Abstract
In all eukaryotes, the unfolded protein response (UPR) relieves endoplasmic reticulum (ER) stress, which is a potentially lethal condition caused by the accumulation of misfolded proteins in the ER. In mammalian and yeast cells, reactive oxygen species (ROS) generated during ER stress attenuate the UPR, negatively impacting cell survival. In plants, the relationship between the UPR and ROS is less clear. Although ROS develop during ER stress, the sources of ROS linked to ER stress responses and the physiological impact of ROS generation on the survival from proteotoxic stress are yet unknown. Here we show that in Arabidopsis thaliana the respiratory burst oxidase homologs, RBOHD and RBOHF, contribute to the production of ROS during ER stress. We also demonstrate that during ER stress RBOHD and RBOHF are necessary to properly mount the adaptive UPR and overcome temporary and chronic ER stress situations. These results ascribe a cytoprotective role to RBOH-generated ROS in the defense from proteotoxic stress in an essential organelle, and support a plant-specific feature of the UPR management among eukaryotes.
Keywords: Unfolded protein response, ER stress, Arabidopsis thaliana, reactive oxygen species, RBOHD, RBOHF
Significance Statement:
Although reactive oxygen species (ROS) are known to be produced during endoplasmic reticulum stress, the source of ROS and their potential effects on the canonical unfolded protein response (UPR) are largely unknown. Here we demonstrate that the respiratory burst oxidase homologs RBOHD and RBOHF positively contribute to plant survival under these conditions.
Introduction
Physiological and stress situations causing insufficiency of the endoplasmic reticulum (ER) to meet cellular demands for secretory protein folding lead to a potentially lethal condition, known as ER stress (Gething et al. 1992). To overcome ER stress and restore homeostasis, protective signaling cascades, collectively called the unfolded protein response (UPR), are originated at the ER and lead to the synthesis of ER chaperones and foldases to attenuate ER stress. If this adaptive UPR fails, such in conditions of unresolved or chronic ER stress, cells commit to programmed cell death (Ron et al. 2007, Ruberti et al. 2015, Angelos et al. 2017). In all eukaryotes, the adaptive phase of the UPR is initiated with the activation of IRE1, an ER-associated membrane kinase and ribonuclease, which catalyzes the unconventional splicing of transcripts of bZIP-transcription factors: yeast Hac1, mammalian Xbp1, or plant bZIP60. This step is necessary for the production of an active transcription factor that controls the expression of UPR target genes (Deng et al. 2011, Nagashima et al. 2011, Moreno et al. 2012). In multicellular eukaryotes, the UPR has expanded to include additional UPR effectors, such as membrane tethered bZIP transcription factors (MTTFs), namely mammalian ATF6 and plant bZIP28. Upon sensing ER stress, these MTTFs translocate to the Golgi, where the cytosol-exposed transcription factor domain is proteolytically removed from the transmembrane anchor and translocated to the nucleus for the transcriptional regulation of UPR target genes (Liu et al. 2007b, Gao et al. 2008, Srivastava et al. 2013, Sun et al. 2013).
In metazoan cells, a third arm of the UPR is activated by the ER associated PKR-like ER kinase (PERK) protein, which oligomerizes and autophosphorylates in conditions of ER stress. PERK activity results in the phosphorylation and inactivation of the eukaryotic translation initiation factors eIF2A thereby promoting a global repression of the rate of the protein translation (Harding et al. 1999, Shen et al. 2002). The transcription factors ATF4 and Nrf2 downstream of PERK are able to escape this translational repression and upregulate an oxidative stress response through the production of antioxidant proteins (Harding et al. 2000, Cullinan et al. 2003). Indeed, mutations in PERK signaling markedly increase the generation of reactive oxygen species (ROS), which negatively affect UPR efficiency and the ability of cells to survive ER stress (Marciniak et al. 2004, Back et al. 2009, Han et al. 2013, Maity et al. 2016). In yeast, which similarly to plants lack PERK, a dysregulated production of H2O2 under ER stress results in a translational attenuation of ER stress genes (Maity et al. 2016). Therefore, in metazoans and yeast, ROS cause attenuation of the cytoprotective functions of the UPR and acceleration of responses leading to cell death. In humans, this process potentiates the development of multiple diseases including diabetes, neurodegenerative diseases, and atherosclerosis (Malhotra et al. 2007).
