Abstract
Species-specific genomic imprinting is an epigenetic phenomenon leading to parent-of-origin-specific differential expression of maternally and paternally inherited alleles. To date, no studies of imprinting have been reported in rapeseed, a tetraploid species. Here, we analysed global patterns of allelic gene expression in developing rapeseed endosperms from reciprocal crosses between inbred lines YN171 and 93275. A total of 183 imprinted genes, consisting of 167 maternal expressed genes (MEGs) and 16 paternal expressed genes (PEGs), were identified from 14,394 genes found to harbour diagnostic SNPs between the parental lines. Some imprinted genes were validated in different endosperm stages and other parental combinations by RT-PCR analysis. A clear clustering of imprinted genes throughout the rapeseed genome was identified, which was different from most other plants. Methylation analysis of 104 out of the 183 imprinted genes showed that 11 genes (7 MEGs and 4 PEGs) harboured differentially methylated regions (DMRs). Unexpectedly, only 1 MEG out of these 11 genes had a DMR that exhibited high CG methylation rate in paternal allele and had big difference between parent alleles. These results extend our understanding of gene imprinting in plants and provide potential avenues for further research in imprinted genes.
Keywords: rapeseed, endosperm, RNA sequencing, imprinting, allele-specific, DNA methylation
1. Introduction
Genomic imprinting is an epigenetic regulatory phenomenon in which there is allele-biased expression of certain genes depending on the parent of origin.1 Imprinted gene expression plays a role in fetal growth regulation and postnatal behaviour in mammals, formation of viable seeds and the inhibition of interspecies hybridization in plants.2–5
In mammals, imprinting occurs in the placenta, embryo and adult tissues, while most imprinting in plants occurs in the endosperm.6–10 The seed of angiosperms comprises three major components, the seed coat, the embryo and the endosperm, which supports embryogenesis.11 There is evidence for cross-regulation between seed components. Seed growth and development require communication and coordination of distinct genetic programs that govern the development of each seed component. Among them, the endosperm plays a central role. The endosperm controls seed growth and allows transfer of maternal nutrients to the embryo. In certain species, including cereals, the endosperm stores reserves in the form of starch, proteins and lipids. In Arabidopsis thaliana and rapeseed (dicot), the endosperm is transient and is consumed by the embryo, which stores the seed reserves.12 Epigenetic regulation and imprinting are known to be vital for proper endosperm development and seed viability because mutations in the components of these regulatory circuits produce unviable seeds.13–17 Although imprinting was first discovered in maize (Zea mays) nearly four decades ago,18 studies of the phenomenon in plants have lagged behind those in animals for some time. Only a few of imprinted genes have been discovered in plants based on parent-of-origin–specific phenotypes associated with mutant alleles.7,8,13,15,19,20 The appreciation of the importance of epigenetic mechanisms in fundamental biological processes has increased recently. With the advent of high-throughput mRNA sequencing (RNA-seq) technologies, genome-wide analysis of parental allele expression has become possible and led to the identification of hundreds of candidate imprinted genes in a number of plant species, including the model plant A. thaliana and several important crops, such as rice, maize, castor bean and sorghum, although the consequences of the imprinting remain largely unknown.21–27
The molecular mechanisms underlying genomic imprinting generally involve epigenetic factors, including DNA methylation, histone modification, long non-coding RNA and sRNA.28 DNA methylation is a classic epigenetic mechanism which is associated with silencing of genes and transposable elements but can also promote gene activity.29–32 It has therefore been considered a major regulator of gene imprinting. Some imprinted genes, such as MEA,13,16FWA,33FIS234 and MPC,35 have been shown to be regulated by DNA methylation or demethylation in Arabidopsis. In recent years, genome-wide allele-specific DNA methylation maps together with the associated imprinted genes have been produced for seeds of Arabidopsis,36–38 rice39,40 and maize.25,41 Many but not all imprinted genes within differentially methylated regions (DMRs) show hypomethylation in maternal alleles and hypermethylation in paternal alleles, in which the hypomethylation of maternal alleles of imprinted genes may be mediated by DEMETER-LIKE activity across species.23,37,42,43 However, there is a great deal of variability between different plant species in terms of the imprinted genes, the proportion of imprinted genes controlled by these epigenetic marks and the role of the epigenetic marks for maternal and paternal alleles.26,27,44 Imprinted genes therefore need to be investigated separately in each species in order to decipher their epigenetic underpinnings.
Rapeseed (Brassica napus L.), like Arabidopsis, belongs to the family Cruciferae. As the third largest source of vegetable oil globally, rapeseed is an important crop plant. Clarification of the genetic regulation involved in endosperm development would help to understand storage accumulation pattern in seed filling. Studies of imprinted genes in the rapeseed endosperm are therefore necessary. In addition, the seed is relatively large compared to Arabidopsis, making the rapeseed endosperm easy to be separated from the seed coat and embryo. Rapeseed thus provides an excellent system for identifying imprinted loci and characterizing parent-of-origin effects of imprinted genes in endosperm development. However, rapeseed is a tetraploid (4×) species derived from B. rapa (AA) and B. oleracea (CC), both of which underwent whole genome triplication. Although the rapeseed genome has been well annotated, the high degree of homology in the genomic sequences increases the difficulty of identifying imprinted genes in the endosperm.45–47
In this study, using the genome of Zhongshuang 11 as reference sequence, we identified rapeseed imprinted genes and analysed their cytosine methylation levels. First, we performed deep mRNA sequencing and identified SNPs between two rapeseed lines, YN171 and 93275. Using YN171 and 93275 as parents, mRNA-seq libraries of reciprocal hybrid endosperm were constructed. Finally, based on the differential expression of parental alleles in the reciprocal endosperms, 183 imprinted genes with parent-of-origin-specific expression were identified. Further genomic DNA methylation sequencing showed that the imprinted genes used for cytosine methylation analysis were not associated with DNA methylation. Our study not only provided an opportunity to understand the mechanisms driving gene imprinting and seed development in rapeseed but also shed some light on the evolution and biological significance of gene imprinting in plants in general.
2. Experimental procedures
2.1. Plant tissue collection
Rapeseed (B. napus L.) lines YN171, 93275, ZY036, 6F313 and 61616 introduced in our previous studies were grown in fields at Wuhan, China.48,49 Reciprocal inter-strain F1 hybrids, YN171 (♀) pollinated with 93275 (♂) (designated Y9) and 93275 (♀) pollinated with YN171 (♂) (designated 9Y) were produced by manual pollination in 2016. Other reciprocal inter-strain F1 hybrids including 93275 and ZY036, 93275 and 9T364, 93275 and 61616 were used for identification of imprinted genes.
Endosperms for RNA-seq and DNA-seq were sampled from immature seeds 30 days after pollination (DAP), from the reciprocal F1 hybrids, both 9Y and Y9, and their parental lines, 93275 and YN171, in two biological replicates. Endosperms for RT-PCR were collected at 15, 20, 25, 30 and 35 DAP separately. Each sample was collected from at least 10 individual plants. Other tissues, including leaf, stem, root, bud, pericarp, embryo and flower, were also sampled from YN171 line in two biological replicates. The endosperm was collected according to the method introduced by Li et al.50 To be specific, the seeds were kept on ice and punched rapidly at the top with a sharp steel needle to make a small hole. The prepared glass micropipette was immediately inserted into the hole and gently aspirated liquid endosperm from the embryo sac. Then the lipid endosperm was quickly transferred into EP tube on ice. All database related to the identification of rapeseed imprinted genes in this MS were uploaded to https://www.ncbi.nlm.nih.gov (BioProject ID: PRJNA470566). A table including detailed information of database was also supplied (Supplementary Table S1).
