Subsequently, (S)-2,3-epoxysqualene is converted to beta-amyrin and camelliol C in the presence of multifunctional (S)-2,3-epoxysqualene cyclase via beta-amyrin synthase and camelliol C synthase, respectively. Similar reports about triterpenoid biosynthesis from (S)-2,3-epoxysqualene cyclases are available for O. basilicum and Catharanthus roseus.41,42
3.7. SSR discovery and analysis
The Illumina HiSeq 2000 system offers the opportunity to analyse molecular markers such as SSRs that are related to terpenoid pathway genes. SSR molecular markers have proven to be a powerful method for understanding genetic variation. Moreover, polymorphic SSR markers are very important for the investigation of related comparative genomics, genetic diversity, evolution, linkage mapping, gene-based association studies and relatedness. Even though SNP markers have become promising, especially for studying complex genetic traits and high-throughput mapping, SSRs provide many advantages compared with other marker systems. Hence, SSRs have become the preferable codominant molecular marker for a construction of linkage maps.43 Therefore, the development of novel SSR molecular markers for S. guaranitica plants could be a valuable tool for breeding studies and genetic applications. Therefore, SSR markers were identified from transcriptome sequencing data using MISA (MIcroSAtellite) (http://pgrc.ipkgatersle-ben.de/misa/misa.html). Of the 61,400 transcripts of S. guaranitica, 5,262 transcripts were observed to have SSRs (Supplementary Table S7). The total number of SSR-containing sequences in S. guaranitica was 5,931, following stringent selection criteria used to identify these SSRs. The analysis data showed that dinucleotide repeats were the most abundant motif type in S. guaranitica (2,787; 45.25%), followed by trinucleotide (1,555; 23.58%), mononucleotide (1,452; 23.58%), tetranucleotide (92; 1.493%), and hexanucleotide (28; 0.454%) types, while the pentanucleotide type was the least abundant motif (17; 0.276%) (Supplementary Table S8 and Fig. S4). Except for the absence of mononucleotide, these results were similar to the previous results obtained from the transcriptome of O. sanctum and O. basilicum (members of the same family), which have dinucleotide repeats as the most abundant motif type, followed by tri-, tetra-, hexa- and pentanucleotide types as the least abundant motif.7 After analysing the data from mono- to hexanucleotide motifs to obtain the number of repeat units, we found that the highest repeat unit of potential SSRs was 10, which accounted for 1,376 SSRs (27.08%), followed by 5 SSRs (1,049; 20.65%), 7 (728; 14.33%), and 6 (573; 11.28%), and the smallest repeat unit of potential SSRs was ≥24 (3; 0.0 5%) (Supplementary Table S9). The AG/CT dinucleotide repeat was the most prevalent motif detected in all SSRs (1,893; 30.73%), followed by A/T as a mononucleotide repeat (1,408; 22.86%). In contrast, the least abundant motif in all SSRs (3; 0.048%) was detected in (AAAAC/GTTTT/AAAAG/CTTTT/AAACC/GGTTT) as pentanucleotide repeat and in (AAACAC/GTGTTT/AAACGG/CCGTTT/AAAGAC/CTTTGT) as hexanucleotide repeat. Finally, several SSR motifs were associated with many unique sequences that encode enzymes (e.g. SgDXS1, SgDXR1, SgMCT, SgHDR9, SgIDI3, SgAACT1, SgHMGS, SgHMGR2, SgHMGR6, SgMVK2, SgGGPSΙΙ2, SgGibberellin 20-oxidase, SgBeta-amyrin synthase, SgSqualene monooxygenase and Sgfarnesyl-diphosphate farnesyltransferase) involved in terpenoid biosynthesis (Supplementary Table S10).
3.8. Validation of the gene expression patterns by quantitative RT-PCR
To determinate the reliability of the Illumina HiSeq 2000 read analysis, 15 candidate genes with a higher differential expression were selected, and their expression profiles were compared within young leaf, old leaf, stem, flower, bud flower and root samples. Quantitative real-time (qRT) PCR was used to determine the ‘transcriptional control’ which indicates the number of mRNA copies of the enzyme that complements the end-product quantity. Therefore, the correlation between the TPS mRNAs with their products and the end-products showed a relationship between the chosen differentially expressed genes (DEGs), monoterpene synthase (SgGPPS; KX893917), sesquiterpene synthase (SgFPPS; KX893918), β-caryophyllene (SgHUMS; KX893973), neomenthol synthase-1 (SgNEOD-1; KX893955), neomenthol synthase-2 (SgNEOD-2; KX893956), neomenthol synthase-3 (SgNEOD-3; KX893957), germacrene-A synthase (SgTPS-1; KX893975), selinene synthase (SgTPS-3; KX893978), germacrene-d synthase (SgTPS-6; KX893977), linalool synthase-1 (SgLINS-1; KX893965), linalool synthase -2 (SgLINS-2; KX893966), (E, E)-geranyl linalool synthase (SgGLNS; KX893967), geraniol isomerase synthase (SgGERIS; KX893968), valencene synthase (SgTPS-V; KX893974), farnesol dehydrogenase (SgFARD; KX893969) and the terpenoid biosynthesis pathway of S. guaranitica. SgACTIN was used as an internal reference gene (Supplementary Table S1). The expression patterns of the 15 selected DEGs in the young leaf, old leaf, stem, flower, bud flower and root samples were examined (Fig. 5) by qRT-PCR, and the results were consistent with the results from the Illumina HiSeq 2000 read analysis. At the current stage, we may be able to answer the question which terpenoid compounds of S. guaranitica accumulated mostly in which tissue. From our results, we found that the next gene, geranyl diphosphate synthase (SgGPPS) gene showed the highest expression levels in the young leaves, followed by roots, stems, old leaves, bud flowers and flowers. These results were nearly compatible with our GC-MS analysis data indicating that the main group of terpenes in roots, bud flowers and young leaves consisted of monoterpene. According to the findings of the GC-MS analysis, we found