Abstract
While actin was discovered in the nucleus over 50 years ago, research lagged for decades due to strong skepticism. The revitalization of research into nuclear actin occurred after it was found that cellular stresses induce the nuclear localization and alter the structure of actin. These studies provided the first hints that actin has a nuclear function. Subsequently, it was established that the nuclear import and export of actin is highly regulated. While the structures of nuclear actin remain unclear, it can function as monomers, polymers, and even rods. Furthermore, even within a given structure, distinct pools of nuclear actin that can be differentially labeled have been identified. Numerous mechanistic studies have uncovered an array of functions for nuclear actin. It regulates the activity of RNA polymerases, as well as specific transcription factors. Actin also modulates the activity of several chromatin remodeling complexes and histone deacetylases, to ultimately impinge on transcriptional programing and DNA damage repair. Further, nuclear actin mediates chromatin movement and organization. It has roles in meiosis and mitosis, and these functions may be functionally conserved from ancient bacterial actin homologs. The structure and integrity of the nuclear envelope and sub-nuclear compartments are also regulated by nuclear actin. Furthermore, nuclear actin contributes to human diseases like cancer, neurodegeneration, and myopathies. Here, we explore the early discovery of actin in the nucleus and discuss the forms and functions of nuclear actin in both normal and disease contexts
Keywords: nuclear actin, nuclear transport, transcription, chromatin organization and movement, nuclear structure
INTRODUCTION
Actin is one of the most abundant and highly conserved proteins in eukaryotes (Dominguez and Holmes, 2011; Pollard, 2017). Actin was initially discovered in rabbit skeletal muscle (Straub, 1943) and is required for muscle contraction (Huxley, 1969). Later studies determined that actin is also an essential component of nonmuscle cells, where it contributes to cell shape, motility, and cytokinesis (Pollard, 2017). We now know of three actin isoforms in vertebrates: skeletal or α-actin, and the nonmuscle β- and γ-actins (Dominguez and Holmes, 2011). Actin monomers, also known as globular-actin (G-actin), can polymerize into filamentous actin (F-actin) (Dominguez and Holmes, 2011). This process is tightly regulated by a vast number of actin binding proteins (Pollard, 2016). Thus, actin polymerization is highly dynamic and this dynamic nature is critical for cellular functions. Because of actin’s abundance, conservation, and essential functions, actin has been extensively studied. While research has primarily focused on understanding the cytoplasmic and cytoskeletal activities of actin, it also localizes and functions within the nucleus (de Lanerolle, 2012; Viita and Vartiainen, 2017; Venit et al., 2018). While early studies reported the presence of actin in the nucleus, this research was not well accepted. Thus, mechanistic study of nuclear actin has lagged behind, as we gained a rich understanding of cytoplasmic actin. Here we explore the discovery of actin in the nucleus, the mechanisms that control actin localization, the structures of nuclear actin, nuclear functions of actin, and how nuclear actin contributes to a number of disease states. We aim to present all of the historical findings on nuclear actin and put them in context with more recent research to give insight into why actin localizes to the nucleus.
EARLY OBSERVATIONS OF ACTIN IN THE NUCLEUS
Coinciding with the finding that actin is not restricted to muscle cells (Ishikawa et al., 1969), actin was reported to be in the nucleus (Ohnishi et al., 1963; Ishikawa et al., 1969; Jockusch et al., 1971). The original studies were largely observational. The first ones described actin in the nuclear subcellular fraction of calf thymus cells (Ohnishi et al., 1963; Ohnishi et al., 1964). Soon after, nuclear actin was described by Nancy Lane (1969), who initially demonstrated using electron microscopy that fibrillar bodies form in the nucleoplasm of Triturus viridescens (newt) oocytes in response to actinomycin D treatment, which inhibits global transcription. These fibrillar bodies, or nuclear bundles (subsequently referred to in the literature as rods), were composed of many individual 5–7 nm filaments – similar in diameter to that of F-actin – suggesting that these rods may be composed of actin (Huxley, 1957; Lane, 1969). Nuclear rods were later observed in chicken sympathetic neurons (Masurovsky et al., 1970), rabbit hypothalamus neurons (Clattenburg et al., 1972), mouse muscle (Miranda and Godman, 1973), and slime mold (Ryser, 1970; Lestourgeon et al., 1975). Thus, these historic studies reveal that nuclear actin is observed across many cell types and species.
While nuclear rods were well characterized by electron microscopy, their molecular composition remained unknown. Subsequent electron microscopy and subcellular fractionation studies indicated the rods were indeed composed of actin (Jockusch et al., 1971; Somosy et al., 1976; Fukui, 1978). Furthermore, nuclear actin rods were observed to form in response to cellular stress. Indeed, dimethyl sulfoxide (DMSO) treatment in Dictyostelium, protists, and cultured mammalian cells (Fukui, 1978; Fukui and Katsumaru, 1979, 1980; Osborn and Weber, 1980; Sanger et al., 1980a) results in nuclear actin rods. Nuclear rods were also observed in cultured cells in response to the ionophore A23187 and magnesium treatment (Osborn and Weber, 1980), heat shock (Welch and Suhan, 1985; Iida et al., 1986), and ATP depletion (Pendleton et al., 2003). Further studies in Dictyostelium demonstrated that these rods formed quickly in response to cellular stress (Fukui and Katsumaru, 1980). Interestingly, these rods were resistant to high concentrations of the actin depolymerizing agent cytochalasin B; this resistance was proposed to be due to interactions with other proteins (Fukui and Katsumaru, 1980). While rods were repeatedly observed under treated conditions they were rarely observed in untreated cells (Lane, 1969). It remains to be determined whether nuclear actin rod formation is an artifact of treatment to various cellular stressors, or a coordinated response to them. Because of the prevalence of nuclear actin rods across many systems and cell types, and the recent observation that rods form in mammalian cells in response to physiologically-relevant stimuli (Plessner et al., 2015), we favor the latter hypothesis.
Soon after the discovery of stress-induced nuclear actin rods, actin was observed in the nucleus under basal conditions by subcellular fractionation in many different cell types including: isolated nuclei of Physarum polycephalum (P. polycephalum) (Jockusch et al., 1974), extracted rat liver nuclei (Douvas et al., 1975), fractionated nuclei of Dictyostelium amoebae (Pederson, 1977), and hand-isolated nuclei of Xenopus laevis (X. laevis) oocytes (Clark and Merriam, 1977). At the time, these findings were met with criticism (Goldstein et al., 1977) and attributed to contamination from actin-rich cytoplasmic fractions. However, recent studies, discussed in the subsequent sections, validate these historical findings and demonstrate that actin, indeed, localizes and functions within the nucleus across organisms and cell types.
NUCLEOCYTOPLASMIC TRANSPORT OF ACTIN
For a long time, it was argued whether an active or passive process controlled the localization of actin to the nucleus. For example, one study contended that the nuclear envelope was not a barrier to actin due to equilibrium of the protein between the cytoplasm and nucleus (Goldstein et al., 1977). Meanwhile, it was observed that the formation of nuclear actin rods in DMSO-treated PtK2 cells coincided with disappearance of cytoplasmic stress fibers, suggesting that actin translocated to the nucleus (Sanger et al., 1980a). It was later found that heat shock led to both the translocation of actin to the nucleus and rod formation in a number of mammalian cell lines (Iida et al., 1986). Together, these findings suggested that actin indeed localizes to the nucleus in response to cellular stress, and the majority hypothesized via an active mechanism.
Subsequently, numerous studies began to uncover the active mechanisms controlling the nuclear localization of actin. Protein sequence analysis revealed that actin does not contain a nuclear localization sequence (NLS) (Vandekerckhove and Weber, 1978; Iida et al., 1992). However, the actin binding protein Cofilin contains a classical bipartite SV40-type NLS that is conserved among higher eukaryotes (Nishida et al., 1984; Matsuzaki et al., 1988; Gunsalus et al., 1995; Munsie et al., 2012). Cofilin is best known for its activity as an actin depolymerizing factor (Bamburg, 1999), where it cooperatively binds regions of F-actin, alters filament rotation, and destabilizes the filament (Hayden et al., 1993; McGough et al., 1997). However, Cofilin also binds G-actin at a 1:1 stoichiometry (Matsuzaki et al., 1988). Additionally, Cofilin, like actin, localizes to the nucleus upon heat shock or DMSO treatment, is a component of nuclear actin rods, and its NLS is required for nuclear actin rod formation (Nishida et al., 1987; Iida et al., 1992; Abe et al., 1993). Further, anti-Cofilin antibodies block the nuclear localization of actin in mast cells (Pendleton et al., 2003). Considering that Cofilin’s NLS is located on the opposite side compared to its actin-binding site and that actin lacks a putative NLS, it was hypothesized that actin localization to the nucleus was dependent on Cofilin (Lappalainen et al., 1997; Pendleton et al., 2003).
More recent studies uncovered that, indeed, actin/Cofilin complexes are transported to the nucleus by a particular import factor (Fig. 1). Fluorescent Recovery After Photobleaching (FRAP) studies and an RNAi screen of a subset of import factors identified Importin 9 as the nuclear importer of actin (Dopie et al., 2012). Importin 9 is a member of the Importin β superfamily, and is highly conserved among higher eukaryotes (Jakel et al., 2002). The Saccharomyces cerevisiae (S. cerevisiae) homolog of Importin 9, Karyopherin 114, is non-essential (Morehouse et al., 1999; Pemberton et al., 1999). Considering this finding, and that the Cofilin NLS is not conserved in yeast (Iida et al., 1993), it is likely that the nuclear import of yeast actin is mediated by another pathway. Alternatively, transgenic disruption of importin 9 in mouse (Blake et al., 2017) and Drosophila (ranbp9) is homozygous lethal (Kelpsch, Jaime, and Tootle, unpublished observation). These findings suggest that nuclear actin is essential for survival. Together, these data establish Importin 9 as the factor mediating the nuclear localization of actin/Cofilin complexes in higher eukaryotes and this translocation is critical for development. However, it remains unclear what other factors are translocated to the nucleus by this importin. Recent data has shown that Importin 9 is sufficient, but not required, for the nuclear localization of the core histones and c-Jun (Muhlhausser et al., 2001; Waldmann et al., 2007).
