Abstract
In many organisms, MutSγ plays a role in meiotic recombination, facilitating crossover formation between homologous chromosomes. Failure to form crossovers leads to improper segregation of chromosomes and aneuploidy, which in humans result in infertility and birth defects. To improve current understanding of MutSγ function, this study investigates the binding affinities and structures of MutSγ in complex with DNA substrates that model homologous recombination intermediates. For these studies, we overexpressed and isolated from Escherichia coli the yeast MutSγ protein Saccharomyces cerevisiae (Sc) Msh4-Msh5. Sc Msh4-Msh5 binds Holliday junction (HJ)-like substrates, 3′ overhangs, single-stranded (ss) forks, and the displacement loop with nanomolar affinity. The weakest binding affinities are detected for an intact duplex and open-junction construct. Similar to the human protein, Sc Msh4-Msh5 exhibits the highest affinity for the HJ with a Kd < 0.4 nM in solution. Energy-transfer experiments further demonstrate that DNA structure is modulated by the binding interaction with the largest changes associated with substrates containing an ss end. Upon binding, Sc Msh4-Msh5 displaces the ss away from the duplex in most of the ss-containing intermediates, potentially enabling the binding of RPA and other proteins. In the case of the junction-like intermediates, Msh4-Msh5 binding either stabilizes the existing stacked structure or induces formation of the stacked X conformation. Significantly, we find that upon binding, Msh4-Msh5 stacks an open-junction construct to the same extent as the standard junction. Stabilization of the junction in the stacked conformation is generally refractory to branch migration, which is consistent with a potential role for MutSγ to stabilize HJs and prevent branch migration until resolution by MutLγ. The different binding modalities observed suggest that Msh4-Msh5 not only binds to and stabilizes stacked junctions but also participates in meiotic recombination before junction formation through the stabilization of single-end invasion intermediates.
Introduction
The mismatch repair (MMR) protein family plays critical roles in postreplication DNA repair and recombination processes, helping to maintain genome integrity and cellular stability (1, 2, 3). In prokaryotes, MutS and MutL act as homodimers, whereas in eukaryotes, MutS homologs (Msh) and MutL homologs (Mlh) participate in MMR as heterodimers (4, 5). The Msh proteins share significant sequence identity and have common structural motifs, such as a θ-shaped structure, but exhibit key differences that enhance their respective function (6, 7, 8). For instance, the Msh2-Msh6 protein complex, also known as MutSα, recognizes and initiates repair of mismatches and small insertion-deletion loops, whereas Msh2-Msh3 or MutSβ is involved in repairing larger loops (9). The Msh2-Msh3 protein complex is also implicated in the propagation of triplet repeat diseases (10, 11). Within this protein family, the MutSγ or Msh4-Msh5 heterodimeric complex plays an important role in meiotic recombination but is not involved in MMR (12). Genetic studies in Saccharomyces cerevisiae (Sc) have shown that Sc Msh4-Msh5 expression is induced in meiosis and that loss of either protein decreases crossover recombination and spore viability (12, 13, 14). Crossover recombination is needed for proper segregation of chromosomes during meiosis, and hMSH4-hMSH5 foci are found at DSB and crossover sites in human oocytes (15). Defective segregation in humans leads to aneuploid offspring, which are typically inviable. Human aneuploidies are associated with and are a major cause of infertility, miscarriages, and congenital birth defects such as Down syndrome (16, 17).
Meiotic recombination begins with programmed double strand breaks (DSB) that are resected to form 3′ single-stranded (ss) tails for invasion into a homologous duplex. Subsequent strand invasion and displacement from duplex DNA form displacement loops (D-loops) and Holliday junction (HJ) precursors (18, 19, 20). Extension of the D-loop coupled with capture of the second DSB end leads to the formation of double HJs (dHJs), which upon resolution form crossovers. Msh4-Msh5 is critical for stabilizing joint molecule DNA recombination intermediates, for promoting crossover recombination, and for proper assembly of the synaptonemal complex in S. cerevisiae meiotic prophase nuclei (17, 20, 21, 22). Recent work has suggested that Sc Msh4-Msh5 stabilizes single-end invasion (SEI) intermediates (21), whereas in other species, such as mice, Caenorhabditis elegans, and plants, genetic and cytological studies have indicated that MutSγ appears at early stages in meiotic recombination before dHJ formation and potentially plays a role in stabilizing HJ precursors (23, 24, 25, 26).
Crossover formation in S. cerevisiae and many other eukaryotes occurs through a homologous recombination pathway involving MutSγ and the MutLγ complex, formed from the Mlh1-Mlh3 heterodimer (5, 25). The endonuclease activity of Mlh1-Mlh3 in coordination with Msh4-Msh5 leads to biased nicking of dHJs, resulting in a crossover upon resolution of the joint molecule. One possible mechanism to explain biased nicking of the dHJ intermediate is that the interaction between Mlh1-Mlh3 and Msh4-Msh5 leads to an asymmetry in the resolution of junctions (5, 27, 28, 29, 30).
Previous in vitro biochemical analysis demonstrated that human MSH4-MSH5 binds to HJs with relatively high affinity and protects all four strands at the junction core as revealed by DNase I footprinting (31, 32). Interaction with the HJ promotes the exchange of ADP for ATP in both subunits and conversion of the ATP-bound protein into a clamp form that can encircle and slide over both duplex DNA arms of a stacked junction, ultimately diffusing away from the junction core (31, 32). This behavior is consistent with that of the other MutS proteins, in which binding to the appropriate substrate stimulates ADP-ATP exchange and induces conformational changes that ultimately lead to recruitment of MutL proteins (5, 33). ATPase activity is a significant component of MutSγ function because mutations in the putative ATP binding region of either subunit conferred a null phenotype (34).
In this study, we report purification of Sc Msh4-Msh5 protein and characterization of the complexes formed with DNA substrates that closely resemble distinct meiotic recombination intermediate structures using steady-state and time-resolved (TR) fluorescence spectroscopic methods at near-physiological conditions. Fluorescence anisotropy measurements, which monitor molecular rotation from which size and shape can be inferred, have been used effectively to measure protein binding to DNA in such diverse systems as CAP-DNA, HU-DNA, and others (35, 36, 37, 38). These methods have been shown to accurately determine dissociation constants in the nanomolar and picomolar ranges (36) while avoiding the caging and dilution effects that sometimes influence gel-based data (39). We also employed Förster resonance energy transfer (FRET) measurements to probe the conformational changes associated with Sc Msh4-Msh5 binding to different DNA substrates that model recombination intermediates. Previously, FRET measurements have effectively measured DNA conformational changes induced by protein binding in MutS and Msh2-Msh6 (40, 41, 42). Importantly, TR fluorescence measurements yield information on local conformational dynamics and can capture any heterogeneities in the system (43, 44, 45, 46). By using these methods to examine Sc Msh4-Msh5 binding interactions, we have determined that the complex binds with high affinity to both SEI and junction-like intermediates and induces conformational changes upon binding. These results shed light on the preferred binding substrates of the Msh4-Msh5 protein; are consistent with Msh4-Msh5 engaging with early, pre-dHJ recombination intermediates; and suggest a role for Msh4-Msh5 at multiple points in the process of meiotic recombination.
Materials and Methods
Purification of S. cerevisiae MutS homologs Msh4 and Msh5
Unless otherwise noted, all chemical and biochemical reagents were obtained from Millipore Sigma (St. Louis, MO). The open reading frame and 75 basepairs (bps) of the 3′ untranslated region of the MSH4 gene were amplified from S. cerevisiae S288C genomic DNA, and NcoI and BamHI sites were incorporated as restriction cut sites. This MSH4 gene sequence matched in full the yeast genome database (YFL003C). To differentiate Msh4 protein from Msh5 protein by molecular weight, we incorporated MSH4 into the pET-42a(+) plasmid to express N-terminal GST-tagged Msh4 protein. The NcoI/BamHI digested PCR product was cloned into the NcoI/BamHI sites of the multiple cloning site of pET-42a(+) plasmid. The insertion was verified by the Keck DNA sequencing lab, Yale University (44637562–44637566). For MSH5 cloning, genomic DNA was prepared from S288C as described above. The open reading frame plus 49 bps of the 3′ untranslated region of the MSH5 gene was amplified using S288C genomic DNA with the N-terminal primer containing an NdeI site (43975553) and the C-terminal primer containing an AatII site (43975554) and cloned into MCS2 of NdeI/AatII sites in pCDFDuetTM-1 vector. This clone was confirmed by sequencing (47075525–47075529, 48500851), and the result matched with the deposited S288C MSH5 gene in the yeast genome database (YDL154W). A detailed list of the primers used for cloning and the sequence verification are provided in Table S1.