In plants, a functional connection between ROS and the UPR management has not yet been clearly defined. During growth and in conditions of stress, several organelles including the mitochondria, chloroplasts, and peroxisomes generate ROS intracellularly (Tripathy et al. 2012). ROS are also produced in the apoplast mainly by the activity of the membrane-bound NADPH Oxidase (NOX) enzymes, known in plants as respiratory burst oxidase homologues (RBOH) (Torres et al. 2002). The NOX/RBOH enzymes generate O2− through electron transfer from NADPH to oxygen (Gapper et al. 2006). O2− is highly reactive and is dismutated to H2O2 either via the superoxide dismutase (SOD) enzymes or spontaneously (Mori et al. 2004). H2O2 is more stable than O2− and moves through membranes via aquaporins (Bienert et al. 2007), and it is therefore considered as a potent signaling ROS in plants (Sadhukhan et al. 2017). Thus far in plants, an increase of soluble H2O2 and lipid peroxidation during ER stress have been reported (Ozgur et al. 2014, Ozgur et al. 2015, Ozgur et al. 2018) but the source of ROS during ER stress and the influence of ER-stress generated ROS on UPR signaling are not yet established. The oxidative function of ERO1 in the ER lumen is a significant source of ER peroxides both in homeostatic conditions of growth and during ER stress; however, an increase in NADPH Oxidase activity has also been observed via biochemical assays during the early UPR (Tu et al. 2004, Sevier et al. 2008, Zito 2015, Ozgur et al. 2018). Under similar conditions, small inductions in transcript levels of RBOHD and RBOHF, two plasma membrane-localized NADPH Oxidases (Torres et al. 2002, Torres et al. 2005), were also noted in tissues subjected to ER stress treatment, leading to the suggestion that increases in ROS levels were due to the respiratory burst oxidase homologs D and F (RBOHD and RBOHF) (Ozgur et al. 2014). Nonetheless, it is yet to be experimentally tested to what extent RBOH activity contributes to the ROS levels during ER stress, and whether these enzymes are necessary for the actuation of effective ER stress responses. More generally, the downstream effects of increased ROS levels during plant ER stress responses are also largely unmapped. Previous reports have shown a variable regulation of the antioxidant defense systems under conditions of ER stress. For example, in conditions of ER stress the O2− scavenging activity of superoxide dismutase is induced in roots, but down-regulated in shoots. Conversely, H2O2 scavenging activities of catalase and ascorbate peroxidase are upregulated in shoots but have no change in roots (Ozgur et al. 2014). However, in both roots and shoots a significantly increased glutathione content and glutathione reductase activity during ER stress lends weight to the conclusion that antioxidant defenses are upregulated to manage the increased ROS production in ER stress (Ozgur et al. 2014). Nonetheless, critical questions remain unanswered. For example, it is yet unknown what effects an increase in ROS levels may have on the adaptive UPR, nor is it understood whether ROS may contribute to life or death decisions in temporary and chronic ER stress. To address these fundamental questions, in this work we explored the functional connection between ROS and the UPR in plants. We demonstrate that O2− and H2O2 are significantly contributed by RBOHD and RBOHF during ER stress. We also show that RBOHD and RBOHF elicit a transcriptional response to ROS during ER stress in the adaptive UPR. Furthermore, we provide evidence for a stringent requirement of RBOHD and RBOHF to prevent the progression of cell death in recovery from temporary ER stress and under chronic ER stress. Together, these findings support a positive role of superoxide signaling in potentiating the cell’s ability to survive temporary and chronic ER stress and ascribe a significant role to two RBOH proteins in this process.