eight monoterpene compound are accumulated in the root, two monoterpene compounds in bud flowers and one monoterpene compound are accumulated in young leaves (Table 3).Therefore, we suggest that the roots are the primer site for monoterpene biosynthesis and accumulation, followed by, bud flower, and young leave. These results are not in agreement with6,44,45 that found that the main monoterpenes in some salvia plant species are formed and accumulated in very young leaves epidermal glands. Because, the formation of most epidermal glands and the accumulation of the monoterpenes, take very short time in young leave tissues. And our S. guaranitica plant has limited number from epidermal gland trichomes on old leave, young leaves and stem. Moreover, Sesquiterpene synthase (FPPS) gene recorded the highest expression levels in the root followed by flower, bud flower, young leave, old leave and stem. On the other hand, these results were not similar with GC-MS analysis data that showed that the main group of sesquiterpenes was mostly accumulated in young leaves. Which have five compounds followed by old leaves have 12 compounds, roots have four compounds, flowers have two and bud flower has one compound (Table 3). Besides, from our study, we found a correlation and linkage between the β-Caryophyllene product and β-Caryophyllene synthase genes expression level in different tissues. For instance, the highest of the β-Caryophyllene synthase gene product and expression level presented in the young leaves followed by old leaves, roots then flowers (Table 3 and Fig. 1). Also, we found a correlation and linkage between the (-)-Germacrene D, Germacrene-A product and Germacrene-D synthase (TPS-6), Germacrene-A synthase (TPS-1) genes expression level in different tissues. Such as the highest of (-)-Germacrene-D, Germacrene-A gene product and expression level present in the young leave followed by old leave. Some of our results are in agreement with those of the previous studies6,44–53 that reported that the terpene quantity levels are thought to be mainly controlled transcriptionally thought producing the different TPS enzymes. (+)-Neomenthol dehydrogenase-1,-2,-3, TPS-3-Selinene synthase, Linalool synthase-1,-2, (E, E)-geranyl linalool synthase, Geraniol isomerase synthase, TPS-Valencene synthase and Farnesol dehydrogenase genes that were detected in the Illumina HiSeq 2000 reads and QRT-PCR but was not detected in the GC-MS analysis data. We suggest that this could be due to the cyclic expression of terpene synthases is under circadian control. Although, changes in transcript levels may not directly determine protein levels or enzyme activities due to possible posttranscriptional, post-translational or enzyme-regulatory mechanisms, the positive correlation between transcript levels and volatile emission suggests that changes in transcript level are an important determinant of scent production. Furthermore, the different rates of protein synthesis and proteolytic turnover and/or differences in protein modifications. And the secondary modification of monoterpene olefins (e.g. oxidation/glycosylation) or sequestration also could contribute to the monoterpene emission profile.54 The combination of the analysed data reads from Illumina HiSeq 2000, qRT-PCR and the GC-MS will pave the way to understand the complex mechanisms of controlling and regulating the diversity of terpene compound production.
Figure 5.
Quantitative RT-PCR validation of expression of terpene synthase genes selected from the DGE analysis in S. guaranitica. Total RNAs were extracted from old leaves, young leaves, stem, flower, bud flower and root samples and the expression of SgGPPS, SgFPPS, SgHUMS, SgNEOD-1, SgNEOD-2, SgNEOD-3, SgTPS-1, SgTPS-3, SgTPS-6, SgLINS-1, SgLINS-2, SgGLNS, SgGERIS, SgTPS-V and SgFARD genes were analysed using quantitative real-time. SgACTIN was used as the internal reference. The values are means ± SE of three biological replicates.
3.9. Functional characterization of terpene synthase genes in transgenic A. thaliana leaves
To test A. thaliana in a transgenic expression system for the production of Salvia terpenes, the following genes were selected from S. guaranitica: farnesyl pyrophosphate synthases (FPPS), geranyl diphosphate synthases (GPPS) and (3S)-linalool synthase (LINS) encoded by SgFPPS, SgGPPS and SgLINS, respectively. Transgenic A. thaliana was carried out by using the Agrobacterium-mediated floral dip method of A. thaliana flowers using A. tumefaciens strain EHA105 carrying pB2GW7-FPPS, pB2GW7-GPPS and pB2GW7-LINS under the control of 35S promoter vector. Fully mature leaves from fifteen 35-day-old putative transgenic plants and wild type plant (Fig. 6A), were collected for semiquantitative RT-PCR to analyse the positive transgenic A. thaliana and assessed the expression levels of terpene genes from the different samples (Fig. 6B). The terpenes were extracted with hexane and analysed by GC-MS. The mono-, sesqui- and di-terpene peaks were clearly detected, and the type and amount of compounds represented by the percentage of peak area (% peak area). Compounds were identified by comparing their mass spectra the compounds with mass spectra libraries. The detected components were also confirmed by comparing them with the published references and extracts of wild-type Arabidopsis which produce different types and amounts of terpenes. Overexpression of the SgFPPS, SgGPPS, and SoLINS genes produced different amounts from mono-, sesqui- and di-terpenes and other terpenoids. Moreover, from the results shown in Table 4 and Supplementary Fig. S5, we found that the transient expression of the different TPS genes from Salvia produced different types and amounts of mono-, sesqui- and di-terpenes and other terpenoid compounds.
