Abstract
Background
Quantification of biofluid cytokines is a rapidly growing area of translational research. However, comparability across the expanding number of available assay platforms for detection of the same proteins remains to be determined. We aimed to directly compare a panel of commonly measured cytokines in plasma of typically aging adults across two high sensitivity quantification platforms, Meso Scale Discovery high performance electrochemiluminiscence (HPE) and single-molecule immunosorbent assays (Simoa) by Quanterix.
Methods
57 community-dwelling older adults completed a blood draw, neuropsychological assessment, and brain MRI as part of a healthy brain aging study. Plasma samples from the same draw dates were analyzed for IL-10, IP-10, IL-6, TNFα, and IL-1β on HPE and Simoa, separately. Reliable detectability (coefficient of variance (CV)<20% and outliers 3 interquartiles above the median removed), intra-assay precision, absolute concentrations, reproducibility across platforms, and concurrent associations with external variables of interest (e.g., demographics, peripheral markers of vascular health, and brain health) were examined.
Results
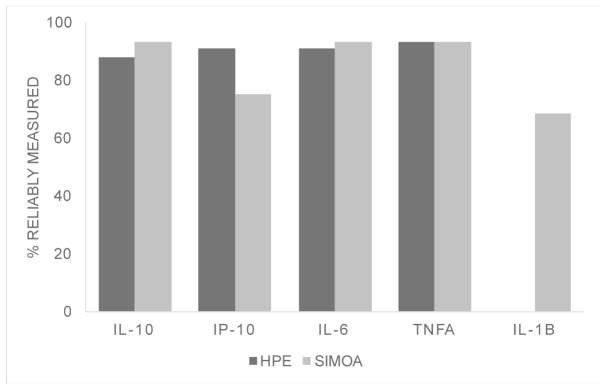
The proportion of cytokines reliably measured on HPE (87.7–93.0%) and Simoa (75.4–93.0%) did not differ (ps>0.32), with the exception of IL-1β which was only reliably measured using Simoa (68.4%). On average, CVs were acceptable at <8% across both platforms. Absolute measured concentrations were higher using Simoa for IL-10, IL-6, and TNFα (ps<0.05). HPE and Simoa shared only small-to-moderate proportions of variance with one another on the same cytokine proteins (range: r=0.26 for IL-10 to r=0.64 for IL-6), though platform agreement did not dependent on cytokine concentrations. Cytokine ratios within each platform demonstrated similar relative patterns of up- and down-regulation across HPE and Simoa, though still significantly differed (ps<0.001). Supporting concurrent validity, all 95% confidence intervals of the correlations between cytokines and external variables overlapped between the two platforms. Moreover, most associations were in expected directions and consistently so across platforms (e.g., IL-6 and TNFα), though with several notable exceptions for IP-10 and IL-10.
Conclusions
HPE and Simoa showed comparable detectability and intra-assay precision measuring a panel of commonly examined cytokine proteins, with the exception of IL-1β which was not reliably detected on HPE. However, Simoa demonstrated overall higher concentrations and the two platforms did not show agreement when directly compared against one another. Relative cytokine ratios and associations demonstrated similar patterns across platforms. Absolute cytokine concentrations may not be directly comparable across platforms, may be analyte dependent, and interpretation may be best limited to discussion of relative associations.
Keywords: Meso Scale Discovery, Quanterix, immune activation, typical aging, plasma markers
Introduction
Proteomic markers are a rapidly growing area of clinical research to help characterize the biologic cascades underlying disease and behaviors, and improve monitoring and diagnostics in humans. Biofluid cytokine concentrations are a particularly widely measured, diverse group of proteins used to estimate immune activation processes in various healthy and diseased populations; a tool that now spans across disciplines (e.g., psychiatry, neurology) and in large multisite clinical studies1. Advancements in technologies have facilitated ease and pragmatics of biofluid marker quantification by allowing for simultaneous analysis of multiple markers via multiplexing, higher analytical sensitivity and lower limits of detection and quantification, and reduction in the amount of sample needed (e.g., compared to standard ELISA)2–4. However, the growing number of available platforms with slightly differing technologies measuring the same protein markers has made it difficult to not only select the most optimal quantification tool, but to compare marker results across platforms and studies. Though tempting to interpret these “biologic data” as free from external biases, the reproducibility and reliability of the same protein across each platform may not be one-to-one5. These issues become especially relevant in the context of multisite (i.e., multiple laboratories) and longitudinal studies, and for cytokine measurement, which are posited to reflect more nuanced and potentially rapidly changing immune cascades with differing levels of detectability, potentially heightening any (even minute) differences in platform technologies. As the field continues forward with these exciting tools to probe into molecular mechanisms in humans, a more comprehensive and empirical characterization of distinct proteomic marker approaches will be critically important.
While a host of studies have examined platform differences in cytokine measurement using pooled control and known cytokine concentration spiked samples under highly controlled protocols, much fewer have conducted these direct comparisons in actual patient populations. Even using the former standardized spiked samples approach, distinct differences across platforms emerge. High performance electrochemiluminiscence (HPE) by Meso Scale Discovery (MSD) and Biosource Luminex appear to demonstrate overall comparable spike recoveries, coefficients of variance, and low limits of detection, though even these findings may be analyte dependent6–10. For instance, in a direct comparison study by Fu and colleagues, in unspiked serum, MSD HPE showed better performances for IL-10, TNFa, and IL-2, IFNy, and CSF-GM, Luminex showed better performances for IL-8, and the platforms appeared comparable for IL-68. Indeed, the authors concluded that no single assay platform can likely be used for all cytokine quantifications, and that optimal assays and platforms must be selected by cytokine of interest. The comparability of cytokine measurements across platforms may be even less clear when examining biofluids in patient who have unknown levels of immune activation and countless potentially contributing factors to cytokine expression. Despite the inherently less controlled nature of patient platform comparison studies, understanding how distinct proteomic platforms perform in such “real world” samples is needed to support the ecologic validity and translation of biofluid marker technologies for human application. In one study, Breen and colleagues examined 13 cytokine analytes on two platforms (MSD HPE and Luminex) across six laboratory sites as part of the Multicenter AIDS Cohort Study and Women Interagency HIV Study1. Every cytokine examined demonstrated at least one significant lab and/or assay lot effect. Only IL-6 was directly compared on the same samples across platforms and demonstrated correlations with standard ELISA varying between r=0.13 to 0.79 in serum and plasma with MSD HPE or Luminex (from Bio-Rad, BioSources, and Linco manufacturers) platforms. Notably, however, there was convergence of the relative patterns of cytokine elevations across platforms, lots, and laboratories in HIV infection, including measurable cytokine increases in pre-/post-HIV viremia and significant differences between seropositive and negative individuals. Interestingly, other studies that have examined serum cytokine concentrations in clinical HIV6 and aging and Alzheimer’s disease5 demonstrate moderate to high correlations across platforms (HIV: IL-6 and IL-8 r’s=0.62 to 0.80; aging/AD r’s=0.60 to 0.97), and fairly consistent diagnostic group differences. Given that anywhere from <2% to 94% of the variance in cytokine concentration may be explained by platform approach that appears to be highly dependent on the analyte measured and the growing number of new platforms, data quantifying when these differences may be expected are needed to make sense of biofluid proteomic data moving forward.