MSH5 in pCDFDuet vector and N-terminal GST-tagged MSH4 in pET-42(a+) vector were co-transformed in BL21CodonPlus(DE3)-RIL expression cell lines (Agilent Technologies, Santa Clara, CA). A separate version of MSH4 without the GST tag was placed in the pCDFDuet vector and expressed using the same conditions as described below to generate the Msh4ΔGST-Msh5 protein. The cells were grown at 37°C and induced with 0.75 mM isopropyl β-D-thiogalactopyranoside when the optical density OD600 reached 0.6. After induction, the cells were grown for 2 h at 30°C. The cells were harvested and resuspended in 10 mM Tris-HCl (pH 8.0), 10% sucrose solutions and stored at −80°C. All subsequent steps were carried out on ice at 4°C. The cells were lysed in buffer A (10 mM Tris-Cl (pH 8.0), 0.01 mM EDTA, 1 mM dithiothreitol (DTT), 0.4 mg/mL lysozyme, 1 mM phenylmethylsulfonylfluoride, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1X protease inhibitor cocktail, 1 M NaCl, and 5% glycerol) followed by three rounds of homogenization using an EmulsiFlex C5 homogenizer (Avestin, Ottawa, Canada). The cell lysate was centrifuged at 6774 × g for 30 min. The clear supernatant was dialyzed overnight against buffer B (10 mM Tris-Cl (pH 8.0), 0.01 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonylfluoride, 1X protease inhibitor cocktail, 100 mM NaCl, and 5% glycerol). The dialyzed supernatant was purified on a sulphopropyl Sepharose Column (GE Healthcare Life Sciences, Waukesha, WI) pre-equilibrated with buffer B and eluted using a linear gradient of 10 column volumes from 100 mM to 1 M NaCl. Msh4 was eluted around 250–300 mM NaCl, and Msh5 was eluted between 200 and 250 mM NaCl. These fractions were pooled and applied to a Heparin Sepharose column (GE Healthcare Life Sciences) pre-equilibrated with buffer B. The proteins were eluted using a linear gradient from 100 mM to 1 M NaCl over 10 column volumes. The heterodimeric complex eluted at ∼225 mM NaCl and was used without further purification. Fractions containing Msh4 (∼250 mM NaCl) and Msh5 (∼200 mM NaCl) were mixed together, and the resulting heterodimer was purified using a size exclusion chromatography (SEC) Superdex 200 Increase GL column (1 cm inner diameter × 24 cm; GE Healthcare Life Sciences) pre-equilibrated with buffer C (10 mM Tris (pH 8.0), 200 mM NaCl, 1 mM DTT, 0.01 mM EDTA, and 5% glycerol) (Fig. S1). We determined a molecular weight of 220 kD for the Msh4-Msh5 heterodimer with GST through comparison with a standard curve, generated using a gel-filtration high molecular weight calibration kit (GE Healthcare Life Sciences) (Fig. S1 B). The identities of the purified proteins were confirmed by liquid chromatography with tandem mass spectrometry (Keck MS & Proteomics Resource; Yale University, New Haven, CT). Concentrations of the purified proteins were determined by Bradford analysis. Aliquots of the pure protein were stored at −80°C in buffer C with 20% glycerol. All experiments were performed with the purified protein obtained from recombined subunits, and as a control, some experiments were also performed with the isolated, purified heterodimer. In all cases, the recombined and purified proteins yielded the same results. For the Msh4ΔGST-Msh5 heterodimer, the Msh4ΔGST was expressed, grown, and purified separately, followed by mixing with Msh5 and SEC purification, as described above. All experiments were performed with the GST-tagged protein except where indicated.
Steady-state nuclease assay
To check for contaminant nuclease activity, we used a steady-state fluorescence assay, in which the presence of nuclease is readily detected by an increase in fluorescence. Approximately 0.5 μM of the Msh4-Msh5 complex was added to 100 nM of 6-MI-incorporated duplex DNA and mixed in the presence of a 5 mM Tris (pH 8.0) buffer containing 1 mM DTT, 5 mM MgCl2, and 200 mM NaCl. The samples were incubated for 30 min, and fluorescence was monitored for 3 h at 30°C using the conditions as described below. Those samples containing protein that did not exhibit a significantly different slope than the DNA-only samples were considered nuclease-free and used for subsequent experiments (Fig. S1 C).
Stability of MutS homologs monitored by CD
Circular dichroism (CD) measurements were performed using a J-810 spectropolarimeter (Jasco, Easton, MD). Thermal melting measurements were performed in a 10 mM sodium phosphate buffer (pH 7.4), 0.01 mM EDTA, 1 mM DTT, 200 mM NaCl, and 5 mM MgCl2 with a minimum of 3 μM of protein. Spectra were obtained from 275 to 190 nm and measured over a temperature range from 10 to 95°C. The amount of folding was determined by monitoring the ellipticity at 210 and 223 nm.
Folding was analyzed in terms of a two-state model, in which we assumed that at low or high temperatures, the protein was either 100% folded or 100% unfolded. The melting temperatures, Tm, were taken as the point at which the protein is 50% unfolded. The Keq was calculated at each temperature and plotted as a function of to determine the enthalpic (ΔH) and entropic (ΔS) parameters using the van ’t Hoff equation (Fig. S1 D; Table S2).
DNA purification and substrate preparation
High performance liquid chromatography (HPLC)-purified DNA strands containing a C6 amino terminal linker were labeled with either 5-carboxyfluorescein, succinimidyl ester (FAM) and/or 5-carboxytetramethylrhodamine, succinimidyl ester and used to prepare donor-only, acceptor-only and doubly labeled DNA substrates (Eurofins Genomics, Louisville KY; Integrated DNA Technologies, Coralville, IA). HPLC-purified 6-methylisoxanthopterin (6-MI)-incorporated DNA strands were obtained from Fidelity Systems (Gaithersburg, MD). Unlabeled DNA strands were obtained from Integrated DNA Technologies and purified using a denaturing 7 M urea, 20% polyacrylamide gel as previously reported (36). DNA sequences used for preparing all substrates are provided in Table S3.
DNA substrates were prepared by adding equimolar amounts of the individual strands in a 10 mM Tris buffer (pH 7.4) with 300 mM NaCl and 0.1 mM EDTA. The samples were heated in a water bath at 65°C for 4 h and then slowly cooled to room temperature (−3.5°C/h). The procedure was the same for duplex DNA except the samples were heated for 5 min at 90°C before slow cooling. Proper formation of all DNA substrates was verified using nondenaturing gel electrophoresis (35, 36) and thermal melting profiles to determine Tm values (Fig. S1 E).
Gel mobility shift assays
Gel mobility shift assays were performed as previously described (35) except a 5% native polyacrylamide gel (29:1) was used with a Tris borate buffer (pH 8.0). The DNA binding buffer consisted of 10 mM Tris (pH 8.0), 0.01 mM EDTA, 1 mM DTT, 200 mM NaCl, 5 mM MgCl2, and 2% Ficoll. Visualization of DNA bands was done using SYBR Green1 (Thermo Fisher, Waltham, MA) and a Typhoon Trio Variable Mode Imager (GE Healthcare Biosciences). Image Quant software (GE Healthcare Biosciences) was used to determine the intensity of the free DNA bands as a function of increasing protein concentration from which an apparent dissociation constant Kd was determined (Fig. S2 A). Analysis was done as previously described, assuming a 1:1 protein/DNA ratio (35).
Fluorescence spectroscopy experiments
All steady-state fluorescence data were generated with a Horiba SPEX Fluoromax-4 spectrofluorometer (Edison, NJ). Fluorescence intensity experiments were performed using 6-MI-incorporated oligomers in HJ, pre-HJ, D-loop with invading single strand (DLIS), single-stranded forks (ss forks), and 3′-overhang DNA substrates (Table S3) (36). The open junction (open HJ) was prepared with FAM-end-labeled strands. 6-MI experiments used an excitation wavelength of 340 nm, and emission was monitored at 430 nm with an integration time of 15 s and a 6 nm bandpass. For the FAM-labeled DNA, the excitation wavelength was 494 nm, and emission was monitored at 518 nm with an integration time of 15 s and 5 nm bandpass. All intensity measurements were performed with the polarizers set to 0° for excitation and 54.7° for emission to eliminate any polarization artifacts. For anisotropy experiments, the same conditions were used, except measurements were performed with the emission polarizer set to either 0 or 90°. The G-factor was measured for each emission wavelength independently and used to calculate the anisotropy. The protein was titrated into a 600 μL volume of either 0.1 nM for HJ DNA, 1 nM for pre-HJ, ss forks, or D-loop DNA or 5 nM for 3′ overhangs, homoduplex, or open HJ DNA. All samples were in a 5 mM Tris (pH 8.0), 0.01 mM EDTA, 1 mM DTT, 5 mM MgCl2, 200 mM NaCl buffer. Samples were incubated for 3 min after each addition of protein. During spectral acquisition, the samples were stirred continuously and maintained at 10°C. Anisotropy and intensity binding curves were analyzed assuming a 1:1 binding interaction with the following equation:
where r is the anisotropy or intensity measured at each protein concentration and rf and ri refer to initial and final anisotropy or intensity values, respectively. The fits were generated and calculated using Origin 9.2.214 (MicroCal, Northampton, MA). The stoichiometry of binding was measured using fluorescence anisotropy by titrating Msh4-Msh5 into a solution of FAM-labeled DNA at a concentration 10-fold greater than the Kd measured for that protein-DNA complex (Fig. 2 C).
Figure 2.