Figure 6.
Overexpression of three S. guaranitica terpenoid genes in transgenic Arabidopsis. (A) Comparison of the phenotypes of the transgenic A. thaliana and wild type A. thaliana. (B) Semiquantitative RT-PCR to confirm the expression of terpenoid genes.
Table 4.
The major chemical compositions in transgenic A. thaliana leaves over-expressing of SgFPPS, SgGPPS and SgLINS
| N | Compound name | Retention time (min.) | Retention time index | Formula | Molecular mass(g mol−1) | Terpene type | W.T |
SgFPPS |
SgGPPS |
SgLINS |
|---|---|---|---|---|---|---|---|---|---|---|
| % Peak area | % Peak area | % Peak area | % Peak area | |||||||
| 1 | alpha-Pinene | 5.942 | 936 | C10H16 | 136.24 | Mono | – | – | 27.63 | – |
| 2 | beta-Pinene | 7.945 | 974 | C10H16 | 136.24 | Mono | – | – | 5.77 | – |
| 3 | Menthol | 20.812 | 1167 | C10H20O | 156.2652 | Mono | 2.75 | – | – | – |
| 4 | Thiourea, tetramethyl- | 22.326 | 1872 | C5H12N2S | 132.227 | 1.56 | – | – | – | |
| 5 | Cadina-1, 4-diene | 26.844 | 1533 | C15H24 | 204.3511 | Sesqui | – | 1.94 | – | – |
| 6 | β-Elemene | 27.314 | 1386 | C15H24 | 204.3511 | Sesqui | – | 2.14 | – | – |
| 7 | β-Caryophyllene | 28.406 | 1417 | C15H24 | 204.3511 | Sesqui | – | 17.71 | 15.4 | 4.64 |
| 8 | Cycloheptasiloxane, tetradecamethyl | 29.621 | 1519 | C14H42O7Si7 | 519.0776 | – | 1.91 | – | – | |
| 9 | (Z)-α-Bisabolene | 29.628 | 1503 | C15H24 | 204.3511 | Sesqui | – | – | 6.76 | – |
| 10 | Germacrene D | 30.516 | 1482 | C15H24 | 204.3511 | Sesqui | – | 71.39 | 6.29 | 10.73 |
| 11 | Norcarane | 31.33 | 796 | C7H12 | 96.1702 | – | – | – | 4.06 | |
| 12 | Germacrene-d-4-ol | 33.499 | 1576 | C15H26O | 222.372 | Sesqui | – | 1.33 | – | – |
| 13 | Cyclooctasiloxane, hexadecamethyl- | 34.487 | 1688 | C16H48O8Si8 | 593.2315 | – | 1.17 | – | – | |
| 14 | Heneicosane | 35.592 | 2100 | C21H44 | 296.5741 | 1.83 | – | – | – | |
| 15 | Allethrin | 38.072 | 2034 | C19H26O3 | 302.4079 | 2.6 | – | – | – | |
| 16 | Nonadecane | 38.456 | 1900 | C19H40 | 268.5209 | 1.92 | – | – | – | |
| 17 | Cyclohexasiloxane, dodecamethyl- | 38.648 | 1342 | C12H36O6Si6 | 444.9236 | – | 0.93 | – | – | |
| 18 | Stearic acid | 40.237 | 2178 | C18H36O2 | 284.4772 | 2 | – | – | – | |
| 19 | Phytane | 41.196 | 1800 | C20H42 | 282.5475 | Diter | 2.56 | – | – | |
| 20 | 1-Monolinoleoylglycerol trimethylsilyl ether | 42.337 | 2780 | C27H54O4Si2 | 498.89 | – | 0.8 | – | – | |
| 21 | Undecane, 4, 8-dimethyl- | 43.781 | 1214 | C13H28 | 184.3614 | 3.26 | – | – | – | |
| 22 | Oleic acid | 44.818 | 2141 | C18H34O2 | 282.468 | 2.5 | – | – | – | |
| 23 | Palmitic acid | 45.31 | 2010 | C16H32O2 | 256.4241 | 22.06 | – | – | 13.31 | |
| 24 | Palmitic acid, trimethylsilyl ester | 46.044 | 2015 | C19H40O2Si | 328.6052 | – | – | – | ||
| 25 | Octadecane | 46.248 | 1792 | C18H38 | 254.4943 | 3.17 | – | – | – | |
| 26 | 1-Butanol, 4-butoxy- | 46.518 | 1705 | C8H18O2 | 146.2273 | – | – | 6.4 | – | |
| 27 | Trimethylsilyl hexadecanoate | 47.316 | 2015 | C19H40O2Si | 328.6052 | 3.81 | – | – | – | |
| 28 | Phytol | 47.772 | 2115 | C20H40O | 296.531 | Diter | – | – | – | 11.08 |
| 29 | N-Hexacosane | 48.608 | 2598 | C26H54 | 366.707 | 3.67 | – | – | – | |
| 30 | dl-Methyltryptamine | 48.695 | 1770 | C11H14N2 | 174.24 | – | – | – | 7.59 | |
| 31 | trans-Elaidic acid | 49.469 | 2123 | C18H34O2 | 282.4614 | 35.41 | – | – | – | |
| 32 | Linoleic acid | 49.842 | 2152 | C21H40O2Si | 352.6266 | – | – | – | 11.52 | |
| 33 | Hexacos-9-ene | 49.993 | 2566 | C26H52 | 364.6911 | – | – | 9.79 | – | |
| 34 | Heptadecane | 50.911 | 1700 | C17H36 | 240.4677 | 3.03 | – | – | – | |
| 35 | 1-chloroeicosane | 51.916 | 2264 | C20H41Cl | 316.993 | – | – | 4.6 | – | |
| 36 | Tetradeca-1, 13-diene | 52.855 | 1385 | C14H26 | 194.356 | – | – | 4.34 | – | |
| 37 | cis-4-tetradecene | 55.024 | 1389 | C14H28 | 196.378 | – | – | 2.4 | – | |
| 38 | Diisooctyl phthalate | 59.952 | 2545 | C24H38O4 | 390.564 | – | – | 10.62 | 21.73 | |
| 39 | 17β-Estradiol, 3-deoxy | 63.773 | 2300 | C18H24O | 256.3826 | – | – | – | 4.9 | |
| 40 | Trichloroacetic acid | 69.121 | 1390 | Cl3CCOOH | 163.39 | – | – | – | 3.96 | |
| 41 | Cholestanol (5α-cholestan-3β-ol), TMS | 74.956 | 3169 | C30H56OSi | 460.8505 | – | – | – | 2.91 | |
| 42 | 1, 2-Epoxyhexane | 77.382 | 768 | C6H12O | 100.1589 | – | – | – | 3.57 | |
| 43 | 13, 23, 27-trimethylhenpentacontane | 78.973 | 5164 | C54H110 | 759.4512 | 7.87 | – | – | – | |
| Total % peak area | % 100 | % 100 | % 100 | % 100 |
Mono, monoterpene; Sesqui, sesquiterpene; Dit, diterpene; –, the terpene and other compounds not detected.