2.2. RNA-seq data generation and processing
Total RNA was extracted using a commercial kit (TaKaRa Mini BEST Plant RNA Extraction Kit). RNA quantity was measured using a NanoDrop (ND-1000, Nano-Drop). RNA-seq was performed in BGI. Specifically, libraries were constructed following the TruSeq RNA Sample Preparation Guide. The Agilent 2100 Bioanaylzer and ABI Step One Plus Real-Time PCR System (11 amplification cycles) were used for qualification and quantification of the sample libraries. Twenty million clean 150 bp paired-end reads from the parental lines and 34 million clean 150 bp paired-end reads from the hybrid lines were obtained, providing sufficient sequencing depth for the imprinting analysis. Low-quality reads were removed from the raw sequences and low-quality nucleotides were clipped from the beginning and the end of sequences using the trimmomatic-0.36 with Sliding Window 4: 15 and MINLEN: 130.51 The reads were mapped to the B. napus genome (http://www.genoscope.cns.fr/brassicanapus/) using TopHat version 2.1.1 (-i 10–read-edit-dist 10).52 To obtain gene expression level in tissues, reads that were not uniquely mapped were excluded. To determine whether the SNPs identified strictly referred to the high similarity of paralogous genes in the tetraploid rape, we compared the diversity between species and paralogous genes by calculating the ratio between the variant sites (mismatch and small Indel) and covered length at both cDNA and DNA level. We note that 94% genes with larger variation between paralogous than between species, and more than 82% genes with significant variation (Fisher exact test, P < 0.05) which has enough sensitivity and specificity in SNP detection. For each gene, the expression level was measured by Reads Per Kilobase exon Model per Million mapped reads (RPKM).
To obtain a more comprehensive list of SNPs between the parental orthologs, the genomes of the two parental samples were also re-sequenced. Reads from the genome samples were mapped to the genome of Zhongshuang 11 using BWA.53 Samtools (1.3.1) was used to exclude reads that were not uniquely mapped and pairs that were split across different chromosomes in all of the RNA and DNA analysis.54 In addition, duplicate reads were removed from the DNA analysis. Alignments from same parent were merged together for SNP identification.
2.3. Identification of SNPs between parental orthologs
A list of SNPs between YN171 (Y) and 93275 (9) was generated using the Samtools command Mpileup and the Bcftools program.54 First, a list of all positions containing potential variants and consistent sites with reference in exon regions for both YN171 and 93275 were generated. SNPs between YN171 and 93275 were filtered using the following criteria: (i) depth >=10 (at least 10 reads covering the SNP site), (ii) exclusion of heterozygous sites in each sample by retaining the sites with the alternative bases accounted for >95% of the reads.
2.4. Identification of imprinted genes
The ratio of maternal to paternal alleles in hybrid endosperm is theoretically 2: 1, so the observed ratio of maternal versus paternal allele counts for each gene in Y9 and 9Y was used to perform a χ2 test. Genes with a parental allele bias differing from 2: 1 (q < 0.05) in reciprocal hybrids were classified as potentially imprinted genes. Two replicates were considered separately and imprinted genes were identified as overlap among replicates. To identify the imprinted genes with greater confidence, more stringent standards were applied. In maternally biased endosperm genes (parental bias greater than 2: 1 at q < 0.05), criteria was set for at least 85% of the informative reads were maternal in both directions of the reciprocal cross. For paternally biased genes (parental bias less than 2: 1 at q < 0.05), criteria was set at least 50% of informative reads in both directions of the reciprocal cross were paternal. Genes that met these criteria in the reciprocal hybrids were defined as allele-specific imprinted genes.22 All statistical analysis was performed using R (version 3.2.1).55 Corrections for multiple testing were performed with Storey’s q-values of 0.05.56
2.5. DNA preparation and locus-specific sequencing
Genomic DNA was extracted from parental leaf tissues (YN171, 93275) using a Qiagen DNeasy Plant Mini Kit according to the manufacturer’s instructions. Primers used for SNP identification were designed manually (Supplementary Table S2). PCR was performed using TaqPlus polymerase (Qiagen). The PCR products were sequenced by Sanger sequencing.
2.6. Gene expression analysis by RT-PCR
Total RNA from endosperms of reciprocal F1 hybrids and their parental lines was used for identification of putative imprinted genes. Approximately 800 ng of total RNA was used for cDNA synthesis using a reverse transcription (RT) kit (TransGen Biotech, Beijing, China) following the manufacturer’s instructions. RT-PCR primers were designed manually (Supplementary Table S2). The PCR products were sequenced by Sanger sequencing.
2.7. Clustering analysis
For clustering analysis, 183 predicted imprinted genes were mapped to the rapeseed chromosomes (except for 4 genes, which could not be mapped to any chromosome). Arabidopsis imprinted genes were collected from the studies published in recent years.21,44,57,58 Sliding windows of 1 MB with step size 0.1 MB were used to compare the number of mapped reference genes and imprinted genes using a hypergeometric distribution. Consecutive significant windows (P < 0.05) were combined to form a cluster. The statistical analysis was performed using R (version 3.2.1).55
2.8. Gene MapMan analysis
MapMan analysis of the imprinted genes was performed with the Classification SuperViewer on the website (http://bar.utoronto.ca/welcome.htm) based on sequence homology with Arabidopsis genes. Categories with P < 0.001 were classified as enriched categories. PEGs and MEGs at each significant level were analysed separately.
2.9. Genomic DNA extraction and bisulphite sequencing
Genomic DNA was extracted from endosperm at 30 DAP by CTAB method and bisulphite sequenced by BGI. Specific for, at least 1 μg of RNase-treated DNA was used for library preparation. The DNA samples were prepared according to the following treatments: The DNA, fragmented to 100–300 by sonication (Covaris), was purified with MiniElute PCR Purification Kit (QIAGEN) and incubate the tube at 20 °C with End Repair Mix. The fragmented DNA was purified and added a single ‘A’ nucleotide to the 3′ ends of the blunt fragments. The purified genomic fragment was added with Methylated Adapter to the 5′ and 3′ ends of each strand and was purificated with QIAquick Gel Extraction kit (QIAGEN) and Then bisulphite treated with an EZ DNA Methylation GoldTM kit (Zymo Research). Finally, a narrow 350–400 bp size range after PCR with uracil-tolerant DNA polymerase for 10 cycles was purified with QIAquick Gel Extraction kit (QIAGEN). Libraries were subjected to QC on a bioanalyser before sequencing on an Illumina HiSeq2000 using 2 × 150 paired-end reads. Low-quality reads were removed and low quality nucleotides were clipped from the beginning and the end of sequences following the same procedure used for the RNA data. Processed reads were then mapped to both the Brassica napus reference genome and chloroplast genome using Bismark v0.16.3 with the following parameters: –non_directional -N 1 -L 32. The generated SAM files were then fed into the deduplicate_bismark subprogram for deduplication and the bismark_methylation_extractor subprogram to extract the methylations.59
The average methylation level of MEG, PEG and non-imp gene were calculated as following method: Gene body regions were separated into 60 bins, and extended 5-kb up- and downstream regions were separated into 50 bins. The average methylation levels for each bin was calculated by the ratio of the methylated Cs/Gs and total number of Cs/Gs and Ts/As from all sites with more than four reads in each bin.60
2.10. Analysis and identification of pDMRs
Reads overlapping SNP were classified as maternal, paternal using custom Perl programs, in which C > T SNPs for forward reads and G > A SNPs for reverse reads were ignored. The reads from forward and reverse strand were merged together. The metilene was used to identify CG, CHG and CHH pDMR.61 Regions containing more than five CGs/CHGs/CHHs supported with at least four reads were further analysed and significant regions were kept (P < 0.05).
