Abstract
Skeletal muscle is generated by the successive incorporation of primary (embryonic), secondary (fetal), and tertiary (adult) fibers into muscle. Conditional excision of Pitx2 function by an MCKCre driver resulted in animals with histological and ultrastructural defects in P30 muscles and fibers, respectively. Mutant muscle showed severe reduction in mitochondria and FoxO3-mediated mitophagy. Both oxidative and glycolytic energy metabolism were reduced. Conditional excision was limited to fetal muscle fibers after the G1–G0 transition and resulted in altered MHC, Rac1, MEF2a, and alpha-tubulin expression within these fibers. The onset of excision, monitored by a nuclear reporter gene, was observed as early as E16. Muscle at this stage was already severely malformed, but appeared to recover by P30 by the expansion of adjoining larger fibers. Our studies demonstrate that the homeodomain transcription factor Pitx2 has a postmitotic role in maintaining skeletal muscle integrity and energy homeostasis in fetal muscle fibers.
Keywords: Skeletal Muscle, Mouse, Pitx2, Mitochondria, Autophagy
1. Introduction
The Pitx2 locus encodes one of the vertebrate set of roughly 200 transcription factors that use the evolutionarily conserved homeodomain (HD) to selectively bind specific DNA sequences at cis-regulatory modules (CRMs) of target loci. HD factors comprise approximately 10% of the sequence specific transcription factors (SSTFs) encoded in the vertebrate genome, are expressed in spatially-restricted, progressively-overlapping patterns throughout embryogenesis, and genetic perturbations at their loci often cause developmental patterning defects characterized by homeosis, or the replacement of one body part by the likeness of another. Progressive deployment of HD-factors acts collectively to specify body part identity during embryogenesis, with each progressive deployment of a new HD-factor applying a new lineage restriction. For example, the onset of Cdx2 expression in outer morula cells represses Nanog and Oct4 RNA, and restricts the trophoectoderm lineage from the inner cell mass (Strumpf et al., 2005). Similarly, the onset of Lbx1 expression in the ventrolateral dermomyotome restricts a subset the default-medial migrating muscle precursors to become lateral-migrating muscle precursors (Gross et al., 2000a). Lineage restriction by a given HD factor in embryos is exquisitely dependent on the lineage of the particular cell in which it is becoming newly expressed. Thus, onset of Lbx1 expression in the developing neural tube does not lead neural tube cells to become laterally migrating muscle cells. Instead, it contributes to lineage restriction, or specification, of progenitors for at least five neuronal subtypes, each in a distinct way that depends on which layer of the neuroepithelium it is emerging from (Gross et al., 2002; Kioussi and Gross, 2008; Kioussi et al., 2006). It is therefore not surprising that HD factors act in specifying muscle progenitor cell types, with different HD-restriction histories acting in different spatial domains to initiate myogenic determination (Harel et al., 2012; Schubert et al., 2018; Shih et al., 2007a; 2007b).
The myogenic progression emerges from classic primary culture-based studies, and determined myoblasts have long been characterized by expression of Myf5, Myf6, or Myod1, rather than by appearance of any particular HD-factor. Removal of FGF extracts, or serum withdrawal, causes myoblasts to pull out of the cell cycle, into a G0 phase, and undergo terminal differentiation. The point at which cells could no longer be induced to proliferate by restoring growth conditions was operationally defined as the point of commitment to terminal differentiation and became associated with Myog expression. Terminal differentiation itself was characterized by observing onset of structural RNA and protein expression and fusion into multinucleated syncytia. When these culture systems were developed, embryological grafting studies indicated that somite-generated muscle cells were plastic and adopted their specific properties from the local environment. Consistent with this view, a TCF4+ prepattern delineates the muscle anlagen pattern of the limb, even in the absence of migratory muscle progenitors from the somite (Kardon et al., 2003). However, newer grafting experiments demonstrate that some predispositions of muscle progenitors, such as Lbx1-dependent lateral migration, are intrinsically encoded in axial subsets of the somites using Hox codes (Alvares et al., 2003; Kessel and Gruss, 1991). The full extent and progression of pre-MRF lineage restrictions induced by HD-factors in each presumptive muscle structure (Kassar-Duchossoy et al., 2005; Lozano-Velasco et al., 2011; Relaix et al., 2005; 2004; Schubert et al., 2001) remains to be determined, yet it remains likely that such restrictions predispose chromatin of muscle progenitor cells at each anatomical location in a slightly different way, so that MRF-entrainment (S. Liu et al., 2018) leads to different flavors of postmitotic muscle nuclei.
Expression of the Pitx2LacZ knock-in allele is observed in the vast majority of fetal muscle anlagen and adult muscles in both the head and trunk (Shih et al., 2007b; 2007a). Somitic Pitx2LacZ expression begins at E10.25 in Pax3+Myod1+ cells of the ventrolateral dermomyotome at forelimb levels. Onset in other somites progresses in both the rostral and caudal direction from this location, and is closely associated with close apposition of a non-somitic Pitx2+ cell population that appears to originate from an early body wall Pitx2 expression domain (Eng et al., 2012a; Shih et al., 2007a). Six hours later, Pitx2+Pax3+Myod1+ cells are also observed intermingled with the central myotome, where Myod1+ cells are known to secondarily colonize the primary myotome (Cinnamon et al., 1999; Gross et al., 2000a; Kahane et al., 1998; Kalcheim et al., 1999; L’honoré et al., 2007; Shih et al., 2007a; Smith et al., 1994).
Early Lbx1+Pax3+Myod1− limb muscle precursors (LMPs) delaminate from forelimb level somites at E9.25 and migrate into the limb. The dorsal and ventral muscle masses of E10.25 forelimbs consist largely of Pax3+Lbx1+; Myod1+ and Myog+ cells (Gross et al., 2000a; Shih et al., 2007a). At this time, Pitx2+ cells start to appear in the forelimb bud along the interior surface of the muscle masses at E10.25. They co-express Pax3 and Myod1, but not Pax7, and by E10.75 they are intermingled with muscle masses (Kioussi et al., 2002; Shih et al., 2007a). At E10.5, a few Pax7+ cells are observed in the forelimb, but by E11.5 the limb muscle masses contain Pitx2+ mixtures of Pax3+Pax7−, Pax3+Pax7+ and Pax3−Pax7+ cells (unpublished). The absence of Pitx2 from early Pax3+Pax7− LMPs and its presence later LMPs is consistent with the idea Pitx2 is expressed in the fetal muscle progenitor (FMP) lineage, but not in the preceding embryonic muscle progenitor (EMP) lineage (Messina and Cossu, 2009). This fetal muscle lineage derives from Pax3+Pax7− progenitors that activate Pax7 expression in the limb bud (Hutcheson et al., 2009; Lepper and Fan, 2010).
The FMP lineage alone appears to be responsive to ß-catenin as myofiber assembly proceeds in the limb muscle anlagen of the fetal period (Hutcheson et al., 2009). ß-catenin signaling increases expression of the Pitx2LacZ knock-in allele in Pitx2-expressing organs (including muscle), acts together with Pitx2 protein to remove deacetylases from Pitx2-bound, repressed promoters, and activates a serial recruitment of coactivators to Pitx2-bound promoters of growth control genes acting at early to mid G1 (Kioussi et al., 2002). Wnt signaling, acting through Pitx2 to boost proliferation (Clevers, 2002), has been demonstrated in chick somites (Abu-Elmagd et al., 2010). The ß-catenin responsiveness in “fetal” but not “embryonic” muscle cells is also consistent with the model that Pitx2 plays a role in former rather than the latter.
Four circumstances complicate the genetic analysis of Pitx2 function in nascent trunk muscle. First, null mutants show early phenotypes at many non-muscle locations and die between E12.5 and E14.5 (Gage et al., 1999; Kitamura et al., 1999; Lin et al., 1999; Lu et al., 1999), precluding evaluation of adult muscle phenotypes. Fetal null mutants appear to have the muscle anlagen in the nearly all of the correct places, but overall deformation of the fetus at this stage distorts the shape and relative position of most anlagen in a way that precludes detailed experimental comparisons (Campbell et al., 2012; Eng et al., 2012a; Lozano-Velasco et al., 2011), with two exceptions, the loss of branchial arch and extraocular musculature by improper specification cranial paraxial mesoderm (Schubert et al., 2018; Shih et al., 2007b) and the loss of body wall musculature that is associated with altered axial specification of lateral plate mesoderm (Eng et al., 2012a). Second, somitic cells initiate Pitx2 expression just at the time when they come near Pitx2+ cells of presumptive lateral plate mesoderm origin, making it uncertain if defects observed in mutants are cell-autonomous in dermomyotome, lateral plate mesoderm, or both. Third, the continued expression of Pitx2 throughout the myogenic progression makes it difficult to separate early and late Pitx2 functions. Fourth, the Pitx3 gene appears to be expressed in a pattern that very similar, but slightly delayed, to that of Pitx2. No apparent muscle phenotype was observed in Pitx3-null mutants, but ectopically upregulation of Pitx2 in E13.5 muscle anlagen, suggested that Pitx2 compensated for Pitx3-dependent defects that occurred prior to this time (L’honoré et al., 2007).