One such newer biofluid protein analysis technology is single-molecule arrays (Simoa) by Quanterix. Simoa are bead-based enzyme-linked immunosorbent assays conducted on high-density microarrays with femtoliter-sized wells each containing an individual bead (single- or multi-plexed). Simoa was designed using this microscopic bead technology with the goal of increasing sensitivity of detection to low concentration proteins. During analysis, beads containing an enzyme-labeled immunocomplex are hydrolyzed into a fluorescent product that can then be quantified as the proportion of wells that generate a light signal3. Though both based on ELISA techniques, this technology differs slightly from the more extensively utilized MSD platform, which utilizes a high-performance electrochemiluminescence (HPE) detection system. In HPE, plate wells are fitted with carbon electrodes that are coated with different capture-antibodies of interest. Following standard sandwich ELISA techniques and using a ruthenium-conjugated secondary antibody, upon electrical stimulation, the ruthenium chemical label emits light at the surface of each analyte-specific electrode. Light intensity is then measured per electrode to estimate analyte concentrations. While MSD HPE has been more comprehensively studied and demonstrates comparable or better sensitivity and intra-assay precision compared to other available platforms, due to its single-bead femtomolar-sized wells technology, Simoa requires less sample and purports to be more sensitive to proteins at very low concentrations. While a handful of studies have demonstrated superiority of Simoa compared to standard ELISA in these respects, very few have compared MSD HPE to Quanterix Simoa. In one study, Myzithras and colleagues found Simoa achieved higher sensitivity detecting a growth factor (GDF11), a 100-fold improvement in the lower limit of quantification compared to MSD, in standardized pooled samples11. Another study demonstrated that use of CSF AB42/Ab40 ratios as quantified on HPE or Simoa were comparably sensitive in classifying abnormal amyloid PET12. However, no clinical studies have systematically compared HPE and Simoa across a panel of inflammatory cytokines.
Given the scientific and clinical potential of proteomic markers, technological availability including MSD HPE (widely validated) and Quanterix Simoa (high sensitivity to otherwise unmeasurable molecules), and yet apparent analyte- and platform-specific quantification effects, pragmatic guidelines regarding platform selection per analyte are needed. We aimed to characterize the comparability of commonly measured inflammatory cytokine markers drawn from typically aging adults analyzed across MSD HPE versus Quanterix Simoa. We directly examined intra-assay precision, reliable detectability, inter-platform reproducibility, and examined concurrent validity via relative profiles of associations with external measures of vascular and brain health across HPE and Simoa techniques.
Methods
Participants
57 typically aging, community-dwelling older adults completed a blood draw that was analyzed on both MSD HPE and Quanterix Simoa platforms, as well as neuropsychological and neurological evaluations and a brain MRI as part of a larger healthy aging study at the University of California, San Francisco (UCSF) Memory and Aging Center (see Table 1). Inclusion criteria at study visit were a neurologic and neuropsychological exam within normative standards per consensus research criteria13, no major memory concerns or diagnosed memory condition, and no major medical/psychiatry condition that may affect cognitive functioning (e.g., schizophrenia, epilepsy)
Table 1.
Demographic and clinical characteristics of typical aging adult study sample (N=57). Mean and standard deviation are reported unless otherwise specified.
| Age, y | 78.2 (6.4), range 64–95 |
|
| |
| Education, y | 17.9 (2.0), range 12–20 |
|
| |
| Sex (%F) | 57.9% (33) |
|
| |
| Race | |
| % White | 93.0% (53) |
| % Asian | 3.5% (2) |
| % Other/Unknown | 3.5% (2) |
|
| |
| MMSE (median, IQR) | 29 (28, 30), range 25–30 |
|
| |
| Modified Trail Making Test (# sequences/second) | 0.64 (0.22) |
|
| |
| Corpus Callosum FA | 0.54 (0.04) |
|
| |
| Body Mass Index | 26.5 (4.2), range 19.9–37.2 |
|
| |
| Triglycerides (mg/L) (n=35) | 84.5 (39.7), range 31–186 |
|
| |
| HOMA-IR (n=38; median, IQR) | 1.9 (1.3, 2.8), range 0.7–8.7 |
|
| |
| C-reactive protein (n=38; median, IQR) | 1.05 (0.38, 1.8), range 0.1–18.6 |
Note. MMSE = Mini-Mental State Examination; IQR = inter-quartile range; FA = fractional anisotropy; HOMA-IR = homeostatic model assessment of insulin resistance.
Cytokine Measurement
After a 12-hour overnight fast, blood was drawn in the morning and centrifuged at 2000x g for 15 minutes at 4°C before being transferred into 500μL polypropylene cryovials for storage. Samples had never been thawed prior to analysis and were gradually brought to room temperature for analysis (described below). Plasma collected from the same participants on the same day were then analyzed for interleukin 10 (IL-10), interferon gamma-induced protein 10 (IP-10; CXCL10), interleukin 6 (IL-6), tumor necrosis factor alpha (TNFα), and interleukin 1 beta (IL-1β) on HPE and Simoa platforms, separately. All analyses were conducted by a board-certified laboratory technician blinded to study design. Both HPE and Simoa analytic procedures employ diluents that aim to reduce interference from rheumatoid factors (RF) and human anti-mouse antibody (HAMA) to reduce noise to signal ratios (i.e., HAMA blocker that contains a specific binder directed against all types of heterphilic interference including HAMA and RF)
High-performance electrochemiluminiscence by MSD
Plasma cytokine concentrations were measured by high-performance electrochemiluminiscence (HPE) using the multiplex 1 V-PLEX Human Proinflammatory (IL-10, IL-6, TNFα, and IL-1β) and Human Chemokine (IP-10) panels. The multiplex arrays were analyzed with a MESO QuickPlex SQ 120 imager (MSD, Rockville, MD) and Discovery Workbench v4.0 software. Concentrations were obtained in duplicate per each sample in accordance with the manufacturer’s protocol. The following reflect the published manufacturer standards for the lower limit of detectability (LoD) and the lower to upper limit of quantification (LoQ) for each of the measured proteins: IL-6: LLoD 0.06pg/mL and LoQ 1.58–488pg/mL; TNFα: LLoD 0.04pg/mL and LoQ 0.69–248pg/mL; IL-10: LLoD 0.03pg/mL and 0.68–233; IP-10: LLoD 0.37pg/mL and LoQ 1.37–500pg/mL; IL-1β: LLoD 0.04pg/mL and LoQ 2.14–375 pg/mL.