Binding and stoichiometry of Sc Msh4-Msh5 with recombination intermediates. (A) EMSA binding analysis of Sc Msh4-Msh5 with different recombination intermediates. The recombined Msh4-Msh5 complex (black) binds to the HJ with comparable affinity as the isolated Msh4-Msh5 complex (red) (also shown in the inset). Original data are shown in Fig. S2. (B) Fluorescence intensity binding analysis of Msh4-Msh5 with different recombination intermediates. Msh4-Msh5 bound HJ DNA with 100-fold higher binding affinity (0.3 ± 0.1 nM) relative to dsDNA (40 ± 15 nM) and the open-junction DNA (46 ± 7 nM) at 200 mM NaCl and 5 mM MgCl2 in a 5 mM Tris (pH 8.0) buffer. Sc Msh4-Msh5 exhibited high affinity binding to recombination intermediates such as 3′ overhangs, ss forks, DLIS, and pre-HJ. All dissociation constants are given in Table 1. (C) Sc Msh4-Msh5 binding stoichiometry determined by fluorescence anisotropy. For all substrates examined, the binding stoichiometry determined from the intersection of the linear binding range with the nonbinding plateau indicates a 1:1 interaction. Data shown are an average from three independent measurements. To see this figure in color, go online.
Steady-state FRET assays
Steady-state FRET experiments were carried out using end-labeled donor-only and donor-acceptor strands that were prepared as described above. All experiments were performed in our binding buffer (5 mM Tris (pH 8.0), 0.01 mM EDTA, 1 mM DTT, 200 mM NaCl, and 5 mM MgCl2) at a concentration of 100 nM DNA. Protein was added to either the donor-only or donor-acceptor sample in stoichiometric amounts. The sample volume was kept constant at 600 μL. Fluorescence emission spectra were obtained by exciting the donor dye at 494 nm and scanning the emission at a rate of 3 nm/pt with an integration time of 1 s. Samples were contained in 5 × 5 mm glass cuvettes and maintained at 10°C with constant stirring.
The efficiency of energy transfer was calculated by the decrease in donor fluorescence in the presence of acceptor relative to the donor-only emission (47). Energy-transfer efficiencies, distances, and R0 values were calculated as described previously (35, 48). R0 values were determined for each DNA substrate, and any changes in intensity as a consequence of protein binding were taken into account when calculating the FRET efficiencies and R0 values. Spectral analysis was performed using Grams AI ver. 9.2 (Thermo Fisher Scientific), and the fits were generated using Origin v.9.2.214 (OriginLab, Northampton, MA).
TR fluorescence lifetime measurements
TR-FRET data were acquired using DNA samples labeled as described above for the steady-state measurements. TR measurements were performed in a 5 mM Tris (pH 8.0), 0.01 mM EDTA, 1 mM DTT, 200 mM NaCl, 5 mM MgCl2 buffer with 100 nM DNA. Stoichiometric amounts of DNA and protein were incubated for 3 min at 10°C, and samples were stirred continuously during spectral acquisition, which typically took 45–50 min. To ensure that any fluorescence quenching associated with Msh4-Msh5-DNA complex formation did not contribute to the measured energy transfer, donor-only spectra were obtained in presence of protein.
TR fluorescence measurements were performed using the time-correlated single-photon counting method (PTI TimeMaster instrument, Horiba, NJ). Samples were excited using a Becker & Hickl 375 nm pulsed picosecond-laser diode (BDL-375-SMC; rep rate = 1 MHz; <1 mW average power). Emission was detected at 520 nm using a 450 nm cutoff filter with a time window of 55 ns. Excitation and emission slits were set to a 15 nm spectral bandpass. The intensity decay data were collected to 20,000 in the peak channel, with the emission polarizer set to 54.7° to avoid any anisotropic bias from the vertically polarized laser light. Analysis of TR fluorescence decays was performed as described previously (48). All fitting and analysis were performed using the FARGOFIT program written by Igor Negrashov (49). To model the FRET distributions without any bias, the decays of the donor-only and the doubly labeled samples were globally fitted using an iterative reconvolution method linking decay terms and amplitudes between donor and donor-acceptor data sets. The χ2 parameter was used to monitor the goodness of the fit, lifetime decay values were determined from a minimum of two separate experiments (Table S4), and the plots were generated using Origin 9.2.214 (OriginLab, Northampton, MA).
Data analysis was also performed using the maximal entropy method (50) utilizing the Shannon-Jaynes entropy function (FelixGX Data Analysis program, PTI, Horiba):
where S is the entropy and αi is the amplitude of each component. The equation has been maximized to recover the least biased set of amplitudes (αi) out of all feasible solutions. Plotting the amplitudes (αi) as a function of lifetime (logτ) reveals the homogeneity in the FRET distributions for each individual substrate. The maximal entropy method and sum of exponential analysis methods yielded essentially the same results for all of the substrates examined (Fig. S3).
Results
Expression and purification of S. cerevisiae Msh4-Msh5
Yeast genetics have provided many details about Msh4-Msh5 function, including its critical role in synaptonemal complex and crossover formation during meiosis (5, 12, 20, 34, 51). Corresponding biochemical studies to fully illuminate the structure-function properties of this protein have been limited by a lack of purified yeast protein. To obtain sufficient protein to study the S. cerevisiae Msh4-Msh5 protein binding to recombination intermediates, including the HJ, in greater biochemical detail, we developed a strategy for overexpressing and purifying the yeast proteins in Escherichia coli (Fig. 1). The MSH4 and MSH5 genes were cloned from the yeast strain S288C and incorporated into the pCDFDuet-1 and pET-42a(+) plasmids. The most successful strategy involved coexpressing the proteins on separate plasmids in the BL21 CodonPlus(DE3)-RIL E. coli strain. To differentiate the Msh4 protein from the Msh5 protein and improve the solubility of the Msh4 protein, we introduced a GST tag at the N-terminus (Fig. 1 A). This tag did not have a significant effect on protein binding to DNA because protein generated without the tag exhibited similar binding affinities (Fig. S2 B). As shown in Fig. 1 B, overexpression of the proteins was sufficient for isolation using a combination of ion exchange and size-exclusion chromatography. The total yield was ∼7 μg/L cells of Msh4-Msh5 recombined from purified subunits and ∼1 μg/L cells for the purified heterodimer.
Figure 1.
Expression, purification, and thermodynamic characterization of Sc Msh4-Msh5. (A) The cloning strategy utilized for Msh4 and Msh5 expression in E. coli BL21-CodonPlus(DE3)-RIL cell lines. An N-terminal GST tag was incorporated before the MSH4 gene to facilitate differentiation between the Msh4 and Msh5 proteins, which otherwise have similar molecular weights. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of the purified Sc Msh4-Msh5 visualized using silver stain or in a Western blot using an anti-GST antibody. Detailed procedures for cloning and purification are provided in Materials and Methods. (C) Thermal melting profiles of the Sc Msh4-Msh5, Sc Msh4ΔGST-Msh5, Sc Msh2-Msh6, and Taq MutS yielded Tm values of 64, 59, 64, and 88°C, respectively. In all cases, cooperative melting profiles are observed, indicative of stable complexes. The Taq MutS Tm is significantly elevated relative to the other proteins, consistent with its thermophilic origin. The thermodynamic parameters of the melts derived through a van ’t Hoff analysis (Fig. S1D) are given in Table S2. To see this figure in color, go online.
The SEC elution profile yielded one peak for the recombined Msh4-Msh5 (Fig. S1 B) corresponding to a molecular weight of 220 kD, as expected for the heterodimer with one GST tag. Electrophoretic mobility shift assays (EMSA) (Fig. S2 A) and fluorescence binding experiments (Fig. 2) with the isolated or recombined Sc Msh4-Msh5 gave the same affinity for HJ DNA (Table 1), indicating that there was no appreciable difference between the two preparations.
Table 1.
Binding Parameters Determined for Msh4-Msh5 DNA Interactions
| DNA Substrate | Dissociation Constant Kd (nM) |
Protein/DNA Stoichiometry | |
|---|---|---|---|
| EMSAa | Fluorescence Intensityb | ||
| Duplex | 120 ± 33 | 40 ± 15 | 1:1 |
| 3′ overhangs | 47 ± 9 | 15 ± 4 | 1:1 |
| ss forks | 25 ± 7 | 5 ± 1 | 1:1 |
| DLIS | 12 ± 2 | 5 ± 2.5 | 1:1 |
| Pre-HJ | 15 ± 5 | 6 ± 1 | 1:1 |
| Open HJ | 88 ± 22 | 46 ± 7 | 1:1 |
| HJ (I) | 12 ± 2 | 0.3 ± 0.1 | 1:1 |
| HJ (R) | 14 ± 2 | 0.3 ± 0.1 | 1:1 |
EMSA Kd values are determined from the fraction of free DNA.
Kd values determined from the change in fluorescence intensity or anisotropy as a function of Msh4-Msh5 concentration, as described in the Materials and Methods. All data shown are an average from at least three independent measurements.