Figure 1:
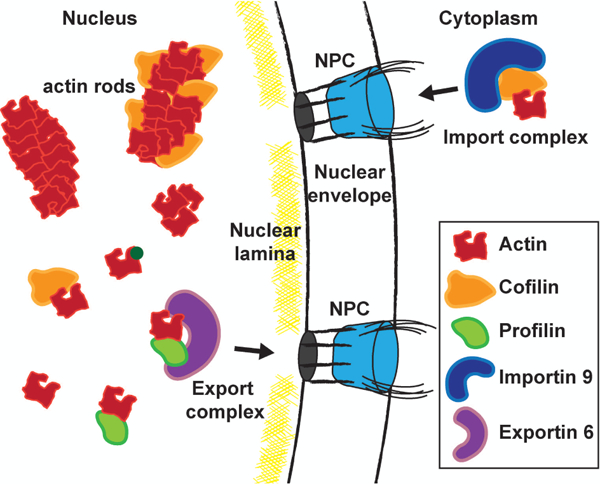
Nucleo-cytoplasmic shuttling of actin and types of nuclear actin. Importin 9 (blue) takes actin (red) and Cofilin (orange) complexes into the nucleus through the nuclear pore complex (NPC). Exportin 6 (purple) mediates the nuclear export of actin and Profilin (green) complexes. Within the nucleus actin takes on many forms. Actin can polymerize into weakly phalloidin-stainable rods and Cofilin-decorated rods. For simplicity, individual actin filaments are shown to represent nuclear actin rods, but rods may be composed of bundles of actin filaments. Additionally, there are short polymers of actin and pools of actin monomers that exist on their own, in an altered conformational state (dark green circle on actin) or complexed with Cofilin or Profilin.
While Importin 9 is responsible for the translocation of actin into the nucleus (Fig. 1), the mechanism controlling the nuclear export of actin has faced controversy over the years. Two functional nuclear export sequences (NESs) have been identified in mammalian α-, β-, and γ-actin as well as S. cerevisiae actin (Wada et al., 1998). Interesting, treatment with Leptomycin B, a specific inhibitor of CRM1 (also known as Exportin 1), enhanced nuclear actin rod formation in response to heat shock in mammalian cells, suggesting that actin is exported by CRM1 (Wada et al., 1998). It should be noted that CRM1 is responsible for the cytoplasmic translocation of many different factors, including ribosomal RNAs – thus, Leptomycin B treatment is also a cellular stressor (Hutten and Kehlenbach, 2007). While nuclear actin rod formation following CRM1 inhibition may indicate CRM1 plays a direct role in the nuclear export of actin, this does not eliminate the possibility that the rod formation was a result of Leptomycin B-induced cellular stress. Supporting this idea, several studies have argued against CRM1-mediated export of nuclear actin (Stuven et al., 2003; Dopie et al., 2012). Indeed, it was found that Exportin 6, a member of the Importin β superfamily, mediates actin’s translocation into the cytoplasm (Stuven et al., 2003) (Fig. 1). Interestingly, Exportin 6 is not capable of binding actin alone; the two proteins only interact in the presence of Profilin (Stuven et al., 2003). Thus, actin is exported from the nucleus in a complex with Profilin. Profilin is a well-studied actin binding protein that canonically binds G-actin at a 1:1 stoichiometry, accelerates the exchange of ADP for ATP on actin, and ultimately promotes F-actin polymerization (Witke, 2004). Exportin 6 is highly conserved among higher eukaryotes and has very few cargos – actin/Profilin complexes, and the actin binding proteins Diaphanous 1, VASP (vasodilator-stimulated phosphoprotein), and Mena (mammalian enabled) (Stuven et al., 2003). However, no Exportin 6 ortholog has been identified in S. cerevisiae, suggesting that the cytoplasmic translocation of actin depends on a different mechanism in yeast (Stuven et al., 2003). Together these data support the model that in higher eukaryotes, actin/Profilin complexes are exported from the nucleus via Exportin 6.
Through decades of research, the mechanisms regulating the transport of actin in and out of the nucleus are now well established (Fig. 1). Specifically, Importin 9 is responsible for the nuclear localization of actin/Cofilin complexes (Dopie et al., 2012), while Exportin 6 mediates the translocation of actin/Profilin complexes to the cytoplasm (Stuven et al., 2003). These results are striking, considering the critical roles of Cofilin and Profilin in F-actin dynamics, and suggests that actin polymerization and depolymerization may also occur in the nucleus. Furthermore, the highly regulated nuclear localization of actin strongly suggests that there is some purpose for actin within the nucleus. However, the presence of actin regulators in the nucleus also raises the questions of what the structures of nuclear actin are and whether actin binding proteins impact the nuclear functions of actin.
FORMS OF NUCLEAR ACTIN
In the cytoplasm, actin is highly dynamic – actin monomers quickly polymerize into filaments and filaments disassemble into actin monomers (Dominguez and Holmes, 2011). These filaments are beautifully labelled by fluorescent phalloidin, can be arranged into higher order structures, and, ultimately, contribute to cellular shape and mobility, among many other cytoplasmic functions (Dominguez and Holmes, 2011; Pollard, 2017). The polymerization and organization of actin is tightly regulated by actin binding proteins (Pollard, 2016). While numerous actin binding proteins localize to the nucleus, the structure and regulation of nuclear actin remains poorly understood.
Nuclear actin exists as monomers, polymers, and rods (Fig. 1). Monomers are canonical G-actin. However, whether nuclear G-actin is post-translationally modified and the identity of its binding partners remain largely unknown. Conversely, as explained throughout this section, filamentous nuclear actin is largely distinct from cytoplasmic F-actin and many terms have been used to describe these nuclear structures that contain actin, including fibrillar bodies (Lane, 1969), nuclear bundles (Lane, 1969), paracrystals (Osborn and Weber, 1980), filaments (Welch and Suhan, 1985), bars (Dopie et al., 2012), and rods (Sanger et al., 1980b). While the literature lacks consensus, we distinguish two major forms of polymerized nuclear actin: polymers and rods. Nuclear actin polymers are oligomers of actin that do not have an obvious filament structure, while nuclear actin rods are larger polymers of actin that resemble either cytoplasmic actin filaments or bundles (Fig. 1).
While dynamic formation of nuclear actin rods was observed in response to various cellular stressors (Fukui, 1978; Fukui and Katsumaru, 1979, 1980; Osborn and Weber, 1980; Welch and Suhan, 1985; Iida et al., 1986; Pendleton et al., 2003), nuclear phalloidin staining was largely absent from the literature (Nishida et al., 1987). Thus, nuclear actin rods were considered to be distinct from the easily phalloidin stainable F-actin observed in the cytoplasm. This difference could be for several reasons. Perhaps the structure of nuclear actin rods is distinct enough from cytoplasmic F-actin that phalloidin cannot bind (Bettinger et al., 2004). Alternatively, many nuclear actin rods are decorated with Cofilin (Bamburg et al., 2010) and this interaction is sufficient to occlude phalloidin binding (Nishida et al., 1987; McGough et al., 1997). While several studies have demonstrated phalloidin stainable nuclear actin rods (Nishida et al., 1987; Baarlink et al., 2013), this weak nuclear phalloidin signal is only observed with high exposure of cytoplasmic phalloidin. This finding supports the concept that the structure of these filaments is distinct from that of cytoplasmic F-actin (Baarlink et al., 2013; Belin et al., 2015; Plessner et al., 2015; Baarlink et al., 2017).
One of the commonly used tools to study nuclear actin are anti-actin antibodies. Interestingly, the use of different antibodies to label nuclear actin suggests that there are distinct pools of nuclear actin (Gonsior et al., 1999; Schoenenberger et al., 2005; Wineland et al., 2018). Specifically, the 2G2 anti-actin antibody, which was raised against actin/Profilin complexes, reveals nuclear structures in myogenic cells and X. laevis oocytes (Gonsior et al., 1999). Alternatively, the 1C7 anti-actin antibody, which recognizes a different epitope on actin from the 2G2 antibody, labels a distinct nuclear actin pool from the 2G2 antibody in mammalian cell lines (Schoenenberger et al., 2005). Further, neither of these antibodies label canonical F-actin in its native state (Gonsior et al., 1999; Schoenenberger et al., 2005), suggesting these antibodies either label G-actin or actin polymers. One tool to specifically label actin monomers is fluorescently-conjugated DNase I (Hitchcock, 1980). DNase I staining is ubiquitous throughout Drosophila oogenesis, demonstrating that every nucleus features nuclear actin monomers (Wineland et al., 2018). In this system, another antibody, C4 anti-actin, which has been used to visualize nuclear actin in several other systems (Parfenov et al., 1995; Gedge et al., 2005; Lenart et al., 2005; Maslova and Krasikova, 2012), identifies a subset of actin monomers and polymers in different nuclei during early oogenesis (Kelpsch et al., 2016; Wineland et al., 2018). Alternatively, the AC15 anti-actin antibody, previously used to visualize nuclear actin in plants (Cruz and Moreno Diaz de la Espina, 2009) and examine it in Drosophila cultured cells (Dopie et al., 2012), labels a distinct, polymeric nuclear actin pool during mid-to-late oogenesis in Drosophila (Wineland et al., 2018). In mammalian cells, the nuclear actin chromobody (ChromoTek) – a genetically encoded NLS-tagged actin nanobody – reveals actin monomers throughout the nucleus and nuclear actin rod formation following serum stimulation or the induction of cellular spreading (Plessner et al., 2015). Notably, this tool fails to recognize actin/Cofilin rods (Plessner et al., 2015). Thus, different anti-actin antibodies identify separate pools of nuclear actin monomers and polymers.
In addition to anti-actin antibodies, a growing number of actin labeling tools are now being utilized to characterize nuclear actin (Melak et al., 2017). Specifically, FRAP and fluorescence correlation spectroscopy (FCS) of fluorescently-tagged actin in mammalian cells indicates that roughly half of the nuclear actin is monomeric and the remaining fraction is polymeric (McDonald et al., 2006). Expression of GFP-actin in the Drosophila germline results in the formation of nuclear actin rods that exhibit regions that are either Cofilin or phalloidin positive (Kelpsch et al., 2016). An NLS-tagged version of LifeAct-GFP, a tool derived from S. cerevisiae Abp140 to label F-actin (Riedl et al., 2008), fails to recognize actin/Cofilin rods (Munsie et al., 2009), but labels a pool of nuclear actin that polymerizes in mammalian cells following serum-stimulation (Baarlink et al., 2013). Expression of LifeAct-GFP in the Drosophila female germline causes female sterility, severe actin defects in the cytoplasm, and nuclear actin rods in the oocyte nucleus that label with phalloidin and LifeAct-GFP (Spracklen et al., 2014). Thus, LifeAct-derived tools are useful for examining phalloidin-positive nuclear actin rods. The expression of a fluorescently labeled NLS-tagged truncation of Utrophin, termed Utr230-EN, identified two different pools of nuclear actin rods in mammalian cells (Belin et al., 2013). When full-length GFP-Utrophin is expressed in the Drosophila female germline, it results in severe cytoskeletal defects and nuclear actin rods in all germ cells that label with Utrophin and phalloidin (Spracklen et al., 2014). Given the difference in nuclear actin rod induction by LifeAct-GFP and GFP-Utrophin in the Drosophila germline (Spracklen et al., 2014), and the different actin binding properties of these tools (Winder et al., 1995; Riedl et al., 2008; Prochniewicz et al., 2009), it is likely that these tools recognize distinct forms of nuclear actin rods. Like the different actin antibodies, the unique patterns of the actin labeling tools support the ideas that nuclear actin takes on a number of different structures, and that multiple pools of nuclear actin exist.