Approximately 0.8–1.0 g of genomic DNA was used for bisulphite conversion using an EZ DNA Methylation GoldTM kit (Zymo Research) according to the manufacturer’s instructions. Primers were designed based on methylated sequences located on the regulatory regions and within rapeseed imprinted genes (Supplementary Table S2). Bisulphite-treated DNA was then amplified using HotStarTaq Plus polymerase (Qiagen). PCR products were cloned into a Peasy-Blunt Zero Cloning Vector (TransGen Biotech) for sequencing. At least 20 randomly chosen clones were sequenced for each gene, and the methylation levels were calculated as a percentage (%) per site for each CG type of cytosine residues.
3. Results
3.1. Genome-wide screening of putative imprinted genes in rapeseed endosperm
During rapeseed silique development, endosperm volume reached a maximum after about 30 DAP and then gradually decreased during seed maturation (Supplementary Fig. S1a). Therefore the developing endosperm tissues (30 DAP) from reciprocal hybrid seeds, Y9 and 9Y, were collected for screening imprinted genes using the method described in Li et al. (Supplementary Figs S1b and S2).50 To discriminate the parental origin of allelic expression in hybrids, a deep high-throughput RNA sequencing of endosperm libraries constructed from two rapeseed parents, YN171 and 93275, was first conducted to discover SNPs between the two varieties, yielding a total of about 80 million 2 × 120–150 bp paired-end clean reads. Additionally, genomic sequence data consisting of about 75 million clean 2 x 120–150 bp paired-end reads from the parents, which covered about 80% of the rapeseed genome, were also used for SNP identification. Although there are multiple homeologs in rapeseed, only 12.18–12.95% reads are matched to multi-places. The multi-matched reads were excluded during gene matching analysis firstly. In order to assess the matching reliability of specific region, the edit distances for both the best sites and the second-best alignment sites for each non-redundant read covered SNPs were compared. In all the reads covering SNPs in hybrid samples, about 37% reads were matched only one location. In the remaining reads, about 85% reads with best alignment showed great differences (edit distance>=3) compared with the second-best alignment. Totally, over 90% reads harbouring diagnostic SNPs between two parental could be matched to the corresponding genes (Supplementary Fig. S3). Finally, 176,464 high-quality SNPs in 25,115 genes between YN171 and 93275, which accounted for about 25% of all the rapeseed genes, were recovered for identifying and measuring allelic expression (Fig. 1A). To validate the accuracy of SNP analysis, 30 genes were chosen for SNP identification. Our analysis revealed the expected SNPs in all these 30 genes, confirming that we could accurately assess allele-specific expression in reciprocal hybrid endosperms (Supplementary Fig. S4).
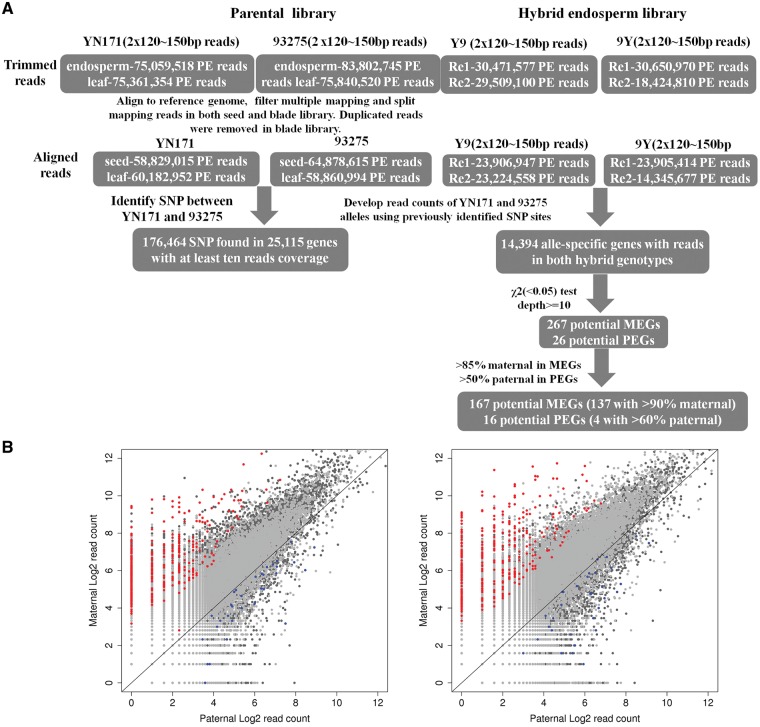
Figure 1.
Flow chart for identification on imprinted genes in rapeseed endosperm. (A) The RNA-seq data and genome sequencing data from YN171 and 93275 were used for identification of allele-specific SNPs The RNA-seq data from the hybrids were used for allele-specific expression analyses to identify imprinted genes. (B) Log 2 normalized read counts for all SNP loci for the endosperms The red dots represent the MEG loci sorted out with >85% read percentage bias and the blue dots represent the PEG loci sorted out with >50% read percentage bias in both reciprocal cross endosperms The black line denotes the 1: 1 ratios (maternal to paternal).
To screen for imprinted genes, about 18–30 million 2 × 120–150 bp paired-end clean reads from hybrid endosperm libraries with two replicates were aligned to reference genomic sequences for calling SNP coverage reads and characterizing the allelic expression of a given hybrid endosperm. (Fig. 1A). Based on saturation curve of rapeseed expression genes in every hybrid endosperm sample, sequence abundance is enough for imprinted gene analysis (Supplementary Fig. S5). The two replicates were considered separately and the imprinted genes were identified as overlap among two replicates. In total, about 14,394 genes covered by at least 10 reads could be assigned to a specific allele in both of the two reciprocal hybrid endosperms (Fig. 1A).
Analysis of gene expression showed that most of genes with SNP sites in the maternal and paternal alleles had a 2: 1 allelic-specific expression ratio in four endosperm samples, indicating unbiased expression (Fig. 1B). A small proportion of genes (∼3.8%) exhibited parent-of-origin differences in the expression of parental alleles in all samples. Among these, 267 genes exhibited maternally biased expression (MEG) and 26 genes displayed paternally biased expression (PEG) (Fig. 1A) in the reciprocal hybrids (χ2 test, q < 0.05). These differentially expressed genes were considered as putative imprinted loci. To obtain a high-confidence list of imprinted genes, stringent standards were applied (for MEGs, >85% of the reads supporting expression originated from the maternal allele; for PEGs, >50% of the reads supporting expression originated from the paternal allele). Using these criteria, 183 candidate imprinted genes were identified. Interestingly, most of the imprinted genes identified in our mRNA-seq analysis exhibited partial imprinting (preferential expression of one allele) rather than complete imprinting (strict monoallelic expression). Of these imprinted genes, 167 genes were maternally expressed and 16 genes were paternally expressed (Fig. 1B, Supplementary Tables S3 and S4). We also found that when the gene was imprinted in one subgenome, the counterpart in another genome was not always imprinted. In all imprinted genes, only seven genes in A subgenome could be found to also be imprinted in C subgenome. Another 30 homologous with imprinted genes which have SNP and expression are not imprinted. Additionally, as an allotetraploid, B. napus also exhibits expression bias between the homoeologous from An and Cn genomes. For example, in the endosperms of 93,275, we found 8,401 gene pairs showing expression bias including 60 imprinted genes (Supplementary Fig. S6; Table S3).