Characterization of Sc Msh4-Msh5 complex
The relative stability of the heterodimer obtained from SEC was evaluated thermodynamically using CD spectroscopy by monitoring the amount of helical content as a function of temperature and using the halfway point or Tm for stability comparisons. As shown in Fig. 1 C, the Msh4-Msh5 heterodimer exhibited cooperative unfolding, as did the protein without the GST tag. The melting profile of Sc Msh4-Msh5 gave a Tm of ∼64°C, which is comparable to that obtained for the Msh2-Msh6 heterodimer (Tm = 59°C). This similarity in thermodynamic stability, as reflected in the Tm values and the calculated enthalpy and entropy values (Table S2), indicates the purified Sc Msh4-Msh5 forms a relatively stable heterodimeric complex. The GST tag confers ∼5°C of stability to the heterocomplex based on the Tm obtained for the Sc Msh4ΔGST-Msh5 protein (Tm = 59°C); the Tm plus the retrieved thermodynamic parameters are consistent with formation of a stable heterodimer in the absence of the GST tag (Table S2). The considerably higher stability (∼30°C) observed for the Taq MutS homodimer is attributed to the fact that the protein is isolated from a thermophilic organism. Melting parameters obtained for Taq MutS are in good agreement with previously reported values for other thermophilic MutS proteins (52).
Upon obtaining purified Msh4-Msh5 heterodimeric protein, we investigated the binding affinity and stoichiometry with different DNA substrates that model intermediates in the recombination process that occur as precursors to dHJs (18) (Fig. 2). We specifically examined the interaction of Msh4-Msh5 with junction-like intermediates, including the HJ and the pre-HJ as well as the interaction of Msh4-Msh5 with SEI-like recombination intermediates such as a 3′ overhang, ss forks, and DLIS (Table S3). Absorption melting profiles verified the formation and relative stability of these constructs (Fig. S1 E). We find that the recombined and isolated heterodimers yield similar binding affinities for the different substrates (Table 1), where the highest affinity is observed for the intact HJs (Fig. 2, A and B). As measured by EMSA, the recombined Msh4-Msh5 heterodimer binds the HJ (14 ± 2 nM) with the same affinity as the purified heterodimer (10 ± 3 nM), and these affinities are comparable to those previously reported for human MSH4-MSH5 (31) (Fig. 2 A; Fig. S2 A). Significantly, the weakest affinities (Kd > 85 nM) are observed for binding to homoduplex DNA and a model of an open HJ, which contains six mismatched bps at the center of the junction (53). We further investigated binding to double-stranded DNA (dsDNA) that contains either a GT mismatch or a 1 bp insertion-deletion loop. EMSA measurements show little to no binding to these substrates at concentrations as high as 3 μM (Fig. S2 A). This finding is in marked contrast to the binding behavior observed for MutSα and MutSβ heterodimers and is consistent with previous reports that Msh4-Msh5 or MutSγ is a meiosis-specific protein and not involved in DNA MMR (12, 51).
Fluorescence intensity experiments performed using internally and externally labeled substrates yielded the same binding trends as the gel shift assays (Fig. 2 B). Notably, binding to a random sequence 34 bp duplex DNA yielded a Kd of 40 ± 15 nM, whereas binding to the HJ yielded a Kd of 0.3 ± 0.1 nM (Table 1), demonstrating that Msh4-Msh5 exhibited a 100-fold higher binding affinity for junction DNA relative to dsDNA in solution. The difference in affinity measured by solution and gel experiments may be caused by dilution effects in the gel; solution methods typically yield higher affinities relative to those determined by EMSA (39, 54). To detect the DNA in the EMSA measurements, the HJ concentration was more than 10-fold higher than the solution-measured Kd, which likely led to significant inaccuracy in the gel-measured Kd value for the HJ substrate. The model HJ used in our studies is based on J3, a stable, nonmigrating four-way junction originally developed and characterized by Lilley, Ha, and others (35, 55, 56, 57, 58) (Table S3). Although the stacked junction (Fig. S4) is the preferred substrate as demonstrated by the subnanomolar binding affinity detected (Fig. 2 B), binding to all other recombination-like substrates was in the nanomolar range, consistent with earlier studies using human MSH4-MSH5, for which preferential binding to HJs over other DNA substrates was observed (31).
Fluorescence anisotropy experiments performed with FAM-labeled DNA substrates confirm protein-DNA complex formation and demonstrate that Msh4-Msh5 forms a 1:1 complex with all substrates (Fig. 2 C). The stoichiometric experiments were performed under conditions in which all added protein binds to DNA. The point of intersection between the linear binding region and saturated plateau gives the binding stoichiometry, whereas the relatively flat plateau suggests that only a small amount of nonspecific binding occurs at higher protein concentrations.
Conformational changes induced by Sc Msh4-Msh5 binding
To elucidate the potential function of Sc Msh4-Msh5 in binding to these different recombination-like substrates, we have examined the conformational changes induced upon binding using a combination of TR and steady-state FRET experiments (Figs. 3 and 4). For all of the FRET measurements performed, we labeled the DNA strands at the 5′ ends with fluorescein as the donor dye and rhodamine as the acceptor dye. Judicious placement of the dyes allowed us to examine the conformational changes upon protein binding, and in some instances, we employed more than one labeling scheme to verify the changes observed. FRET efficiencies were determined from the behavior of doubly labeled molecules relative to donor-only molecules and were quantified by either a change in donor lifetime or a change in donor fluorescence intensity of the FRET substrate. We compare donor fluorescence in singly and doubly labeled constructs because the TR measurements measure FRET efficiency in terms of donor lifetime (47). To account for any fluorescence quenching caused by protein binding, donor-only measurements were performed in the presence of protein when appropriate. We used the fluorescence binding and stoichiometry information (above) to ensure the maximal amount of complex formation under our measurement conditions. The gel shift insets (Figs. 3 and 4), obtained with samples postirradiation, demonstrate both the presence of the protein-DNA complex and the absence of any substantive damage to the sample as a consequence of the measurement.
Figure 3.
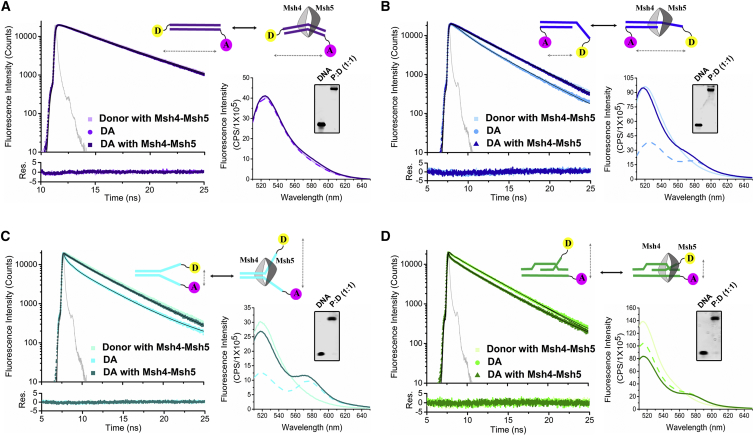
Conformational analysis of Sc Msh4-Msh5 binding to duplex and ss-containing intermediates. TR fluorescence decays of donor-only and donor-acceptor DNA substrates, which are labeled as shown and used for measuring energy transfer with and without protein. Insets depict steady-state fluorescence and EMSA data of the same samples. In the inset, steady-state donor-acceptor fluorescence spectra obtained in the absence of protein are shown with a dashed line. (A) FRET analysis of Sc Msh4-Msh5 binding to dsDNA suggests a minimal amount of induced bending, based on the slightly shorter lifetime and reduced donor intensity observed in TR and steady-state spectra, respectively. (B) FRET analysis of Msh4-Msh5 binding to 3′-overhang DNA resulted in an increase in donor fluorescence and longer lifetimes. This reduction in transfer efficiency suggests Msh4-Msh5 straightens the 3′ overhang. (C) Sc Msh4-Msh5 binding to ss forks led to an increase in donor fluorescence and a longer lifetime in the steady-state and TR spectra, respectively, consistent with a reduction in FRET, indicative of protein-induced opening of the fork. (D) Sc Msh4-Msh5 binding to the DLIS-like intermediate led to a decrease in donor fluorescence and a shorter lifetime in the steady-state and TR spectra, respectively, consistent with an increase in FRET efficiency. This suggests Sc Msh4-Msh5 binding brings the single end closer to the duplex in this DLIS-like substrate. The instrument response function is shown in gray. Bottom traces depict the residuals obtained from fitting the decays. Buffer conditions are as in Fig. 2. Data were acquired and analyzed as described in the Materials and Methods. To see this figure in color, go online.
Figure 4.
For a Figure360 author presentation of Fig. 4, see the figure legend at https://doi.org/10.1016/j.bpj.2018.10.029.
Conformational analysis of Msh4-Msh5 binding to joint molecule intermediates (pre-HJ, open HJ, and HJ). Data are presented as in Fig. 3. (A) Sc Msh4-Msh5 binding to pre-HJds minimally perturbed lifetime or donor fluorescence intensity in TR and steady-state data, respectively, signifying little to no change in energy transfer when the probes are placed on the intact junction side, as shown. (B) Placement of probes on the incomplete junction side (pre-HJss) led to an increase in donor fluorescence and a longer lifetime in the steady-state and TR spectra, respectively, indicating a reduction in transfer efficiency, consistent with the ss moving away from the junction. (C) FRET analysis of the open HJ. In the absence of protein, donor fluorescence intensity and the relatively long lifetime observed in the steady-state and TR spectra, respectively, are indicative of low energy transfer and are consistent with an open-junction conformation. Protein binding led to a pronounced reduction in donor fluorescence intensity and a shorter lifetime, which is consistent with an increase in FRET efficiency. These findings suggest the protein induces the junction to adopt a stacked conformation upon binding. A similar result is obtained if the labels are placed to assess the other possible stacked conformation (Figs. S4 and S5B). (D) FRET analysis of the stacked HJ. Msh4-Msh5 binding led to minimal changes in donor lifetime and fluorescence intensity in the TR and steady-state spectra, respectively. The relatively constant FRET efficiency observed is consistent with a stacked conformation of the junction that is not significantly altered upon protein binding. The instrument response function is shown in gray. Bottom traces depict the residuals obtained from fitting the decays. Data were acquired and analyzed as described in the Materials and Methods. To see this figure in color, go online.