The putative functions of TPS genes isolated from S. guaranitica were initially predicted according to the conserved motifs using the InterPro protein sequence analysis and classification (http://www.ebi.ac.uk/interpro/) database. The SgGPPS protein with a 418-aa length has a metal-binding domain (IPR005630) from 74-418 aa; inside this domain are two motifs: both are DDxxD motif (DDVLD) one motif starting at 177 aa, and the other one is starting at 304 aa. Additionally, the SgFPPS protein is 349-aa length has a metal-binding domain (IPR005630) from 6-349 aa; inside this domain are two motifs: both are DDxxD motif one is (DDIMD) starting at 100 aa, and the other one is a (DDYLD) starting at 239 aa. On the other hand, the SgLINS protein is 541-aa in length, this protein has an N-terminal domain (IPR001906) from 69-279 aa and a metal-binding domain (IPR005630) from 270-540 aa, and inside the latter domain are DDxxD conserved motifs (DDIFD) starting at 347 aa Supplementary Fig. S6. Finally, the protein sequences contaned one or two of this domain belong to the terpene synthase family.
Furthermore, Croteau and coworkers shed light on the carbocationic reaction mechanism for all monoterpene synthases by reporting that the reaction was initiated by the divalent metal ion-dependent ionization of the substrate. The resulting cationic intermediate undergoes a series of hydride shifts or other rearrangements and cyclizations until the reaction was terminated by the addition of a nucleophile or proton loss. Croteau and coworkers illustrated this reaction mechanism by studying the native enzymes with substrate inhibitors, analogues and intermediates.55,56 Moreover, Rodney Croteau et al.56 elucidated the preliminary conversion of the geranyl cation to the tertiary linalyl cation to facilitate cyclization to a six-membered ring. Afterward, the linalyl cation provides the cyclic α-terpinyl cation; this is an important branching point intermediate in the formation of all cyclic monoterpenes because multiple terpene products can be obtained through electrophilic attack of C1 on the C6–C7 linalyl cation double bond and from the α-terpinyl cation. From the previous discussion, the reaction mechanisms of monoterpene synthases are highly reticulate. The individual intermediate may have multiple fates, which suggests the explanation for the ability of terpene enzymes to make various terpene products.57–60 On the other hand, the carbocationic reaction mechanism that uses sesquiterpene synthase to form sesquiterpenes by catalysing FPP recycling is similar to the reaction mechanism by those monoterpene synthases. Moreover, the larger carbon skeleton of FPP and the presence of three double bonds instead of two suggest a rationale for increases of the structural diversity of the sesquiterpene products. Furthermore, the initial cyclization reactions for sesquiterpene synthases can be divided into two types. Type one involves cyclization of the initially formed farnesyl cation to yield 11-membered ((E)-humulyl cation) rings of large size and a C2–C3 double bond (this type has no barrier to cyclization). The second type involves cyclization that proceeds after the tertiary nerolidyl cation produced from preliminary isomerization of the C2–C3 double bond. This isomerization mechanism is directly analogous to the isomerization of GPP to yield a linalyl cation in monoterpene synthesis. The nerolidyl cation is considered an intermediate in the sesquiterpene synthase mechanism.61–65 Collectively, we can state that the ability of TPS genes to convert a prenyl diphosphate substrate into diverse products during different reaction cycles is one of the unique traits of this type of enzyme. As described above, this property is found in the majority of all characterized monoterpene and sesquiterpene synthases. However, some monoterpene and sesquiterpene synthases can catalyse substrates into a single product, and the proteins may have specific methods for multiple product formations. For example, γ-humulene synthase from A. grandis has two DDxxD motifs located on opposite sides and can generate 52 different sesquiterpenes. This protein is able to bind substrates with two different conformations and resulting in different sets of products.66 In another example regarding the first monoterpene synthase cloned from Salvia officinalis, (+)-sabinene synthase produces 63% (+)-sabinene but also 21% γ-terpinene, 7.0% terpinolene, 6.5% limonene and 2.5% myrcene in in vitro assays.67 These additional monoterpene products or their immediate metabolites are also found in the monoterpene-rich essential oil of the S. guaranitica plant.