At forelimb levels Pitx3 is not expressed in the LPM-derived domain of Pitx2, and it is not expressed in the ventrolateral dermomyotome at E10.5, where Pitx2 expression begins in paraxial mesoderm derivatives (somites). A floxed Pitx2 allele has been reported (Ai et al., 2006), yet it appears that phenotypic analyses using trunk skeletal muscle drivers has only been reported when Pitx3 was also excised (L’honoré et al., 2014). Double conditional Pitx2/3 mutants excised either with MSD-CRE during early somite formation, or with Pax3Cre during dermomyotome formation, form variably reduced fetal muscle anlagen by E13.5 that express RNA for the commitment marker Myog. MSDCre excision in all somitic cells leads to robust apoptosis in thoracic somites at E12.5, whereas HSACre excision in the differentiated cells of those somites does not (L’honoré et al., 2014), demonstrating that Pitx2/3 plays an essential role early role prior to terminal differentiation.
Taken together, these observations suggest that onset of Pitx2 expression in cells of the ventrolateral dermomyotome at E10.5, restricts the lineage at that location, from one which is forming progenitors for “embryonic”-type, primary muscle fibers (EMFs), into one that will form progenitors for “fetal”-type, secondary muscle fibers (FMFs). Functional ablation of Pitx2 would therefore be expected to lead to a cellular neoteny, in which the embryonic muscle precursors (EMPs) in the ventrolateral dermomyotome at E10.5, failing to normally deploy Pitx2 and become fetal muscle precursors (FMPs), merely remain as supernumerary EMPs that eventually undergo apoptosis. The massive apoptosis observed in E12.5 somites in MSD-excised Pitx2/3 conditionals begins at E11.5 (L’honoré et al., 2014) and appears similar to massive apoptosis observed shortly after specification failure in the dorsal horn of Lbx1 mutants (Gross et al., 2000b). In timing and location it appears appropriate for languishing, supernumerary EMPs. However, double null mutants apparently still have limb muscle that can express Myog at E13.5 (L’honoré et al., 2014). These must arise from the LMPs that have arrived in the limb bud before onset of Pitx2 expression in the ventrolateral dermomyotome. Independent onset of Pitx2 in some of these cells is likely to restrict lineage in much the same way it does in the somite, from EMP to FMP.
Here we report the development of a new floxed allele at the Pitx2 locus and demonstrate that conditional loss of Pitx2 function in postmitotic nuclei of terminally differentiating myofibers of trunk skeletal muscle, driven by an MCKCre transgene (Brüning et al., 1998), leads to an adult phenotype. Two key determinants of skeletal muscle function, anatomical structure and energy state, were defective in Pitx2MCK muscle, indicating that Pitx2 plays a key role in maintaining muscle homeostasis by governing postmitotic gene expression.
2. Materials and methods
2.1. Generation and functional confirmation of a new conditional Pitx2 allele
A 1.8 kb PGK-neo (PGKN) cassette, flanked by FLP sites, was inserted between upstream (3.5kb SalI/NheI) and downstream (3kb KpnI/KpnI) flanking regions so that it was transcribed in the same direction as the locus. LoxP sites and a diagnostic Bam HI site were inserted around exon 5, which encodes the N-end of the homeodomain (Fig. 1A). Sal1 linearized targeting vector (25μg) was electroporated into 2×107 embryonic stem cells (R1), grown in DMEM supplemented with 15% fetal calf serum, 0.1mM ß-mercaptoethanol, 4mM glutamine and 103 U/ml rLIF (Chemicon, Tokyo, Japan). ES cells were plated onto NeoR embryonic fibroblasts, grown in the presence of 0.3 mg/ml G418 for two weeks, and 300 resistant clones isolated. Homologous recombinant clones were identified by Southern blot analyses (Fig 1B) and chromosome analyses were performed to ensure euploidy (data not shown). Three clones were microinjected into C57BL/6 blastocysts, and then transferred to pseudopregnant females. Mice homozygous for the floxed allele (Pitx2FL) bred true, and were crossed with mice heterozygous for the null lacZ knock-in allele Pitx2Z (Lin et al., 1999), to the generate the heterozygote Pitx2FL/Z mice that were crossed with mice bearing the MCKCre transgenic driver (Brüning et al., 1998).
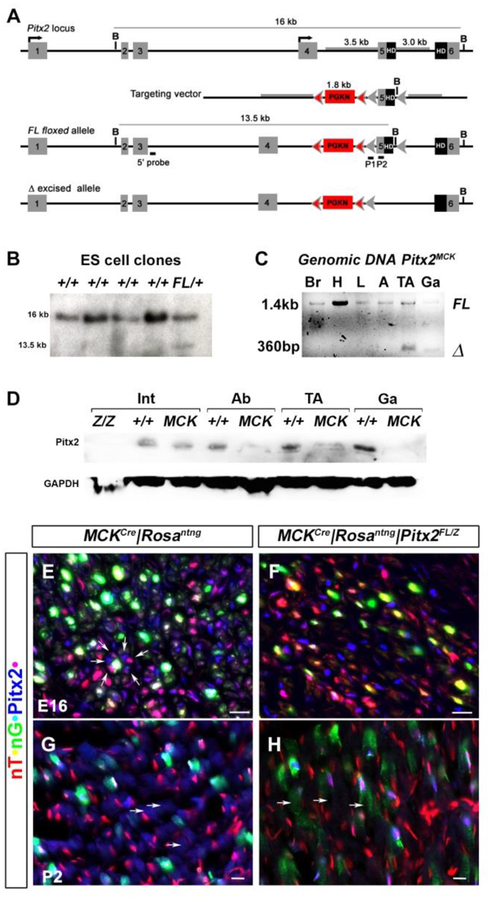
Figure 1. Generation of new conditional allele and functional confirmation of the Pitx2 muscle specific knockout mouse.
(A) Schematic representations of the endogenous Pitx2 locus, targeting vector, Pitx2FL allele, and the Cre-recombined allele. Exon 5, encoding the upstream end of the homeobox, is flanked by loxP sites (grey arrowheads), while the PGKNeo (PGKN) cassette is flanked by FRT sites (pink arrowheads). Tissue-specific recombination with a Cre driver results in the Pitx2Δ allele (bottom). (B) Homologous recombination in ES cells. Genomic DNA from ES cell clones was digested with BamH1 and probed with the 5’ probe shown above to identify homologous recombination with a13.5 kb fragment (C) Demonstration of Cre recombination of genomic DNA from tissues. The floxed (Pitx2FL) and deleted (Pitx2Δ) form of the allele are detected as 1400 and 360bp amplicons, respectively, in PCR amplifications of genomic DNA isolated from brain (Br), cardiac outflow track (H), liver (L), white adipose tissue (A), or muscles (TA, Ga) of P30 conditional null mice (Pitx2MCK). Excision was only detected tibialis anterior (TA) and gastrocnemius (Ga). (D) Western blot of total protein extracts from the organs of normal and conditional null mice at P30. Pitx2 protein levels were reduced in the abdominal body wall (Ab) and muscles (TA, Ga) of conditional mutants but were unaffected in intestine (I). Intestinal Pitx2 protein levels are reduced in complete (Pitx2Z/Z) but not conditional (Pitx2MCK) mutants. (E-H) Triple labelling immunohistochemistry of E16 hindlimb muscle (E, F) and P2 tibialis anterior (G, H) of control MCKcre|RosanT-nG (E, G) and mutant MCKcre|RosanT-nG |Pitx2FL/Z (F, H) mice. Onset of Cre excision is indicated by the onset of green signal in red nuclei (yellow). Loss of Pitx2 protein signal (blue) was observed in these cells.
Functional confirmation of excision in target tissues was demonstrated by PCR analysis of genomic DNA isolated from P30 tissues of Pitx2FL/+ mice (Fig 1C). External PCR primers were designed so that the 1.4 kb amplicon of the Pitx2FL allele becomes reduced to a 360 bp amplicon when excision generates the tissue specific Pitx2Δ allele. The smaller amplicon was observed with genomic DNA from two muscles (TA, Ga) but was not detected with genomic DNA from brain, cardiac outflow tract, liver, or white adipose tissue, indicating that MCKCre mediated tissue specific genome recombination is found preferentially in muscle, and specifically occurred in the tibialis anterior (TA) and gastrocnemius (Ga) muscles on which we perform much of our adult analysis.
The ability of the MCKCre driver to selectively reduce Pitx2 protein in muscle tissue was demonstrated by western blot (Fig 1D). Protein extracts from P30 wild type, conditional, and complete null mice were compared using an antibody derived against the C-terminus of the Pitx2 protein (Kioussi et al., 2002). Pitx2 protein levels were reduced below detection in extracts of the abdominal wall (Ab), or the two muscles (TA, Ga), but appeared unaffected in extracts of the intestine (I), where expression is clearly reduced in complete null (Pitx2Z/Z) extracts.
To monitor conditional Pitx2 excision in our system at the cellular level, we a made use of dual-color nuclear lineage tracing allele (RosanT-nG) designed and tested by the Schmidt group (Prigge et al., 2013). The constitutive, endogenous ROSA promoter drives expression of red nuclear Tomato (nT) protein until Cre excision makes it drive expression of green nuclear GFP (nG). Cryostat sections of fetal hindlimb muscle (E16), or the postnatal (P2) TA muscle, from either normal (MCKCre|RosanT-nG) or conditional null (MCKCre|RosanT-nG|Pitx2Z/FL) mice, were compared using triple labeling immunohistochemistry with anti-Pitx2, anti-GFP, and anti-Tomato antibodies (Fig 1E–H). Onset of excision by the MCKCre transgene driver, indicated by nT+nG+ nuclei, occurred in both normal and conditional muscles, at both E16 and P2. Pitx2 protein was readily observed in nT+nG+ nuclei (indicated by white) of normal, but not conditional null muscle. The excision observed at E16 indicates that the MCKCre driver is already active in differentiating cells of the fetal anlagen, consistent with the association of MCK expression with fetal muscle cells (Ferrari et al., 1997). Excision continued in the postnatal period (see results; Fig 3A,B).