3.2. Validation of imprinted rapeseed genes
To determine whether the imprinted genes in rapeseed are conserved in other species, the rapeseed imprinted genes were compared with the known imprinted genes identified from other plants. The results showed there were 16, 2, 2, 1 and 1 rapeseed imprinted genes which were also imprinted in Arabidopsis, sorghum, castor bean, maize and rice, respectively (Table 1).
Table 1.
Overlaps between rapeseed imprinted genes and those of Arabidopsis, sorghum, castor bean, maize and rice.
| Query | Query length | Query coverage | Subject | Subject length | Subject covrage | Identity | Mapping length | Mismatch | Gap | Evalue | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis | BnA08g0333300 | 346 | 100 | AT1G09540 | 366 | 560 | 82.065 | 368 | 41 | 8 | 0 |
| BnC08g0884370 | 329 | 100 | AT1G09540 | 366 | 489 | 73.098 | 368 | 57 | 9 | 4.50E-175 | |
| BnA04g0174740 | 1258 | 92.61 | AT1G42470 | 1272 | 91.59 | 90.129 | 1165 | 111 | 2 | 0 | |
| BnA07g0301490 | 205 | 100 | AT1G77960 | 420 | 117 | 38.565 | 223 | 92 | 7 | 3.76E-31 | |
| BnUnng1014900 | 175 | 82.29 | AT2G01300 | 156 | 251 | 83.333 | 144 | 24 | 0 | 4.63E-87 | |
| BnA03g0108770 | 502 | 100 | AT2G32750 | 509 | 827 | 78.571 | 504 | 103 | 4 | 0 | |
| BnA04g0178370 | 748 | 100 | AT2G34880 | 806 | 973 | 64.942 | 773 | 221 | 16 | 0 | |
| BnA04g0178970 | 600 | 100 | AT2G34880 | 806 | 791 | 64.263 | 624 | 168 | 11 | 0 | |
| BnC04g0675620 | 736 | 100 | AT2G34880 | 806 | 985 | 65.876 | 759 | 217 | 15 | 0 | |
| BnC05g0699760 | 405 | 54.32 | AT2G35670 | 755 | 195 | 46.818 | 220 | 112 | 4 | 1.17E-55 | |
| BnC03g0551690 | 133 | 66.92 | AT3G06860 | 725 | 12.28 | 59.551 | 89 | 21 | 1 | 5.46E-24 | |
| BnA01g0031940 | 196 | 95.92 | AT3G20750 | 208 | 112 | 48.404 | 188 | 87 | 3 | 3.07E-31 | |
| BnA01g0031980 | 205 | 96.1 | AT3G20750 | 208 | 112 | 47.716 | 197 | 84 | 4 | 3.67E-31 | |
| BnA08g0309250 | 239 | 94.98 | AT4G12900 | 231 | 323 | 70.485 | 227 | 58 | 5 | 3.31E-113 | |
| BnA02g0056250 | 900 | 100 | AT5G21150 | 896 | 100 | 79.868 | 909 | 160 | 8 | 0 | |
| BnA06g0253460 | 406 | 99.75 | AT5G23340 | 405 | 100 | 91.852 | 405 | 33 | 0 | 0 | |
| Sorghum | BnA05g0188930 | 245 | 94.69 | Sobic.003G027000 | 270 | 85.56 | 61.538 | 234 | 85 | 3 | 8.65E-94 |
| BnA02g0058320 | 330 | 45.76 | Sobic.005G073500 | 481 | 29.31 | 25.166 | 151 | 103 | 3 | 1.62E-09 | |
| Castor bean | BnA06g0231270 | 503 | 93.64 | 29792.m000624 | 507 | 92.5 | 50.317 | 473 | 229 | 5 | 8.07E-174 |
| BnA07g0273570 | 456 | 98.9 | 29905.m000439 | 458 | 97.6 | 41.612 | 459 | 248 | 10 | 4.45E-108 | |
| Maize | BnA03g0137530 | 112 | 77.68 | GRMZM2G107839 | 157 | 57.32 | 52.222 | 90 | 40 | 1 | 1.21E-25 |
| Rice | BnC09g0906740 | 152 | 98.68 | LOC_Os04g39150 | 158 | 98.73 | 27.044 | 159 | 104 | 5 | 0.000000115 |
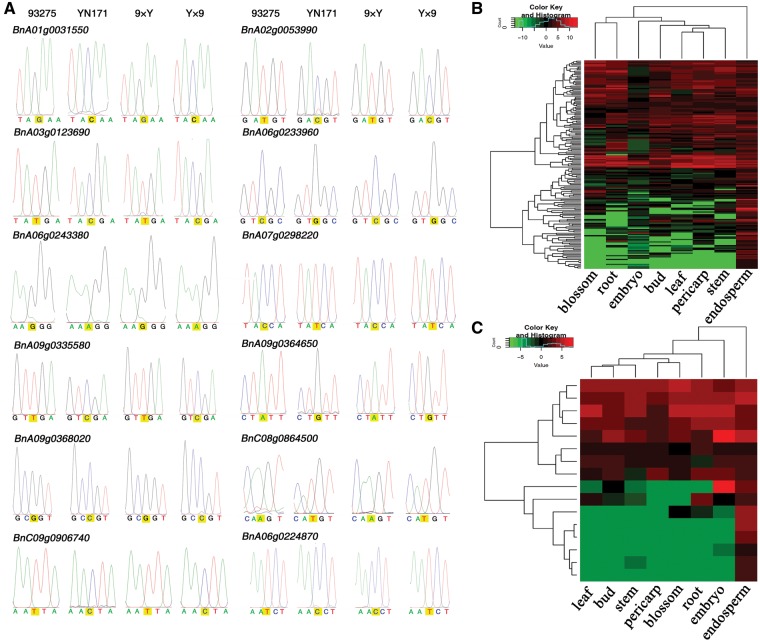
In addition to the imprinted genes which have been reported in other species, it is necessary to confirm the accuracy of some other imprinted genes identified in rapeseed. However, because of high homology between the An subgenome and Cn subgenome in rapeseed, it is very difficult to separate the imprinted genes from its homologs by designing specific primers for most of rapeseed genes. By analysing differences in the sequences and expression of the imprinted genes and their homologs, 11 MEGs and 1 PEGs with nearly complete imprinting were screened for confirmation of the putative imprinting by measuring their allelic-specific expression patterns. RT-PCR and sequencing analysis with the total RNA from 30 DAP hybrid endosperm showed that the expression patterns of these 12 tested imprinted genes were consistent with the mRNA-seq results (Fig. 2A).
Figure 2.
Analysis of imprinted genes in rapeseed endosperm. Allele-specific expression validation of the imprinted genes by RT-PCR sequencing (A); heat map analysis of MEGs (B) and PEGs (C) based on their expression in vegetative tissues and seeds. Each row represents a gene, and each column represents a tissue type. Tissue types are: blossom, root, embryo, bud, leaf, pericarp, stem and endosperm. Red or green indicate tissues in which a particular gene is highly expressed or repressed, respectively.
These 12 imprinted genes were also used to assess whether imprinting could be detected in other reciprocal hybrid combinations (9T364 and 93275; 61,616 and 93,275; zy036 and 93,275). The genes which had the same SNPs that were found between YN171 and 93275 were identified to be imprinted in the other reciprocal hybrid combinations (Supplementary Fig. S7), leading to the conclusion that the predicted imprinted genes were subjected to genomic imprinting and, furthermore, that mRNA-seq analysis was a reliable method to identify parent-of-origin-specific gene expression.