4. Conclusion
In this study, a large, high-quality transcriptome database was established for S. guaranitica leaves using NGS technology to characterize and identify genes that are related to the terpenoid biosynthesis pathway. Using de novo sequencing and analysis of the S. guaranitica transcriptome data via the Illumina HiSeq 2000 system, we identified many genes that encode enzymes involved in terpenoid biosynthesis. The purpose of identifying these genes is not only to facilitate functional studies but also to develop biotechnology for improving the production of medicinal ingredients through metabolic engineering. We profiled terpenoids from six tissues of S. guaranitica and used qRT-PCR to determine the correlation between the expression levels of TPS genes and the end-products. By combining the transcriptome and metabolome analysis with RNA-Seq or qRT-PCR with GC-MS approaches, this study paves the way for understanding the complex metabolic genes for the production of the diverse terpene compounds in blue anise sage. The results from our study will allow us to understand the specific activities of TPS in S. guaranitica for the production of interesting compounds and to develop new technology for utilization.
To our knowledge, this is the first study used Illumina HiSeq 2000 PE sequencing technology to investigate the global transcriptome of S. guaranitica. The valuable genetic resource in salvia will provide the foundation for future genetic and functional genomic research on S. guaranitica or closely related species. We further studied the functions of various S. guaranitica TPS genes, including SgFPPS, SgGPPS and SoLINS, by expressing these genes in A. thaliana transgenic plants. SgFPPS, SgGPPS and SoLINS were successfully expressed in the leaves of A. thaliana, and these transgenes altered the levels of terpenoids, as confirmed by GC-MS analysis of extracted transgenic A. thaliana leaves. The GC-MS analysis revealed that these S. guaranitica terpenoid synthases isolated from S. guaranitica can convert a prenyl diphosphate substrate into diverse products, which is one of the unique traits of this type of enzyme. Our study provides new insights into our understanding of plant terpenoid biosynthesis and the potential for biotechnology application.
Acknowledgements
The authors thank Associate Professor. Hazem Abdelnabby, Dr Jin Huanan, Dr Mohammed ayaad, Dr El sayed Elnishawy and Dr Mahmoud Kalil for proof-reading the manuscript. Also, the authors thank Dr Hongbo Lin for assistance for running the GC-Ms and data analysis. We also owe thanks to Prof. Zhang Yan sheng to give us the S. guaranitica seedlings. Also, the authors thank Dr Mohamed Hamdy Amar and Dr Wael Moussa, for their constructive comments and help. We also owe thanks to all Soybean Molecular Genetic lab and Rapeseed Lipid Biology lab members for facilitating the practical work at all levels of the experiments; the first author would like to thank the China Scholarship Council (CSC) and Huazhong Agricultural University for the scholarship.
Accession numbers
KX869088, KX869089, KX869090, KX869091, KX869092, KX869093, KX869094, KX869095, KX869096, KX869097, KX869098, KX869099, KX869100, KX869101, KX869102, KX869103, KX869104, KX869105, KX869106, KX869107, KX869108, KX869109, KX869110, KX869111, KX869112, KX869113, KX869114, KX869115, KX869116, KX869117, KX869118, KX869119, KX869120, KX869121, KX869122, KX869123, KX869124, KX869125, KX893913, KX893914, KX893915, KX893916, KX893917, KX893918, KX893925, KX893926, KX893919, KX893920, KX893921, KX893927, KX893922, KX893923, KX893924, KX893928, KX893929, KX893930, KX893931, KX893932, KX893933, KX893934, KX893935, KX893936, KX893937, KX893938, KX893939, KX893940, KX893941, KX893942, KX893943, KX893944, KX893945, KX893946, KX893947, KX893948, KX893949, KX893950, KX893951, KX893952, KX893953, KX893954, KX893955, KX893956, KX893957, KX893958, KX893959, KX893960, KX893961, KX893962, KX893963, KX893964, KX893965, KX893966, KX893967, KX893968, KX893969, KX893970, KX893971, KX893972, KX893973, KX893974, KX893975, KX893976, KX893977, KX893978, KX893979, KX893980, KX893981, KX893982, KX893983, KX893984, KX893985, KX893986, KX893987, KX893988, KX893989, KX893990, KX893991, KX893992, KX893993, KX893994, KX893995, KX893996, KX893997, KX893998, KX893999, KX894000, KX894001, KX894002, KX894003, KX894004, KX894005, KX894006, KX894007, KX894008, KX894009, KX894010, KX894011, KX894012, KX894013, KX894014, KX894015, KX894016, KX894017
Ethics approval and consent to participate
No investigations were undertaken using humans/human samples in this study. No experimental animals were used to conduct any of the experiments reported in this manuscript. Our study did not involve endangered or protected species. No specific permits were required from Wuhan Botanical Garden, China, for obtaining the seedlings of Salvia guaranitica L and Prof. Qingfeng Wang and Zhang Yan sheng should be contacted for future permissions.