Together, different tools can be used to identify actin monomers, polymers, and rods. The use of different antibodies and actin labeling tools has identified distinct pools of nuclear actin, which likely feature different conformations. The mechanisms that define the conformational states of actin within the nucleus remain unknown. However, it is appealing to consider roles for post-translational modifications of actin (Terman and Kashina, 2013). For example, actin oxidation occurs in the nucleus, reduces nuclear actin levels, and alters gene expression (Lundquist et al., 2014). Alternatively, actin interacts with a large number of proteins. These interactions may reveal antigens that define specific pools of nuclear actin monomers, polymers, and rods. For example, the 2G2 anti-actin antibody was raised against the actin/Profilin complex (Gonsior et al., 1999). Further, LifeAct-GFP and the nuclear actin chromobody identify nuclear actin rods that do not colocalize with Cofilin (Munsie et al., 2009; Plessner et al., 2015). Whether and how post-translational modifications or protein/protein interactions define the distinct pools of and/or contribute to specific functions of nuclear actin remains to be fully explored.
FUNCTIONS OF NUCLEAR ACTIN
The findings that nuclear actin exists in numerous forms, the structure of actin is known to control its activities, and its nuclear localization is highly regulated, suggest that actin has important functions within the nucleus. Indeed, nuclear actin has been widely implicated in regulating transcription (Fomproix and Percipalle, 2004; Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004), chromatin remodeling (Zhao et al., 1998; Galarneau et al., 2000; Shen et al., 2000; Serebryannyy et al., 2016a), DNA-damage repair (Andrin et al., 2012; Belin et al., 2015; Serebryannyy et al., 2017; Wang et al., 2017), and nuclear structure (Sasseville and Langelier, 1998; Bohnsack et al., 2006; Feric and Brangwynne, 2013) (Fig. 2). These nuclear actin activities are discussed below. However, as the body of literature on nuclear actin grows, so do the reported functions of nuclear actin. For example, more recent evidence suggests nuclear actin has roles in apoptosis (Grzanka et al., 2010a; Grzanka et al., 2010b; Grzanka et al., 2011), DNA replication (Parisis et al., 2017), and viral infection (Fuchsova et al., 2015).
Figure 2:
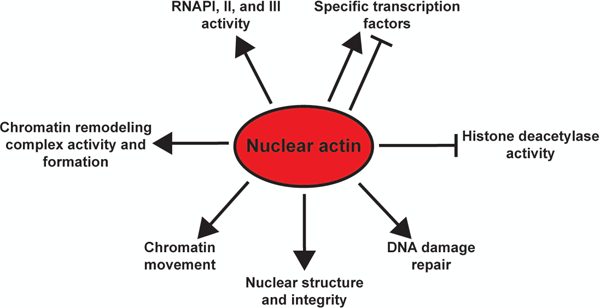
Summary of nuclear actin functions. Nuclear actin positively regulates RNA polymerase activity, chromatin remodeling complex activity and formation, chromatin movement, nuclear structure and integrity, and DNA damage repair. Histone deacteylase activity is negatively regulated by nuclear actin, while several transcription factors are positively or negatively regulated by nuclear actin.
Transcription
The early finding that nuclear actin rods form in response to treatment with a potent inhibitor of transcription spurred research into the roles of nuclear actin in transcription (Lane, 1969). Studies in Chironomus tentans (C. tentans) found that an anti-actin antibody labels transcriptionally active regions of chromatin and that actin specifically interacts with a subset of proteins that bind new transcripts as they are synthesized (Percipalle et al., 2001; Percipalle et al., 2002). Disruption of this actin interaction blocks global transcription in both C. tentans (Percipalle et al., 2003) and human cells (Kukalev et al., 2005). These data suggest that nuclear actin plays an essential role in transcription. Recent studies support that this is a widely conserved function of nuclear actin, as Drosophila actin interacts with regions of euchromatin, as observed via DNA adenine methyltransferase ID (DamID) technology, (Filion et al., 2010), and actin is required for transcription, as blocking the nuclear import of actin via RNAi knockdown of Importin 9 or Cofilin inhibits global transcription (Dopie et al., 2012). Thus, disruption of nuclear actin in many different species perturbs transcription. However, the mechanisms that underlie the regulation of transcription by nuclear actin remain to be fully identified.
A large body of evidence supports that nuclear actin regulates transcription by directly affecting RNA Polymerase (RNAP) activity (Fomproix and Percipalle, 2004; Hofmann et al., 2004; Hu et al., 2004; Philimonenko et al., 2004). Early studies identified that a molecule, presumed to be actin, co-purified with RNAPII from P. polycephalum (Weaver, 1976; Smith et al., 1979), S. cerevisiae (Bell et al., 1977), and calf thymus (Hodo and Blatti, 1977). These studies suggested that nuclear actin directly interacts with transcription machinery but were widely criticized, as actin was thought to be a contaminant of the purification process. However, more recent studies support the validity of this early research, as they identified that actin not only directly interacts with transcription machinery, but also regulates the process. Indeed, studies of Pleurodeles waltlii (salamander) oocytes demonstrated that the injection of anti-actin antibodies into the nucleus blocked RNAPII-dependent transcription (Scheer et al., 1984). Further, actin co-purified with the RNAPII pre-initiation complex from mammalian cells (Egly et al., 1984). In purified systems, actin was sufficient to stimulate transcription, while actin antibodies blocked transcription (Egly et al., 1984; Hofmann et al., 2004). Together, these data indicate that nuclear actin regulates the initiation of transcription. Nuclear actin also contributes to elongation as actin monomers recruit a kinase, Positive Transcription Elongation Factor b (P-TEFb), to RNAPII to activate it via phosphorylation of its C-terminal domain (Qi et al., 2011). Thus, nuclear actin directly interacts with transcription machinery and influences transcriptional initiation and elongation (Fig. 3). Further, this role of nuclear actin is conserved amongst higher eukaryotes.
Figure 3:
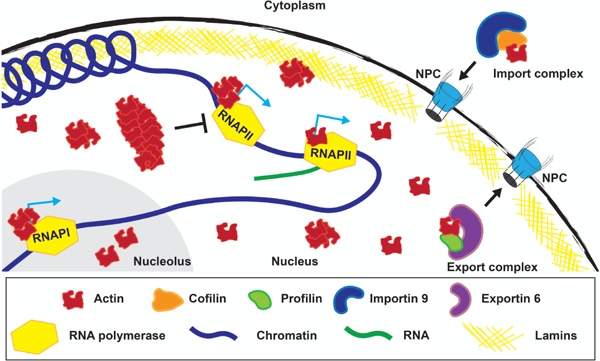
Nuclear actin regulation of transcription. Nuclear import and export of actin (red) is mediated by Importin 9 (blue) and Exportin 6 (purple), respectively, through the nuclear pore complex (NPC). Nuclear actin monomers and polymers are found throughout the nucleus and make up a gel-like matrix at the periphery with the nuclear lamina (yellow lines). Both actin monomers and polymers regulate the activity of RNA polymerase II (RNAPII; yellow hexagons), while nuclear actin rods inhibit RNAPII activity. For simplicity, individual actin filaments are shown to represent nuclear actin rods, but rods may be composed of bundles of actin filaments. Nuclear actin monomers are also found in the nucleolus, where polymerization is required for RNAPI (yellow hexagon) activity.
While the exact mechanisms that underlie nuclear actin’s regulation of RNAPII have yet to be identified, recent studies have begun to explore the structure of the nuclear actin involved. Specifically, the formation of nuclear actin rods blocks RNAPII-dependent transcription, and this is rescued by expression of a non-polymerizable NLS-tagged actin (R62D mutation) (Serebryannyy et al., 2016b). This finding suggests that nuclear actin monomers are essential for RNAPII activity. However, other studies have implicated polymeric actin in transcription. Specifically, a number of studies have determined that nuclear myosin I localizes to the nucleus where, like actin, it interacts with RNAPII (Pestic-Dragovich et al., 2000; Hofmann et al., 2006). Further, nuclear myosin I antibodies block RNAPII activity in vitro (Pestic-Dragovich et al., 2000; Hofmann et al., 2006). Canonically, myosins are molecular motors that interact with F-actin (Sellers, 2000). Considering this, and that both nuclear actin and nuclear myosin I interact with RNAPII and are required for RNAPII activity, it seems likely that nuclear myosin I and polymerized nuclear actin interact to contribute to transcription (Philimonenko et al., 2004; Hofmann et al., 2006; Ye et al., 2008; Philimonenko et al., 2010). Thus, it is likely that dynamic polymerization of nuclear actin is required for RNAPII activity (Fig. 3).
While nuclear actin is critical for RNAPII-dependent transcription, many studies have also implicated roles for actin in regulating the other RNA polymerases. Actin binds to RNAPIII and is essential for basal transcriptional activity (Hu et al., 2004). While actin antibody injection in salamander oocyte nuclei did not block RNAPI-dependent transcription (Scheer et al., 1984), actin was later found to associate with RNAPI in mammalian cells (Fomproix and Percipalle, 2004; Philimonenko et al., 2004). Specifically, actin was found in the nucleolus of HeLa cells, where it physically associates with RNAPI and is required for RNAPI activity (Fomproix and Percipalle, 2004; Philimonenko et al., 2004). Thus, nuclear actin regulates RNAPI activity in mammalian cells, but not in salamander oocytes, suggesting that this process could be specific to mammals or nuclear actin regulation of RNAPI is not required in the salamander oocyte or, potentially, oocytes in general. Together, these data suggest that nuclear actin regulates RNAPI and RNAPIII, in addition to RNAPII.
Considering its interactions with RNAPI and RNAPIII, nuclear actin likely participates in ribosome biosynthesis in the nucleolus. Supporting this idea, immunogold-labeling and electron microscopy demonstrated that actin localized to the nucleolus at sites of ribosomal DNA (rDNA) transcription (Kysela et al., 2005). Further, mouse embryonic fibroblasts lacking β-actin have reduced rDNA transcription, resulting in impaired cell growth and proliferation (Almuzzaini et al., 2016). While reintroduction of wild-type NLS-actin rescues these phenotypes, the expression of polymerization-deficient or -promoting mutant actins only partially rescues. Further, recent studies demonstrated that actin polymers promote RNAPI activity, as non-polymerizable actin fails to bind to the complex (Ye et al., 2008). Thus, dynamic nuclear actin polymerization is a critical for regulating nucleolar function through its interaction with and regulation of RNAPI and RNAPIII in mammalian cells (Fig. 3). Recent data from Drosophila suggests the nucleolar functions of actin are conserved as actin localizes to this subnuclear compartment (Wineland et al., 2018). However, actin’s functions in the nucleolus during development have yet to be explored.