The effects of protein binding on the amplitude-weighted mean lifetimes and energy-transfer efficiencies for all of the recombination intermediates examined are shown in Fig. 5. Generally, analysis of the TR decays yields two lifetime components, one of which is relatively long-lived (4.5–5 ns) and is comparable to the lifetime of the FAM monomer. For most of the substrates, this component is unchanged with protein binding. The second lifetime obtained is shorter (1–3 ns) and is more dependent on the DNA substrate monitored (Table S4). The amplitude-weighted mean lifetime takes into account the populations associated with each lifetime and can be used to determine the amount of energy transfer (47), in which shorter mean lifetimes of the donor-acceptor molecule correspond to higher energy-transfer efficiencies. We further verified the relative populations of each component using the maximal entropy analysis method, which does not assume a specific kinetic model, and resolved similar distributions (Fig. S3). Additionally, transfer efficiencies obtained from the steady-state and TR fluorescence experiments are in excellent agreement in terms of the trends observed upon protein binding as well as the values obtained. The correlation between these different measuring and analytical methods provide high confidence in the protein-induced conformational changes observed.
Figure 5.
Mean lifetime (τmol) and EFRET analysis of Msh4-Msh5 binding to model recombination intermediates. (A) Amplitude-weighted mean lifetimes obtained from the analysis of the individual fluorescence decays for donor and donor-acceptor DNA substrates in the presence and absence of Sc Msh4-Msh5. Fit parameters for analysis of TR fluorescence decays are given in Table S4. (B) A comparison of FRET efficiencies determined from TR and steady-state measurements. FRET efficiencies obtained from both types of measurements are in excellent agreement, demonstrating that the differences observed are consistent with conformational changes induced by protein binding. The most dramatic protein-induced conformational changes are associated with the 3′ overhang, ss forks, pre-HJss, and DLIS substrates. Raw data are shown in Figs. 3 and 4. Efficiencies are given in Table 2. To see this figure in color, go online.
As shown in Fig. 3 A, Msh4-Msh5 binding to duplex DNA led to slight decreases in donor fluorescence lifetime and donor fluorescence intensity in the donor-acceptor-labeled molecule as measured by TR and steady-state fluorescence spectroscopy, respectively. These changes are indicative of a slight increase in FRET efficiency, which signifies a small amount of bending induced upon binding. Since the length of the duplex is at the limit of the FRET efficiency range for the dyes used, there is a considerable degree of uncertainty in this measurement (Table 2). Nevertheless, the distances obtained suggest that the amount of bending observed is less than that detected for Msh2-Msh6 protein binding to mismatched DNA (6, 8, 59). Given the considerable flexibility introduced by the mismatch (60, 61, 62, 63), it is not surprising that a greater degree of bending is observed in the Msh2-Msh6 mismatch complex. Nevertheless, the absence of a significant bend induced by Msh4-Msh5 binding suggests that the protein interacts with duplex DNA differently than Msh2-Msh6, as would be expected given the considerable lack of homology in the N-terminal DNA binding domains of the two protein complexes and the absence of clearly defined N-terminal DNA binding domains in Msh4-Msh5 (32, 64).
Table 2.
Steady-State and TR FRET Efficiencies for Msh4-Msh5-DNA Complex
| DNA or Protein-DNA Complex | EFRETa |
End-to-End Distance R (Å)b | R0 (Å)c | |
|---|---|---|---|---|
| Steady State | TR | |||
| dsDNA (high salt) | 0.01 ± 0.01 | 0.02 ± 0.02 | – | 52.0 |
| Msh4-Msh5-dsDNA | 0.02 ± 0.01 | 0.07 ± 0.02 | 98 ± 52 | 51.1 |
| 3′ overhangs (high salt) | 0.53 ± 0.01 | 0.50 ± 0.01 | 50 ± 9 | 51.3 |
| Msh4-Msh5-3′ overhangs | 0.07 ± 0.01 | 0.02 ± 0.02 | 79 ± 18 | 51.1 |
| Forks (high salt) | 0.74 ± 0.01 | 0.71 ± 0.02 | 43 ± 7 | 51.1 |
| Msh4-Msh5-forks | 0.04 ± 0.01 | 0.07 ± 0.02 | 81 ± 24 | 47.7 |
| Forks (high salt)d | 0.00 ± 0.00 | 0.05 ± 0.05 | 100 ± 24e | 50.8 |
| Msh4-Msh5-forksd | 0.14 ± 0.01 | 0.16 ± 0.02 | 65 ± 13 | 48.3 |
| DLIS (high salt) | 0.20 ± 0.03 | 0.19 ± 0.01 (0.11 ± 0.01)f | 63 ± 16 (71 ± 16)g | 49.8 |
| Msh4-Msh5-DLIS | 0.41 ± 0.03 | 0.47 ± 0.07 (0.48 ± 0.07)f | 53 ± 13 (51 ± 13)g | 50.2 |
| pre-HJss (high salt) | 0.24 ± 0.01 | 0.25 ± 0.03 (0.28 ± 0.03)f | 62 ± 10 (60 ± 10)g | 51.2 |
| Msh4-Msh5-pre-HJss | 0.04 ± 0.01 | 0.03 ± 0.03 (0.0 ± 0.03)f | 85 ± 29 (100 ± 29)e,g | 50.0 |
| pre-HJds | 0.41 ± 0.01 | 0.38 ± 0.02 (0.61 ± 0.02)f | 54 ± 08 (47 ± 08)g | 50.6 |
| Msh4-Msh5-pre-HJds | 0.40 ± 0.01 | 0.40± 0.02 (0.60 ± 0.02)f | 54 ± 9 (47 ± 9)g | 50.6 |
| Open HJ316 | 0.10 ± 0.01 | 0.11 ± 0.05 (0.12 ± 0.05)f | 73 ± 15 (70 ± 15)g | 50.6 |
| Msh4-Msh5-open HJ316 | 0.25 ± 0.02 | 0.29 ± 0.03 (0.55 ± 0.03)f | 61 ± 12 (49 ± 12)g | 50.4 |
| Open HJ317d | 0.08 ± 0.01 | 0.06 ± 0.01 (0.17 ± 0.01)f | 76 ± 15 (65 ± 15)g | 50.3 |
| Msh4-Msh5-open HJ317d | 0.22 ± 0.01 | 0.34 ± 0.01 (0.64 ± 0.01)f | 62 ± 12 (46 ± 12)g | 50.5 |
| HJ (high salt) | 0.29 ± 0.05 | 0.31 ± 0.02 (0.61 ± 0.02)f | 60 ± 17 (48 ± 17)g | 51.9 |
| Msh4-Msh5-HJ | 0.27 ± 0.01 | 0.32 ± 0.03 (0.60 ± 0.03)f | 63 ± 14 (50 ± 14)g | 53.6 |
FRET efficiency (EFRET) was calculated from the decrease in donor fluorescence intensity, donor, or amplitude-weighted lifetime in the presence of the acceptor as described in the Materials and Methods. The data shown are an average from at least three independent measurements. Fit parameters obtained from analysis of TR fluorescence decays are given in Table S4.
The end-to-end distances (R) were calculated from the steady-state efficiencies as described in the Materials and Methods. The larger error in the distances relative to efficiencies arises from consideration of the steady-state fluorescence anisotropy values of the dyes (80).
R0 values were calculated for each donor-acceptor pair as described previously (35).
Because the measured efficiency is 0, we assume that the distance is at least 2R0.
Efficiencies are calculated from one donor lifetime (τ2), which changes upon protein binding (Table S4).
Distances are calculated based on efficiencies determined from the individual donor lifetimes.
ss intermediates
In contrast to dsDNA, we find that Msh4-Msh5 significantly distorted the 3′ overhangs (Fig. 3 B) and the ss forks (Fig. 3 C) upon binding. For both substrates, we observed an increase in donor lifetime and fluorescence intensity upon binding signifying a loss in FRET efficiency. In the absence of protein, the 3′-overhang is bent, exhibiting a relatively high transfer efficiency between the ss end and the 5′ end of the duplex (Table 2). The reduction in efficiency upon Msh4-Msh5 binding is consistent with a straightening of the 3′-overhang or the ss end moving away from the duplex region. Similarly, we find that Msh4-Msh5 binding led to the two ends of the ss forks moving farther away from each other, resulting in a significant decrease in transfer efficiency. To confirm the opening of the fork upon protein binding, we employed an alternate labeling scheme, in which one of the labels was located at the duplex end and one at an ss end as opposed to placing both labels at the ss ends (Fig. S5). In this alternate labeling scheme, a slight decrease in mean fluorescence lifetime is observed upon protein binding, consistent with the ss end moving closer to the duplex 5′ end. Thus, both labeling schemes are suggestive of an opening of the fork upon binding. We speculate that this protein-induced separation of the ssDNA from the dsDNA in both the 3′ overhang substrate and the ss fork substrate potentially facilitates the binding of proteins needed to stabilize the ss such as RPA (discussed below) (26).