Results
ER stress induces accumulation of superoxide by NADPH Oxidases
We first aimed to test whether ER stress activates NADPH Oxidases by establishing the levels of O2−, the product of NADPH Oxidases, in seedlings subjected to induced ER stress conditions. To do so, we followed O2− accumulation in situ using nitrotetrazolium blue (NBT), a chromogenic substrate for oxidases commonly applied to the study of NADPH Oxidase activity in vivo (Dunand et al. 2007). We analyzed roots of wild type (WT) and a double RBOHD and RBOHF knockout seedlings (herein dubbed rbohd rbohf (Torres et al. 2002)). RBOHD and RBOHF belong to a ten-member family of proteins and are mainly expressed in the shoot and vascular tissue (Morales et al. 2016) After 7 days of growth on solid ½ LS media, the seedlings were transferred to solid ½ LS media containing the well-established ER stress inducer tunicamycin (Tm) for 24 or 48 hours to induce the adaptive UPR in a plate system (Iwata et al. 2005, Liu et al. 2007a, Chen et al. 2013).. We found that at 24 hours, the levels of NBT staining were only slightly increased, mainly at the root tip, both in Tm-treated WT and rbohd rbohf compared to the respective DMSO controls (Tm solvent) (Figure 1A; root tip indicated with Δ). However, at 48 hours of TM treatment, in WT and in the rbohd rbohf line the levels of NBT staining had dissipated at the root tip and increased along the maturation zone and mature root tissues (Figure 1A; mature zone and maturation zone indicated with † and ‡, respectively). The NBT staining was higher in the mature zone of WT roots compared to rbohd rbohf. These observations were validated by measurements of the relative levels of NBT staining along the root length using ImageJ (Figure 1B). These verified differences in NBT staining at the tissue level are consistent with the notion that RBOHD and RBOHF are expressed largely in shoot tissues and mature root tissues but not at the root tip in Arabidopsis (Morales et al. 2016). We next tested whether the observed NBT staining in rbohd rbohf could be due to other RBOH activity. Therefore, we supplemented the Tm-treated samples with diphenyleneiodonium (DPI), a broad spectrum flavoprotein inhibitor that is commonly applied in the study of RBOH proteins (Ogasawara et al. 2008). We found that DPI-treatment largely ablated the Tm-induced NBT-staining in WT and rbohd rbohf (Figure 1A,B). These results indicate that the remaining Tm-induced NBT staining found at the root tips may be sourced from the activity of RBOH other enzymes functioning redundantly to RBOHD and RBOHF (Huang et al. 2016). Together, these results indicate that RBOHD, RBOHF, and potentially other RBOH enzymes are involved in ER stress-induced O2− production, which at least for RBOHD and RBOHF is with the previously described expression pattern at the tissue level.
Figure 1. NADPH oxidase dependent O2- is generated during ER Stress partially through RBOHD and RBOHF activity.
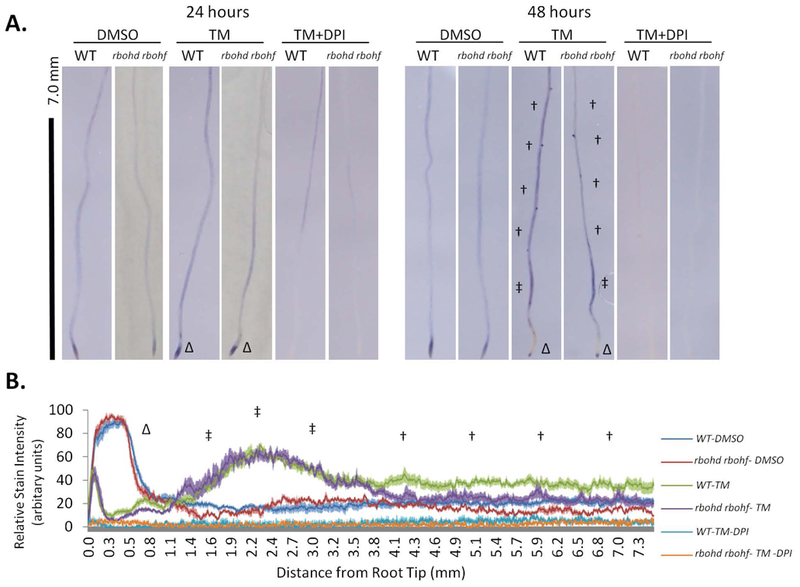
In situ detection and semi-quantification of superoxide in root tissues by staining with nitro-tetrazolium blue (NBT) A. WT or rbohd rbohf seedlings treated with 1.0 μg/ml Tm, 1.0 μg/ml TM + 2.5 μM DPI, or DMSO were stained with NBT after 24 or 48 hours. †: mature zone; ‡: maturation zone; Δ: root tip. B. Tissue-specific differences in superoxide are shown by relative pixel intensities of NBT stain from the root tip ± SE.