Nuclear actin, in addition to controlling global transcription through its interactions with all three RNAPs, also binds to and regulates specific transcription factors. Specifically, actin regulates the localization and activity of Myocardin-related transcription factor A (MRTF-A; also referred to as MAL), a serum response factor (SRF) coactivator (Vartiainen et al., 2007). Another study, using breast cancer cells, found that β-actin interacts with Estrogen receptor α (ERα) following its activation and nuclear localization; it is suspected that this interaction is functionally relevant for the expression of ERα target genes (Ambrosino et al., 2010). Additionally, polymerized nuclear actin recruit Coronin 2A, a component of the nuclear receptor co-repressor (NCoR) complex, away from Toll-like receptors to de-repress transcription (Huang et al., 2011). Whether nuclear actin binds and regulates the activity of other transcription factors remains to be determined.
Several signaling pathways impinge on transcription via modulating nuclear actin. For example, the induced expression of the Homeobox B (HoxB) gene cluster by retinoic acid treatment of mammalian cells is dependent on nuclear actin polymerization (Ferrai et al., 2009). Furthermore, extracellular cues, like fibronectin, stimulate nuclear actin polymerization via the Linker of the Nucleoplasm and Cytoskeleton (LINC) complex proteins, Sun1 and Sun2 (Plessner et al., 2015). This pathway is suspected to impinge on transcriptional regulation by MAL/SRF, although other transcriptional regulators may also be affected. Recent research also identified that the loss of the extracellular cue Laminin-111 reduces Exportin 6 activity and thereby, increases nuclear actin levels to activate transcriptional networks promoting growth and proliferation (Fiore et al., 2017). Thus, nuclear actin is a key mediator of the transcriptional responses downstream of a variety of signal transduction cascades.
Decades of research indicate that nuclear actin regulates transcription at many levels, including regulating RNAPII initiation (Egly et al., 1984) and elongation (Qi et al., 2011), regulating the activities of RNAPI (Fomproix and Percipalle, 2004; Philimonenko et al., 2004) and RNAPIII (Hu et al., 2004), modulating activity of specific transcription factors (Vartiainen et al., 2007; Ambrosino et al., 2010; Huang et al., 2011), and being a downstream target of signaling pathways that impinge on transcription (Plessner et al., 2015; Fiore et al., 2017). However, many questions remain. While nuclear actin is essential for RNAP activity, the structure – monomers versus polymers – involved remains debated and the mechanisms of its actions are unknown. While nuclear actin polymerization state influences the activity of several transcription factors, the proteins that control actin polymerization in the nucleus remain to be defined. Additionally, how the activity of nuclear actin binding proteins is regulated by various signaling pathways to alter nuclear actin-dependent transcription is not yet understood. Finally, transcriptional regulation by nuclear actin has largely been explored in cell lines and a limited number of organisms. Thus, it is unclear how universal the roles of nuclear actin are in regulating transcription, specifically in in vivo, multicellular contexts.
Chromatin organization
Like its regulation of transcription, it is now well-recognized that nuclear actin modulates chromatin organization. A prominent study demonstrated that injection of anti-actin antibodies into the X. laevis oocyte nucleus blocked chromosome condensation, while injection into the cytoplasm had no effect (Rungger et al., 1979). Another study injecting anti-actin antibodies and actin binding proteins showed similar results, and suggested actin monomers and polymers likely play roles in chromatin organization (Scheer et al., 1984). While these data implicated nuclear actin in chromatin organization, the mechanisms remained unknown. More recent studies have revealed that actin is a component of some, but not all, chromatin remodeling complexes. Here we will briefly discuss the interaction of actin with several of these complexes (for more detailed insight see (Venit et al., 2018)).
One chromatin remodeling complex that contains actin is the highly conserved BAP (Brahma-associated proteins) complex in Drosophila, which is referred to as SWI/SNF (switching/sucrose nonfermenting) in S. cerevisiae and BAF (BRG1-associated factors) complex in mammals. Specifically, protein interaction and genetic studies discovered that an actin-related protein (ARP) associated with the BAP complex (Papoulas et al., 1998). Subsequently β-actin monomers were found to not only interact with the BAF ATPase subunit, Brg1 (Brahma-related gene 1; Brahma in Drosophila; SWI2/SNF2 in S. cerevisiae), but are required for optimal activity and, therefore, proper chromatin remodeling (Zhao et al., 1998). Further showing the functional role of nuclear actin in chromatin remodeling, knockout of β-actin in mouse embryonic fibroblasts results in transcriptional reprogramming via loss of Brg1 binding to chromatin, and increased Histone 3 lysine 9 trimethylation (H3K9me3) and Heterochromatin protein (HP)1α levels (Xie et al., 2018). These effects were partially rescued by the expression of NLS-tagged β-actin (Xie et al., 2018). Together these data demonstrate that nuclear actin is essential for BAP complex activity.
Actin is also a defined component of the S. cerevisiae inositol requiring (INO80) complex (Shen et al., 2000). It was shown through mutagenesis of INO80 components that Arp4, Arp8, and actin monomers bind to the N-terminus of the INO80 ATPase, Ino80. Loss of this interaction impairs chromatin remodeling activity and reduces survival in a similar manner to loss of Ino80 itself (Shen et al., 2003). Further, subdomain 2 of actin is essential for INO80 binding to chromatin (Kapoor et al., 2013). Thus, actin is directly required for INO80 activity, as it is involved in the binding of this complex to chromatin.
Actin is also a component of the TIP60 (Tat-interactive protein 60 kDa) complex in S. cerevisiae and the homologous NuA4 (Nucleosome acetyltransferase of histone 4) complex in mammals (Galarneau et al., 2000; Ikura et al., 2000); this type of chromatin remodeling complex promotes transcription. Interestingly, actin is not required for the ATPase activity of this complex but is suspected to contribute its structure (Ikura et al., 2000).
TIP60 is also essential for the response to γ-radiation-induced DNA lesions. Specifically, loss of complex activity in HeLa cells results in apoptotic escape (Ikura et al., 2000). Considering that actin is likely a structural component of this complex, it is suspected to have a role in the response to DNA damage (Ikura et al., 2000). Supporting this idea, studies expressing non-polymerizable nuclear actin (G13R) and utilizing F-actin depolymerizing drugs revealed that nuclear actin polymerization is necessary for DNA damage repair (Andrin et al., 2012). Indeed, DNA damage induced Formin2- and Spire1/2-dependent nuclear actin polymerization is required for proper repair (Belin et al., 2015). Additionally, α-catenin localizes to sites of DNA damage via a mechanism that depends on nuclear actin polymerization and β-catenin; although the function of these proteins at sites of damage remains enigmatic (Serebryannyy et al., 2017). More recent studies found DNA damage rapidly induced nuclear actin rod formation and recruits ATR (Ataxia telangiectasia and Rad3-related) for DNA repair (Wang et al., 2017). Thus, nuclear actin is likely critical for DNA damage responses, both in general and those mediated by TIP60.
Actin is a component of specific remodeling complexes. While it does not interact with imitation switch (ISWI) containing chromatin remodeling complexes (Zhao et al., 1998), it is present in the BAF, INO80, and TIP60 complexes. However, it is still unclear how actin differentially influences the functions of each of these complexes. As Arp4, and several other ARPs, directly bind to core histones (Harata et al., 1999; Kapoor et al., 2013) and actin (Schafer and Schroer, 1999), it suggests that actin/Arp4 complexes bind nucleosomes directly and serve as a scaffold for binding different subsets of ARPs to recruit specific chromatin remodeling complexes (Kapoor et al., 2013). Therefore, actin likely functionally regulates the activity of BAF, INO80, and TIP60 via interactions with specific ARPs.
More recently, actin was identified as a functional binding partner of histone deacteylase (HDAC) 1 and 2 of the nucleosome remodeling histone deacetylase (NuRD) complex and the corepressor for element-1-silencing transcription factor (CoREST) complex (Serebryannyy et al., 2016a). Nuclear actin monomers bind HDACs and inhibit their function, whereas stimulation of nuclear actin polymerization alleviates this suppression (Serebryannyy et al., 2016a). HDACs remove acetyl-moieties from histones, relaxing chromatin and promoting transcription (Seto and Yoshida, 2014). Thus, actin monomers negatively regulate transcription, at least through their interactions with HDACs (Serebryannyy et al., 2016a). Interestingly, HDAC2 interacts with Arp4 (Joshi et al., 2013). This interaction is similar to that of Arp4 with other chromatin remodeling complexes and suggests a potential mechanism of regulation of HDACs by nuclear actin.
More recent studies have explored the role of nuclear actin in regulating cell fate specific transcriptional programs. Transplanting somatic nuclei into the nuclei of X. laevis oocytes, which are rich in nuclear actin, is an efficient system to directly study transcriptional reprogramming (Gurdon and Melton, 2008; Miyamoto et al., 2011). This reprogramming requires nuclear actin polymerization. Specifically, nuclear actin polymerization at the Oct4 locus recruits the BAF chromatin remodeling complex to induce the expression of this pluripotency gene (Miyamoto et al., 2011). Similarly, nuclear actin polymerization is necessary for the osteogenic differentiation of mesenchymal stem cells (Sen et al., 2015). Surprisingly, treatment with cytochalasin D, an inhibitor of actin polymerization (Casella et al., 1981), increased the levels of and caused polymerization of nuclear actin; this, in turn, induced the transcriptional activation of genes that define an osteogenic fate (Sen et al., 2015). However, the chromatin remodeling complexes mediating this nuclear actin-dependent process remain undefined. Together these studies strongly implicate nuclear actin, in the context of modulating chromatin remodeling complexes, as a key regulator of transcriptional reprogramming.
Numerous studies have demonstrated that nuclear actin binds to and regulates the activity of a variety of chromatin remodeling complexes. This interaction functionally alters complex activity, ultimately controlling transcriptional output. Indeed, recent studies indicate that nuclear actin regulates transcriptional programs controlling cell fate (Miyamoto et al., 2011; Sen et al., 2015). Notably, chromatin remodeling is just one of the means by which nuclear actin regulates transcription; as discussed above, nuclear actin also directly regulates all three RNAPs and affects specific transcription factors.
Chromatin movement
Nuclear actin has also been implicated in the movement of chromatin over long distances. Live imaging of fluorescently-labelled transgenes demonstrated that loci move away from the nuclear periphery upon transcriptional activation (Chuang et al., 2006). This movement was delayed by the expression of non-polymerizable NLS-actin (G13R mutation), while it was accelerated by the expression of F-actin stabilizing NLS-actin (S14C mutation) (Chuang et al., 2006). Similarly, polymerization of nuclear actin is critical for repositioning transgenic loci to Cajal bodies (Dundr et al., 2007) or nuclear speckles (Khanna et al., 2014) for subsequent expression. While these studies have been incredibly informative, they utilized complex transgenes that may not represent endogenous conditions. A more recent study utilized fluorescent in situ hybridization (FISH) probes to label endogenous chromatin territories to demonstrate that perturbing actin polymerization via latrunculin A or jasplakinolide alters chromatin movement (Mehta et al., 2010). As these drugs alter actin polymerization throughout the entire cell, it is possible that the defects are due to perturbing cytoplasmic, nuclear, or both pools of actin. Together these studies suggest that polymeric actin, and likely nuclear actin polymerization, contributes to the movement of chromatin.