Fig. 3 D depicts the fluorescence changes induced by Msh4-Msh5 binding to our substrate that models a DLIS intermediate. In this case, the doubly labeled DLIS fluorescence intensity and lifetime were close to that of the donor, only indicative of a relatively low transfer efficiency (Table 2). This low efficiency implies that the ss end is relatively distant from the labeled duplex end. The observation of some energy transfer is consistent with the fact that the strand is tethered to the substrate through base pairing of the first nine bases in the D-loop (Fig. 3 D). Interestingly, protein binding led to decreases in the lifetime and intensity indicative of an increase in energy transfer, which suggests that protein binding constrains the ss end closer to the duplex end. The efficiency almost doubles with protein binding, and the distance between the probes decreases by more than 10 Å (Table 2). This behavior is in marked contrast to what we observed with the 3′ overhang and ss forks, for which protein binding separated the ss from the duplex. We speculate that for the DLIS intermediate, the protein-induced changes in conformation facilitate formation of a HJ by stabilizing the position of the ss (Fig. 3 D).
Junction-like intermediates
We have further explored the effect of Sc Msh4-Msh5 binding to HJs through examination of a number of junction-like constructs (Fig. 4). We have specifically studied the protein-induced conformations of a pre-HJ in which one arm is ss, an open HJ in which the central six bases of each arm are mismatched, and a canonical HJ that is immobile. For the junction-like substrates, primarily one decay component changes upon protein binding; therefore, we have also used this lifetime to calculate FRET efficiencies, revealing the conformational changes associated with the population undergoing energy transfer, which are typically larger than the population-averaged values (Table 2).
For the pre-HJ, we have introduced two different labeling schemes to probe the conformation upon protein binding. On the side of the junction with two intact duplex arms (pre-HJds) (Fig. 4 A), the intensity and lifetime measurements yield moderate transfer efficiencies (Table 2), suggesting the junction arms are stacked in the absence of protein. Sc Msh4-Msh5 binding did not alter the conformation of this end of the junction and preserved the stacked arrangement. In contrast, a labeling scheme that monitored the ss end of the junction (pre-HJss) (Fig. 4 B) suggested that this end of the junction is more “open,” where the ss is displaced from the duplex end, as observed previously in the DLIS-like intermediate. For pre-HJss, however, protein binding led to further displacement of the ss end away from the duplex, as shown by the pronounced increase in fluorescence lifetime and intensity indicating a loss of transfer efficiency. Binding to the pre-HJ supports a model in which Sc Msh4-Msh5 exhibits two different binding modes: one in which the protein induces or retains the stacked configuration of the junction and a second modality in which the ss strand is displaced from the junction, possibly to facilitate binding of RPA, initiation of the homology search, and formation of SEI intermediates.
In the case of the open HJ (Fig. 4 C), donor lifetimes and intensities were consistent with low levels of energy transfer, signifying that the arms of the junction are relatively distant from each other as expected. Interestingly, upon protein binding, we observed that the lifetimes and intensities decreased, indicative of an increase in energy transfer. This finding suggests that protein binding brings the arms into closer proximity consistent with that of a stacked conformation. In fact, the transfer efficiencies and calculated distances (Table 2) suggest that the amount of stacking observed is the same as that detected for the intact junction (see below) despite the presence of mismatched bases in the center. We have also investigated an alternate labeling scheme for this construct (open HJ317) (Fig. S5) and obtained the same results for both labeling schemes, consistent with all four arms forming a stacked conformation (Table 2). The TR fluorescence measurements yielded equivalent populations for both possible conformations (Table S4), indicating that the protein does not preferentially stack the junction in one conformation over the other (Fig. S4).
In contrast to our open-junction construct, under our binding conditions (200 mM NaCl, 5 mM MgCl2), the intact junction adopts a stacked conformation (35, 56, 58, 65). Lilley and co-workers have previously characterized the populations of junction 3 conformers in detail and concluded that the IsoII conformer, in which the R and X arms are proximal, dominates in solution (this study, IsoII: 70%; IsoI: 30%) (Fig. S4) (56, 57, 58). Based on this previous finding, we labeled the R and X strands to monitor junction conformation with and without protein. We found that neither the fluorescence intensity nor lifetime changed significantly upon protein binding (Fig. 4 D), indicating that protein binding did not alter junction conformation. The TR fluorescence measurements further showed that binding did not lead to any shift in the relative populations of stacked conformers (Table S4). These findings suggest that Sc Msh4-Msh5 binding stabilizes the stacked X junction conformation with existing junction populations. In this conformation, the arms are ∼48 Å apart, and their relative proximity is consistent with the relatively high concentrations of Na+ and Mg2+ present in the buffer (35, 66). In addition, we find that all of the junction-like complexes exhibit a similar degree of stacking in the protein-bound state, in which the distance between arms for all the protein-bound-stacked junctions and junction precursors is 48 ± 2 Å (Table 2).
Examination of the energy-transfer efficiencies as a group yields some notable trends (Fig. 5 B). Specifically, we observe that recombination intermediates with ss ends exhibit the largest changes in transfer efficiencies upon protein binding, in which binding decreases the efficiency, suggesting that the ss end is displaced from the duplex end and becomes more labile. A notable exception to this trend is the DLIS intermediate, which exhibits an increase in energy transfer upon protein binding, consistent with the protein constraining the ss end closer to the duplex end (Fig. 5 B). These induced changes observed for the DLIS-like intermediate are comparable to the conformational changes observed upon protein binding to junction-like intermediates, in which Sc Msh4-Msh5 binding either induced or stabilized the stacked conformation of the junction. The transfer efficiencies obtained with the protein-open HJ complex demonstrated that the protein stacked the junction arms upon binding, as binding increased the efficiency to the same level detected with the intact junction, consistent with Sc Msh4-Msh5 bringing the open HJ duplex arms closer together (Figs. 4 C and 5 B). Interestingly, Sc Msh4-Msh5 binding to the pre-HJ substrate exhibited both types of binding behavior, in which the side with the intact junction was stabilized in a stacked configuration, whereas binding to the other side led to displacement of the ss strand away from the duplex (Figs. 4, A and B and Fig. 5 B).
Discussion
Sc Msh4-Msh5 binds with high affinity to ss-containing recombination intermediates, signifying a possible role in engaging pre-dHJ intermediates
The fundamental cellular process of meiosis occurs via many different steps requiring a complex machinery of proteins to ensure accurate segregation of homologous chromosomes to form haploid cells. In several different eukaryotic organisms, MutSγ has been shown to be crucial for the formation of crossovers, arising from the asymmetric resolution of dHJs (20, 25, 67, 68). Deletion of MutSγ leads to a reduced number of crossovers (34), which is attributed in part to the role MutSγ plays in stabilizing HJs. An interesting finding from these steady-state and TR fluorescence results is that Sc Msh4-Msh5 binds with high affinity to 3′ overhangs and ss forks that model pre-HJ recombination intermediates (Figs. 3, B–D). These forks and overhangs recapitulate some of the DNA structures that appear in the early stages of prophase I, after the introduction of DSB (69) and 3′ end resection but before dHJ formation (18). Importantly, we observe that Sc Msh4-Msh5 not only binds with high affinity to these intermediates but also induces conformational changes upon binding, intimating that the observed binding has functional implications. These findings are consistent with a model in which MutSγ participates in meiotic recombination at time points distinct from those associated with dHJ formation and resolution (Fig. 6).
Figure 6.
Model for possible role for MutSγ in meiotic recombination. Based on our in vitro results, we propose a model in which MutSγ binds to and stabilizes early recombination intermediates, possibly as soon as 3′ ends are resected after the introduction of DSB. Stabilization by MutSγ prevents repair by other pathways and promotes the formation of HJs and dHJs, ultimately leading to crossover formation. To see this figure in color, go online.
Our results indicate that for structures containing an ss end (except for the DLIS as discussed below), Sc Msh4-Msh5 binding induces the ss end to move away from the duplex (Figs. 3 and 5). We propose that this displacement of the ssDNA end facilitates the binding of RPA, which prevents the ss strand from forming any secondary structure. In C. elegans, a recent structured illumination microscopy study examining the different stages of meiotic recombination revealed Msh5 foci at resected DSB ends flanked by populations of RPA. Interestingly, as meiosis progressed toward the formation of dHJs, the appearance of a second MutSγ population along with a loss of the RPA populations was detected. From this observation, the authors inferred that the loss of RPA was associated with a loss of recombination intermediates containing ssDNA, as would be expected with the formation of dHJs (26). Moreover, in budding yeast, loss of Sc Msh4-Msh5 resulted in defects in formation of SEI intermediates (21), and in Arabidopsis, mutation of Atmsh4 results in a delay in proceeding to the first meiotic division, which was attributed to repair of the recombination intermediates before dHJ formation (23). In that case, MutSγ was inferred to stabilize these intermediates, which promoted dHJ formation and ultimately crossovers. The central elements and processes of meiosis are largely conserved across organisms (68), suggesting common mechanisms of action. Thus, our results supporting an expanded role for Sc Msh4-Msh5 are consistent with a number of genetic, cytological and biochemical studies in multiple organisms in which MutSγ function has been implicated at earlier stages in meiotic recombination (Fig. 6).