ER stress induces the accumulation of hydrogen peroxide dependent upon RBOHD and RBOHF activity and intact UPR signaling
Because O2− is converted to H2O2, we next aimed to establish the levels of accumulation of H2O2 in seedlings undergoing ER stress. To do so, we first set up an assay to measure reliably H2O2 levels in untreated tissues. We utilized a sensitive enzymatic fluorimetric assay based on the stoichiometric oxidization of non-fluorescent Amplex Ultra Red (AUR) by H2O2 by exogenous horseradish peroxidase (HRP) to brightly fluorescent resorufin (Queval et al. 2008, Zhu et al. 2010, Chakraborty et al. 2016). We expected that this assay would lead to the detection of resorufin fluorescence in the presence of HRP and to reduced levels of resorufin fluorescence in the absence of this enzyme. We first conducted tissue extraction using potassium phosphate buffer (Chakraborty et al. 2016, Le et al. 2016), and found no significant differences in oxidization of non-fluorescent AUR by added HRP compared to the control (Figure 2A). These results indicate either that the assay was not sensitive enough or that the AUR oxidation was saturated in the absence of HRP. To test this, we implemented a 10% trichloroacetic acid extraction (neutralized: NTCA) to precipitate possibly interfering enzymatic reactions, and then assayed the levels of H2O2. Consistent with our original hypothesis, we found that, in the presence of HRP, the levels of resorufin fluorescence were significantly higher than in the absence of the enzyme compared to the control (Figure 2A). As a further control for the validity of our assay, we treated the samples with catalase, which specifically dismutates H2O2 in the extract and should therefore further lower the levels of resorufin fluorescence in HRP-treated samples. Conversely, we expected no differences in catalase treatment of samples extracted in potassium phosphate buffer compared the untreated control. In samples extracted with NTCA, we found that the levels of resorufin fluorescence were significantly lower in catalase-treated samples compared untreated samples (Figure 2C). As expected also, the addition of catalase did not alter the levels of fluorescence of potassium phosphate buffer-extracted samples compared to untreated samples (Figure 2B). These results support that the resorufin fluorescence levels detected in the potassium phosphate buffer-extracted samples are due to H2O2-independent oxidation of AUR. Importantly, these results also indicate that we have established a quantitative approach to track specifically H2O2 levels in tissues.
Figure 2. ER Stress induced H2O2 are controlled by RBOHD and RBOHF as well as intact UPR signalling.
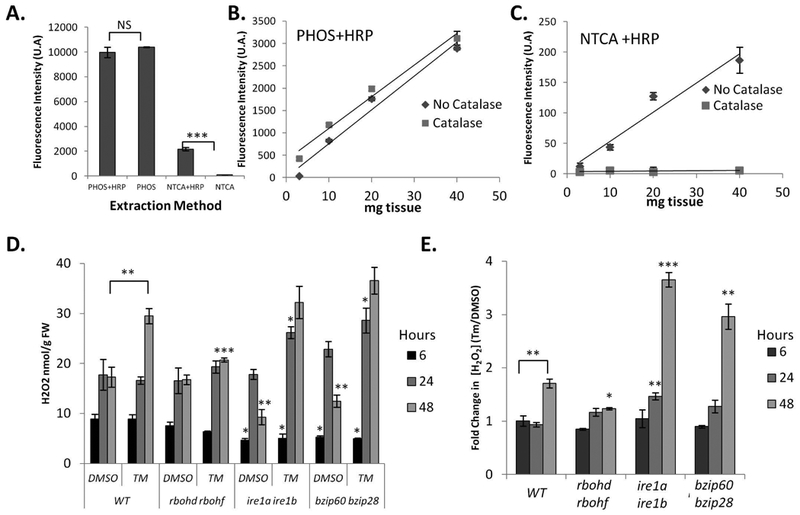
Development and application of an Amplex Ultra Red protocol for H2O2 quantification from seedling tissues subjected to ER stress. A. Comparison of H2O2 extraction methods using potassium phosphate buffer (PHOS) or 10% TCA neutralized with NaHCO3 (NTCA) incubated in the presence or absence of horseradish peroxidase (HRP). B. Evaluation of tissue loading during PHOS extraction and catalase treatment. C. Evaluation of tissue loading during NTCA extraction and catalase treatment. D. WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 seedlings were treated in a plate system with Tm or DMSO for the indicated time and H2O2 quantified; see materials and methods. Data represent the mean concentration ± SE. E. From the values in (D) average fold change ± SE (Tm/DMSO) was determined from biological replicates in the order that they were measured. Statistical significance compared to equivalent WT value, unless a bracket is used to indicate comparison. Statistical significance determined using Student’s unpaired t-test and indicated by: *=p<0.05, **=p<0.005 ***=p<0.0005, NS=not significant.