In addition to regulating chromatin organization in interphase cells, actin has also been implicated in chromosome segregation during meiosis and mitosis. Actin has been observed on meiotic spindles in plants (Forer and Jackson, 1979; Forer et al., 1979), insects (Silverman-Gavrila and Forer, 2000; Fabian and Forer, 2007), and mice (Bogolyubova and Ginzburg, 2013). It was later shown that actin filaments are required for proper chromosome alignment and segregation during meiosis in multiple mammalian oocytes (Mogessie and Schuh, 2017). Considering that improper meiotic chromosome segregation results in aneuploidy (Nagaoka et al., 2012), actin plays a protective role in meiosis (Mogessie and Schuh, 2017). Actin has also been reported to be a part of the mitotic spindle and regulate spindle length in X. laevis embryos (Woolner et al., 2008). Supporting this finding, we recently identified a pool of actin that localizes to mitotic spindles in Drosophila follicle cells (Wineland et al., 2018). These studies suggest a conserved, in vivo role for actin in chromosome movement, including during mitosis and meiosis.
The roles of nuclear actin in regulating chromatin organization and chromosome separation are not surprising given the functions of actin in prokaryotes. While bacterial MreB only features 15% identity to S. cerevisiae actin, its 3D structure is strikingly similar to monomeric actin and it forms filaments comparable to F-actin (van den Ent et al., 2001). MreB not only contributes to bacterial shape but has been implicated in chromosome segregation. Specifically, MreB is required for chromosome movement to the opposite sides of dividing bacteria (Soufo and Graumann, 2003; Gitai et al., 2005; Kruse and Gerdes, 2005). Additionally, MreB interacts with RNAP to mediate chromosome separation but may also influence transcription (Wachi and Matsuhashi, 1989; Kruse and Gerdes, 2005). Another bacterial actin, ParM, also strongly resembles actin monomer structure and forms filaments reminiscent of F-actin (van den Ent et al., 2002). ParM has a well-defined role in the segregation of a bacterial plasmid in vivo and in vitro (Moller-Jensen et al., 2002). Interestingly, ParM filaments are incredibly unstable unless bound to plasmids on both ends (Garner et al., 2004). Thus, the functions of bacterial actins in the regulation of cell shape and chromosome segregation mirrors that observed for eukaryotic cytoplasmic and nuclear actin, respectively, and represents an excellent example of functional conservation.
Together, these studies implicate that actin’s contributions to chromatin movement during transcriptional activation, meiosis, and mitosis may have derived from an ancient, ancestral function of bacterial actins. Further studies are needed to delineate the mechanisms by which actin mediates chromatin movement.
Nuclear structure
Early studies of nuclear actin also proposed a structural role for actin in the giant nuclei of amphibian oocytes. Analysis of hand-dissected nuclei from X. laevis oocytes revealed that the majority of nuclear actin is diffusible and not filamentous (Clark and Merriam, 1977). The remaining filamentous actin is randomly oriented and makes up a gel-like matrix (Clark and Merriam, 1977; Clark and Rosenbaum, 1979). More recent studies demonstrated that X. laevis oocytes lack Exportin 6, which results in the high level of nuclear actin (Bohnsack et al., 2006). When Exportin 6 was added back to the oocytes, the nuclei became very fragile; this finding strongly supports a structural role for nuclear actin (Bohnsack et al., 2006). Further, stabilization of the nuclear F-actin network blocks nuclear envelope breakdown, a process necessary for meiosis, in X. laevis and Patiria miniata (starfish) (Okada et al., 2012; Mori et al., 2014). Nuclear actin is thought to contribute to the structure of oocyte nuclei because of their large size. However, polymerized nuclear actin is observed in bovine lymphocytes and is thought to contribute to their nuclear structure as well (Nakayasu and Ueda, 1983). Furthermore, in cultured cells, when cell spreading is induced by fibronectin, the forces on the nucleus change and transient nuclear actin rods are observed (Li et al., 2015; Plessner et al., 2015). Nuclear actin polymerization is also essential for the nuclear expansion following mitotic exit. Specifically, as cultured mammalian cells exit mitosis, their nuclear volume expands and the chromatin reorganizes; these processes depend on the transient formation of nuclear actin rods (Baarlink et al., 2017). Together, these findings suggest that nuclear actin contributes to the structure, dynamics, and stability of the nuclear envelope.
While it is unknown how nuclear actin contributes to the stability of the nuclear envelope, actin has been found to interact with nuclear proteins that contribute to nuclear structure. Specifically, nuclear actin likely regulates the structure of the nucleus and nuclear envelope, at least in part, through interactions with lamins, intermediate filament proteins that are a key component of the nucleoskeleton (de Leeuw et al., 2018). In vitro studies uncovered that actin binds to the C-terminus of lamin A, where it may have a role in chromatin organization and nuclear structure (Sasseville and Langelier, 1998). Further, the C-terminus of both A- and B-type lamins can bind and bundle F-actin in vitro (Simon et al., 2010). Supporting that this interaction extends to a cellular context, an RNAi based screen in Drosophila cultured cells suggested that lamins regulate nuclear actin polymerization (Dopie et al., 2015). Interestingly, loss of A-type lamins in Drosophila larval muscle results in phalloidin stainable nuclear actin rods (Schulze et al., 2009). These data suggest a conserved role for lamins in the regulation of nuclear actin polymerization. Actin also interacts with another nuclear envelope protein, Emerin, in mammalian myoblasts (Fairley et al., 1999; Lattanzi et al., 2003). Interestingly, Emerin promotes actin polymerization in vitro (Holaska et al., 2004) and this polymerized actin is expected to mediate the nuclear envelope localization of various chromatin remodeling complexes and respond to mechanical tension (Holaska and Wilson, 2007). Thus, nuclear actin polymers are likely critical for maintaining nuclear envelope shape and the overall structure of the nucleus via their interactions with Lamins and Emerin.
Nuclear actin also regulates nuclear organization. Specifically, the nuclear actin matrix in amphibian and avian oocytes is essential for nuclear organization (Maslova and Krasikova, 2012; Feric and Brangwynne, 2013). Interestingly, a number of studies have demonstrated that both the nucleolus and silenced chromatin undergo phase separation to defined sub-nuclear compartments (Feric et al., 2016; Larson et al., 2017; Strom et al., 2017). The proper formation and organization of the nucleolus by phase separation is essential for ribosomal processing, and therefore cell survival (Feric et al., 2016; Banani et al., 2017). Because perturbation of nuclear actin can induce chromatin and nucleolar coalescence, this suggests that nuclear actin may, by unknown mechanisms, regulate phase separation.
Together, these data demonstrate that nuclear actin contributes to the structure and organization of the nucleus, and this process is highly conserved among higher eukaryotes. Specifically, nuclear actin is essential for nuclear envelope integrity and this may be accomplished via interactions with Emerin and lamins. Further, nuclear actin may contribute to the organization of nuclear subcompartments. However, the exact mechanisms by which nuclear actin regulates nuclear and nuclear subcompartment organization remain to be defined.
NUCLEAR ACTIN IN DISEASE
Considering the vast functions of actin in the nucleus, it is not surprising that nuclear actin has been implicated in a number of diseases including cancer (Spencer et al., 2011; Fiore et al., 2017), neurodegeneration (Bamburg et al., 2010; Munsie et al., 2011), and myopathies (Costa et al., 2004; Bathe et al., 2007; Domazetovska et al., 2007) (Fig. 4).
Figure 4:
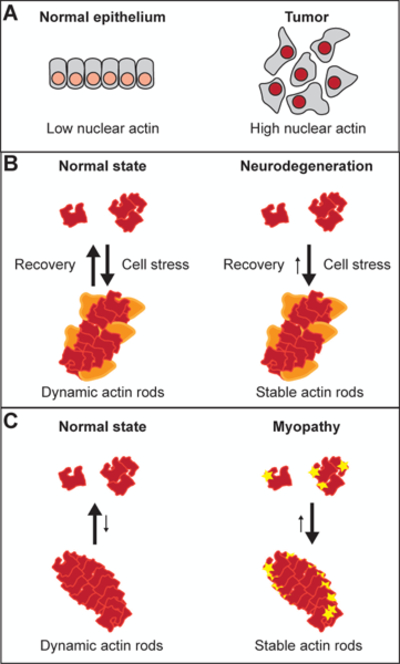
Nuclear actin in disease states. (A) Model for the role of nuclear actin in cancer. Extracellular cues maintain low levels of nuclear actin (light red nuclei in grey cells) in the normal epithelium. Changes in the microenvironment induce a tumor-like phenotype by increasing nuclear actin (dark red nuclei in grey cells) and thereby, causing transcriptional reprogramming (adapted from (Fiore et al., 2017)). (B) Model for the role of nuclear actin in neurodegeneration. When normal cells are stressed, nuclear actin transiently polymerizes into Cofilin-decorated rods (actin in red; Cofilin in orange). In cases of neurodegeneration, the actin/Cofilin rods are stabilized. (C) Model for the role of nuclear actin in myopathy. Under standard conditions, nuclear actin rod formation is limited. However, mutations in actin (yellow star on red actin) promotes polymerization and stabilization of nuclear actin rods. For simplicity, individual actin filaments are shown to represent nuclear actin rods, but rods may be composed of bundles of actin filaments.
While each of the functions of nuclear actin discussed above could easily be connected to cancer, it was recently demonstrated that there is a link between cellular senescence, the escape from it (i.e. tumorigenesis), and nuclear actin levels (Spencer et al., 2011). Specifically, mammary epithelial cells maintain their senescence through Laminin-111 induced signaling. Perturbing this pathway increases nuclear actin levels, transcription, and eventually proliferation (Spencer et al., 2011). Further, the expression of an NLS-tagged non-polymerizable actin (R62D) allowed cells to escape senescence regardless of the presence of Laminin-111, suggesting that maintaining a low level of nuclear actin monomers is essential for senescence (Spencer et al., 2011). A more recent study found that Laminin-111 promotes the cytoplasmic localization of actin by positively regulating Exportin 6 via phosphoinositide 3-kinase (PI3K) signaling (Fiore et al., 2017). Malignant cells are insensitive to Laminin-111 and exhibit high levels of nuclear actin. Notably, the malignant phenotype can be replicated by knocking down Exportin 6 in previously quiescent cells (Fiore et al., 2017). Thus, high levels of nuclear actin contribute to a tumor-like phenotype (Fig. 4A). As perturbing Laminin-111-induced signaling ultimately causes transcriptional reprogramming of these cells, it is tempting to hypothesize that nuclear actin regulates chromatin remodeling complexes to drive tumor formation and progression. Future studies are needed to explore this and other potential mechanisms of nuclear actin activity in cancer.