Sc Msh4-Msh5 stabilizes HJs in the stacked X conformation, preventing branch migration
In our studies, Sc Msh4-Msh5 exhibited the highest affinity for the stacked X configuration of the HJ (Fig. S4), and the FRET measurements demonstrated that the protein does not alter the junction conformation upon binding (Figs. 4 D and 5). This behavior differs significantly from that of the MutSα homolog Msh2-Msh6, which induces formation of an open junction upon binding (70, 71) (Fig. S5 C). Stabilization of the stacked X form is consistent with the findings of Fishel and co-workers, who observed similar preferential binding and found that hMSH4-hMSH5 binding to HJ stimulated ADP to ATP exchange, leading to formation of a sliding clamp that embraces both duplex arms of the junction (31, 32). Sc Msh4-Msh5 binding to the DLIS-like intermediate also supports a model in which the protein stabilizes the stacked X form of the junction (Fig. 3 D). Our experiments reveal that Sc Msh4-Msh5 binding brings the ssDNA closer to the duplex, which we infer is in preparation for forming a stacked HJ. We speculate that because the D-loop has already formed in this intermediate, the interaction with Sc Msh4-Msh5 differs from earlier ss-containing intermediates in which the protein separates ssDNA from the duplex end. MutSγ was previously proposed to stabilize nascent D-loops in the mycelial fungus Sordaria macrospora based on the behavior of Msh4 deletion mutants, which affected the pairing of chromosomes (67). Our FRET results with the protein-open HJ complex have shown that Sc Msh4-Msh5 not only stabilizes but also induces formation of the stacked X form of the junction (Fig. 4 C; Fig. S5 B), adding to the previous finding that hMSH4-MSH5 binds to stacked junctions (31, 32). Significantly, when Sc Msh4-Msh5 induced the open HJ to adopt the stacked X conformation, there was no bias in the stacked conformers obtained (Table S4), suggesting that conformational bias induced by Sc Msh4-Msh5 binding is not a possible pathway for asymmetric resolution of dHJs.
In the later stages of prophase I, extension of the D-loop and capture of the second DSB end lead to the formation of a dHJ, which can be resolved asymmetrically to form crossovers (Fig. 6). In many organisms, MutSγ is required for crossover formation, and cytologically, Msh4 and Msh5 foci have been observed at dHJs (23, 24, 25, 26), although in yeast, some crossover formation occurs through a different pathway (29, 72). In budding yeast, the endonuclease Mlh1-Mlh3 interacts with Sc Msh4-Msh5 to resolve dHJs, although how the asymmetry in resolution occurs is not understood, particularly because the junctions form hundreds of bps apart from each other. One proposed model for MutSγ function is the stabilization of dHJs to prevent dissolution before resolution by MutLγ (20, 31, 73). Fishel and co-workers have proposed that multiple MutSγ complexes form sliding clamps around the junction site, potentially facilitating asymmetric resolution of the junctions (31), whereas others have suggested that the polymerization of Mlh1-Mlh3 enables asymmetric junction resolution (27). We note that Sc Msh4-Msh5 stabilization of the stacked X configuration is refractory to branch migration and prevents the dissolution of junctions without the need for multiple copies of the protein, as demonstrated by the 1:1 binding interaction observed (Fig. 2 C). Interestingly, Mlh1-Mlh3 has been shown to preferentially bind to open junctions (53), although MutSγ stabilizes the stacked X form (31 and this work). When and how the junction changes conformation after interacting with Sc Msh4-Msh5 remains an open area of investigation. In other MutS proteins, ADP-ATP exchange induces changes in protein and DNA conformation, creating a binding site for MutL and turning the MutS protein into a sliding clamp (41, 74, 75, 76). We speculate a similar mechanism may be occurring in MutSγ, in which ATP binding facilitates changes in MutSγ conformation and the formation of an open junction, which is more amenable to MutLγ binding. In our study, MutSγ conformational changes are not monitored by our FRET experiments and will need to be addressed in future studies.
Our experiments with the pre-HJ junction-like substrate support a model in which Sc Msh4-Msh5 exhibits two different binding modes depending on the structure of the recombination intermediate. Specifically, we suggest that Sc Msh4-Msh5 binds to SEI intermediates and stabilizes these substrates in preparation for forming junctions (Fig. 6). Importantly, our results also indicate that the protein actively displaces the ssDNA from the main duplex, potentially to facilitate the binding of RPA. Our results also show that Sc Msh4-Msh5 stabilizes junction-like intermediates and our DLIS-like substrate in the stacked X conformation, which is not capable of branch migration. These two different protein-binding modes are observed in the complex formed with the pre-HJ, where both displacement of an ssDNA end and stacking of junction arms is observed. We propose that Sc Msh4-Msh5 stabilization of the stacked X conformer effectively inhibits the dissolution of junctions until they are processed by other proteins such as Mlh1-Mlh3. Distinct from this function of binding to HJs, which has both enzymatic and structural components, we propose that the function of Msh4-Msh5 binding to SEI-like substrates is an architectural one, primarily involving positioning of the ssDNA end for optimal binding by ss binding proteins, such as RPA, DMC1, and Rad51. The binding of these proteins is key for maintaining the ssDNA for homology searching and strand invasion. A mainly architectural role for MutSγ in meiotic recombination provides an attractive explanation as to why it can be functionally replaced by other proteins in organisms lacking MutSγ such as Drosophila and fission yeast (68, 77, 78).
In summary, our results point to a broad role for MutSγ in regulating meiotic recombination intermediates in which the protein complex conformationally modifies SEI intermediates to facilitate binding of RPA and other proteins. This function occurs in addition to its role in stabilizing HJs to position them in a manner that facilitates processing by MutLγ as a precursor to crossover formation. These results make clear that Sc Msh4-Msh5 stabilizes HJs and junction-like intermediates in a stacked X conformation that inhibits branch migration and dissolution of the junctions. These findings suggest that MutSγ may play more of an architectural role in meiotic recombination, although future investigations analyzing MutSγ ATPase activity in conjunction with DNA binding are needed to fully address this point. Yet to be elucidated is how MutSγ, working with MutLγ, facilitates crossover formation and the asymmetric nicking of a dHJ.
Author Contributions
Y.L. and S.L. created the clones. S.L. and I.M. designed the research. S.L. performed all the experiments. S.L. and I.M. analyzed and interpreted the data. I.M. and M.M.H. directed and oversaw the project. S.L., I.M., and M.M.H. wrote and reviewed the manuscript.
Acknowledgments
We thank Miho Sakato for help with cloning Msh4-Msh5. We thank Amy J. MacQueen, Donald B. Oliver, and Tithi Banerjee for helpful discussions.
This work was supported by the National Science Foundation (MCB-0843656 to I.M.), the National Institutes of Health (R15GM114743 to M.M.H.), and Wesleyan University.
Editor: Jason Kahn.
Footnotes
Five figures and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31218-9.
Supporting Citations
Reference (79) appears in the Supporting Material.