Single molecule arrays by Quanterix
Automated plasma cytokine measurements by the single molecule array (Simoa) principle were conducted with an HD1-Analyzer (Quanterix, Lexington, MA). IL-10, IL-6 and TNFα were measured in multiplex assays using the Cytokine 3-Plex A panel. IP-10 and IL1β were measured with single-analyte assays. Analyses were performed in duplicate, according to manufacturer’s protocol. The following reflect the published manufacturer standards for the lower limit of detectability (LoD) and lower limit of quantification (LLoQ) for each of the measured proteins: IL-6: LoD 0.006pg/mL and LoQ 0.01pg/mL; TNFα: LoD 0.02pg/mL and LoQ 0.03pg/mL; IL-10: LoD 0.004pg/mL and LoQ 0.02pg/mL; IP-10: 0.05pg/mL and LoQ 0.18pg/mL; IL-1β: LoD 0.02pg/mL and LoQ 0.08pg/mL.
Demographic and Clinical Variables of Interest
Data were additionally gathered on several external variables of interest to serve as objective indicators of relative construct validity and comparability of cytokine marker associations across platforms. Given this analysis was conducted within the context of a healthy brain aging study, we selected to examine the relationship between cytokine markers across MSD and Simoa with age, sex, education, cognition, white matter integrity, and several peripheral measures of vascular health.
Cognition
Participants completed the Mini-Mental State Examination as a global indicator of cognition14. Additionally, speeded executive control was measured utilizing a modified version of the Trail Making Test which required participants to serially alternate between numbers and days of the week. The total number of sequences completed was divided by the time to complete, such that higher values indicate better cognitive control15. We selected a measure that taps into both processing speed and executive functions given that these abilities are most strongly and consistently associated with inflammatory markers among both healthy older adults and in neurodegenerative syndromes16.
White Matter Integrity
Given established animal studies and growing clinical literature implicating the role of immune activation in white matter health (via both potentially vascular pathways and primary neurodegenerative pathologies)17,18, we aimed to determine the associations between cytokine concentrations measured via MSD HPE or Quanterix Simoa platforms, and a white matter microstructure tract, corpus callosum, quantified by diffusion tensor imaging. We selected the corpus callosum as our region of interest to reflect a global metric of white matter integrity, given that it comprises tracts from many cortical regions, is relatively free of cross-fibers, and appears to be a robust marker of diffuse white matter changes.
MRI acquisition
Magnetic resonance imaging (MRI) scans were obtained using a Siemens 12-channel head coil and were performed on a 3 Tesla Siemens Tim TrioMR scanner at the UCSF Neuroscience Clinical Research Unit. Whole brain T1 images were acquired using Magnetization-prepared rapid gradient echo (MPRAGE) in the axial plane: TR=2300ms; TE=3.43ms; TI=900 ms; flip angle=9; slice thickness=1 mm; FOV=256*224 mm; voxel size=1 mm*1mm; matrix size=256*224; and number of slices=176. Diffusion-Weighted Images (DTI) were acquired using single-short spin-echo sequence with the following parameters: TR=5300 ms; TE=88 ms; TI=2500 ms; flip angle=90; FOV=256*256 mm; two diffusion values of b=0 and 1000 s/mm; 12 diffusion directions; four repeats; 40 slices; matrix size=128*128; voxel size=2 mm*2 mm; slice thickness=3 mm; and GRAPPA=2.
Image processing and analysis
We analyzed fractional anisotropy (FA) as the primary parameter of interest from diffusion-weighted images as a marker of white matter tract microstructure. DTI processing began with denoising.19 FSL software co-registered the diffusion direction images with the b=0 image, then applied a gradient direction eddy current and distortion correction.20 Diffusion tensors were calculated using a non-linear least-squares algorithm from Dipy.21 After quality control, patients’ tensor (four-dimensional image) were registered linearly and non-linearly into a common space using DTI-TK. At baseline, patients’ tensors were moved into a group template. DTI FA maps were calculated from the patients’ tensor in the group template space. White matter tracts were masked using the ICBM-DTI-81 white matter labels and tract atlas22, and mean FA was averaged across callosal subregions.
Peripheral Vascular Health
Blood collected from the fasting morning draw was also analyzed for clinical labs in a subset of individuals (n=38), including serum levels of C-reactive protein, insulin, glucose, and triglycerides using standard laboratory protocols. C-reactive protein (CRP) was selected as a comparison of a widely used clinical indicator of “pro-inflammatory state.”23,24 A homeostatic model assessment of insulin resistance (HOMA-IR) was calculated in mass units [(glucose*insulin)/405] to provide a global estimate of insulin resistance at the time of blood draw. Body mass index was also calculated at the time of blood draw. These clinical markers of vascular health were selected given their consistently reported association with pro-inflammatory cytokine elevations in serum and plasma25,26, and to provide more direct comparison between peripheral cytokines and peripheral indicators of clinical health (versus cognition or brain structure, which reflect central processes).