Nuclear actin has also been implicated in neurodegenerative disorders. These disease states are associated with cytoplasmic and/or nuclear actin/Cofilin rods (Minamide et al., 2000; Bamburg et al., 2010; Munsie et al., 2011). Interestingly, in the context of Huntington’s disease, mutant huntingtin protein localizes to the nucleus of cultured neurons and is covalently crosslinked to nuclear actin/Cofilin rods (Munsie et al., 2011). It is suspected that this protein interaction stabilizes nuclear actin/Cofilin rods and thereby, contributes to disease progression (Munsie et al., 2011) (Fig. 4B). Considering nuclear actin rod formation is a common response to cellular stress (Fukui, 1978; Fukui and Katsumaru, 1979, 1980; Osborn and Weber, 1980; Welch and Suhan, 1985; Iida et al., 1986; Pendleton et al., 2003; Plessner et al., 2015), it is likely these cells experienced some type of stress, nuclear actin rod formation was induced, and these rods were stabilized by mutant huntingtin. Interestingly, similar actin/Cofilin rods have been observed in the cytoplasm and nucleus of neurons from Alzheimer’s patients and cultured cells induced to mimic the disease (Minamide et al., 2000; Huang et al., 2008; Whiteman et al., 2009; Bamburg et al., 2010; Munsie et al., 2011). Thus, actin/Cofilin rods are associated with a number of neurodegenerative disorders. Rods have also been observed at an increased frequency with age in normal samples (Masurovsky et al., 1970; Feldman and Peters, 1972; Fiori, 1987; Bamburg et al., 2010). The mechanisms that contribute to rod formation in the contexts of aging and Alzheimer’s disease have yet to be explored. Together, these data suggest that persistent nuclear actin rods occur in neurodegenerative states. We suspect that the persistence of the rods contributes to these pathologies. Indeed, persistent rod formation results in disruption but not death of neurites in culture, mimicking disease pathogenesis (Minamide et al., 2000).
A subset of actin myopathies are referred to as intranuclear rod myopathies, so named because they exhibit actin rods in the nuclei of muscle cells (Clarkson et al., 2004). Specifically, several patient myopathy-linked mutations of α-actin result in nuclear actin rod formation in cultured myoblasts and myotubes (Costa et al., 2004; Bathe et al., 2007) (Fig. 4C). The formation of these rods happens in a similar manner to that of wild-type α-actin nuclear actin rod formation in response to cellular stressors (Domazetovska et al., 2007). Further, a specific mutation of α-actin induces nuclear actin rods and reduces the mitotic index of cultured myoblasts, suggesting that persistent nuclear actin rods are detrimental to muscle development (Domazetovska et al., 2007). The exact mechanisms by which nuclear actin rods contribute to myopathies remains unknown. However, as persistent nuclear actin rods alter HDAC activity and, ultimately, transcription (Serebryannyy et al., 2016b), it is appealing to hypothesize the involvement of altered nuclear actin-dependent transcription in intranuclear rod myopathies.
These studies strongly support the model that the level and structure of nuclear actin have to be tightly regulated for normal cellular function and human health; misregulation contributes to diseases such as cancer (Spencer et al., 2011; Fiore et al., 2017), neurodegeneration (Bamburg et al., 2010; Munsie et al., 2011), and myopathies (Costa et al., 2004; Bathe et al., 2007; Domazetovska et al., 2007). Given the broad functions of nuclear actin, we suspect that misregulation of nuclear actin also contributes to other disease. Indeed, it is particularly intriguing to consider the roles of nuclear actin in diseases that are associated with cellular stress, such as metabolic diseases including diabetes, heart disease, fatty liver, and obesity.
CONCLUSION
The initial observations of nuclear actin were >50 years ago. While early studies hinted at function, they were met with skepticism; this caused the study of nuclear actin to lag behind its cytoplasmic counterpart. The discovery that the nuclear localization of actin is an active and regulated process strongly suggested that there was a purpose for actin to translocate to the nucleus. We now know that nuclear actin has many functions, including regulating all three RNA polymerases, specific transcription factors, chromatin remodeling complex activity and formation, histone deacetylases, chromatin movement, DNA damage repair, and nuclear structure/integrity (Fig. 2). However, the precise mechanisms that underlie actin’s contribution to each of these functions remain to be fully defined and must be further explored. Multiple pools of nuclear actin monomers and polymers have also been identified – the regulation and functions of these pools are largely unknown. Nuclear actin also contributes to human health and disease, although to what extent needs to be fully explored. In order to overcome these knowledge gaps, we must identify tools to specifically probe the nuclear functions of actin separately from its cytoplasmic activities. Further, the vast majority of nuclear actin studies have been performed in cultured cells or large oocyte nuclei. The use of in vivo, genetic models will greatly enhance our understanding of nuclear actin. Such studies will build on the groundwork that has been laid – actin localizes to the nucleus and its functions there are essential for key cellular processes.
Acknowledgements
We thank members of the Tootle Lab for careful review of the manuscript. Information Technology Services – Research Services, provided data storage support. This project is supported by National Institutes of Health R01GM116885. D.J.K. is partially supported by NINDS T32NS045549.
Grant sponsors: NIH/NIGMS, NIH/NINDS; Grant numbers: R01GM116885, T32NS045549
Literature cited
- Abe H, Nagaoka R, Obinata T. 1993. Cytoplasmic localization and nuclear transport of cofilin in cultured myotubes. Exp Cell Res 206:1–10. [DOI] [PubMed] [Google Scholar]
- Almuzzaini B, Sarshad AA, Rahmanto AS, Hansson ML, Von Euler A, Sangfelt O, Visa N, Farrants AK, Percipalle P. 2016. In beta-actin knockouts, epigenetic reprogramming and rDNA transcription inactivation lead to growth and proliferation defects. FASEB J 30:2860–2873. [DOI] [PubMed] [Google Scholar]
- Ambrosino C, Tarallo R, Bamundo A, Cuomo D, Franci G, Nassa G, Paris O, Ravo M, Giovane A, Zambrano N, Lepikhova T, Janne OA, Baumann M, Nyman TA, Cicatiello L, Weisz A. 2010. Identification of a hormone-regulated dynamic nuclear actin network associated with estrogen receptor alpha in human breast cancer cell nuclei. Mol Cell Proteomics 9:1352–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrin C, McDonald D, Attwood KM, Rodrigue A, Ghosh S, Mirzayans R, Masson JY, Dellaire G, Hendzel MJ. 2012. A requirement for polymerized actin in DNA double-strand break repair. Nucleus-Austin 3:384–395. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Plessner M, Sherrard A, Morita K, Misu S, Virant D, Kleinschnitz EM, Harniman R, Alibhai D, Baumeister S, Miyamoto K, Endesfelder U, Kaidi A, Grosse R. 2017. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat Cell Biol 19:1389–1399. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Wang H, Grosse R. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340:864–867. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol 15:185–230. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW, Davis RC, Flynn KC, Goldsbury C, Jensen JR, Maloney MT, Marsden IT, Minamide LS, Pak CW, Shaw AE, Whiteman I, Wiggan O. 2010. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr Alzheimer Res 7:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe FS, Rommelaere H, Machesky LM. 2007. Phenotypes of myopathy-related actin mutants in differentiated C2C12 myotubes. BMC Cell Biol 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. 2013. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell 24:982–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Lee T, Mullins RD. 2015. DNA damage induces nuclear actin filament assembly by Formin −2 and Spire-(1/2) that promotes efficient DNA repair. [corrected]. Elife 4:e07735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI, Valenzuela P, Rutter WJ. 1977. Phosphorylation of yeast DNA-dependent RNA polymerases in vivo and in vitro. Isolation of enzymes and identification of phosphorylated subunits. J Biol Chem 252:3082–3091. [PubMed] [Google Scholar]
- Bettinger BT, Gilbert DM, Amberg DC. 2004. Actin up in the nucleus. Nat Rev Mol Cell Biol 5:410–415. [DOI] [PubMed] [Google Scholar]
- Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ , the Mouse Genome Database G. 2017. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res 45:D723–D729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogolyubova N, Ginzburg A. 2013. Actin Colocalization with Metaphase Chromosomes of the Second Meiosis in Ovulated Mouse Oocytes. Developmental Biology Journal 2013:6. [Google Scholar]
- Bohnsack MT, Stuven T, Kuhn C, Cordes VC, Gorlich D. 2006. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol 8:257–263. [DOI] [PubMed] [Google Scholar]
- Casella JF, Flanagan MD, Lin S. 1981. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 293:302–305. [DOI] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16:825–831. [DOI] [PubMed] [Google Scholar]
- Clark TG, Merriam RW. 1977. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell 12:883–891. [DOI] [PubMed] [Google Scholar]
- Clark TG, Rosenbaum JL. 1979. An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell 18:1101–1108. [DOI] [PubMed] [Google Scholar]
- Clarkson E, Costa CF, Machesky LM. 2004. Congenital myopathies: diseases of the actin cytoskeleton. J Pathol 204:407–417. [DOI] [PubMed] [Google Scholar]
- Clattenburg RE, Singh RP, Montemurro DG. 1972. Intranuclear filamentous inclusions in neurons of the rabbit hypothalamus. J Ultrastruct Res 39:549–555. [DOI] [PubMed] [Google Scholar]
- Costa CF, Rommelaere H, Waterschoot D, Sethi KK, Nowak KJ, Laing NG, Ampe C, Machesky LM. 2004. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J Cell Sci 117:3367–3377. [DOI] [PubMed] [Google Scholar]
- Cruz JR, Moreno Diaz de la Espina S. 2009. Subnuclear compartmentalization and function of actin and nuclear myosin I in plants. Chromosoma 118:193–207. [DOI] [PubMed] [Google Scholar]
- de Lanerolle P 2012. Nuclear actin and myosins at a glance. J Cell Sci 125:4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw R, Gruenbaum Y, Medalia O. 2018. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol 28:34–45. [DOI] [PubMed] [Google Scholar]
- Domazetovska A, Ilkovski B, Cooper ST, Ghoddusi M, Hardeman EC, Minamide LS, Gunning PW, Bamburg JR, North KN. 2007. Mechanisms underlying intranuclear rod formation. Brain 130:3275–3284. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. 2011. Actin structure and function. Annu Rev Biophys 40:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Rajakyla EK, Joensuu MS, Huet G, Ferrantelli E, Xie T, Jaalinoja H, Jokitalo E, Vartiainen MK. 2015. Genome-wide RNAi screen for nuclear actin reveals a network of cofilin regulators. Journal of Cell Science 128:2388–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. 2012. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A 109:E544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvas AS, Harrington CA, Bonner J. 1975. Major nonhistone proteins of rat liver chromatin: preliminary identification of myosin, actin, tubulin, and tropomyosin. Proc Natl Acad Sci U S A 72:3902–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179:1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egly JM, Miyamoto NG, Moncollin V, Chambon P. 1984. Is actin a transcription initiation factor for RNA polymerase B? EMBO J 3:2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L, Forer A. 2007. Possible roles of actin and myosin during anaphase chromosome movements in locust spermatocytes. Protoplasma 231:201–213. [DOI] [PubMed] [Google Scholar]
- Fairley EA, Kendrick-Jones J, Ellis JA. 1999. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci 112 ( Pt 15):2571–2582. [DOI] [PubMed] [Google Scholar]
- Feldman ML, Peters A. 1972. Intranuclear rods and sheets in rat cochlear nucleus. J Neurocytol 1:109–127. [DOI] [PubMed] [Google Scholar]