Funding
M.A. was supported by the China Scholarship Council (CSC). This study was supported by the National Basic Research Development Program of China (973 Program, Grant 2013CB127001), and collaboration fund between Huazhong Agricultural University and Anhui Agricultural University.
Availability of data and materials
All data supporting my findings can be available and found in the Supplementary data.
Conflict of interest
None declared.
Supplementary Material
References
- 1. Alziar G. 1988–1993, Catalogue synonymique des Salvia L. dumonde (Lamiaceae). I.–VI. Biocosme Mesogéen., 5 (3–4): 87–136; 6(1–2, 4): 79–115, 163–204; 7(1–2): 59–109; 9(2–3): 413–497; 10(3–4): 33–117.
- 2. Takano A., Okada H.. 2011, Phylogenetic relationships among subgenera, species, and varieties of Japanese Salvia L. (Lamiaceae), J. Plant Res., 124, 245–52. [DOI] [PubMed] [Google Scholar]
- 3. Carretero-Paulet L., Ahumada I., Cunillera N. M., Rodríguez C., Ferrer A., Boronat N.. 2002, Campos, expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol-4-phosphate pathway, Plant Physiol., 129, 1581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao J., Lawrence C. D., Robert V.. 2005, Elicitor signal transduction leading to production of plant secondary metabolites, Biotechnol. Adv., 23, 283–333. [DOI] [PubMed] [Google Scholar]
- 5. Ward J. A., Ponnala L., Weber C. A.. 2012, Strategies for transcriptome analysis in nonmodel plants, Am. J. Bot., 2, 267–76. [DOI] [PubMed] [Google Scholar]
- 6. Mohammed A., Penghui L., Guangbiao S., Daofu C., Xiaochun W., Jian Z.. 2017, Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis), Sci. Rep., 7, 16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shubhra R., Seema M., Ankita B.. 2014, De novo sequencing and comparative analysis of holy and sweet basil transcriptomes, BMC Genomics, 15, 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hua W. P., Zhang Y., Song J., Zhao L. J., Wang Z. Z.. 2011, De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients, Genomics, 98, 272–9. [DOI] [PubMed] [Google Scholar]
- 9. Meena S., Kumar S. R., Venkata Rao D. K., et al. 2016, De novo sequencing and analysis of lemongrass transcriptome provide first insights into the essential oil biosynthesis of aromatic grasses, Front. Plant Sci., 7, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hyun T. K., Rim Y., Jang H.-J., et al. 2012, De novo transcriptome sequencing of Momordica cochinchinensis to identify genes involved in the carotenoid biosynthesis, Plant Mol. Biol., 79, 413–27. [DOI] [PubMed] [Google Scholar]
- 11. Huang H. H., Xu L. L., Tong Z. K., et al. 2012, De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis, BMC Genomics, 13, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi C. Y., Yang H., Wei C. L., et al. 2011, Deep sequencing of the Camellia sinens is transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds, BMC Genomics, 12, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monica R. L., Federica M., Rosa T., et al. 2010, Comparative chemical composition and antiproliferative activity of aerial parts of Salvia leriifolia Benth. and Salvia acetabulosa L. Essential oils against human tumor cell in vitro models, J. Med. Food., 13, 62–9. [DOI] [PubMed] [Google Scholar]
- 14. Aziz R. A., Hamed F. k., Abdulah N. A.. 2008, Determination of the main components of the essential oil extracted from Salvia fruticosa by sing GC and GC-MS DAMASCUS, J. Agric. Sci., 24, 223–36. [Google Scholar]
- 15. Su-Fang E., Zeti-Azura M., Roohaida O., Noor A. S., Ismanizan I., Zamri Z.. 2014, Functional characterization of sesquiterpene synthase from polygonum minus, Sci. World J., doi: 10.1155/2014/840592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aharoni A., Giri A. P., Deuerlein S., et al. 2003, Terpenoid metabolism in wild-type and transgenic Arabidopsis plants, Plant Cell, 15, 2866–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim H. A., Lim C. J., Kim S., et al. 2014, High-throughput sequencing and de novo assembly of Brassica oleracea var. capitata L. for transcriptome analysis, PLoS One, 9, e92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A.. 2011, Full-length transcriptome assembly from RNA-Seq data without a reference genome, Nat. Biotechnol., 29, 644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anders S., Huber W.. 2010, Differential expression analysis for sequence count data, Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak K. J., Schmittgen T. D.. 2001, Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method, Methods, 25, 402–8. [DOI] [PubMed] [Google Scholar]
- 21. Hongmei L., Luo H., Zhu Y., et al. 2014, Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation, Physiol. Plant, 152, 241–55. [DOI] [PubMed] [Google Scholar]
- 22. Fateme A. M., Mohammad H. F., Abdolhossein R., Ali Z., Maryam S.. 2013, Volatile constituents of Salvia compressa and Logochilus macranthus, two labiatae herbs growing wild in Iran, Res. J. Recent Sci., 2, 66–8. [Google Scholar]