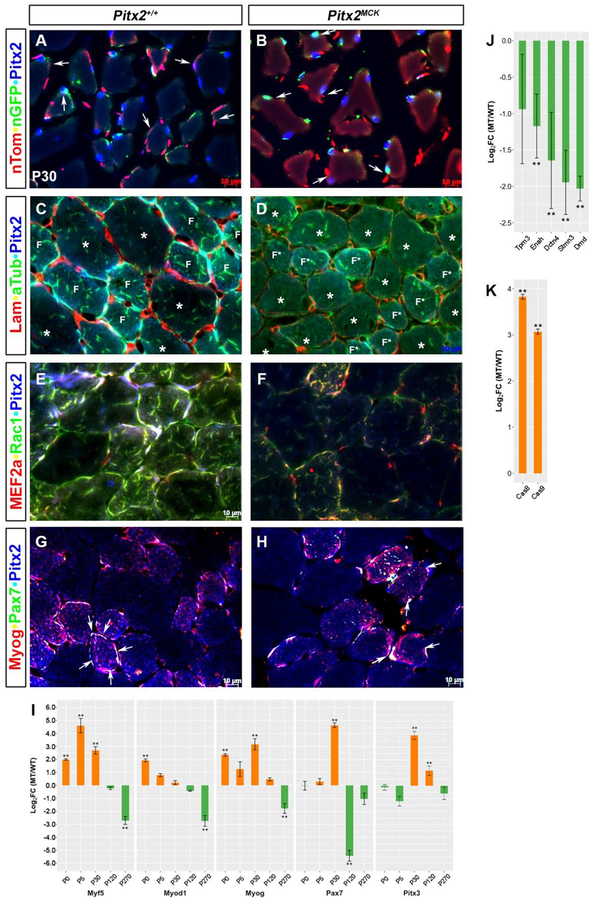
Figure 3. Cytoskeletal Defects Localized to Smaller Fibers in Pitx2MCK muscles.
Immunohistochemical and qPCR comparison of P30 TA muscle Triple labelling to monitor (A, B) docking, conditional allele excision, and Pitx2 expression in myofibers; (C, D) Laminin, alpha-tubulin, and Pitx2 proteins; (E, F) MEF2a, Rac1 and Pitx2 proteins; (G, H) Myog, Pax7, and Pitx2 proteins. Note that smaller (F, F*) and larger(*) fibers respond differently to mutation (see C, D). (I) qPCR analysis of total RNA from TA muscles at P0, P5, P30, P120, and P270. using primers for RNAs encoding muscle specific SSTFs. (J, K) qPCR analysis of P30 TA muscle RNA for cytoskeletal components Tpm3 (Tropomyosin), Enah (Enabled homolog), Dctn4 (Dynactin), Stmn3 (Stathmin), Dmd (Dystrophin), and apoptosis proteins (Caspases 8 and 9).
2.2. Mice
All animal experiments were performed in accordance to institutional and National Health and Medical Research Council guidelines. The experimental protocol was approved by the Institutional Animal Care and Use Committee at Oregon State University. Mice were housed in 12 light/12 dark cycle environment and fed the standard PicoLab Rodent Diet 20, 5053*, a managed formulation that delivers constant nutrition.
2.2. Quantitative RT-qPCR
Skeletal muscle biopsies (100mg) from wild type and Pitx2MCK mice were collected and homogenized with Bullet Blender (Next Advance, Averill Park, NY, USA). RNA from selected tissues was extracted (Qiagen, Helden, Germany) and transcribed to cDNA with High Capacity cDNA Reverse Transcription Kit (AB, Foster City, CA, USA). Both RNA and cDNA quality and quantity were tested by NanoDrop (Thermo Scientific, Waltham, MA, USA). 100 ng of cDNA, 1 pmol of primer and SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) were applied in each well in 7500 Real Time PCR System (AB). Each sample was tested in technical triplicates. Expression levels of 18S rRNA were used as internal quantity and quality control. Ct value of each well was calculated. Relative gene expression was calculated using the ddCt method. All primers are listed in Table S1.
2.3. Histology and Immunohistochemistry
Eosin-Hematoxylin (H&E) staining: Muscle biopsies were fixed in 3.7% paraformaldehyde for 30 min, dehydrated with ethanol and xylene and embedded in paraffin. Five micron thick serial cross-sections were mounted on lysine coated slides and incubated at 65°C overnight to removal of paraffin. Slides were rehydrated and incubated in hematoxylin solution (Fisher Scientific, Hampton, NH) for 90s, washed with running tap water, Blueing reagent (Fisher Scientific. Hampton, NH), Clarification reagent (EMD Millipore, Burlington, MA) and 95% ethanol for 1min for each step. Slides were stained with eosin for 30s, sequentially washed twice with 100% ethanol and xylene for 1 min for each step, and mounted with DPX (VWR, Radnor, PA).
Immunohistochemistry: Cytoskeleton staining: Muscle biopsies isolated, fixed in 4% paraformaldehyde and immunohistochemistry was performed as described in (Shih et al., 2007c). Sections were incubated with antibodies against GFP (rat anti-GFP, (Shih et al., 2007a)), RFP (1:3,000, Abcam, Cambridge, United Kingdom ab62341), alpha-Tubulin (, 1:100, Sigma, St. Louis, MO T6074), Laminin A (1:300, Sigma, St. Louis, MO L9393), MYOG (1:100, Santa Cruz Biotechnology, Santa Cruz CA SC576), PAX7 (1:50, DHSB Iowa City IA), Rac1 (1:200, Biosciences 610650), MEF2a (1:100, Santa Cruz Biotech SC10794;), mitochondria (1:50, EMD Millipore MAB1273), or PITX2 (anti-Pitx2a, (Kioussi et al., 2002)), and Cy™2-conjugated AffiniPure Donkey Anti-Mouse IgG (H+L) 715-225-151, Jackson ImmunoResearch, West Grove, PA) and Cy™5-conjugated AffiniPure Donkey Anti-Rabbit IgG (H+L) 711-175-152, Jackson ImmunoResearch). Myosin Heavy Chain (MHC) staining: Muscle biopsies were snap frozen in liquid nitrogen-chilled isopentane and embedded in OCT. Sections of 10 μ were rehydrated with PBS, permeabilized with PBST (0.5% Triton X-100 in PBS) for 30 min and blocked with BSA (5% in PBS) for 30 min at room temperature. Sections were incubated with 4° C overnight with MYH2 (1:100, DHSB SC-71) and MYH4 (1:100, DHSB BF-F3). After washing with 3 times in PBS, slides were incubated with secondary antibody cocktail: Alexa Fluor 594 IgG1 (y1) Goat anti-mouse (1:200, Invitrogen Carsbad, CA A21125) and Alexa Fluor 488 IgM Goat anti-mouse (1:200, Invitrogen Carsbad, CA A21042). After washing with 3 times in PBS, slides were mounted with glycerol (90%glycerol in 10× PBS). Slides were observed and photographed in Axiolmager Z1 Zeiss microscope.
2.4. Transmission Electron Microscopy
Skeletal muscle biopsies from control and mutant mouse TA were fixed immediately in 2.5% gluteraldehyde, 1% paraformaldehyde in 0.1M cacodylate buffer, pH7.4. Tissue was washed in 0.1M cacodylate buffer, post-fixed with 1% osmium tetraoxide, washed and dehydrated with serial acetone concentrations, followed by araldite and embedded in araldite. Ultrathin sections were observed in a FEI Titan 80–200 TEM electron microscope.
2.5. Myoblast Cultures
Wild type and mutant P30 TA myofibers were ioslated, rinsed with a HEPES-buffered salt solution (HBSS) containing 140mM NaCl, 5.4mM KCl, 0.2mM Na2HPO4 and 0.2mM KH2PO4, pH 7.2–7.4 and treated with 2% collagenase (Worthington Biochem, Lakewood, NJ) in serum-free DMEM/F12 for 90min as described by (Shefer and Yablonka-Reuveni, 2005). Healthy myofibers were selected and cultured in DMEM/F12 supplement with 20% FBS, sodium pyruvate, and antibiotics. After 7 days in culture, cells were trypsinized and platted again in presence of DMEM/F12 (Gibco, Carlsbad, CA), 15% horse serum (Sigma, St. Louis, MO) supplemented with sodium pyruvate and antibiotics at 37 °C with 5% CO2. For FoxO3 silencing cells were transfected with si-scramble (AM6011, Ambion, Austin, TX) and si-FoxO3 (4390771, Ambion) (Kannike et al., 2014). Three days after transfection, cells were harvested for RNA preparation and quantitation assays. For immunocytochemistry, cells were cultured on gelatin (1 g/L) coated coverslips, fixed with 4% paraformaldehyde, treated with methanol, and stained for antibodies against Myogenin and Mitochondria (1:100, MAB1273, EMD Millipore, Burlington, MA) as previously described by (Chang et al., 2013). Slides were observed and photographed in an Axiolmager Z1, Zeiss microscope.
2.6. Chromatin Immunoprecipitation
ChIP was performed as previously described by (Eng et al., 2012b).