- Feric M, Brangwynne CP. 2013. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol 15:1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Naum-Ongania G, Longobardi E, Palazzolo M, Disanza A, Diaz VM, Crippa MP, Scita G, Blasi F. 2009. Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol Biol Cell 20:3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A, Spencer VA, Mori H, Carvalho HF, Bissell MJ, Bruni-Cardoso A. 2017. Laminin-111 and the Level of Nuclear Actin Regulate Epithelial Quiescence via Exportin-6. Cell Rep 19:2102–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori MG. 1987. Intranuclear inclusions in Schwann cells of aged fowl ciliary ganglia. J Anat 154:201–214. [PMC free article] [PubMed] [Google Scholar]
- Fomproix N, Percipalle P. 2004. An actin-myosin complex on actively transcribing genes. Exp Cell Res 294:140–148. [DOI] [PubMed] [Google Scholar]
- Forer A, Jackson WT. 1979. Actin in spindles of Haemanthus katherinae endosperm. I. General results using various glycerination methods. J Cell Sci 37:323–347. [DOI] [PubMed] [Google Scholar]
- Forer A, Jackson WT, Engberg A. 1979. Actin in spindles of Haemanthus katherinae endosperm. II. Distribution of actin in chromosomal spindle fibres, determined by analysis of serial sections. J Cell Sci 37:349–371. [DOI] [PubMed] [Google Scholar]
- Fuchsova B, Serebryannyy LA, de Lanerolle P. 2015. Nuclear actin and myosins in adenovirus infection. Exp Cell Res 338:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y 1978. Intranuclear actin bundles induced by dimethyl sulfoxide in interphase nucleus of Dictyostelium. J Cell Biol 76:146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Katsumaru H. 1979. Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp Cell Res 120:451–455. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Katsumaru H. 1980. Dynamics of nuclear actin bundle induction by dimethyl sulfoxide and factors affecting its development. J Cell Biol 84:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell 5:927–937. [DOI] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Mullins RD. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306:1021–1025. [DOI] [PubMed] [Google Scholar]
- Gedge LJ, Morrison EE, Blair GE, Walker JH. 2005. Nuclear actin is partially associated with Cajal bodies in human cells in culture and relocates to the nuclear periphery after infection of cells by adenovirus 5. Exp Cell Res 303:229–239. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341. [DOI] [PubMed] [Google Scholar]
- Goldstein L, Rubin R, Ko C. 1977. The presence of actin in nuclei: a critical appraisal. Cell 12:601–608. [DOI] [PubMed] [Google Scholar]
- Gonsior SM, Platz S, Buchmeier S, Scheer U, Jockusch BM, Hinssen H. 1999. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci 112 ( Pt 6):797–809. [DOI] [PubMed] [Google Scholar]
- Grzanka D, Grzanka A, Izdebska M, Gackowska L, Stepien A, Marszalek A. 2010a. Actin reorganization in CHO AA8 cells undergoing mitotic catastrophe and apoptosis induced by doxorubicin. Oncol Rep 23:655–663. [DOI] [PubMed] [Google Scholar]
- Grzanka D, Marszalek A, Gagat M, Izdebska M, Gackowska L, Grzanka A. 2010b. Doxorubicin-induced F-actin reorganization in cofilin-1 (nonmuscle) down-regulated CHO AA8 cells. Folia Histochem Cytobiol 48:377–386. [DOI] [PubMed] [Google Scholar]
- Grzanka D, Marszalek A, Izdebska M, Gackowska L, Andrzej Szczepanski M, Grzanka A. 2011. Actin cytoskeleton reorganization correlates with cofilin nuclear expression and ultrastructural changes in cho aa8 cell line after apoptosis and mitotic catastrophe induction by doxorubicin. Ultrastruct Pathol 35:130–138. [DOI] [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. 1995. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol 131:1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Melton DA. 2008. Nuclear reprogramming in cells. Science 322:1811–1815. [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U. 1999. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell 10:2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden SM, Miller PS, Brauweiler A, Bamburg JR. 1993. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32:9994–10004. [DOI] [PubMed] [Google Scholar]
- Hitchcock SE. 1980. Actin deoxyroboncuclease I interaction. Depolymerization and nucleotide exchange. J Biol Chem 255:5668–5673. [PubMed] [Google Scholar]
- Hodo HG 3rd, Blatti SP. 1977. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry 16:2334–2343. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol 6:1094–1101. [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Vargas GM, Ramchandran R, Stojiljkovic L, Goodrich JA, de Lanerolle P. 2006. Nuclear myosin I is necessary for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II. J Cell Biochem 99:1001–1009. [DOI] [PubMed] [Google Scholar]
- Holaska JM, Kowalski AK, Wilson KL. 2004. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol 2:E231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL. 2007. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry 46:8897–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N. 2004. A role for beta-actin in RNA polymerase III transcription. Genes Dev 18:3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TY, Minamide LS, Bamburg JR, Bokoch GM. 2008. Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell 15:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, Rosenfeld MG, Glass CK. 2011. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature 470:414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17:193–201. [DOI] [PubMed] [Google Scholar]
- Huxley HE. 1957. The double array of filaments in cross-striated muscle. J Biophys Biochem Cytol 3:631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. 1969. The mechanism of muscular contraction. Science 164:1356–1365. [PubMed] [Google Scholar]
- Iida K, Iida H, Yahara I. 1986. Heat shock induction of intranuclear actin rods in cultured mammalian cells. Exp Cell Res 165:207–215. [DOI] [PubMed] [Google Scholar]
- Iida K, Matsumoto S, Yahara I. 1992. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct 17:39–46. [DOI] [PubMed] [Google Scholar]
- Iida K, Moriyama K, Matsumoto S, Kawasaki H, Nishida E, Yahara I. 1993. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-M(r) actin-binding and depolymerizing protein. Gene 124:115–120. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–473. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Bischoff R, Holtzer H. 1969. Formation of arrowhead complexes with heavy meromyosin in a variety of cell types. J Cell Biol 43:312–328. [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. 2002. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 21:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Becker M, Hindennach I, Jockusch H. 1974. Slime mould actin: homology to vertebrate actin and presence in the nucleus. Exp Cell Res 89:241–246. [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Brown DF, Rusch HP. 1971. Synthesis and some properties of an actin-like nuclear protein in the slime mold Physarum polycephalum. J Bacteriol 108:705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. 2013. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol 9:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P, Chen M, Winkler DD, Luger K, Shen X. 2013. Evidence for monomeric actin function in INO80 chromatin remodeling. Nat Struct Mol Biol 20:426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelpsch DJ, Groen CM, Fagan TN, Sudhir S, Tootle TL. 2016. Fascin regulates nuclear actin during Drosophila oogenesis. Mol Biol Cell 27:2965–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. 2014. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol 24:1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Gerdes K. 2005. Bacterial DNA segregation by the actin-like MreB protein. Trends Cell Biol 15:343–345. [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol 12:238–244. [DOI] [PubMed] [Google Scholar]
- Kysela K, Philimonenko AA, Philimonenko VV, Janacek J, Kahle M, Hozak P. 2005. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol 124:347–358. [DOI] [PubMed] [Google Scholar]
- Lane NJ. 1969. Intranuclear fibrillar bodies in actinomycin D-treated oocytes. J Cell Biol 40:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. 1997. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J 16:5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattanzi G, Cenni V, Marmiroli S, Capanni C, Mattioli E, Merlini L, Squarzoni S, Maraldi NM. 2003. Association of emerin with nuclear and cytoplasmic actin is regulated in differentiating myoblasts. Biochem Biophys Res Commun 303:764–770. [DOI] [PubMed] [Google Scholar]
- Lenart P, Bacher CP, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. 2005. A contractile nuclear actin network drives chromosome congression in oocytes. Nature 436:812–818. [DOI] [PubMed] [Google Scholar]
- Lestourgeon WM, Forer A, Yang YZ, Bertram JS, Pusch HP. 1975. Contractile proteins. Major components of nuclear and chromosome non-histone proteins. Biochim Biophys Acta 379:529–552. [PubMed] [Google Scholar]
- Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, Dickinson RB. 2015. Moving Cell Boundaries Drive Nuclear Shaping during Cell Spreading. Biophys J 109:670–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist MR, Storaska AJ, Liu TC, Larsen SD, Evans T, Neubig RR, Jaffrey SR. 2014. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell 156:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslova A, Krasikova A. 2012. Nuclear actin depolymerization in transcriptionally active avian and amphibian oocytes leads to collapse of intranuclear structures. Nucleus 3:300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masurovsky EB, Benitez HH, Kim SU, Murray MR. 1970. Origin, development, and nature of intranuclear rodlets and associated bodies in chicken sympathetic neurons. J Cell Biol 44:172–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F, Matsumoto S, Yahara I, Yonezawa N, Nishida E, Sakai H. 1988. Cloning and characterization of porcine brain cofilin cDNA. Cofilin contains the nuclear transport signal sequence. J Biol Chem 263:11564–11568. [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. 2006. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 172:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. 1997. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol 138:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta IS, Amira M, Harvey AJ, Bridger JM. 2010. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol 11:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melak M, Plessner M, Grosse R. 2017. Actin visualization at a glance. J Cell Sci 130:525–530. [DOI] [PubMed] [Google Scholar]
- Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. 2000. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol 2:628–636. [DOI] [PubMed] [Google Scholar]
- Miranda AF, Godman GC. 1973. The effects of cytochalasin D on differentiating muscle in culture. Tissue Cell 5:1–22. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Pasque V, Jullien J, Gurdon JB. 2011. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Gene Dev 25:946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogessie B, Schuh M. 2017. Actin protects mammalian eggs against chromosome segregation errors. Science 357. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Jensen RB, Lowe J, Gerdes K. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J 21:3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse H, Buratowski RM, Silver PA, Buratowski S. 1999. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc Natl Acad Sci U S A 96:12542–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Somogyi K, Kondo H, Monnier N, Falk HJ, Machado P, Bathe M, Nedelec F, Lenart P. 2014. An Arp2/3 nucleated F-actin shell fragments nuclear membranes at nuclear envelope breakdown in starfish oocytes. Curr Biol 24:1421–1428. [DOI] [PubMed] [Google Scholar]