- 23. Daniel J. S. 2004, Localization of salvinorin A and related compounds in glandular trichomes of the psychoactive sage Salvia divinorum, Ann. Bot., 93, 763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takano A., Okada H.. 2014, Volatile profiling of aromatic traditional medicinal plant, Polygonum minus in different tissues and its biological activities, Molecules, 19, 19220–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z., Fang B., Chen J., et al. 2010, De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of ISSR markers in sweet potato (Ipomoea batatas), BMC Genomics, 11, 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang C., Liu X., Yiu S.-M., Lim B. L.. 2013, De novo assembly and characterization of Camelina sativa transcriptome by paired-end sequencing, BMC Genomics, 14(1), 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Annadurai R. S., Neethiraj R., Jayakumar V., et al. 2013, De Novo transcriptome assembly (NGS) of Curcuma longa L. rhizome reveals novel transcripts related to anticancer and antimalarial terpenoids, PLoS One, 8, e56217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. An J., Shen X., Ma Q., Yang C., Liu S., Chen Y.. 2014, Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.), PLoS One, 9, e99940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang L. L., Yang X., Sun P., Tong W., Hu S. Q.. 2012, The first Illumina-based de novo transcriptome sequencing and analysis of safflower flowers, PLoS One, 7, e38653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yan W., Yan P., Zhe L., et al. 2013, De novo transcriptome sequencing of radish (Raphanus sativus L.) and analysis of major genes involved in glucosinolate metabolism, BMC Genomics, 14, 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gahlan P., Singh H. R., Shankar R., et al. 2012, De novo sequencing and characterization of Picrorhiza kurrooa transcriptome at two temperatures showed major transcriptome adjustments, BMC Genomics, 13, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang L., Ding G., Lin H., et al. 2013, Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis, PLoS One, 8, e80464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie F., Burklew C. E., Yang Y., et al. 2012, De novo sequencing and a comprehensive analysis of purple sweet potato (Impomoea batatas L.) transcriptome, Planta, 236, 101–13. [DOI] [PubMed] [Google Scholar]
- 34. Hao D. C., MA P., Mu J., et al. 2012, De novo characterization of the root transcriptome of a traditional Chinese medicinal plant Polygonum cuspidatum, Sci. China Life Sci., 55, 452–66. [DOI] [PubMed] [Google Scholar]
- 35. Kanehisa M., Goto S.. 2000, KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Virginie V. D., Germaine S., Yamina O., et al. 2001, Crystal structure of isopentenyl diphosphate: dimethylallyl diphosphate isomerase, EMBO J., 20, 1530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dorothea T. 2006, Terpene synthases and the regulation, diversity and biological roles of terpene metabolism, Curr. Opin. Plant Biol., 9, 297–304. [DOI] [PubMed] [Google Scholar]
- 38. Douglas J. M.-G., Rodney C.. 1995, Terpenoid metabolism, Plant Cell, 7, 1015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nagegowda D. A. 2010, Plant volatile terpenoid metabolism: biosynthetic genes, transcriptional regulation and subcellular compartmentation, FEBS Lett., 584, 2965–73. [DOI] [PubMed] [Google Scholar]
- 40. Razborsek M. I., Voncina D. B., Dolecek V., Voncina E.. 2008, Determination of oleanolic, betulinic and ursolic acid in lamiaceae and mass spectral fragmentation of their trimethylsilylated derivatives, Chromatographia, 67, doi: 10.1365/s10337-008-0533-6. [Google Scholar]
- 41. Misra R. C., Maiti P., Chanotiya C. S., Shanker K., Ghosh S.. 2014, Methyl jasmonate-elicited transcriptional responses and pentacyclic triterpenoid biosynthesis in sweet basil, Plant Physiol., 164, 1028–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang L., Li J., Ye H., Li C., et al. 2012, Molecular characterization of the pentacyclic triterpenoid biosynthetic pathway in Catharanthus roseus, Planta, 236, 1571–81. [DOI] [PubMed] [Google Scholar]
- 43. Verma P., Shah N., Bhatia S.. 2013, Development of an expressed gene catalogue and molecular markers from the de novo assembly of short sequence reads of the lentil (Lens culinaris Medik.) transcriptome, Plant Biotechnol. J., 11, 894–905. [DOI] [PubMed] [Google Scholar]
- 44. Sabine G.-G., Corinna S., Ralf S., Johannes N.. 2012, Seasonal influence on gene expression of monoterpene synthases in Salvia officinalis (Lamiaceae), J. Plant Physiolol., 169, 353–9., [DOI] [PubMed] [Google Scholar]