Statistical Analyses
We determined the intra-assay precision of cytokine measurements in each platform by determining their reliability. To identify cytokine values that were reliably measured, we first removed values with coefficient of variation (CV) >20%. We then removed outliers defined as 3 times the sample upper interquartile range for each marker, and remaining values were determined to be reliable. The reproducibility of the assays was determined by comparing the proportions of “reliably detectable” values across MSD HPE and Quanterix Simoa via McNemar’s test of symmetry for paired nominal data. For subsequent analyses, all cytokines and CV values were log-transformed to allow for parametric analyses. Matched-pairs t-tests were then conducted to examine the comparability of CVs and absolute values (pg/mL) across platforms. Pearson correlations were calculated to estimate the agreement, and Bland-Altman plots were developed to examine differences between MSD HPE and Quanterix Simoa measurement techniques across each cytokine marker. Additionally, to inform future studies using pathway analyses examining relative cytokine concentrations across a panel of proteins, we explored relative ratios of cytokine concentrations within each platform; given that some markers had negative log values, we added an integer (log+3) to all values to depict positive ratios. Finally, Pearson correlations were also used to examine the relative effect sizes of each cytokine marker with external variables of interest across MSD HPE and Quanterix Simoa.
Results
Reliable Detectability, Intra-assay Precision, and Absolute Concentrations
IL-1β
While IL-1β had reliable detectability (CV<20% and outliers 3*upper IQR removed) of 68.4% on the Quanterix Simoa platform, none of the IL-1β values met our pre-specified parameters of reliable detectability and CVs ranged from 35.0 to 131.4% on MSD HPE. Therefore, our subsequent platform comparison analyses were restricted to IL-10, IP-10, IL-6, and TNFα.
IL-10, IP-10, IL-6, TNFα
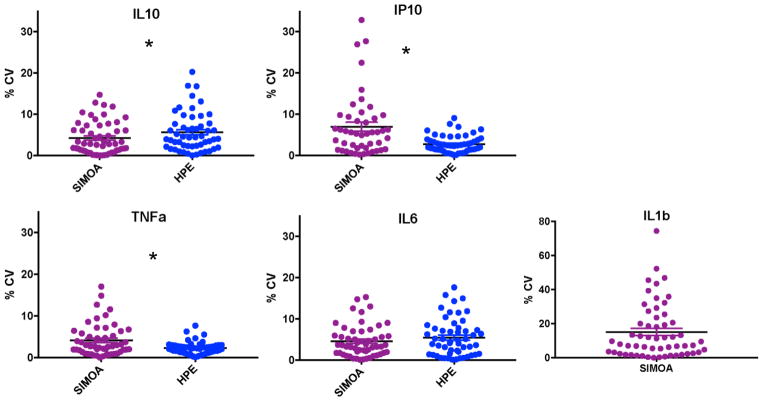
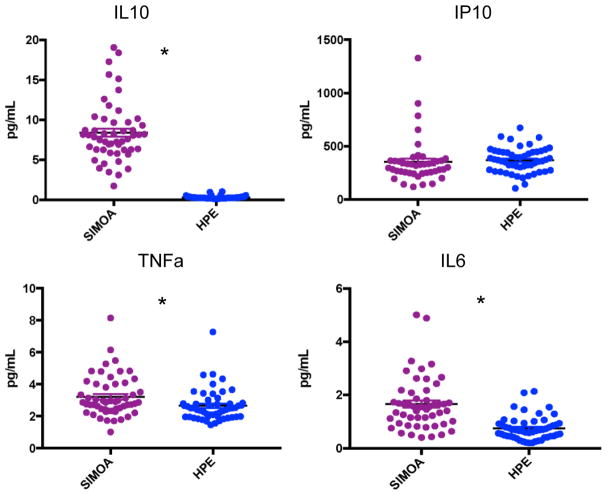
Applying the reliability parameters outline above, of the 57 older adult samples on which we attempted to measure cytokine concentrations, MSD HPE resulted in 87.7–93.0% reliable detectability while Quanterix Simoa resulted in 75.4–93.0% reliable detectability (Figure 1). The two platforms did not significantly differ in the amount of reliable data produced (McNemar’s test ps>0.32). Regarding intra-assay precision, on average, both platforms demonstrated CVs within an acceptable range <8%. However, Quanterix Simoa demonstrated a 259% significantly higher CV for IP-10 (t(43)=3.4, p=0.001), while MSD HPE demonstrated a 133% significantly higher CV for IL-10 (t(48)=−2.6, p=0.01). CVs were statistically comparable for IL-6 (t(49)=0.72, p=0.48) and TNFα (t(49)=1.6, p=0.11) (Figure 2). To increase comparability of interpretation, all subsequent analyses were conducted among participants with reliably detectable data on both Quanterix Simoa and MSD HPE (IL-10 n=47; IP-10 n=39; IL-6 n=48; TNFα=49). The Quanterix Simoa platform resulted in higher absolute levels (pg/mL) of IL-10 (t(46)=40.9, p<0.001), IL-6 (t(47)=11.2, p<0.001)), and TNFα (t(48)=2.8, p=0.007) compared to the MSD HPE platform (Figure 3). Quanterix Simoa and MSD HPE did not significantly differ in absolute measurement of IP-10 (t(38)=1.9, p=0.07).
Figure 1.
Proportion of samples reliably measured across cytokine markers on high performance electrochemiluminescence (HPE) and single molecule array (SIMOA) platforms did not differ, with the exception of IL1β.
Figure 2.
Intra-assay precision of plasma cytokine measurements by high performance electrochemiluminescence (HPE) and single molecule array (SIMOA) platforms.
Note. CV = coefficient of variance; *p<0.05.
Figure 3.
Absolute concentrations (pg/mL) differ between high performance electrochemiluminescence (HPE) and single molecule array (SIMOA) platform measurements for IL-10, IL-6, and TNFα
Note. *p<0.05.
Inter-platform Reproducibility
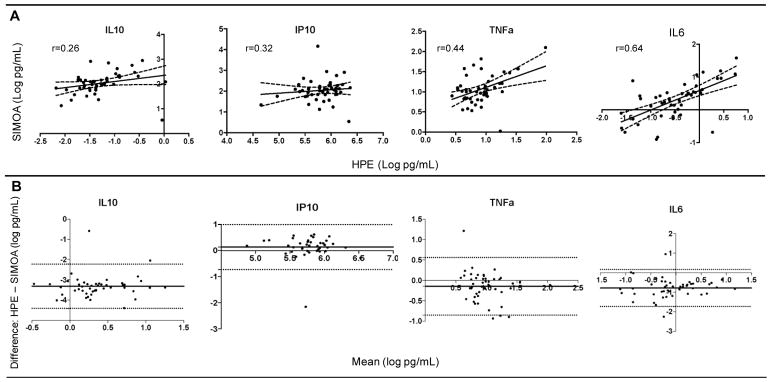
Cytokine concentrations of the same protein marker demonstrated only small-to-medium associations across platforms. Though all statistically significant (ps<0.05), IL-6 demonstrated the strongest (r=0.64) and IL-10 the weakest correlation (r=0.26) between MSD HPE and Quanterix Simoa platform measurements (Figure 4a). Given concerns that restricted range in a healthy cohort may artificially contribute to weaker correlation values, we also examined correlation coefficients among participants with outliers included (IL-10 n=49, IP-10 n=40, IL-6 n=50, TNFα n=50), but CVs>20% removed; however, the range of Pearson correlation coefficients remained comparable (r range = 0.29 to 0.56).