- Muhlhausser P, Muller EC, Otto A, Kutay U. 2001. Multiple pathways contribute to nuclear import of core histones. EMBO Rep 2:690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie L, Caron N, Atwal RS, Marsden I, Wild EJ, Bamburg JR, Tabrizi SJ, Truant R. 2011. Mutant huntingtin causes defective actin remodeling during stress: defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet 20:1937–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsie LN, Caron N, Desmond CR, Truant R. 2009. Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat Methods 6:317. [DOI] [PubMed] [Google Scholar]
- Munsie LN, Desmond CR, Truant R. 2012. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci 125:3977–3988. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 13:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu H, Ueda K. 1983. Association of actin with the nuclear matrix from bovine lymphocytes. Exp Cell Res 143:55–62. [DOI] [PubMed] [Google Scholar]
- Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, Sakai H. 1987. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci U S A 84:5262–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E, Maekawa S, Sakai H. 1984. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 23:5307–5313. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Kawamura H, Tanaka Y. 1964. [Actin and Myosin-Like Proteins in the Calf Thymus Cell Nucleus]. J Biochem 56:6–15. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Kawamura H, Yamamoto T. 1963. [Extraction of a Protein Resembling Actin from the Cell Nucleus of the Calf Thymus]. J Biochem 54:298–300. [DOI] [PubMed] [Google Scholar]
- Okada I, Fujiki S, Iwase S, Abe H. 2012. Stabilization of actin filaments prevents germinal vesicle breakdown and affects microtubule organization in Xenopus oocytes. Cytoskeleton (Hoboken) 69:312–323. [DOI] [PubMed] [Google Scholar]
- Osborn M, Weber K. 1980. Dimethylsulfoxide and the ionophore A23187 affect the arrangement of actin and induce nuclear actin paracrystals in PtK2 cells. Exp Cell Res 129:103–114. [DOI] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955–3966. [DOI] [PubMed] [Google Scholar]
- Parfenov VN, Davis DS, Pochukalina GN, Sample CE, Bugaeva EA, Murti KG. 1995. Nuclear actin filaments and their topological changes in frog oocytes. Exp Cell Res 217:385–394. [DOI] [PubMed] [Google Scholar]
- Parisis N, Krasinska L, Harker B, Urbach S, Rossignol M, Camasses A, Dewar J, Morin N, Fisher D. 2017. Initiation of DNA replication requires actin dynamics and formin activity. Embo Journal 36:3212–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 1977. Isolation and characterization of chromatin from the cellular slime mold, Dictyostelium discoideum. Biochemistry 16:2771–2777. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. 1999. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol 145:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A. 2003. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem 278:14394–14400. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N. 2003. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci U S A 100:6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Jonsson A, Nashchekin D, Karlsson C, Bergman T, Guialis A, Daneholt B. 2002. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res 30:1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol 153:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, Shabanowitz J, Hunt DF, Hozak P, de Lanerolle P. 2000. A myosin I isoform in the nucleus. Science 290:337–341. [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Janacek J, Harata M, Hozak P. 2010. Transcription-dependent rearrangements of actin and nuclear myosin I in the nucleolus. Histochem Cell Biol 134:243–249. [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 6:1165–1172. [DOI] [PubMed] [Google Scholar]
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. 2015. Nuclear F-actin Formation and Reorganization upon Cell Spreading. Journal of Biological Chemistry 290:11209–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 2016. Actin and Actin-Binding Proteins Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 2017. What We Know and Do Not Know About Actin In: Jockusch BM, editor The Actin Cytoskeleton. Cham: Springer International Publishing. p 331–347. [Google Scholar]
- Prochniewicz E, Henderson D, Ervasti JM, Thomas DD. 2009. Dystrophin and utrophin have distinct effects on the structural dynamics of actin. Proc Natl Acad Sci U S A 106:7822–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi TY, Tang W, Wang L, Zhai L, Guo LJ, Zeng XL. 2011. G-actin Participates in RNA Polymerase II-dependent Transcription Elongation by Recruiting Positive Transcription Elongation Factor b (P-TEFb). Journal of Biological Chemistry 286:15171–15181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. Lifeact: a versatile marker to visualize F-actin. Nat Methods 5:605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungger D, Rungger-Brandle E, Chaponnier C, Gabbiani G. 1979. Intranuclear injection of anti-actin antibodies into Xenopus oocytes blocks chromosome condensation. Nature 282:320–321. [DOI] [PubMed] [Google Scholar]
- Ryser U 1970. [The ultrastructure of mitotic nuclei in plasmodia of Physarum polycephalum]. Z Zellforsch Mikrosk Anat 110:108–130. [PubMed] [Google Scholar]
- Sanger JW, Gwinn J, Sanger JM. 1980a. Dissolution of cytoplasmic actin bundles and the induction of nuclear actin bundles by dimethyl sulfoxide. J Exp Zool 213:227–230. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Sanger JM, Kreis TE, Jockusch BM. 1980b. Reversible translocation of cytoplasmic actin into the nucleus caused by dimethyl sulfoxide. Proc Natl Acad Sci U S A 77:5268–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasseville AM, Langelier Y. 1998. In vitro interaction of the carboxy-terminal domain of lamin A with actin. FEBS Lett 425:485–489. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Schroer TA. 1999. Actin-related proteins. Annu Rev Cell Dev Biol 15:341–363. [DOI] [PubMed] [Google Scholar]
- Scheer U, Hinssen H, Franke WW, Jockusch BM. 1984. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell 39:111–122. [DOI] [PubMed] [Google Scholar]
- Schoenenberger CA, Buchmeier S, Boerries M, Sutterlin R, Aebi U, Jockusch BM. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. Journal of Structural Biology 152:157–168. [DOI] [PubMed] [Google Scholar]
- Schulze SR, Curio-Penny B, Speese S, Dialynas G, Cryderman DE, McDonough CW, Nalbant D, Petersen M, Budnik V, Geyer PK, Wallrath LL. 2009. A comparative study of Drosophila and human A-type lamins. PLoS One 4:e7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. 2000. Myosins: a diverse superfamily. Biochim Biophys Acta 1496:3–22. [DOI] [PubMed] [Google Scholar]
- Sen B, Xie Z, Uzer G, Thompson WR, Styner M, Wu X, Rubin J. 2015. Intranuclear Actin Regulates Osteogenesis. Stem Cells 33:3065–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy LA, Cruz CM, de Lanerolle P. 2016a. A Role for Nuclear Actin in HDAC 1 and 2 Regulation. Sci Rep 6:28460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy LA, Parilla M, Annibale P, Cruz CM, Laster K, Gratton E, Kudryashov D, Kosak ST, Gottardi CJ, de Lanerolle P. 2016b. Persistent nuclear actin filaments inhibit transcription by RNA polymerase II. J Cell Sci 129:3412–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebryannyy LA, Yemelyanov A, Gottardi CJ, de Lanerolle P. 2017. Nuclear alpha-catenin mediates the DNA damage response via beta-catenin and nuclear actin. J Cell Sci 130:1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Yoshida M. 2014. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541–544. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12:147–155. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila RV, Forer A. 2000. Evidence that actin and myosin are involved in the poleward flux of tubulin in metaphase kinetochore microtubules of crane-fly spermatocytes. J Cell Sci 113 ( Pt 4):597–609. [DOI] [PubMed] [Google Scholar]
- Simon DN, Zastrow MS, Wilson KL. 2010. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus 1:264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SS, Kelly KH, Jockusch BM. 1979. Actin co-purifies with RNA polymerase II. Biochem Biophys Res Commun 86:161–166. [DOI] [PubMed] [Google Scholar]
- Somosy Z, Csuka O, Sugar J. 1976. Electron microscopic examination of nuclear inclusions induced by Vinca alkaloids. Exp Cell Res 101:429–434. [DOI] [PubMed] [Google Scholar]
- Soufo HJ, Graumann PL. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr Biol 13:1916–1920. [DOI] [PubMed] [Google Scholar]
- Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ. 2011. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci 124:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklen AJ, Fagan TN, Lovander KE, Tootle TL. 2014. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol 393:209–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub F 1943. Actin, ii. Stud Inst Med Chem Univ Szeged 3:23–37. [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, Gorlich D. 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 22:5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman JR, Kashina A. 2013. Post-translational modification and regulation of actin. Curr Opin Cell Biol 25:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 21:6935–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J, Weber K. 1978. Actin amino-acid sequences. Comparison of actins from calf thymus, bovine brain, and SV40-transformed mouse 3T3 cells with rabbit skeletal muscle actin. Eur J Biochem 90:451–462. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316:1749–1752. [DOI] [PubMed] [Google Scholar]
- Venit T, Xie X, Percipalle P. 2018. 15 - Actin in the Cell Nucleus A2 - Lavelle, Christophe In: Victor J-M, editor Nuclear Architecture and Dynamics. Boston: Academic Press; p 345–367. [Google Scholar]
- Viita T, Vartiainen MK. 2017. From Cytoskeleton to Gene Expression: Actin in the Nucleus. Handb Exp Pharmacol 235:311–329. [DOI] [PubMed] [Google Scholar]
- Wachi M, Matsuhashi M. 1989. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol 171:3123–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, Nishida E. 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J 17:1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann I, Walde S, Kehlenbach RH. 2007. Nuclear import of c-Jun is mediated by multiple transport receptors. J Biol Chem 282:27685–27692. [DOI] [PubMed] [Google Scholar]
- Wang YH, Hariharan A, Bastianello G, Toyama Y, Shivashankar GV, Foiani M, Sheetz MP. 2017. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun 8:2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver RF. 1976. Structural comparison of deoxyribonucleic acid-dependent ribonucleic acid polymerases I and II from the slime mold Physarum polycephalum. Arch Biochem Biophys 172:470–475. [DOI] [PubMed] [Google Scholar]
- Welch WJ, Suhan JP. 1985. Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat-shock treatment. J Cell Biol 101:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman IT, Gervasio OL, Cullen KM, Guillemin GJ, Jeong EV, Witting PK, Antao ST, Minamide LS, Bamburg JR, Goldsbury C. 2009. Activated actin-depolymerizing factor/cofilin sequesters phosphorylated microtubule-associated protein during the assembly of alzheimer-like neuritic cytoskeletal striations. J Neurosci 29:12994–13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, Critchley DR, Kendrick-Jones J. 1995. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci 108 ( Pt 1):63–71. [DOI] [PubMed] [Google Scholar]
- Wineland DM, Kelpsch DJ, Tootle TL. 2018. Multiple pools of nuclear actin. The Anatomical Record. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W 2004. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14:461–469. [DOI] [PubMed] [Google Scholar]
- Woolner S, O’Brien LL, Wiese C, Bement WM. 2008. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol 182:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Almuzzaini B, Drou N, Kremb S, Yousif A, Farrants AO, Gunsalus K, Percipalle P. 2018. beta-Actin-dependent global chromatin organization and gene expression programs control cellular identity. FASEB J:fj201700753R. [DOI] [PubMed] [Google Scholar]
- Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. 2008. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Gene Dev 22:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625–636. [DOI] [PubMed] [Google Scholar]