Supporting Material
References
- 1.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel T.A., Erie D.A. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015;49:291–313. doi: 10.1146/annurev-genet-112414-054722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manhart C.M., Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst.) 2016;38:84–93. doi: 10.1016/j.dnarep.2015.11.024. Published online December 2, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren J.J., Pohlhaus T.J., Beese L.S. Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S., Gellert M., Yang W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2011;19:72–78. doi: 10.1038/nsmb.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obmolova G., Ban C., Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 9.Reyes G.X., Schmidt T.T., Hombauer H. New insights into the mechanism of DNA mismatch repair. Chromosoma. 2015;124:443–462. doi: 10.1007/s00412-015-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt M.H., Pearson C.E. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst.) 2016;38:117–126. doi: 10.1016/j.dnarep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Polyzos A.A., McMurray C.T. Close encounters: moving along bumps, breaks, and bubbles on expanded trinucleotide tracts. DNA Repair (Amst.) 2017;56:144–155. doi: 10.1016/j.dnarep.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross-Macdonald P., Roeder G.S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 13.Hollingsworth N.M., Ponte L., Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 14.Pochart P., Woltering D., Hollingsworth N.M. Conserved properties between functionally distinct MutS homologs in yeast. J. Biol. Chem. 1997;272:30345–30349. doi: 10.1074/jbc.272.48.30345. [DOI] [PubMed] [Google Scholar]
- 15.Kneitz B., Cohen P.E., Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb N.E., Sherman S.L., Hassold T.J. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet. Genome Res. 2005;111:250–255. doi: 10.1159/000086896. [DOI] [PubMed] [Google Scholar]
- 17.Reichman R., Alleva B., Smolikove S. Prophase I: preparing chromosomes for segregation in the developing oocyte. Results Probl. Cell Differ. 2017;59:125–173. doi: 10.1007/978-3-319-44820-6_5. [DOI] [PubMed] [Google Scholar]
- 18.Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 19.Zickler D., Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Börner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 22.Lynn A., Soucek R., Börner G.V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 23.Franklin F.C., Higgins J.D., Jones G.H. Control of meiotic recombination in Arabidopsis: role of the MutL and MutS homologues. Biochem. Soc. Trans. 2006;34:542–544. doi: 10.1042/BST0340542. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Tang D., Cheng Z. Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics. 2014;198:1447–1456. doi: 10.1534/genetics.114.168732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray S., Cohen P.E. Control of meiotic crossovers: from double-strand break formation to designation. Annu. Rev. Genet. 2016;50:175–210. doi: 10.1146/annurev-genet-120215-035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woglar A., Villeneuve A.M. Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell. 2018;173:1678–1691.e16. doi: 10.1016/j.cell.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manhart C.M., Ni X., Alani E. The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol. 2017;15:e2001164. doi: 10.1371/journal.pbio.2001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santucci-Darmanin S., Neyton S., Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 2002;11:1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- 29.Zakharyevich K., Tang S., Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenzi M.L., Smith J., Cohen P.E. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am. J. Hum. Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snowden T., Acharya S., Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 32.Snowden T., Shim K.S., Fishel R. hMSH4-hMSH5 adenosine nucleotide processing and interactions with homologous recombination machinery. J. Biol. Chem. 2008;283:145–154. doi: 10.1074/jbc.M704060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modrich P. Mechanisms in E. coli and human mismatch repair (Nobel Lecture) Angew. Chem. Int. Ed. Engl. 2016;55:8490–8501. doi: 10.1002/anie.201601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishant K.T., Chen C., Alani E. Genetic analysis of baker’s yeast Msh4-Msh5 reveals a threshold crossover level for meiotic viability. PLoS Genet. 2010;6:e1001083. doi: 10.1371/journal.pgen.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitoc C.I., Mukerji I. HU binding to a DNA four-way junction probed by Förster resonance energy transfer. Biochemistry. 2011;50:1432–1441. doi: 10.1021/bi1007589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno A., Knee J., Mukerji I. Applying 6-methylisoxanthopterin-enhanced fluorescence to examine protein-DNA interactions in the picomolar range. Biochemistry. 2012;51:6847–6859. doi: 10.1021/bi300466d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson B.J., Larkin C., Schildbach J.F. Using fluorophore-labeled oligonucleotides to measure affinities of protein-DNA interactions. Methods Enzymol. 2008;450:253–272. doi: 10.1016/S0076-6879(08)03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen B., McMurray C. Rapid method for measuring DNA binding to protein using fluorescence anisotropy. Protoc. Exch. 2009 [Google Scholar]
- 39.Fried M.G., Liu G. Molecular sequestration stabilizes CAP-DNA complexes during polyacrylamide gel electrophoresis. Nucleic Acids Res. 1994;22:5054–5059. doi: 10.1093/nar/22.23.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeRocco V.C., Sass L.E., Erie D.A. Dynamics of MutS-mismatched DNA complexes are predictive of their repair phenotypes. Biochemistry. 2014;53:2043–2052. doi: 10.1021/bi401429b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu R., DeRocco V.C., Weninger K.R. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 2012;31:2528–2540. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristóvão M., Sisamakis E., Friedhoff P. Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic Acids Res. 2012;40:5448–5464. doi: 10.1093/nar/gks138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty S., Steinbach P.J., Ansari A. Enhanced spontaneous DNA twisting/bending fluctuations unveiled by fluorescence lifetime distributions promote mismatch recognition by the Rad4 nucleotide excision repair complex. Nucleic Acids Res. 2018;46:1240–1255. doi: 10.1093/nar/gkx1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno A., Knee J.L., Mukerji I. Photophysical characterization of enhanced 6-methylisoxanthopterin fluorescence in duplex DNA. J. Phys. Chem. B. 2016;120:12232–12248. doi: 10.1021/acs.jpcb.6b07369. [DOI] [PubMed] [Google Scholar]
- 45.Kalinin S., Valeri A., Seidel C.A. Detection of structural dynamics by FRET: a photon distribution and fluorescence lifetime analysis of systems with multiple states. J. Phys. Chem. B. 2010;114:7983–7995. doi: 10.1021/jp102156t. [DOI] [PubMed] [Google Scholar]
- 46.Kalinin S., Peulen T., Seidel C.A. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 47.Lakowicz J.R. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 48.Zhang Q., Lahiri S., Mukerji I. Alignment of the protein substrate hairpin along the SecA two-helix finger primes protein transport in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2017;114:9343–9348. doi: 10.1073/pnas.1702201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agafonov R.V., Negrashov I.V., Nesmelov Y.E. Structural dynamics of the myosin relay helix by time-resolved EPR and FRET. Proc. Natl. Acad. Sci. USA. 2009;106:21625–21630. doi: 10.1073/pnas.0909757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livesey A.K., Brochon J.C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys. J. 1987;52:693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novak J.E., Ross-Macdonald P.B., Roeder G.S. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukui K., Bessho Y., Kuramitsu S. Thermostable mismatch-recognizing protein MutS suppresses nonspecific amplification during polymerase chain reaction (PCR) Int. J. Mol. Sci. 2013;14:6436–6453. doi: 10.3390/ijms14036436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranjha L., Anand R., Cejka P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 2014;289:5674–5686. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wojtuszewski K., Hawkins M.E., Mukerji I. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 55.Duckett D.R., Murchie A.I., Lilley D.M. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 56.Murchie A.I., Clegg R.M., Lilley D.M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989;341:763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- 57.McKinney S.A., Déclais A.C., Ha T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 58.Joo C., McKinney S.A., Ha T. Exploring rare conformational species and ionic effects in DNA Holliday junctions using single-molecule spectroscopy. J. Mol. Biol. 2004;341:739–751. doi: 10.1016/j.jmb.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 59.Wang H., Yang Y., Erie D.A. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guest C.R., Hochstrasser R.A., Millar D.P. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991;30:3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- 61.Imhof P., Zahran M. The effect of a G:T mispair on the dynamics of DNA. PLoS One. 2013;8:e53305. doi: 10.1371/journal.pone.0053305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isaacs R.J., Rayens W.S., Spielmann H.P. Structural differences in the NOE-derived structure of G-T mismatched DNA relative to normal DNA are correlated with differences in (13)C relaxation-based internal dynamics. J. Mol. Biol. 2002;319:191–207. doi: 10.1016/S0022-2836(02)00265-6. [DOI] [PubMed] [Google Scholar]
- 63.Isaacs R.J., Spielmann H.P. Insight into G[bond]T mismatch recognition using molecular dynamics with time-averaged restraints derived from NMR spectroscopy. J. Am. Chem. Soc. 2004;126:583–590. doi: 10.1021/ja037333r. [DOI] [PubMed] [Google Scholar]
- 64.Rakshambikai R., Srinivasan N., Nishant K.T. Structural insights into Saccharomyces cerevisiae Msh4-Msh5 complex function using homology modeling. PLoS One. 2013;8:e78753. doi: 10.1371/journal.pone.0078753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duckett D.R., Murchie A.I., Lilley D.M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990;9:583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Litke J.L., Li Y., Mukerji I. Probing the Ion Binding Site in a DNA Holliday Junction Using Förster Resonance Energy Transfer (FRET) Int. J. Mol. Sci. 2016;17:366. doi: 10.3390/ijms17030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zickler D., Espagne E. Sordaria, a model system to uncover links between meiotic pairing and recombination. Semin. Cell Dev. Biol. 2016;54:149–157. doi: 10.1016/j.semcdb.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Youds J.L., Boulton S.J. The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 69.Keeney S., Giroux C.N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 70.Alani E., Lee S., Kolodner R.D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J. Mol. Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 71.Marsischky G.T., Lee S., Kolodner R.D. 'Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes. J. Biol. Chem. 1999;274:7200–7206. doi: 10.1074/jbc.274.11.7200. [DOI] [PubMed] [Google Scholar]
- 72.Argueso J.L., Wanat J., Alani E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 2004;168:1805–1816. doi: 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Sweel N., Raghavan V., Alani E. mlh3 mutations in baker’s yeast alter meiotic recombination outcomes by increasing noncrossover events genome-wide. PLoS Genet. 2017;13:e1006974. doi: 10.1371/journal.pgen.1006974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeong C., Cho W.K., Lee J.B. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat. Struct. Mol. Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma A., Doucette C., Hingorani M.M. Slow conformational changes in MutS and DNA direct ordered transitions between mismatch search, recognition and signaling of DNA repair. J. Mol. Biol. 2013;425:4192–4205. doi: 10.1016/j.jmb.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Groothuizen F.S., Winkler I., Sixma T.K. MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife. 2015;4:e06744. doi: 10.7554/eLife.06744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blanton H.L., Radford S.J., Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Osman F., Dixon J., Whitby M.C. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 79.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 80.Ivanov V., Li M., Mizuuchi K. Impact of emission anisotropy on fluorescence spectroscopy and FRET distance measurements. Biophys. J. 2009;97:922–929. doi: 10.1016/j.bpj.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.