Figure 4.
Cytokine concentrations are significantly, but not strongly correlated with each other and demonstrate significant differences as quantified by high performance electrochemiluminiscence (HPE) versus single molecule array (Simoa) platforms.
Note. A) Scatterplots depicting agreement between HPE (x-axis) and SIMOA (y-axis); B) Bland-Altman plots illustrating platform differences. The y-axis represents the mean differences between HPE and SIMOA, while the x-axis depicts the average between the platforms. The solid horizontal lines indicate the average difference and the dashed horizontal lines represent the 95% confidence intervals.
To more directly examine platform differences and bias, Bland-Altman plots were developed. Quanterix Simoa produced overall higher quantifications than MSD HPE as indicated by the negative solid mean difference lines (Figure 4b), with the exception of IP-10, regardless of overall concentration levels (e.g., Quanterix Simoa was not biased by those with overall higher values). However >95% of the sample fell within the 95% confidence intervals (dotted lines) for all markers examined suggesting that there was not significant bias (difference) on one of the platforms, and there did not appear to be a systematic bias in directionality. That is, most data points fell within and closely outside the 95% confidence intervals with about the same amount falling above and below the mean difference line.
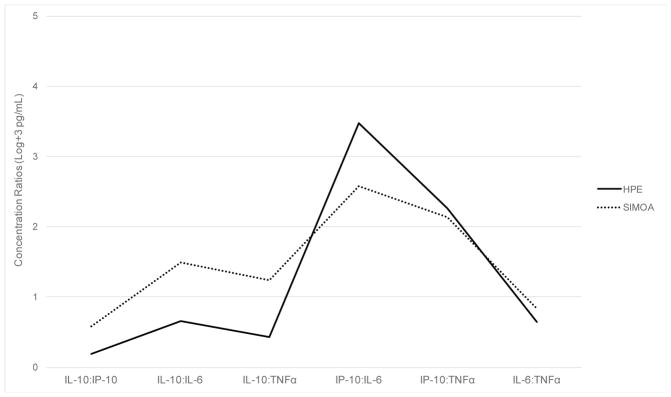
Additionally, given that panels of proteomic markers are commonly examined relative to one another to indicate aberrant up- or down-regulation within a postulated biological pathway, we explored the pattern of relative cytokine concentration ratios within each platform. Figure 5 demonstrates that, relative to the other measured cytokines, MSD HPE and Quanterix Simoa demonstrate a similar pattern of within-platform cytokine increases and decreases across the panel examined. However, when statistically compared, each of the cytokine ratio values significantly differed in paired analyses on MSD HPE compared to Quanterix Simoa (t values= −6.79 to 46.0, ps<0.001).
Figure 5.
Relative ratios of cytokine concentrations demonstrate a similar pattern of increases and decreases, but still significantly differ across platforms.
Note. Y-axis represents cytokine concentration ratios (log transformed + 3 in pg/mL). HPE = high performance electrochemiluminiscence; Simoa = Quanterix single molecule array. All ratios values are significantly different between MSD HPE and Quanterix Simoa (ps<0.001).
Inter-platform Correlations with Clinical and Demographic Variables
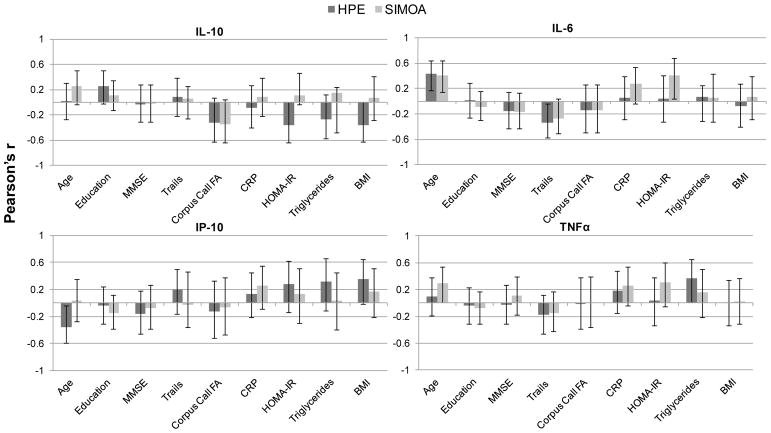
Lastly, we examined the relative effect sizes of each cytokine marker with variables of interest (e.g., age, cognition, peripheral measures of vascular health) across the two platforms. As a whole, 95% confidence interval bars of the associations between the cytokines and external variables overlapped between the two platforms for all variables examined (Figure 6). While the majority of associations between external variables and the cytokine markers demonstrated the same (and expected) directionality across MSD and Simoa, there were several notable exceptions. IL-10 demonstrated the most variability of this nature across platforms. For example, though IL-10 is generally considered to promote anti-inflammatory cascades, it demonstrated a positive association with age across both MSD (r=0.01) and Simoa (r=0.25); additionally, though the error bars overlapped and associations were small, IL-10 measured via MSD showed negative associations with HOMA-IR (r=−0.36), triglycerides (r=−0.27), and BMI (r=−0.36), but positive associations with these variables as measured via Simoa (r’s range=0.07–0.15). Notably, IL-6 again appeared to demonstrate the most consistent associations with variables of interest across the two platforms, both in terms of magnitude and direction of the effect, followed by TNFα. IP-10 generally demonstrated larger effect sizes with external variables when measured via MSD compared to Simoa; however, some associations were in unexpected directions. For example, IP-10 is implicated in the acute pro-inflammatory phase but demonstrated a significant negative association with age (r=−0.35) and a positive association with an executive functioning task (Trails r=0.20) when measured on the MSD platform. None of the cytokine markers across either platform were significantly associated with sex (t-ratio range = 0.4 to 1.6, ps>0.12) or education (r’s range = −0.08 to 0.25, ps>0.08) in our sample.