2.7. Seahorse Assay
For in vitro mitochondrial respiration and glycolysis studies, 20,000 cells/well were grown onto 24-well plates (Seahorse Bioscience, Billerica, MA) under normal conditions for 24 hours. The assay was performed by Seahorse XF Analyzer (Seahorse Bioscience) according to user manual. Mitochondrial function parameters were evaluated by glycolysis, basal respiration, ATP production, proton leakage, and maximal and spare respiratory capacity measurements. The mitochondria analysis was measured by serial injections of oligomycin (1μM ATP, a synthase inhibitor, Seahorse Bioscience), carbonyl cyanide 4-trifluoromethoxy-phenylhydrazone (FCCP 1μM, a protonphore and un-coupler of oxidative phosphorylation in mitochondria, Seahorse Bioscience), and rotenone (1μM, electron transporter inhibitor in mitochondria complex I, Seahorse Bioscience). The cellular glycolysis and respiration profiles were measured by extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), respectively. The OCR and ECAR values were determined from 3 wells per genotype.
3. Results
3.1. Reduced myofiber size and number
A conditional null floxed allele (Pitx2FL) was generated and used to ask if Pitx2 plays a cell autonomous role in muscle development that cannot be compensated by Pitx3. Generation and functional confirmation of the new allele (Fig 1) is described in the methods section. A transgenic MCKCre driver allele that confers skeletal muscle and heart specific expression of Cre RNA in adults (Brüning et al., 1998), was used to generate the conditional Pitx2 mutants (Pitx2MCK) (see methods). This Kahn version of MCKCre driver has been used to analyze over 90 conditional alleles (Tg(Ckmm-cre)5Khn; MGI), mostly in adult muscle. It consists of a 6.5 kb MCK upstream genomic fragment (promoter and enhancer 1, noncoding exon 1, 3kb of intron 1 including the enhancer 2 region, 16bp exon 2) driving expression of nuclear Cre. The 6.5 kb fragment appears to drive transgene expression in all fibers, rather than selectively into fast fibers as some shorter fragments do (Tai et al., 2011)
Pitx2MCK mutant animals were viable without gross morphological defects for at least 3 months. Histological analyses were performed on the TA muscle at P5 and P30 (Fig2 A-D). Mutant TA myofibers were smaller and irregular at both P5 and P30, with half of the normal cross-sectional area (Fig 2E). Mutant myofibers also showed far more central nuclei, an indicator of pathological fiber repair. The ratio of nuclei per myofiber was reduced by 20% at P5 and 35% at P30 (Fig 2F). At P30, myofibers appear to be interlaced with more abundant connective tissue. These results clearly demonstrate that Pitx2 is essential for proper muscle formation in a way that cannot be compensated by the fully wild type Pitx3 locus. The defect is likely cell autonomous in the skeletal muscle lineage. However, Tg(Ckmm-cre)5Khn, used in conjunction with similar Pitx2 conditional alleles, also gives rise to defects in heart (Li et al., 2018) and extraocular muscle (Zhou et al., 2009). We have not yet investigated the heart and extraocular muscles with our allele, yet it stands to reason that all three defects could be either the source, recipient, or both, of secondary physiological consequences in these animals.
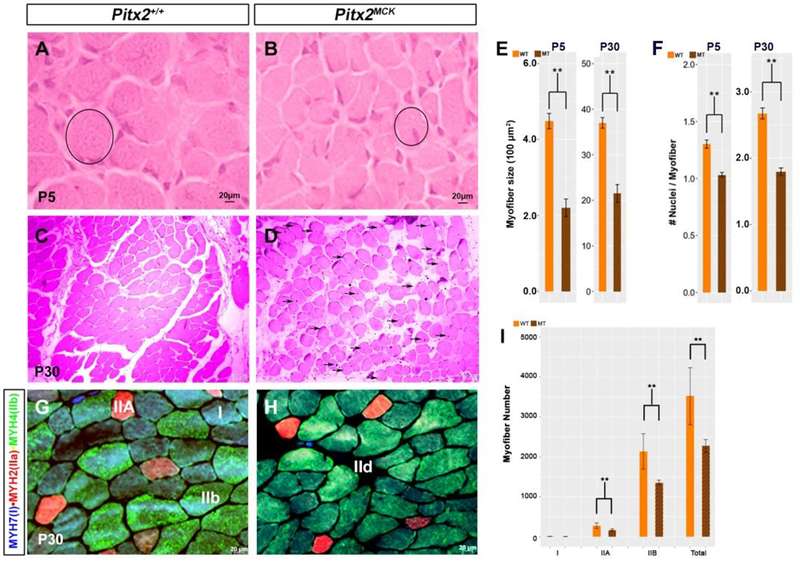
Figure 2. Atrophying muscle in Pitx2MCK mice.
(A-D) Hematoxylin-eosin staining for TA muscle at P5 (A, B) and P30 (C, D). Myofibers became smaller in diameter (circle), with increased inter-myofiber space, and showed centralized nuclei (arrows) (D). (E) Quantitative comparison of myofiber cross sectional area at P5 and P30. Myofibers of H&E stained cross sections were analyzed using ImageJ. Myofibers of ten sequential sections of the entire TA for both genotypes were counted (n=3). (F) Quantitative analysis of nuclei per myofiber cross section at P5 and P30. (G, H) Triple labelling immunohistochemistry of cross sectioned TA at P30 for MYH7, MYH2 and MYH4 to determine fiber types (I) Quantitative analysis of number of myofibers per area of TA at P30. Three sequential sections of the entire TA for both genotypes were used for the quantitation (n=3).
3.2. Altered myosin heavy chain profiles in P30 myofibers
Myofiber type distributions were compared using immunohistology on TA muscle from WT and Pitx2MCK mice (Fig 2G, H). TA normally contains a mix of myofiber types that can be revealed by myosin heavy chain (MYH) isoforms. MHY7 (blue) is expressed in type I slow twitch fibers of embryonic origin with oxidative activity and high mitochondrial density. MYH2 (red) is expressed in type IIA moderate twitch fibers of fetal or postnatal origin with oxidative/glycolytic activity and high mitochondria density. MYH4 (green) is expressed in type IIB faster twitch fibers of early postnatal origin with glycolytic activity and low mitochondria density. Significant changes in MHC expression profiles were observed in conditional mutants at P30. In control and mutant muscle, MYH7 and MYH4 antibodies co-label most fibers. However, the intensity of MYH4 staining declines while the intensity of MYH7 increases in mutants. The MYH2 was detected in far fewer fibers and shows similar intensity in normal and mutant muscle. The fraction of fibers rich in IIA (bright red) and IIB (bright green) was significantly reduced in mutants, by 37% and 40%, respectively (Fig 2I). This is very close to the 36% loss measured for all fiber types, and suggests that mutants fibers express fewer of the myosin heavy chains normally associated with fast, glycolytic and fetal muscle.
3.3. Reduced Pitx2 Protein and Dysmorphic Syncytia at E16 and P2
In muscle cell cultures, both Myog and endogenous MCK RNA expression begin shortly after the G1–G0 transition (Chamberlain et al., 1985). The expression of these two RNAs, as well as those of the myogenic late promoters, depend on Myod1 facilitating deacetylation at their chromatin contexts (Berkes and Tapscott, 2005; Tapscott, 2005). Rising Myog protein levels replace Myod1 at the same regulatory regions at the same time that HDACs are lost from them. This leads to the recruitment of an ATP-dependent chromatin remodeling enzyme at roughly the same time that MEF2D appears ((Ohkawa et al., 2006; Tai et al., 2011). Postnatal muscle biopsies (P2) from controls and Pitx2MCK mutants show green staining that marks the onset of Pitx2 function loss (Fig 1G, H). The green stain marks nuclei, but also appears to spill into the nascent syncytium next to those nuclei. A range of red/green co-labeling is observed in nuclei, with high red/low green, or low red/high green, representing cells that are earlier, or later, in the excision/detection process, respectively. Pitx2 protein was observed in nuclei along this red to green progression, in both control and mutant samples. Pitx2 protein in mutants was only observed early in the color progression (magenta to white), while Pitx2 protein was observed throughout the color progression in controls (magenta to white to cyan). The nascent syncytia of control samples appear to contain Pitx2 protein (dull blue glow; arrows) while those of mutant samples do not. Few, if any, green unassociated cells were observed, indicating that green excision events are always associated with cells docked onto the surface of a nascent syncytium. These results are consistent with the idea that Pitx2 expression in mutants is abolished by conditional excision just as cells are fusing onto the syncytium. It is therefore unlikely that proliferative, Pitx2+, FMPs would excise the conditional allele. Consequently, these would be expected to be unaffected, and could proliferate, until they fuse to the syncytium. Magenta (Pitx2+, high red) or blue (Pitx2+, low red) nuclei, that showed no hint of incipient green, were absent or rare at P2, suggesting that proliferative Pitx2+ cells were rare at this stage. Instead, many pure red (Pitx2−) nucleii were observed both inside and outside of the syncytium at P2, suggesting that some Pitx2− cells proliferate and enter the syncytium without activating MCK.