- 45. Croteau R., Felton M., Karp F., Kjonaas R.. 1981, Relationship of camphor biosynthesis to leaf development in sage Salvia officinalis, Plant Physiol., 67, 820–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dudareva N., Cseke L., Blanc V. M., Pichersky E.. 1996, Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower, Plant Cell., 8, 1137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McConkey M. E., Gershenzon J., Croteau R. B.. 2000, Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint, Plant Physiol., 122, 215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mahmoud S. S., Croteau R. B.. 2003, Menthofuran regulates essential oil biosynthesis in peppermint by controlling a downstream monoterpene reductase, Proc. Natl. Acad. Sci. U.S.A., 100, 14481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mahmoud S. S., Williams M., Croteau R.. 2004, Cosuppression of limonene-3-hydroxylase in peppermint promotes accumulation of limonene in the essential oil, Phytochemistry, 65, 547–54. [DOI] [PubMed] [Google Scholar]
- 50. Xie Z., Kapteyn J., Gang D. R.. 2008, A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes, Plant J., 54, 349–61. [DOI] [PubMed] [Google Scholar]
- 51. Lane A., Boecklemann A., Woronuk G. N., Sarker L., Mahmoud S. S.. 2010, A genomics resource for investigating regulation of essential oil production in Lavandula angustifolia, Planta, 231, 835–45. [DOI] [PubMed] [Google Scholar]
- 52. Schmiderer C., Grausgruber-Gröger S., Grassi P., Steinborn R., Novak J.. 2010, Influence of gibberellin and daminozide on the expression of terpene synthases in common sage (Salvia officinalis), J. Plant Physiol., 167, 779–86. [DOI] [PubMed] [Google Scholar]
- 53. Kampranis S. C., Ioannidis D., Purvis A., et al. 2007, Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function, Plant Cell, 19, 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dudareva N., Martin D., Kish C. M., et al. 2003, (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon, Plant Cell, 15, 1227–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodney C., Mark F., Robert C. R.. 1980, Biosynthesis of monoterpenes – conversion of the acyclic precursor’s geranyl pyrophosphate and nerylpyrophosphate to the rearranged monoterpenes fenchol and fenchone by a soluble enzyme preparation from fennel (Foeniculum vulgare), Arch. Biochem. Biophys., 200, 524–33. [DOI] [PubMed] [Google Scholar]
- 56. Rodney C. 1987, Biosynthesis and catabolism of monoterpenoids, Chem. Rev., 87, 929–54. [Google Scholar]
- 57. Wise M. L., Rodney C.. 1999, Comprehensive Natural Products Chemistry, Isoprenoids Including Caroteinoids and Steroids, vol. 2, pp. 97–135. Elsevier: Amsterdam. [Google Scholar]
- 58. Lücker J., El-Tamer M. K., Schwab W., et al. 2002, Monoterpene biosynthesis in lemon (Citrus Limon) – cDNA isolation and functional analysis of four monoterpene synthases, Eur. J. Biochem., 269, 3160–71. [DOI] [PubMed] [Google Scholar]
- 59. Takehiko S., Tomoko E., Hiroshi F., et al. 2004, Molecular cloning and functional characterization of four monoterpene synthase genes from Citrus unshiu Marc, Plant Sci., 166, 49–58. [Google Scholar]
- 60. Dezene P. W. H., Ryan N. P., Kimberley-Ann G., Rona N. S., Jörg B.. 2005, Characterization of four terpene synthase cDNAs from methyl jasmonateinduced Douglas-fir, Pseudotsuga menziesii, Phytochemistry, 66, 1427–39. [DOI] [PubMed] [Google Scholar]
- 61. Martin D. M., Bohlmann J.. 2004, Identification of Vitis vinifera (-)-alpha-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase, Phytochemistry, 65, 1223–9. [DOI] [PubMed] [Google Scholar]
- 62. David E. C., Stephen S., Pushpalatha P. N. M.. 1981, Trichodiene biosynthesis and the enzymatic cyclization of farnesyl pyrophosphate, J. Am. Chem. Soc., 103, 2136–8. [Google Scholar]
- 63. David E. C., Guohan Y.. 1994, Trichodiene synthase – stereochemical studies of the cryptic allylic diphosphate isomerase activity using an anomalous substrate, J. Org. Chem., 59, 5794–8. [Google Scholar]
- 64. David E. C., Manish T.. 1995, Epicubenol synthase and the stereochemistry of the enzymatic cyclization of farnesyl and nerolidyl diphosphate, J. Am. Chem. Soc., 117, 5602–3. [Google Scholar]
- 65. Alchanati I., Patel J. A. A., Liu J., et al. 1998, The enzymatic cyclization of nerolidyl diphosphate by delta cadinene synthase from cotton stele tissue infected with Verticillium dahlia, Phytochemistry, 47, 961–7. [Google Scholar]
- 66. Steele C. L., Crock J., Bohlmann J., Croteau R.. 1998, Sesquiterpene synthases from grand fir (Abies grandis) – Comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of delta-selinene synthase and gamma-humulene synthase, J. Biol. Chem., 273, 2078–89. [DOI] [PubMed] [Google Scholar]
- 67. Wise M. L., Savage T. J., Katahira E., Croteau R.. 1998, Monoterpene synthases from common sage (Salvia officinalis) – cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1, 8-cineole synthase, and (+)- bornyl diphosphate synthase, J. Biol. Chem., 273, 14891–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting my findings can be available and found in the Supplementary data.