Figure 6.
Correlations (r and 95% error bars) between each measured cytokine and external variables of interest across high performance electrochemiluminiscence (HPE) and single molecule array (SIMOA) platforms.
Note. MMSE = Mini Mental State Examination; Corpus Call FA = corpus callosum fractional anisotropy; CRP = C-reactive protein; HOMA-IR = homeostatic model assessment of insulin resistance; BMI = body mass index.
Discussion
Ours are the first data directly comparing cytokine measurement in clinical samples between high performance electrochemiluminiscence (HPE) by Meso Scale Discovery and single molecule arrays (Simoa) by Quanterix with the aim of helping to guide platform selection and future cross-study interpretations for several commonly examined immune activation markers. Although both predicated on ELISA techniques, distinct differences in cytokine quantification emerged. While the overall ability of each platform to detect our cytokines of interest was largely comparable (with the exception of IL1β), absolute concentrations significantly differed, and only moderate to minimal variance was shared across the platforms when measuring the same markers in the same blood samples. However, patterns of relative cytokine ratios and external associations within platform demonstrated greater similarities and there was not a significant bias across platforms, suggesting that relative (versus absolute) cytokine concentration relationships may be most appropriate when examining across measurement platforms. As interest in biofluid proteins continues to expand, despite representing postulated biological processes, it is important to appreciate that these markers may not be entirely “objective” and are in fact subject to common pitfalls of any indirect assessment approach.
With the exception of IL-1β, both HPE and Simoa demonstrated acceptable rates of quantification and intra-assay precision. Overall, the panel of cytokines was reliably measured (i.e., CVs<20% and outliers excluded) for ~90% of samples attempted with overall CVs <8% on both platform; though not significant, it is notable that IP-10 was only reliably measured in 75% of the sample using Simoa (versus 91% on HPE). The major exception to these comparable results was IL-1β, which only resulted in values for 3 out of the 57 attempted samples using HPE, none of which met our operationalization of reliability (CVs>34%). On the other hand, Simoa reliably measured IL-1β in 68% of samples attempted. Given that IL-1β may be present only at very low concentrations, particularly among healthy individuals, detectability is a clear challenge. Our results demonstrating the relatively poor sensitivity of HPE for IL-1β are consistent with previous studies examining this cytokine in patient samples, including HIV infection1,6, multiple sclerosis10, and kit control samples8. IL-1β is a key mediator in the inflammatory response and is involved in a host of cell functions (proliferation, differentiation, apoptosis), including propagating immune responses to the point of damage during states chronic disease and injury27. Given its ubiquity in immune activation, it is promising that Simoa was able to quantify IL-1β in >60% of the samples and perhaps represents advancement of biofluid technologies moving forward. However, validation that these concentrations are reliably measured across time and indeed represent the molecule of interest is still needed.
The major differences in HPE versus Simoa emerged when directly comparing cytokine concentration measurements head-to-head. Overall, Simoa quantified significantly higher concentrations particularly of IL-10 (27-fold higher), but also for IL-6 and TNFα (1–2 fold higher). Though it is unclear which platform represents “true” biologic levels, these findings are consistent with Simoa’s purported goal of increasing overall measurement sensitivity (i.e., detection of “subfemtomolar concentrations” via femtomolar-sized wells). These differences in absolute cytokine values are consistent with almost all previously reported platform comparisons studies found, such that regardless of platforms examined, the resulting pg/mL concentrations do not appear directly comparable1,10,28. Additionally, directly correlating on the same cytokine markers, though all statistically significant, HPE and Simoa demonstrated only small-to-moderate effect sizes. Notably, IP-10 and IL-10 demonstrated the weakest (rs<0.35), while IL-6 and TNFα demonstrated the strongest (rs>0.40) shared variance. However, Bland-Altman plots did not suggest significant bias across the platforms (95% of data-points fell within the 95% confidence interval) overall, or any bias dependent on concentration levels (i.e., Simoa did not demonstrate disproportionately higher quantifications for individuals with the highest concentrations).
The reason(s) for these quantification differences is difficult to determine, though it is important to keep in mind that HPE and Simoa do differ in their quantification approaches. That is, while HPE utilizes electrodes coated in capture-antibodies that measure fluoresce signal produced by an electric-chemical reaction, Simoa consists of a bead-based enzyme-labeled immunocomplex that is chemically broken down into a measured fluorescent product. HPE quantifies the intensity of the fluorescence signal and Simoa counts the proportion of wells with a fluorescence signal. Additionally, differences in the company-supplied antibodies, dilution protocols, and overall sensitivity and reliability of instruments may have contributed to our findings. Another likely reason for the different absolute concentrations across the platforms is that the assays have not been standardized to each other using common calibrators. It is also possible that the biology underpinning of these cytokines play a role in their detection reliability. That is, IL-6 and TNFα can be considered as widereaching master regulators of immune activation involved in the initiation of both adaptive and innate inflammatory pathways29–31. As such, perhaps it is not surprising that they are among the most commonly reported cytokines indicating the most robust, reliably measured results across patient samples, in parallel to our findings here. These signaling proteins may therefore be particularly helpful to indicate an overall “pro-inflammatory state” but are less nuanced in their ability to implicate specific immune pathways. On the other hand, IP-10 and IL-10 represent relatively more specific immune activation pathways in the patient population or construct of interest, including acutely heightened immune responses and anti-angiogenic processes (IP-10) and anti-inflammatory clearance (IL-10), including inhibiting activity of helper T cells, natural killer cells, and macrophages32–35. Perhaps due to the greater subtleties of these pathways, IP-10 and IL-10 were more sensitive to even the small measurement approach differences examined here and may therefore be less reliable overall when compared across differing platform techniques. Notably, ours is among the few studies that have reported such direct platform comparisons in a clinical sample (i.e., same plasma on two different platforms) and the only one that has done so on HPE versus Simoa, heightening the ecological relevance of our findings. Of those that have, correlations have ranged from 0.1 to 0.85,6, paralleling our findings and indicating that direct platform comparability of biofluid markers may be highly analyte specific.