Endogenous MCK RNA in skeletal muscle is first detected at E13 and is maintained throughout adulthood. Its expression appears to be absent from “embryonic”, or primary, muscle cells, but is very strong in “fetal”, or secondary, muscle cells (Ferrari et al., 1997). These considerations suggested that the MCKCre driver also functions at fetal stages. E16 muscle sections from the hindlimb bear this out (Fig 1E, F). Both mutant and control sections show robust spots of green excision marker that appear far larger than the surrounding single nuclei. Green staining also labels syncytia in controls but not in mutants. The large green nuclei/syncytia of controls are surrounded by florets of magenta or blue nuclei (Fig 1E, arrows). Closer examination reveals that the nuclei in these florets all contained similar levels of blue (Pitx2), while varying drastically in their level of red (inconsistent with the idea that ROSA is expressed equivalently in all cells). Equivalent magenta and blue nuclei exist in mutants, but in more spindle-like associations with the green nuclei. The blue and magenta nuclei appear to be free of syncytia and could belong to proliferative cells. Taken together, these observations indicate that the population of presyncytial cells changes from a Pitx2+ population to a Pitx2− population between E16 and P2, consistent with the idea that adult muscle progenitors (AMPs) begin to dock onto fetal myofibers at this stage, and suggests how muscle fibers are recovered somewhat by P30 (Fig 2; Fig 3A, B).
3.4. Molecular Defects Restricted to Pitx2+ Fibers
One month after birth, the myofibers of mutants are finally recognizable as such (Fig 3A, B). The syncytia of myofibers appear dull blue in controls but dull red in mutants, and mutant myofibers appeared to have less fusion events around their circumference. An osmotic shock during immunohistochemistry allowed docked nuclei to be distinguished from those inside the syncytium. Docking cells of mutants and controls generally lack Pitx2 (pure red) and fuse with very little green signal. Unlike E16 and P2, the green signal is weak and first appears within the syncytium, rather than already being robust in the docking cell. The nuclei associated with these weak green signals within the syncytium are generally Pitx2+, indicating that MCK-driven excision has not worked on these nuclei. The ability of both control and mutant fibers to express Pitx2 within nuclei of the myotubes indicates that the Pitx2FL allele escaped excision in a Pitx2− nucleus that entered the syncytium without experiencing MCKCre, and that Pitx2 nuclear protein levels rose after fusion.
Laminin is a major component of the basal lamina that surrounds individual myofibers. It also surrounds the early embryonic myotome before early Myf5+ cells, delaminating from the dermomyotome, use integrins to organize onto it (Bajanca et al., 2006). Laminin staining in P30 muscle appeared to be localized to cells or basal laminae between the myofibers (Fig 3C, D). However, the intensity and extent of laminin staining was greatly reduced in mutants. The loss of laminin was surprising because the cells that normally express it are outside the syncytia, where conditional gene excision was not observed. Consequently, the observed lamination defect is likely a non-cell autonomous effect of defective myofibers, and suggests that mutant fibers do not appropriately stimulate or organize basal lamina deposition. Sub-sarcolemmal dystrophin interacts with extracellular laminin through the dystroglycan molecule. Mutant muscle expresses dystrophin RNA at fourfold lower levels (Fig 3J), indicating that defects already exist on the inside of the sarcolemma.
The microtubule cytoskeleton undergoes reorganization during muscle cell differentiation into a fixed array of paraxial microtubules that serves as a template for contractile sarcomere formation (Mogessie et al., 2015). Alpha-Tubulin (aTub) tethers nuclei and sarcolemma to the contractile sarcomere (Boudriau et al., 1996). Tubulin staining was observed on the inside of all myofibers in both mutants and controls (Fig 3C, D), yet it was far more intense in the smaller fibers (F, F*), which show intense tubulin expression in and around the Pitx2+ nuclei in their periphery. Both large (asterisk) and small fibers showed less tubulin, and Pitx2, staining in mutants, indicating defects in the assembly of contractile sarcomeres. A 3–4 fold reduction of RNAs encoding the microtubule interacting proteins dynactin (Dctn4) and stathmin (Stmn3) is consistent with this idea (Fig 3J).
Rac1 is an intracellular signal transducer that regulates actin cytoskeleton dynamics (Guo et al., 2006), and mediates fusion in developing muscle fibers (Demonbreun et al., 2015). In embryonic and fetal muscle, M-Cad and Neogenin, activate Rac1 as cells actively fuse. Loss of Rac1 function results in severe muscle phenotypes (Vasyutina et al., 2005). As with the tubulin, the Rac1 signal was higher in smaller Pitx2high syncytia than in larger Pitx2low ones, suggesting that fusion to the larger fibers may use a Rac1 independent method, such as perhaps Tmem8c or CDO. Rac1 staining was particularly high around the peripheral Pitx2+ nuclei of normal small fibers and was severely reduced both there and in the center of mutant small fibers. Rac1 appeared similar in larger fibers of controls and mutants. The selective loss of Rac1 signal from the periphery of smaller Pitx2high syncytia, indicates that Rac1 mediated fusion is abnormal, while the loss from the center of these fibers suggests that other cytoskeletal processes may also be defective. The RNAs encoding actin-binding Tpm3 and actin-regulating Enah declined two fold in mutants (Fig. 3J).
MEF2c, MEF2a, and MEF2d are MADS SSTFs that are expressed sequentially in somites and limbs (Subramanian and Nadal-Ginard, 1996). MEFs function after terminal differentiation to execute late muscle gene expression. MEF2a was selectively expressed in smaller Pitx2high syncytia and is severely reduced and disorganized in mutants (Fig 3E,F), indicating that muscle differentiation is defective in these fibers. MEF2a was not observed in the larger fibers, in either mutant or control, suggesting that another MEF was operational.
3.5. Postnatal Muscle Restoration
At P16 and P2, myofibers of mutants were severely disrupted, yet by P30 they appear relatively normal, even though there is a general reduction in fiber size in mutants. Caspase-dependent pathways are involved in apoptotic nuclear loss during atrophy in skeletal muscle. And caspase levels are 8–16 fold higher in mutant muscle (Fig 3K). The smaller, normally Pitx2+, fibers, become less frequent in mutants as larger Pitx2− fibers in become more frequent, suggesting that a rescue process functioning between E16 and P30. RNAs encoding the muscle determination (Myf5 or Myod1) and commitment (Myog) factors remain abnormally high between P0 and P30, at which time they become abnormally low (Fig 3I). Myf5 and Myog RNA levels remain abnormally high between P0 and P30, while the excessive Myod1 RNA becomes gradually reduced as Pax7 and Pitx3 RNA levels rise. Either satellite cells or Pitx3+Pitx2− AMPs could be fusing with mutant syncytia to rescue them. Pax7 marks muscle stem and satellite cells, while Myog marks committed and terminally differentiating myocytes. Both were readily observed in the in smaller Pitx2high syncytia, but were rare or absent in larger Pitx2low syncytia, in both mutants and controls. Pax7+ MyoG+ syncytia of mutants had fewer and less symmetrically arranged nuclei around them (arrows). The lack of extra Pax7+ cells on mutant large fibers indicates that they are unlikely to drive the rescue. Pitx3 RNA, which expressed at normal levels at P0 and P5, and could mark the AMPs, becomes 16 fold higher than controls at P30, when Myod1 levels are no longer excessive (Fig. 3I). Taken together, the data strongly that Pitx3+Pitx2− AMPs rescue muscle by a Myod1-independent mechanism that does not involve MCK upregulation at fusion. This allows the Pitx2FL allele to enter the syncytium without being inactivated by Cre so that it can be upregulated inside.
3.6. Defects in Ultrastructure and Metabolism
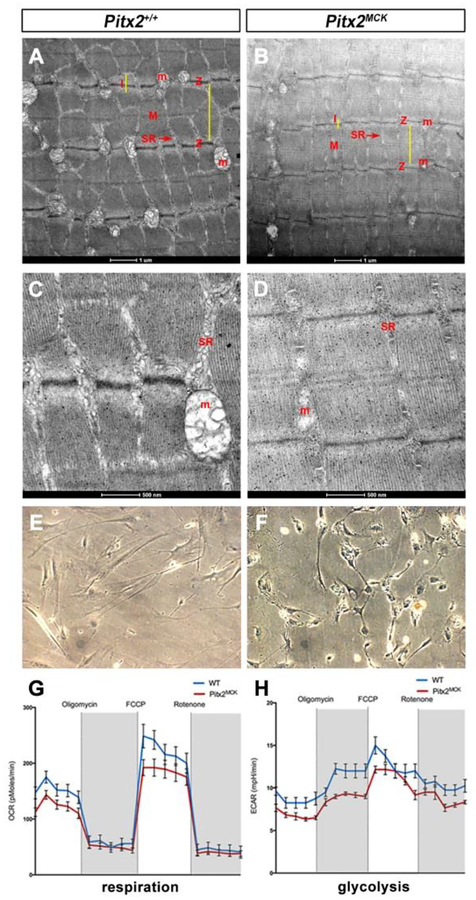
The ultrastructure of the contractile apparatus was compared in P30 biopsies (Fig 4 A–D). Mutant sarcomeres appeared to form a contractile apparatus, but they were abnormal in a variety of features. The distances between Z-lines (anchoring points of actin filaments) were shorter, while the width of myofibrils was larger. The longitudinal striations of sarcomeres were less electron-dense. The lower electron density of the Z-lines themselves may be related to the widening of myofibrils. M-lines (overlay of actin/myosin) of controls appear as single electron dense lines in controls, but 3 lower density lines were observed in mutants. I-bands (thin filament zone) were narrower and less reticulated. Clearly, the contractile apparatus of mutants was abnormal. However, the most striking difference was the emaciated sarcoplasmic reticulum of mutants, in which mitochondria were either vestigial or absent (Fig 4A–D).
Figure 4. Mitochondrial Loss and Respiration Defects in Pitx2MCK Muscle.