3.3. Characterization of imprinted genes in rapeseed
To further characterize the imprinted genes in rapeseed, the allele-specific expression pattern of 5 of those 16 imprinted genes were analysed at earlier and later stages of endosperm development based on RNA-seq analysis. The five imprinted genes maintained their imprinting pattern at the different developmental stages, at five different time points, 15, 20, 25, 30 and 35 DAP. This result suggested that imprinted genes maintained stable regulation of expression during the development of rapeseed (Supplementary Fig. S8).
Furthermore, previous studies have found that most imprinted genes in plants show specific or preferential expression in the endosperm.24 To test whether this is true in rapeseed, transcriptome sequencing data was used to analyse the expression pattern of the validated rapeseed imprinted genes in other tissues of YN171, including the leaf, stem, root, bud, pericarp, embryo and flower. Figure 2B and C shows the results of clustering analysis of MEGs and PEGs based on their expression in eight tissues. Unlike some earlier studies, only about 13% (22 of 167) of the maternally imprinted genes and about 37.5% (6/16) of the paternally imprinted genes were expressed specifically or preferentially in the endosperm in rapeseed, and in most cases the expression was elevated in the endosperm but not endosperm specific (Supplementary Table S5). No obvious difference was found between MEGs and PEGs in terms of expression patterns. These findings suggested that most of the rapeseed imprinted genes might not be functionally restricted to the endosperm.
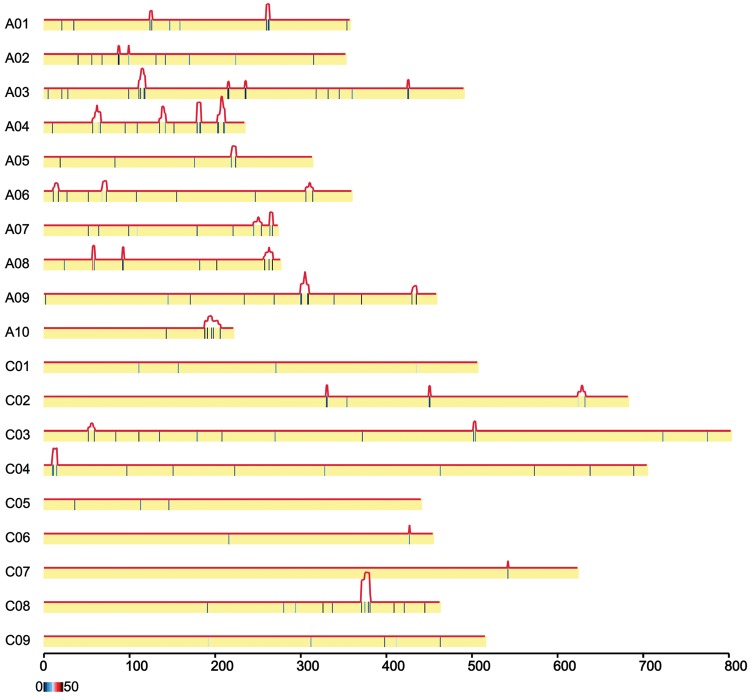
In mammals, imprinted genes often lie in clusters and expression is controlled by an imprinting control region.62 To further investigate whether the imprinted loci in rapeseed were located within clusters, the 183 imprinted genes were mapped onto the 19 rapeseed chromosomes. Sliding windows of 1 MB with step size 0.1 MB were used to compare the number of mapped reference genes and imprinted genes using a hypergeometric distribution. Consecutive significant windows (P < 0.05) were combined to form a cluster. Altogether, 34 clusters were identified to be located into the chromosome. A total of 89 imprinted genes accounting 48.6% (89/183) of the total imprinted genes were clustered suggesting that nearly a half of the imprinted genes may have regional regulation (Fig. 3). Each cluster contained two to six genes (Supplementary Table S6). We showed the representative genome browser shots of two large clusters, c12 and c34 (Supplementary Fig. S9). Detailed information including imprinted genes and non-imprinted genes in every cluster was showed in Supplementary Table S6. These clusters might represent genes controlled by common cis epigenetic regulatory elements. To compare the clustered imprinted regions between rapeseed and Arabidopsis, we analysed 593 Arabidopsis imprinted genes and found that 405 genes were located within 14 clusters. By collinearity analysis to the cluster regions between rapeseed and Arabidopsis, 16 rapeseed clusters were overlapped with 11 Arabidopsis clusters, which suggests that high consistency of imprinted regions exist in rapeseed and Arabidopsis (Supplementary Table S7).
Figure 3.
Chromosomal location of rapeseed imprinted genes. A total of 34 clusters (protrusions in red lines) containing 2–6 imprinted genes were identified.
3.4. Functional annotation analysis of the imprinted genes
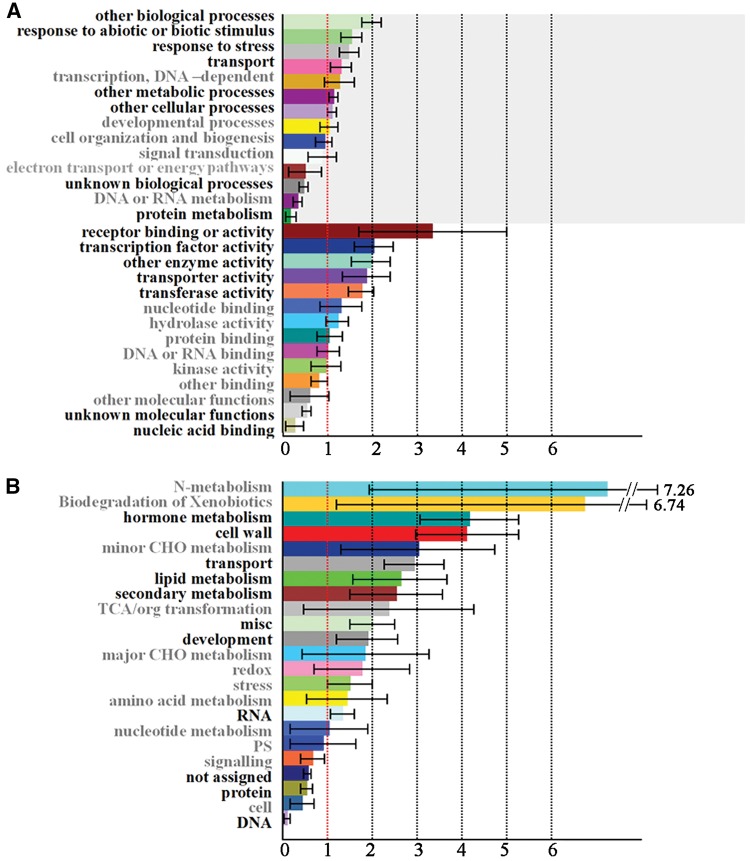
To annotate the function of the rapeseed imprinted genes, the protein sequences encoded by these genes were used to search the homologous Arabidopsis proteins by BLASTP. When the GO biological process annotations of the imprinted genes were assessed, the 183 rapeseed imprinted genes (q < 0.001) were allocated into several biological processes, including other biological processes, response to abiotic or biotic stimulus, response to stress, transport, other metabolic processes, other cellular processes and protein metabolism (Fig. 4A). The molecular functions were enriched in receptor binding or activity, transcription factor activity, other enzyme activity, transporter activity and transferase activity (q < 0.001). MapMan analysis showed that genes related to the hormone metabolism, cell wall, transport, lipid metabolism, secondary metabolism, misc and development were enriched (P < 0.001) (Fig. 4B). Additionally, over 50 imprinted genes were predicted to localize in nucleus and showed binding activities and regulation of transcription functions suggesting that many targets of rapeseed imprinting might have important regulatory roles. Further functional analyses of rapeseed imprinted genes will be necessary to shed light on the functional consequences on seed development.