However, cytokine ratios and associations with external variables within each platform demonstrated similar patterns, supporting relative interpretations of cytokine concentrations across platforms. Regarding the cytokine ratios, our data support the development of proteomic profiles within a targeted pathway as useful application of biofluid markers to help characterize a molecular process of interest, and may be less susceptible to the interpretation limitations inherent when examining a single marker, particularly across platforms and/or studies. That is, characterization of relative patterns of marker up- or down-regulation may be the most prudent interpretation of biofluid markers, rather than attempting to interpret the value of a single marker in the absence of the relative proteomic milieu. Additionally, by exploring the variance explained by cytokines as measured across platforms on the same clinical variables, we aimed to support the concurrent validity of HPE and Simoa. Though the absolute relationship between the measured cytokines and each external variable are still empirical questions currently under investigation (e.g., cognition and IL-6), we posited that the relative direction and size of the relationships should be consistent across the two platforms for the same cytokine marker to support convergence. Indeed, 95% confidence intervals of all associations examined overlapped between HPE and Simoa, though it is notable that these confidence intervals were fairly large (e.g., spanning r<0.2 to r>0.6 even for significant associations) and commonly crossed zero (non-significant). Both IL-6 and TNFα demonstrated consistent associations with external variables across platforms particularly in terms of direction, and neither HPE nor Simoa demonstrated consistently stronger or weaker associations overall. Regarding construct and ecological validities of the cytokine markers overall (i.e., degree to which each platform marker was associated with similar constructs and/or real-world status), IL-6 and TNFα demonstrated associations with external variables of interest in the expected directions, including positive associations with age and C-reactive protein, a clinically obtained marker of immune activation, and negative associations with an executive functions task. On the other hand, perhaps less surprising given their relative discordance, IP-10 and, particularly, IL-10 demonstrated some differences across platforms. For IL-10, a generally anti-inflammatory cytokine, HPE demonstrated consistent negative associations with measures of cardiovascular disease and immune activation (C-reactive protein), whereas Simoa demonstrated small, but consistently positive associations with these factors. Similarly, regarding age, IP-10 (pro-inflammatory) demonstrated a significant negative association via HPE but a positive association via Simoa. Perhaps the two measurement platforms are recognizing and capturing slightly different antibodies on each of these proteins, resulting in the less consistent relationships. Taken as a whole, our data largely supports the relative interpretation of cytokine concentrations when examining markers across platform techniques, though reliability of these relationships may still vary depending on analyte,
Our data are not without limitations. Most notably, though blood was drawn on the same day from the same participants, never underwent a freeze-thaw cycle, and was analyzed by the same lab technician, the analyses were not conducted on the same day (HPE aliquots were analyzed before Simoa aliquots). Though less likely to contribute significant measurement error, length of freeze time and human factors that can differ from day-to-day may have introduced additional noise not related to the platform quantifications themselves. Additionally, our data were collected in a fairly homogeneous patient sample of healthy, relatively well-educated, largely White older adults. Especially in the absence of disease, the range of cytokine concentrations may be limited (ceiling/floor effects) leading to smaller associations overall (including across platforms) simply due to limited variability. Additionally, due to these limitations, we were not able to examine effects of other demographic factors such as race or low education on the cytokine markers, which may be of importance interpreting patient samples in diseased groups in the future. On the other hand, the uniformity of our clinical sample will help provide a baseline understanding not only of platform-specific nuances in cytokine measurement, but also the potential relative demographic influences for individuals similar to our sample.
Conclusions
Our study is among the first to closely characterize a common cytokine panel across HPE and Simoa platforms in a clinical sample. Consistent with previous studies examining comparability of other biofluid platforms, our data suggest that 1) platform selection matters, and 2) examination of relative, but not absolute values of cytokine concentrations may be the most prudent interpretation of these markers. However, the degree to which differences quantification approach impacts measured concentrations may differ by analyte. While IL-6 and TNFα appeared relatively robust, IL-10 and IP-10 were much more sensitive to platform differences. As more diverse quantification approaches and studies emerge, more work is needed to describe biofluid protein measurements across platform techniques, across time, and especially in diverse clinical samples; our conceptual findings will ultimately only be as meaningful as our measurement tools.
Highlights.
MSD HPE and Quanterix Simoa are analytic platforms to detect biofluid cytokines
Detectability and reliability of the same cytokines did not differ across platforms
Absolute cytokine concentrations were higher using Simoa vs. HPE
Cross-platform correlations were low, but cytokine ratios showed similar patterns
Cytokine measurement is platform-specific and is best interpreted in relative terms
Acknowledgments
This study was supported by NIH-NIA grants NIA 1R01AG032289 (PI: Kramer), R01AG048234 (PI: Kramer), 2R01AG038791(PI: Boxer), U54NS092089 (PI: Boxer), and UCSF ADRC P50 AG023501. Our work was also supported by Larry L. Hillblom Network Grant (PI: Kramer; 2014-A-004-NET) and Fellowship Grant (PI: Casaletto; 2017-A-004-FEL), and the University of California Cures Alzheimer’s Disease program.
Footnotes
Disclosures
The authors have the following disclosures:
KBC, FEE, RF, SW, EF, AMS, BB, AK, JCR, ALB, and JHK have no disclosures to report.