(A, D) Transmission electron microscopy for TA at P30 indicated distorted and smaller sarcomeres. Z, I, and M bands show differences. Mitochondria (M) and sarcoplasmic reticulum (SR) are vestigial. (E-H) Seahorse assay on primary TA muscle cell cultures (E, F) from WT and Pitx2MCK mice (n=3). (G) OCR and (H) ECAR measure mitochondrial respiration and glycolysis, respectively.
Due to decreased number of mitochondria, 54% less mitochondria in mutants, the metabolic activity of TA was measured. The Seahorse assay was applied on P30 cultured TA myocytes (Fig. 4E, F), to determine both oxygen consumption rate (OCR), an indicator of mitochondrial respiration, and extracellular acidification rate (ECAR), which is an indicator of glycolysis (Fig. 4G, H). Mutant cells were shorter, more rounded and behaved differently. Mutant cells were viable up to 2 weeks in culture. Viability was tested by 18S RNA levels. Real-time measurements of OCR and ECAR indicated reduced respiration and glycolytic rate of mutant myocytes, suggesting reduced energy production in both cytoplasm (anaerobic respiration) and mitochondria (aerobic respiration).
2.4. Muscle-specific Loss of Pitx2 Activates FoxO3-mediated Mitophagy
Degradation of skeletal muscle protein during wasting is accomplished through the ubiquitin proteasome system (Tawa et al., 1997) and the autophagy pathways. In skeletal muscle, FoxO3 coordinates a variety of stress-response genes by controlling two major systems in protein breakdown, the ubiquitin-proteasome, and the autophagy-lysosome pathways (Mammucari et al., 2007; Milan et al., 2015). To determine if FoxO3 mediated autophagy in the Pitx2MCK muscle, gene expression profiling by RT-qPCR was performed in TA biopsies from wild type and mutant P30 mice. FoxO3 RNA expression increased 20-fold in mutants (Fig 5A) and RNA encoding the mitophagosome-associated proteins, Bnip3 (BCL2 Interacting Protein 3) and Parkin also increased significantly (Fig 5B),
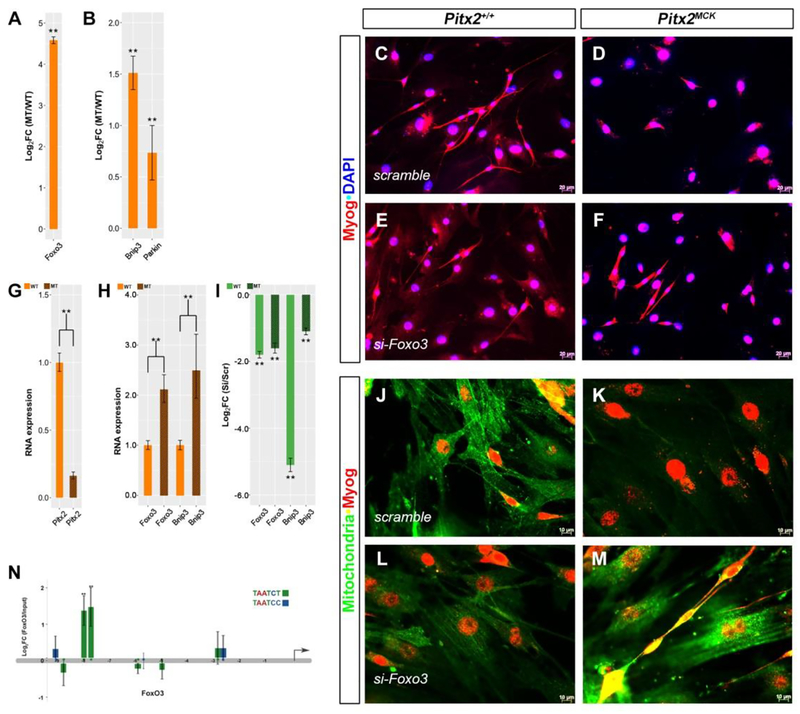
Figure 5. Pitx2 Suppresses FoxO3-dependent Mitophagy.
(A, B) RT-qPCR of total RNA from P30 TA muscle of WT and Pitx2MCK mice. (C-F) Cultured myocytes from P30 TA myofibers from normal (C, E, J, L) and mutant (D, F, K, M) were transfected with scramble siRNA (C, D, J, K) and Foxo3 siRNA (E, F, L, M). Cells were stained for Myog and DAPI (C-F) or Myog and Mitochondria (J-M). (G, H) qPCR of total RNA from scramble treated cell cultures, indicated reduced Pitx2 and increased Foxo3 and Bnip3 levels in mutant cultures (I) qPCR analysis comparing expression between scramble and si-FoxO3 treatments. Levels of FoxO3 and Bnip3 RNAs decline with si-FoxO3 treatment. (N) RT-qPCR ChIP for Pitx2 occupancy on 10 Kb upstream of the FoxO3 transcription start site. Pitx2 bicoid binding domains TAATCT (green) and TAATCC (blue) indicated Pitx2 occupancy (TAATCY; −9170, −8876, −8102, −7816, −6013, −5877, −5099, −2950, −2739). Pitx2 occupancy was greatest at positions −8102, −7816.
To test if repression of FoxO3 expression can reverse the mitochondrial loss observed in ultrastructure, muscle primary cultures from TA of P30 wild type and Pitx2MCK mutant mice were compared by transfection with FoxO3 siRNA (Fig 5). Cells were grown on a glass coverslip for up to nine days, allowing them to proliferate, fuse and become multinucleated fibers. Cultures from normal muscle expressed roughly five fold more Pitx2 RNA (Fig 5G), while cultures from mutants expressed significantly more FoxO3 and Bnip3 RNA (Fig 5H). Both FoxO3 and Bnip3 RNA levels declined significantly with si-FoxO3 treatment compared to scramble controls (Fig 5I). Fused fibers could readily be derived from normal muscle (Fig 5C, J), but little fusion was observed in cultures derived from Pitx2MCK muscle (Fig 5 D, K), despite their expression of Myog (Fig. 5D, F). Treatment of mutant cultures with si-FoxO3 appeared to rescue fusion (Fig. 5F, M), compared to scramble controls (Fig 5D, K), and lead to a dramatic increase in mitochondrial staining (Fig 5 M).
To determine whether Pitx2 could be directly involved in the transcriptional regulation of the FoxO3 gene, Pitx2 protein occupancy was tested using chromatin immunoprecipitation. Nine bicoid sites were identified in the 10kb fragment upstream of the FoxO3 transcriptional start site. Strong Pitx2 occupancy was detected at positions −8102 and −7816 (Fig. 5N). Taken together, the observations suggest that Pitx2 protein represses the FoxO3 locus, and thereby reduces mitophagy. Fibers that import Pitx2+ nuclei, or those that can turn on Pitx2 after nuclear import would likely shift the homeostatic balance of the fiber in favor of more abundant mitochondria.
4. Discussion
4.1. Pitx2-dependent gene regulation in progenitors and fibers
In previous reports we examined the effect of a Pitx2 null allele on GFP+ LMPs sorted from E12.5 limb buds of Lbx1GFP mice using expression microarrays (Campbell et al., 2012). These reports identified genetic targets of Pitx2 that encode proteins with functions in the cytoarchitecture such as cell adhesion-, actin-, and tubulin-related proteins. The relatively small fold changes in target gene RNA levels that were observed in these studies do not resemble the onset of mRNAs encoding muscle structural genes in muscle cell cultures. Recent lineage tracing studies indicate show that LMPs present in the forelimb bud at E10.5 are the last to enter the limb by migration from the somite. All subsequent muscle cells must therefore be derived from these cells by a system that is not yet fully understood. The onset of expression of Pitx2 in LMPs between E10.25 and E11 suggests that a Pitx2+ lineage of muscle cells is restricted from the Lbx1+ LMP lineage during this period. If Pitx2 onset represses Lbx1 expression during lineage restriction, as it does in the E10.5 ventrolateral dermomyotome, then the onset of Pitx2 should quell the Lbx1GFP signal, and Pitx2 expressing cells should not have be isolated in the sort mentioned above. However, the GFP protein itself can persist after the Lbx1 gene is turned off and could have led to the co-purification of the small pool of newly minted Pitx2+ cells from normal E12.5 limb buds. These would not exist in the mutant because the lineage restricting factor, Pitx2, was missing. The small fold changes in Pitx2-dependent RNA expression levels that we observed may merely reflect the small portion of Pitx2+ cells in the isolated pool. They may also indicate that target loci are becoming de-repressed and poised, waiting to be activated when myofibers assemble after commitment. While there clearly are effects of Pitx2 on E12.5 LMP precursor function, the role of Pitx2 on target genes expression after commitment has not yet been explored. The observation of a phenotype with the MCK-driver on the Pitx2 conditional allele indicates that such studies would yield functionally relevant results.
4.2. Tertiary Fibers Outgrow Secondary Fibers to Rescue of Pitx2MCK Muscle
The MCKCre driver is generally used as a universal adult muscle driver. Our studies indicate that the driver is on at least as early as E16 and that it appears to work in only the small subset of fibers at P30 (Fig 3). The onset GFP signal after Cre excision should match the onset of Pitx2Δ allele usage, and therefore mark the time when Pitx2 protein levels are expected to begin to decline without being replaced. Our observations indicate that GFP turns on in nuclei docked onto syncytia, and spills into them. Any residual Pitx2 protein at the time of docking is likely to be spilled into the syncytium, but the allele is likely already excised at this time. The observed phenotypes are therefore unlikely to result from effects on proliferating myoblasts, and must arise from reduced Pitx2 protein levels in myofibers. The selective effect of the MCKCre driver on the smaller fibers, suggests that the many of the conditional analyses reported on muscle should be re-evaluated as a fetal muscle specific knockouts rather than as adult, or differentiated muscle specific knockouts.