Figure 4.
Functional classification analysis of the 183 rapeseed imprinted genes. (A) GO analysis. (B). MapMan analysis. The x-axis shows normed to Freq. in Arabidopsis set (± bootstrap StdDev). Terms in black on the y-axis: P-value < 0.001 and terms in grey: P-value > 0.001.
3.5. Association analysis between allele-differential cytosine methylation and imprinted gene expression in rapeseed
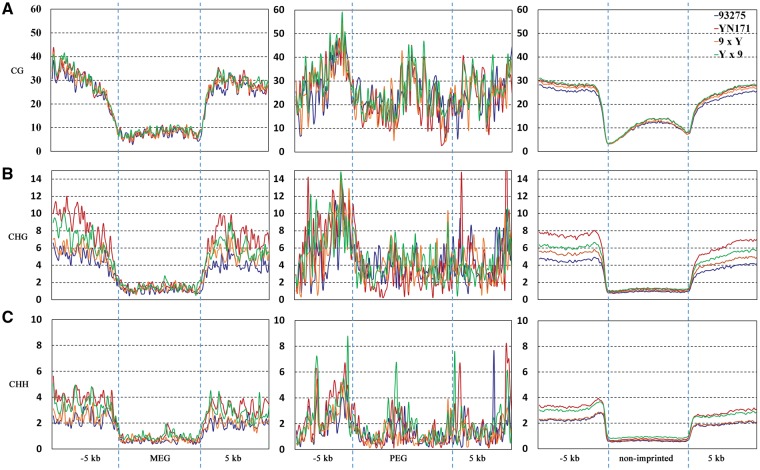
To analyse the association of allele-differential cytosine methylation and imprinted gene expression in rapeseed, bisulphite sequencing to the genomic DNA from endosperm at 30 DAP was performed to assess the patterns of DNA methylation of both maternal and paternal alleles of imprinted loci. The chloroplast genome was used to estimate the bisulphite conversion rate. We found the average methylation levels of CG, CHG and CHH were 1.39%, 0.625% and 0.51%, which showed the bisulphite conversion rate of genome sequence exceeded more than 98%. The methylation level from 5 kb upstream to 5 kb downstream of genes was analysed to investigate the DNA methylation pattern around CG, CHG and CHH residues. No significant difference was found in DNA methylation patterns of MEGs, PEGs and non-imprinted genes in CG methylation levels in the four endosperm samples originating from YN171, 93275 and the reciprocal hybrids. The overall CG methylation level was higher in PEGs than that in MEGs. However, the CG methylation level of PEGs was not significantly different from that of non-imprinted genes, although the overall levels of DNA methylation in the upstream regions of PEGs were slightly higher (Fig. 5A). At CHG and CHH residues, the methylation levels showed obvious difference in parents. CHG methylation levels showed similar in MEGs and PEGs and both of them exhibited slightly but statistically significant lower levels than non-imprinted genes, while CHH methylation levels had no significant difference between imprinted genes and non-imprinted genes (Fig. 5B and C). To eliminate the effects of non-additive gene expression in polyploidy rapeseed, we screened the homoeologous gene pairs showing significant differences in CG methylation and found 1294 gene pairs showing an inverse correlation between degree of methylation and gene expression. Among them, although 10 imprinted genes are found, most of them showed low methylation level in both of An and Cn homoeologs (Supplementary Fig. S10, Table S8).
Figure 5.
DNA methylation analysis in endosperms from the reciprocal hybrids between YN171 and 93275. (A) Average DNA methylation levels of MEGs, PEGs and non-imprinted genes (non-imp) for endosperm in CG context throughout the gene body and its 5-kb upstream and downstream regions. (B) Average DNA methylation levels of MEGs, PEGs and non-imprinted genes (non-imp) in CHG context throughout the gene body and its 5-kb upstream and downstream regions.
In order to detect allele-specific methylation of a gene, it must have sufficient read coverage of both the maternal and paternal genomic alleles in regions with at least one SNP between YN171 and 93275. A total of 104 genes (93 MEGs, 11 PEGs) met this criterion. Of these, seven MEGs and four PEGs showed coherent differential CG methylation between the two parental alleles in reciprocal hybrid endosperm DNAs (Supplementary Table S9). Of the 11 genes with differential CG methylation, 6 DMRs were located within the gene, 4 located at upstream region and 1 located at downstream. The identified DMRs ranged from 72 to 493 bp in length. Surprisingly, contrary to the studies in other plants, seven identified DMRs in rapeseed imprinted genes exhibited low CG methylation rates no matter in maternal alleles or in paternal alleles and had no big difference between parent alleles. Among the other four genes with higher CG methylation rates (BnC04g0675620, BnA09g0355640, BnA04g0165340 and BnA10g0421660), only BnC04g0675620 showed obvious differential methylation levels. To further confirm the reliability of methylation analysis, two genes including 1 MEG (BnA03g0127610) and 1 PEG (BnC08g0868840) were chose to investigate allele-specific methylation patterns. The results indicated that 2 genes showed difference in methylation states between parental alleles, which was coincident with the results from bisulphite sequencing data (Supplementary Fig. S11). Taken together, we found 1 gene (BnC04g0675620) showing high methylation levels in at least 104 imprinted genes. Additionally, lower methylation levels of CHG and CHH were found in some imprinted genes, which showed no great difference between parent alleles (Supplementary Table S9).
4. Discussion
As a result of the increasing focus on the importance of epigenetic mechanisms in fundamental biological processes together with the rapid advancement of RNA-seq technologies, studies of imprinting in diverse plant species, especially in several important crops, have intensified in recent years.25–27,63 At present, with the exception of Arabidopsis and Capsella rubella, there have been no studies on gene imprinting in the endosperms of Cruciferae species.64 Rapeseed is an important oil crop, and a genome-wide survey of imprinted genes in rapeseed endosperm may improve our understanding of storage accumulation and offer another model system for seed development. The first step toward understanding the biological functions of imprinting is to identify a large set of imprinted genes in a given species, although imprinted genes may differ between species and even between different strains within a species.57
In this study, we applied high-throughput mRNA-seq analysis with highly stringent selection criteria and identified 167 MEGs and 16 PEGs from hybrid rapeseed endosperms originating from YN171 and 93275. Although imprinted genes are well conserved in mammals, it is obvious that the overlap of imprinted genes in different plant species is rather limited.23,27 Even between Capsella rubella and Arabidopsis, which share a common ancestor ∼10–14 million years ago, only about 14% of the MEGs and 29% of the PEGs were found to overlap.64 In our study, although limited overlap of identified rapeseed imprinting genes with other plants supports less conserved imprinting in plants, among the 16 genes overlapped with Arabidopsis imprinted genes, BnC05g0699760, the homolog of Arabidopsis FIS2, was the most well-known imprinted gene in Arabidopsis.65FIS2 in the endosperm is initiated and established by DME-mediated active DNA demethylation in the central cell, while the paternal alleles remain methylated and silenced.34 Unfortunately, we could not confirm if the homolog of FIS2 on A05 chromosome is also imprinted because of SNP absence. BnA04g0178370, BnA04g0178970 and BnC04g0675620 are homologs of Arabidopsis JMJ15. JMJ15 is closely related to JMJ14, which is thought to demethylate trimethylated lysine 4 of histone H3 (H3K4me3) and is involved in DRM2-mediated maintenance of DNA methylation.21
Previous studies have not conclusively resolved whether imprinting of genes is conserved across the different stages of seed development. Xin et al.66 showed that MEGs and PEGs in maize exhibited distinct patterns of gene imprinting during endosperm development, with PEGs predominantly detected at 7 DAP and MEGs predominantly detected at 10 DAP. In castor bean, most of the 19 imprinted genes (17 MEGs and 2 PEGs) were found to maintain their imprinting pattern throughout endosperm development.26 In this study, the imprinting status of 5 rapeseed imprinted genes across the 5 developmental stages of endosperm from 15 to 35 DAP were highly similar, suggesting that imprinting occurs early and persists in the rapeseed endosperm.