HZ is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg, has served at advisory boards of Eli Lilly and Roche Diagnostics and has received travel support from TEVA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breen EC, Reynolds SM, Cox C, et al. Multisite comparison of high-sensitivity multiplex cytokine assays. Clin Vaccine Immunol. 2011;18(8):1229–1242. doi: 10.1128/CVI.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hottenstein C, Szapacs M, Fuller K, Evans C. Platforms and techniques used for biomarker assays: where are we now? Bioanalysis. 2017;9(14):1029–1031. doi: 10.4155/bio-2017-0107. [DOI] [PubMed] [Google Scholar]
- 3.Rissin DM, Kan CW, Campbell TG, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivnak AJ, Rissin DM, Kan CW, et al. A fully-automated, six-plex single molecule immunoassay for measuring cytokines in blood. J Immunol Methods. 2015;424:20–27. doi: 10.1016/j.jim.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Bryant SE, Lista S, Rissman RA, et al. Comparing biological markers of Alzheimer’s disease across blood fraction and platforms: Comparing apples to oranges. Alzheimer’s Dement Diagnosis, Assess Dis Monit. 2016;3:27–34. doi: 10.1016/j.dadm.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabitao D, Margolick JB, Bream J. Performance evaluation of two multiplex technologies for the measurement of serum cytokines in HIV-infected individuals. 2010;134:17. [Google Scholar]
- 7.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex?? and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340(1):55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56(2):314–318. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masliah E, Mallory; M, Alford; M, et al. Altered expression of synaptic proteins occurs early during progression of Alzheimer’s disease. Neurology. 2001;56:127–129. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 10.Malekzadeh A, Twaalfhoven H, Wijnstok NJ, Killestein J, Blankenstein MA, Teunissen CE. Comparison of multiplex platforms for cytokine assessments and their potential use for biomarker profiling in multiple sclerosis. Cytokine. 2017;91:145–152. doi: 10.1016/j.cyto.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Myzithras M, Li H, Bigwarfe T, et al. Development of an ultra-sensitive Simoa assay to enable GDF11 detection: a comparison across bioanalytical platforms. Bioanalysis. 2016;8(6):511–518. doi: 10.4155/bio.16.17. [DOI] [PubMed] [Google Scholar]
- 12.Janelidze S, Zetterberg H, Mattsson N, et al. CSF A β 42/A β 40 and A β 42/A β 38 ratios: better diagnostic markers of Alzheimer disease. Ann Clin Transl Neurol. 2016;3(3):154–165. doi: 10.1002/acn3.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7(3):270–279. doi: 10.1016/J.JALZ.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folstein MF. The Mini-Mental State Examination. Arch Gen Psychiatry. 1983;40(7):812. doi: 10.1001/archpsyc.1983.01790060110016. [DOI] [PubMed] [Google Scholar]
- 15.Kramer, Joel H, PsyD, Jurik Jennifer MA, Sha Sharon J, MS, Rankin Kate P, PhD, Rosen Howard J, MD, Johnson Julene K, PhD, Miller BLM. Distinctive Neuropsychological Patterns in Frontotemporal Dementia, Semantic Dementia, And Alzheimer Disease. [Accessed January 7, 2018];Cogn Behav Neurol. 2003 16(4):211–218. doi: 10.1097/00146965-200312000-00002. http://ovidsp.uk.ovid.com/sp-3.27.2b/ovidweb.cgi?QS2=434f4e1a73d37e8c6d5cc3ea7a7100e09ed58082c7a5078041ec5dd93115ee3c4d1685b49b33ca59e684eab23d8ab323358d6fcb2ffb60478ac06f45f37b6625f91b6afcebec9099709edcdc1fe0ebeca549a70b5bce8d76d1de1a064d20884f6e13e08d63. [DOI] [PubMed] [Google Scholar]
- 16.Ownby RL. Neuroinflammation and cognitive aging. Curr Psychiatry Rep. 2010 doi: 10.1007/s11920-009-0082-1. [DOI] [PubMed] [Google Scholar]
- 17.Hinman JD, Abraham CR. What’s Behind the Decline? The Role of White Matter in Brain Aging. Neurochem Res. 2007;32(12):2023–2031. doi: 10.1007/s11064-007-9341-x. [DOI] [PubMed] [Google Scholar]
- 18.Raz N, Yang Y, Dahle CL, Land S. Volume of white matter hyperintensities in healthy adults: Contribution of age, vascular risk factors, and inflammation-related genetic variants. Biochim Biophys Acta - Mol Basis Dis. 2012;1822(3):361–369. doi: 10.1016/J.BBADIS.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veraart J, Fieremans E, Novikov DS. Diffusion MRI noise mapping using random matrix theory. Magn Reson Med. 2016;76(5):1582–1593. doi: 10.1002/mrm.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Garyfallidis E, Brett M, Amirbekian B, et al. Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform. 2014;8:8. doi: 10.3389/fninf.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori S, Oishi K, Jiang H, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridker PM, Hennekens CH, Buring JE, Rifai N. C-Reactive Protein and Other Markers of Inflammation in the Prediction of Cardiovascular Disease in Women. N Engl J Med. 2000;342(12):836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 24.Danesh J, Wheeler JG, Hirschfield GM, et al. C-Reactive Protein and Other Circulating Markers of Inflammation in the Prediction of Coronary Heart Disease. N Engl J Med. 2004;350(14):1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 25.Salvioli S, Capri M, Valensin S, et al. Inflamm-Aging, Cytokines and Aging: State of the Art, New Hypotheses on the Role of Mitochondria and New Perspectives from Systems Biology. Curr Pharm Des. 2006;12(24):3161–3171. doi: 10.2174/138161206777947470. [DOI] [PubMed] [Google Scholar]
- 26.Ungvari Z, Csiszar A, Kaley G. Vascular Inflammation in Aging. Herz. 2004;29(8):733–740. doi: 10.1007/s00059-004-2625-x. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011;22(4):189–195. doi: 10.1016/j.cytogfr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Bryant SE, Mielke MM, Rissman RA, et al. Blood-based biomarkers in Alzheimer disease: Current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimer’s Dement. 2017;13(1):45–58. doi: 10.1016/J.JALZ.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat Immunol. 2006;7(3):247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- 30.Feuerstein GZ, Liu T, Barone FC. Cytokines, inflammation, and brain injury: role of tumor necrosis factor-alpha. Cerebrovasc Brain Metab Rev. 1994;6(4):341–360. [PubMed] [Google Scholar]
- 31.Norris JG, Tang LP, Sparacio SM, Benveniste EN. Signal transduction pathways mediating astrocyte IL-6 induction by IL-1 beta and tumor necrosis factor-alpha. [Accessed December 17, 2017];J Immunol. 1994 152(2):841–850. http://www.ncbi.nlm.nih.gov/pubmed/7506738. [PubMed] [Google Scholar]
- 32.Howard M, O’garra A, Ishida H, De R, Malefyt W, De Vries J. Biological Properties of Interleukin 10. [Accessed December 17, 2017];J Clin Immunol. 1992 12(4) doi: 10.1007/BF00918147. https://link.springer.com/content/pdf/10.1007/BF00918147.pdf. [DOI] [PubMed] [Google Scholar]
- 33.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174(5):1209–1220. doi: 10.1084/JEM.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luster AD, Ravetch JV. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10) J Exp Med. 1987;166(4):1084–1097. doi: 10.1084/JEM.166.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strieter RM, Kunkel SL, Arenberg DA, Burdick MD, Polverini PJ. Interferon γ-Inducible Protein-10 (IP-10), a Member of the C-X-C Chemokine Family, Is an Inhibitor of Angiogenesis. Biochem Biophys Res Commun. 1995;210(1):51–57. doi: 10.1006/BBRC.1995.1626. [DOI] [PubMed] [Google Scholar]