Spillage of Pitx2 protein into fibers at the time of fusion and commitment was not the only way Pitx2MCK muscle fibers could possibly retain some Pitx2 function. We observed that fibers, both large and small, can assimilate some Pitx2+ nuclei without activating the MCKCre driver (Fig. 3; nT-nG tracer stays red). These “red assimilations” are associated with the Pitx2+ nuclei within the syncytia in P30 mutant muscle (Fig 3A), but are scarce or absent at P2 (Fig 1). The small amount of green they were associated with was also within the syncytium rather than filling the docked cell. Individual fibers therefore dock and load different types of nuclei, at sites with or without MCK onset, suggesting that cells of different lineages contribute to individual fibers and that different modes of assimilation are used.
The conditional mutant animals appeared to grow normally into adults, and we did not identify a phenotype until P30 muscles were compared by histology (Fig 2) and EM (Fig 4). Surprisingly, subsequent examinations of E16 and P2 muscle (Fig 1) showed what appeared to be a more severe phenotype than the one initially observed at P30. Immunohistochemistry at P30 demonstrates that there are two types of fibers present (Fig 3). A smaller type, with high Pitx2 and MyoG levels in the syncytium, with Pax7+Pitx2+MyoG+ nuclei docked at its periphery, and which expresses high levels of Rac1, MEF2a, and alpha-tubulin around these nuclei, appears to be defective in mutants. A larger type, which lacks Pitx2 and the other markers and which appears unaffected by Pitx2 loss at P30, appears to be rescuing the animal’s muscle between P2 and P30.
The abnormal rise of Pitx3 RNA expression in mutant muscle during that interval (Fig 3I) may indicate a gene regulatory event within the smaller fibers that is trying to rescue them, or it could indicate that Pitx3 restricts its own lineage of adult muscle progenitors (AMPs) from the Pitx2+ FMP lineage early in development. No apparent muscle phenotype was observed in Pitx3 null mutants, but ectopic upregulation of Pitx2 in E13.5 muscle anlagen was described, and it was suggested that Pitx2 compensated for Pitx3 loss (L’honoré et al., 2007). The time of compensating upregulation of Pitx2 implies that the “Pitx3-dependent processes that need compensating for” have already happened at that early stage. If the Pitx2 and Pitx3 indeed do mark parallel lineages, then these lineages may have different modes of feeding nuclei into syncytia. If the red assimilations are AMPs and the green assimilations are FMPs, then both AMPs and FMPs are assimilated by the smaller fetal muscle fibers (FMFs), while only AMPs are assimilated by the larger adult muscle fibers (AMFs). AMFs therefore receive fewer or no defective, Pitx2-excised nuclei and gradually overgrow FMFs in mutants. If Pitx3 levels are indeed rising within the smaller fibers, then the source of the larger fibers may be the Twist2-dependent progenitor cells that contribute to adult muscle. Twist2-dependent progenitors contribute selectively to fast type IIb/X myofibers and would therefore be expected to produce fibers with low mitochondrial density (N. Liu et al., 2017).
4.3. Postmitotic Excision Effects
The onset of excision after commitment indicates that Pitx2 function in the progenitor pool was unaffected and that normal lineage restriction would occur. As cells enter transient amplification prior to fusion and excision their constellation of SSTFs should be entirely normal. It is only after commitment that the effect of Cre excision can begin. The EMFs should form normally, while altered expression of Pitx2-dependent genes in FMFs is expected to produce phenotypes that represent lost functions of FMFs. The shift to MHC expression profiles to those resembling slower fiber types at P30 is consistent with the loss of fetal character (Fig 2). Selective Pitx2-dependent expression of Rac1, alpha-tubulin, and MEF2a in FMFs indicates that these genes are fiber-specific targets involved in fusion, sarcomere assembly, and differentiated gene expression (Fig 3). The disruption of Laminin expression occurs in cells outside of the syncytia and indicates that mutant myofibers have disrupted signaling in their environment.
In muscle cell cultures, the respiration rate and mitochondrial cell number rise appreciably as myoblasts fuse into myotubes (Remels et al., 2010). Blocking mitochondrial biogenesis inhibits myoblast proliferation and differentiation (Rochard et al., 2000). The loss of mitochondria in EM studies (Fig 4) and the repression of the mitophagy pathway by Pitx2 (Fig 5), both suggest that one normal Pitx2 function is to increase mitochondria levels by preventing their destruction. The reduction in both glycolysis and respiration (Fig 4) suggests that defects in both major energy production pathways occur, rather than a switch from one to another.
5. Conclusions
Postmitotic loss of Pitx2 in myofibers causes defects in FMFs that are rescued by the expanded growth of another distinct fiber type, presumptively the AMFs, so that mutant animals appear normal for at least 3 months after birth. The MCKCre driven Pitx2 perturbation resulted in myofibers with dysfunctional architecture, gene expression, mitochondrial density, and energy metabolism. Pitx2 therefore plays a critical role in muscle gene expression after the G1–G0 transition.
Supplementary Material
Highlights.
Pitx2 regulates fetal myofiber gene expression after G1–G0 transition.
Pitx2 loss in muscle disrupts muscle anatomy and energy metabolism.
Pitx2 represses FoxO3-mediated skeletal muscle mitophagy.
Acknowledgments
The authors thank Greg Dressler for proving the pKSFlox-FLP-Neo vector, Theresa Sawyer for Transmitted Electron Microscopy imaging (NSF 1040588 Murdock Charitable Trust, Oregon Nanoscience and Microtechnologies Institute). The work was supported by the College of Pharmacy Oregon State University, NIH-NIAMS 1R01AR054406 (CK) and March of Dimes FY2005–829 (CK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
No competing interests declared.
REFERENCES
- Abu-Elmagd M, Robson L, Sweetman D, Hadley J, Francis-West P, Münsterberg A, 2010. Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev. Biol 337, 211–219. doi:10.1016/j.ydbio.2009.10.023 [DOI] [PubMed] [Google Scholar]
- Ai D, Liu W, Ma L, Dong F, Lu M-F, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, Black BL, Brown NA, Martin JF, 2006. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Developmental Biology 296, 437–449. doi:10.1016/j.ydbio.2006.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S, 2003. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. DEVCEL 5, 379–390. [DOI] [PubMed] [Google Scholar]
- Bajanca F, Luz M, Raymond K, Martins GG, Sonnenberg A, Tajbakhsh S, Buckingham M, Thorsteinsdóttir S, 2006. Integrin alpha6beta1-laminin interactions regulate early myotome formation in the mouse embryo. Development 133, 1635–1644. doi:10.1242/dev.02336 [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ, 2005. MyoD and the transcriptional control of myogenesis. Seminars in Cell and Developmental Biology 16, 585–595. doi:10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Boudriau S, Côté CH, Vincent M, Houle P, Tremblay RR, Rogers PA, 1996. Remodeling of the cytoskeletal lattice in denervated skeletal muscle. Muscle Nerve 19, 1383–1390. doi:10.1002/(SICI)1097-4598(199611)19:11<1383::AID-MUS2>3.0.CO;2-8 [DOI] [PubMed] [Google Scholar]
- Brüning JC, Michael MD, Winnay JN, Hayashi T, Hörsch D, Accili D, Goodyear LJ, Kahn CR, 1998. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Molecular Cell 2, 559–569. [DOI] [PubMed] [Google Scholar]
- Campbell AL, Shih H-P, Xu J, Gross MK, Kioussi C, 2012. Regulation of motility of myogenic cells in filling limb muscle anlagen by Pitx2. PLoS ONE 7, e35822. doi:10.1371/journal.pone.0035822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JS, Jaynes JB, Hauschka SD, 1985. Regulation of creatine kinase induction in differentiating mouse myoblasts. Mol Cell Biol 5, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-N, Feng M-J, Chen Y-L, Yuan R-H, Jeng Y-M, 2013. p15(PAF) is an Rb/E2F-regulated S-phase protein essential for DNA synthesis and cell cycle progression. PLoS ONE 8, e61196. doi:10.1371/journal.pone.0061196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnamon Y, Kahane N, Kalcheim C, 1999. Characterization of the early development of specific hypaxial muscles from the ventrolateral myotome. Development 126, 4305–4315. [DOI] [PubMed] [Google Scholar]
- Clevers H, 2002. Inflating cell numbers by Wnt. Molecular Cell 10, 1260–1261. [DOI] [PubMed] [Google Scholar]
- Demonbreun AR, Swanson KE, Rossi AE, Deveaux HK, Earley JU, Allen MV, Arya P, Bhattacharyya S, Band H, Pytel P, McNally EM, 2015. Eps 15 Homology Domain (EHD)-1 Remodels Transverse Tubules in Skeletal Muscle. PLoS ONE 10, e0136679. doi:10.1371/journal.pone.0136679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng D, Ma H-Y, Xu J, Shih H-P, Gross MK, Kioussi C, Kiouss C, 2012a. Loss of abdominal muscle in Pitx2 mutants associated with altered axial specification of lateral plate mesoderm. PLoS ONE 7, e42228. doi:10.1371/journal.pone.0042228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng D, Ma H-Y, Xu J, Shih H-P, Gross MK, Kioussi C, Kiouss C, 2012b. Loss of abdominal muscle in Pitx2 mutants associated with altered axial specification of lateral plate mesoderm. PLoS ONE 7, e42228. doi:10.1371/journal.pone.0042228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Molinari S, Melchionna R, Cusella-De Angelis MG, Battini R, De Angelis L, Kelly R, Cossu G, 1997. Absence of MEF2 binding to the A/T-rich element in the muscle creatine kinase (MCK) enhancer correlates with lack of early expression of the MCK gene in embryonic mammalian muscle. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research 8, 23–34. [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA, 1999. Dosage requirement of Pitx2 for development of multiple organs. Development 126, 4643–4651. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M, 2002. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron 34, 535–549. [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M, 2000a. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127, 413–424. [DOI] [PubMed] [Google Scholar]
- Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M, 2000b. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127, 413–424. [DOI] [PubMed] [Google Scholar]
- Guo F, Debidda M, Yang L, Williams DA, Zheng Y, 2006. Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J. Biol. Chem 281, 18652–18659. doi:10.1074/jbc.M603508200 [DOI] [PubMed] [Google Scholar]
- Harel I, Maezawa Y, Avraham R, Rinon A, Ma H-Y, Cross JW, Leviatan N, Hegesh J, Roy A, Jacob-Hirsch J, Rechavi G, Carvajal J, Tole S, Kioussi C, Quaggin S, Tzahor E, 2012. Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc. Natl. Acad. Sci. U.S.A 109, 18839–18844. doi:10.1073/pnas.1208690109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G, 2009. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes & Development 23, 997–1013. doi:10.1101/gad.1769009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahane N, Cinnamon Y, Kalcheim C, 1998. The cellular mechanism by which the dermomyotome contributes to the second wave of myotome development. Development 125, 4259–4271. [DOI] [PubMed] [Google Scholar]
- Kalcheim C, Cinnamon Y, Kahane N, 1999. Myotome formation: a multistage process. Cell Tissue Res 296, 161–173. [DOI] [PubMed] [Google Scholar]
- Kannike K, Sepp M, Zuccato C, Cattaneo E, Timmusk T, 2014. Forkhead transcription factor FOXO3a levels are increased in Huntington disease because of overactivated positive autofeedback loop. J. Biol. Chem 289, 32845–32857. doi:10.1074/jbc.M114.612424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon G, Harfe BD, Tabin CJ, 2003. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. DEVCEL 5, 937–944. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, Tajbakhsh S, 2005. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes & development 19, 1426–1431. doi:10.1101/gad.345505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessel M, Gruss P, 1991. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. CELL 67, 89–104. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG, 2002. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. CELL 111, 673–685. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Gross MK, 2008. How to build transcriptional network models of mammalian pattern formation. PLoS ONE 3, e2179. doi:10.1371/journal.pone.0002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Shih H-P, Loflin J, Gross MK, 2006. Prediction of active nodes in the transcriptional network of neural tube patterning. Proc. Natl. Acad. Sci. U.S.A 103, 18621–18626. doi:10.1073/pnas.0609055103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M, 1999. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 126, 5749–5758. [DOI] [PubMed] [Google Scholar]
- L’honoré A, Commère P-H, Ouimette J-F, Montarras D, Drouin J, Buckingham M, 2014. Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Developmental Cell 29, 392–405. doi:10.1016/j.devcel.2014.04.006 [DOI] [PubMed] [Google Scholar]
- L’honoré A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, Camper S, Drouin J, 2007. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Developmental Biology 307, 421–433. doi:10.1016/j.ydbio.2007.04.034 [DOI] [PubMed] [Google Scholar]
- Lepper C, Fan C-M, 2010. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424–436. doi:10.1002/dvg.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tao G, Hill MC, Zhang M, Morikawa Y, Martin JF, 2018. Pitx2 maintains mitochondrial function during regeneration to prevent myocardial fat deposition. Development dev 168609. doi:10.1242/dev.168609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG, 1999. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282. doi:10.1038/45803 [DOI] [PubMed] [Google Scholar]
- Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez-Ortiz E, Chen B, Shelton JM, Jaichander P, Bassel-Duby R, Olson EN, 2017. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol 19, 202–213. doi:10.1038/ncb3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen H, Ronquist S, Seaman L, Ceglia N, Meixner W, Chen P-Y, Higgins G, Baldi P, Smale S, Hero A, Muir LA, Rajapakse I, 2018. Genome Architecture Mediates Transcriptional Control of Human Myogenic Reprogramming. iScience 6, 232–246. doi:10.1016/j.isci.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Velasco E, Contreras A, Crist C, Hernández-Torres F, Franco D, Aránega AE, 2011. Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev. Biol 357, 165–178. doi:10.1016/j.ydbio.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF, 1999. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276–278. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M, 2007. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metabolism 6, 458–471. doi:10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Messina G, Cossu G, 2009. The origin of embryonic and fetal myoblasts: a role of Pax3 and Pax7. Genes & Development 23, 902–905. doi:10.1101/gad.1797009 [DOI] [PubMed] [Google Scholar]
- Milan G, Romanello V, Pescatore F, Armani A, Paik J-H, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, Blaauw B, DePinho RA, Sandri M, 2015. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat Comms 6, 6670. doi:10.1038/ncomms7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogessie B, Roth D, Rahil Z, Straube A, 2015. A novel isoform of MAP4 organises the paraxial microtubule array required for muscle cell differentiation. Elife 4, e05697. doi:10.7554/eLife.05697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CGA, Imbalzano AN, 2006. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. The EMBO journal 25, 490–501. doi:10.1038/sj.emboj.7600943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge JR, Wiley JA, Talago EA, Young EM, Johns LL, Kundert JA, Sonsteng KM, Halford WP, Capecchi MR, Schmidt EE, 2013. Nuclear double-fluorescent reporter for in vivo and ex vivo analyses of biological transitions in mouse nuclei. Mamm. Genome 24, 389–399. doi:10.1007/s00335-013-9469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M, 2005. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435, 948–953. doi:10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M, 2004. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes & development 18, 1088–1105. doi:10.1101/gad.301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remels AHV, Langen RCJ, Schrauwen P, Schaart G, Schols AMWJ, Gosker HR, 2010. Regulation of mitochondrial biogenesis during myogenesis. Mol. Cell. Endocrinol 315, 113–120. doi:10.1016/j.mce.2009.09.029 [DOI] [PubMed] [Google Scholar]
- Rochard P, Rodier A, Casas F, Cassar-Malek I, Marchal-Victorion S, Daury L, Wrutniak C, Cabello G, 2000. Mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression and activity of myogenic factors. J. Biol. Chem 275, 2733–2744. [DOI] [PubMed] [Google Scholar]
- Schubert FR, Singh AJ, Afoyalan O, Kioussi C, Dietrich S, 2018. To roll the eyes and snap a bite - function, development and evolution of craniofacial muscles. Seminars in Cell and Developmental Biology doi:10.1016/j.semcdb.2017.12.013 [DOI] [PubMed] [Google Scholar]
- Schubert FR, Tremblay P, Mansouri A, Faisst AM, Kammandel B, Lumsden A, Gruss P, Dietrich S, 2001. Early mesodermal phenotypes in splotch suggest a role for Pax3 in the formation of epithelial somites. Dev. Dyn 222, 506–521. doi:10.1002/dvdy.1211 [DOI] [PubMed] [Google Scholar]
- Shefer G, Yablonka-Reuveni Z, 2005. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol. Biol 290, 281–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H-P, Gross MK, Kioussi C, 2007a. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr. Patterns 7, 441–451. doi:10.1016/j.modgep.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Shih H-P, Gross MK, Kioussi C, 2007b. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc. Natl. Acad. Sci. U.S.A 104, 5907–5912. doi:10.1073/pnas.0701122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H-P, Gross MK, Kioussi C, 2007c. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr. Patterns 7, 441–451. doi:10.1016/j.modgep.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Smith TH, Kachinsky AM, Miller JB, 1994. Somite subdomains, muscle cell origins, and the four muscle regulatory factor proteins. J Cell Biol 127, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao C-A, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J, 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. doi:10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Subramanian SV, Nadal-Ginard B, 1996. Early expression of the different isoforms of the myocyte enhancer factor-2 (MEF2) protein in myogenic as well as non-myogenic cell lineages during mouse embryogenesis. Mechanisms of development 57, 103–112. [DOI] [PubMed] [Google Scholar]
- Tai PW, Fisher-Aylor KI, Himeda CL, Smith CL, Mackenzie AP, Helterline DL, Angello JC, Welikson RE, Wold BJ, Hauschka SD, 2011. Differentiation and fiber type-specific activity of a muscle creatine kinase intronic enhancer. Skeletal muscle 1, 25. doi:10.1186/2044-5040-1-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695. doi:10.1242/dev.01874 [DOI] [PubMed] [Google Scholar]
- Tawa NE, Odessey R, Goldberg AL, 1997. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J. Clin. Invest 100, 197–203. doi:10.1172/JCI119513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C, 2005. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes & Development 19, 2187–2198. doi:10.1101/gad.346205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cheng G, Dieter L, Hjalt TA, Andrade FH, Stahl JS, Kaminski HJ, 2009. An altered phenotype in a conditional knockout of Pitx2 in extraocular muscle. Invest. Ophthalmol. Vis. Sci 50, 4531–4541. doi:10.1167/iovs.08-2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.