In plants, the proportions of MEGs and PEGs showing preferential expression in endosperm were dramatically different in maize (61.5% MEGs and 76.5% PEGs) and sorghum (96.7% MEGs and 93.7% PEGs).25,27 However, our analysis showed that most of the rapeseed imprinted genes (66% MEGs and 67% PEGs) were also expressed in other tissues, rather than endosperm specific. The difference between our findings and previous studies might be due to the number of tissues analysed. Only four other tissues were analysed in maize and five in sorghum, while we used seven other tissues in rapeseed. This conjecture was further supported by our finding that about 30% of the imprinted genes were expressed in only two or three tissues. In castor bean, where measurements were made in a range of tissues, including the leaf, stem, root, male and female flower, embryo and endosperm, the expression of 67% of the MEGs and 17% of the PEGs was observed in other tissues.26 These results suggest that most imprinted genes also play important roles in the development of other tissues.
Gene expression within a cluster is often orchestrated by a common imprinting control region.67 Although physical clustering is a hallmark of imprinted genes in mammals, this does not appear to be a consistent characteristic in plants. The proportion of clustered genes in plants is highly variable. In Arabidopsis, 21 of 208 (10%) and 18 of 66 (27%) imprinted genes showed evidence of clustering.44,68 In maize, only 2 clusters including 4 genes were found in a set of 100 imprinted genes (4%),23 however, clear clustering of 74 of 217 (34%) imprinted genes and non-coding RNAs was observed from the same reciprocal hybrids between the two parental lines (B73 and Mo17).68 An analysis of 100 sorghum imprinted genes revealed that only 28 genes (28%) were grouped in 12 clusters. Unlike the results reported in other plant species, we found that 48.6% of the identified imprinted genes in rapeseed formed 34 distinct clusters, although the clustering criteria we used were similar to those in other studies. This suggested that clustering might play a role in imprinted gene regulation in rapeseed as it is in mammals.
In some other plant species, most of DMRs identified are uniformly hypomethylated in maternal alleles and hypermethylated in paternal alleles.26–68 In maize, the identified DMRs have an average of 14% CG methylation in maternal alleles and 94% in paternal alleles.68 In castor bean, the average is 9.6% in maternal alleles and 86% in paternal alleles.24 In this study, among the 104 genes (93 MEGs, 11 PEGs) that cytosine methylation levels could be analysed, 4 PEGs showed differential CG sites with the average CG methylation rates of over 30% in maternal alleles and about 3% in paternal alleles. Only one MEG showing differential methylation levels exhibited high CG methylation rates. Additionally, we also found over 5000 non-coding RNAs in 93275 and YN171. By analysing the locations and expression levels of the non-coding RNA, we found 1 RNA showing exonic overlap on the opposite strand with BnC03g0568040 gene and 14 RNAs locating near 9 imprinted genes (the distance is about 1∼40 kb) (Supplementary Table S10). However, all the RNAs nearly do not affect the expression of imprinted genes. Because DNA methylation is a classic epigenetic mechanism and has been considered a major regulator of gene imprinting, the methylation mechanisms of imprinting in rapeseed imprinted genes is worth being investigated further.
Authors’ contributions
J.L. and W.H. conceived the research plans; H.L. analysed the sequence data; S.F. performed the experiments; S.S., X.Z. and Z.H. provided technical assistance; J.L. and J.L. wrote the article.
Supplementary Material
Acknowledgements
This study was supported by the Ministry of Science and Technology of China (2016YFD0100500), the National Key Basic Research Program of China (2015CB150200) and National High Technology Research and Development Program of China (2013AA102602).
Conflict of interest
None declared.
References
- 1. Feil R., Berger F.. 2007, Convergent evolution of genomic imprinting in plants and mammals, Trends Genet., 23, 192–9. [DOI] [PubMed] [Google Scholar]
- 2. Tycko B., Morison I. M.. 2002, Physiological functions of imprinted genes, J. Cell. Physiol., 192, 245–58. [DOI] [PubMed] [Google Scholar]
- 3. Jiang H., Köhler C.. 2012, Evolution, function, and regulation of genomic imprinting in plant seed development, J. Exp. Bot., 63, 4713–22. [DOI] [PubMed] [Google Scholar]
- 4. Kradolfer D., Wolff P., Jiang H., Siretskiy A., Köhler C.. 2013, An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana, Dev. Cell., 26, 525–35. [DOI] [PubMed] [Google Scholar]
- 5. Kawashima T., Berger F.. 2014, Epigenetic reprogramming in plant sexual reproduction, Nat. Rev. Genet., 15, 613–24. [DOI] [PubMed] [Google Scholar]
- 6. Renfree M. B., Suzuki S., Kaneko-Ishino T.. 2012, The origin and evolution of genomic imprinting and viviparity in mammals, Philos. Trans. R. Soc. Biol. Sci., 368, 20120151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huh J. H., Bauer M. J., Hsieh T. F., Fischer R. L.. 2008, Cellular programming of plant gene imprinting, Cell, 132, 735–44. [DOI] [PubMed] [Google Scholar]
- 8. Jullien P. E., Berger F.. 2009, Gamete-specific epigenetic mechanisms shape genomic imprinting, Curr. Opin. Plant Biol., 12, 637–42. [DOI] [PubMed] [Google Scholar]
- 9. Gehring M. 2013, Genomic imprinting: insights from plants, Annu. Rev. Genet., 47, 187–208. [DOI] [PubMed] [Google Scholar]
- 10. Gehring M., Choi Y., Fischer R. L.. 2004, Imprinting and seed development, Plant Cell, 16, S203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berger F., Chaudhury A.. 2009, Parental memories shape seeds, Trends Plant Sci., 14, 550–6. [DOI] [PubMed] [Google Scholar]
- 12. Li J., Berger F.. 2012, Endosperm: food for humankind and fodder for scientific discoveries, New Phytol., 195, 290–305. [DOI] [PubMed] [Google Scholar]
- 13. Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B.. 1998, Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis, Science, 280, 446–50. [DOI] [PubMed] [Google Scholar]
- 14. Kinoshita T., Yadegari R., Harada J. J., Goldberg R. B., Fischer R. L.. 1999, Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm, Plant Cell, 11, 1945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A.. 2000, Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds, Proc. Natl. Acad. Sci. USA, 97, 10637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U.. 2003, The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1, Genes Dev., 17, 1540–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jullien P. E., Katz A., Oliva M., Ohad N., Berger F.. 2006, Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis, Curr. Biol., 16, 486–92. [DOI] [PubMed] [Google Scholar]
- 18. Kermicle J. L. 1970, Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission, Genetics, 66, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Springer N. M., Gutierrez-Marcos J.. 2009, Imprinting in maize, In: Bennetzen J.L., Hake S.C. (eds.), The Maize Handbook, Springer, New York, NY, pp. 429–40. [Google Scholar]
- 20. Raissig M. T., Baroux C., Grossniklaus U.. 2011, Regulation and flexibility of genomic imprinting during seed development, Plant Cell., 23, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsieh T. F., Shin J., Uzawa R.. 2011, Regulation of imprinted gene expression in Arabidopsis endosperm, Proc. Natl. Acad. Sci. USA, 108, 1755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo M., Taylor J. M., Spriggs A.. 2011, A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm, PLoS Genet., 7, e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Waters A. J., Makarevitch I., Eichten S. R., et al. 2011, Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm, Plant Cell, 23, 4221–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waters A. J., Bilinski P., Eichten S. R., et al. 2013, Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species, Proc. Natl. Acad. Sci. USA, 110, 19639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang M., Xie S. J., Dong X. M., et al. 2014, Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize, Genome Res., 24, 167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu W., Dai M. Y., Li F., Liu A. Z.. 2014, Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean, Nucleic Acids Res., 42, 6987–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang M. S., Li N., He W. N., Zhang H. K., Yang W., Liu B.. 2016, Genome-wide screen of genes imprinted in sorghum endosperm, and the roles of allelic differential cytosine methylation, Plant J., 85, 424–36. [DOI] [PubMed] [Google Scholar]
- 28. Rodrigues J. A., Zilberman D.. 2015, Evolution and function of genomic imprinting in plants, Gene Dev., 29, 1837–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makarevich G., Villar C. B. R., Erilova A., Köhler C.. 2008, Mechanism of PHERES1 imprinting in Arabidopsis, J. Cell Sci., 121, 906–12. [DOI] [PubMed] [Google Scholar]
- 30. Rigal M., Kevei Z., Pélissier T., Mathieu O.. 2012, DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns, EMBO J., 31, 2981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deng S. L., Chua N. H.. 2015, Inverted-repeat RNAs targeting FT intronic regions promote FT expression in Arabidopsis, Plant Cell Physiol., 56, 1667–78. [DOI] [PubMed] [Google Scholar]
- 32. Williams B. P., Pignatta D., Henikoff S., Gehring M.. 2015, Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat, PLoS Genet., 11, e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kinoshita T., Miura A., Choi Y., et al. 2004, One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation, Science, 303, 521–3. [DOI] [PubMed] [Google Scholar]
- 34. Jullien P. E., Kinoshita T., Ohad N., Bergera F.. 2006, Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting, Plant Cell, 18, 1360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tiwari S., Schulz R., Ikeda Y., et al. 2008, Maternally expressed PAB C-terminal, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins, Plant Cell, 20, 2387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gehring M., Bubb K. L., Henikoff S.. 2009, Extensive demethylation of repetitive elements during seed development underlies gene imprinting, Science, 324, 1447–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsieh T. F., Ibarra C. A., Silva P., et al. 2009, Genome-wide demethylation of Arabidopsis endosperm, Science, 324, 1451–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moreno-Romero J., Jiang H., Santos-González J., Köhler C.. 2016, Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm, EMBO J., 35, 1298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zemach A., Kim M. Y., Silva P., et al. 2010, Local DNA hypomethylation activates genes in rice endosperm, Proc. Natl. Acad. Sci. USA, 107, 18729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodrigues J. A., Ruan R., Nishimura T., et al. 2013, Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm, Proc. Natl. Acad. Sci. USA, 110, 7934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang P. F., Xia H., Zhang Y., et al. 2015, Genome-wide high-resolution mapping of DNA methylation identifies epigenetic variation across embryo and endosperm in maize (Zea may), BMC Genomics, 16, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gutiérrez-Marcos J. F., Costa L. M., Dal Prà M., Scholten S., Kranz E., Perez P., Dickinson H. G.. 2006, Epigenetic asymmetry of imprinted genes in plant gametes, Nat. Genet., 38, 876–8. [DOI] [PubMed] [Google Scholar]
- 43. Du M., Luo M., Zhang R., Finnegan E. J., Koltunow A. M.. 2014, Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites, Plant J., 79, 232–42. [DOI] [PubMed] [Google Scholar]
- 44. Wolff P., Weinhofer I., Seguin J., et al. 2011, Highresolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm, PLoS Genet., 7, e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chalhoub B., Denoeud F., Liu S., et al. 2014, Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome, Science, 345, 950–3. [DOI] [PubMed] [Google Scholar]
- 46. Sun F. M., Fan G. Y., Hu Q., et al. 2017, ZS11’ reveals the introgression history in semi-winter morphotype. The high-quality genome of Brassica napus cultivar, Plant J., 92, 452–68. [DOI] [PubMed] [Google Scholar]
- 47. Bayer P. E., Hurgobin B., Golicz A. A., et al. 2017, Assembly and comparison of two closely related Brassica napus genomes, Plant Biotechnol. J., 15, 1602–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu Z. Y., Hua W., Zhang L., et al. 2013, Seed structure characteristics to form ultrahigh oil content in rapeseed, PLoS One, 8, e62099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu J., Hua W., Yang H. L., et al. 2014, Effects of specific organs on seed oil accumulation in Brassica napus L, Plant Sci., 227, 60–8. [DOI] [PubMed] [Google Scholar]
- 50. Li J., Fan S. H., Sun X. C., et al. 2016, Effective method for isolating liquid endosperm and improved procedure for subsequent DNA extraction in Brassica napus, Oil Crop Sci., 2, 20–5. [Google Scholar]
- 51. Bolger A. M., Lohse M., Usadel B.. 2014, Trimmomatic: a flexible trimmer for Illumina Sequence Data, Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L.. 2013, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H., Handsaker B., Wysoker A., et al. 2009, The sequence alignment/map format and SAM tools, Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. RC Team. 2015, A language and environment for statistical computing, Version 32 1 [Software], R Foundation for Statistical Computing, Vienna, Austria.
- 56. Storey J. D., Tibshirani R.. 2003, SAM thresholding and false discovery rates for detecting differential gene expression in DNA microarrays, In: Parmigiani G., Garrett E. S., Irizarry R. A., Zeger S. L. (eds.), The Analysis of Gene Expression Data Statistics for Biology and Health, pp. 272–90. [Google Scholar]
- 57. Pignatta D., Erdmann R. M., Scheer E., Picard C. L., Bell G. W., Gehring M.. 2014, Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting, Elife, 3, e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gehring M., Missirian V., Henikoff S.. 2011, Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds, PLoS One, 6, e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krueger F., Andrews S. R.. 2011, Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications, Bioinformatics, 27, 1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schultz M. D., Schmitz R. J., Ecker J. R.. 2012, ‘Leveling’ the playing field for analyses of single-base resolution. DNA methylomes, Trends Genet., 28, 583–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jühling F., Kretzmer F., Bernhart S. H., Otto C., Stadler P. F., Hoffmann S.. 2016, metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data, Genome Res., 26, 256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Edwards C. A., Ferguson-Smith A. C.. 2007, Mechanisms regulating imprinted genes in clusters, Curr. Opin. Cell Biol., 19, 281–9. [DOI] [PubMed] [Google Scholar]
- 63. Klosinska M., Picard C. L., Gehring M.. 2016, Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus, Nat. Plants, 2, 16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hatorangan M. R., Laenen B., Steige K., Slotte T., Köhler C.. 2016, Rapid evolution of genomic imprinting in two species of the Brassicaceae, Plant Cell., 28, 1815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luo M., Bilodeau P., Koltunow A., Dennis E. S., Peacock W. J., Chaudhury A. M.. 1999, Genes controlling fertilization-independent seed development in Arabidopsis thaliana, Proc. Natl. Acad. Sci. USA, 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xin M., Yang R., Li G., et al. 2013, Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development, Plant Cell, 25, 3212–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bartolomei M. S., Ferguson-Smith A. C.. 2011, Mammalian genomic imprinting, CSH Perspect. Biol., 3, a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang M., Zhao H. N., Xie S. J., et al. 2011, Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm, Proc. Natl. Acad. Sci. USA, 108, 20042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.