We then used this assay to measure the levels of H2O2 in seedlings experiencing ER stress during the adaptive phase of the UPR. Using the same plate system detailed earlier (Figure 1), we compared the effects of Tm treatment for 6, 24, and 48 hours against seedlings growing on plates containing DMSO as control. We tested WT, rbohd rbohf, a mutant lacking the two IRE1 isoforms (ire1a ire1b; (Chen et al. 2012)) and a double mutant lacking functional bZIP60 and bZIP28 transcription factors (bzip60 bzip28; (Deng et al. 2013)). We found differences in H2O2 concentrations among the various backgrounds and their controls (Figure 2D), in support of a significant bearing of RBOHD and RBOHF activity as well as intact UPR on H2O2 production. To better illustrate such differences we estimated the fold change in concentration between Tm/DMSO (Figure 2E). Specifically, we found that in all backgrounds at 6-hours of treatment there were no differences in fold change of H2O2 concentrations (TM/DMSO; Figure 2E). At 24 hours, we established that Tm-treatment led to a small but significant increase in H2O2 levels in the ire1a ire1b mutant background only. However, at 48 hours, coincident with significant increases in O2− in mature WT tissues (Figure 1B), we found a significant increase in H2O2 levels in WT that were not observed in the rbohd rbohf mutant (Figure 2E), indicating that RBOHD and RBOHF are required for the accumulation of H2O2 under ER stress conditions. Noticeably, the increase of H2O2 verified in WT but not rbohd rbohf was significantly greater in ire1a ire1b and bzip60 bzip28 mutants (Figure 2E), supporting a requirement of an intact UPR signaling for the management of H2O2 levels in conditions of ER stress. Together, these results indicate that H2O2 accumulates in response to ER stress in the adaptive phase of the UPR dependent on RBOHD and RBOHF activity and also influenced by the integrity of UPR signaling.
UPR regulators influence RBOHD and RBOHF expression during adaptive UPR
Having established that RBOHD and RBOHF activity is required for ER stress-induced O2− and H2O2 production (Figures 1, 2), we next aimed to determine if the canonical UPR arms affected RBOHD or RBOHF expression at the transcriptional level. Therefore, we tested whether ER stress modulated RBOHF or RBOHD transcript levels as it occurs for other ER stress responsive genes, such as bZIP60, BiP3, ERdj3A and ERdj3B (Chen et al. 2012, Ruberti et al. 2018). To do so, a subset of seedlings from the H2O2 quantification experiments (Figure 2) were used to follow the changes in gene transcript levels by quantitative RT-PCR (qRT-PCR) during Tm treatment in the plate system (Iwata et al. 2005, Liu et al. 2007a, Chen et al. 2013). We found no changes in the RBOHF transcript levels in the WT, ire1a ire1b, or bzip60 bzip28 lines at 6 hours of treatment. However, at 24 and 48 hours of TM treatment, the bzip60 bzip28 line had slightly lower levels of the RBOHF transcripts compared to WT (Figure 3A). Conversely, in the WT and ire1a ire1b, the RBOHD transcripts were transiently induced at 24 hours and then restored to basal levels at 48 hours. We also observed that the RBOHD induction was significantly higher in ire1a ire1b compared to WT (Figure 3B). The RBOHD transcript levels in the bzip60 bzip28 line we found to be slightly repressed at 6 hours of treatment (with a TM/DMSO ratio of ~0.8; Figure 3B). Noticeably, however, at 24 hours in the bzip60 bzip28 line the RBOHD transcript levels were significantly lower compared to WT and ire1a ire1b. Together these results demonstrate that although the integrity of UPR signaling is required for maintaining homeostatic levels of RBOH expression during ER stress, the timing of the observed changes does not directly correlate with RBOHD and RBOHF dependent O2− (Figure 1) or H2O2 production (Figure 2) in WT plants. This indicates that other factors, such as post translational modifications, protein-protein interactions, or altered endomembrane trafficking of the protein product may contribute to these outcomes.
Figure 3. Intact UPR signaling is required to maintain homeostasis of RBOH transcript levels and ROS signaling, while RBOH activity affects UPR homeostasis.
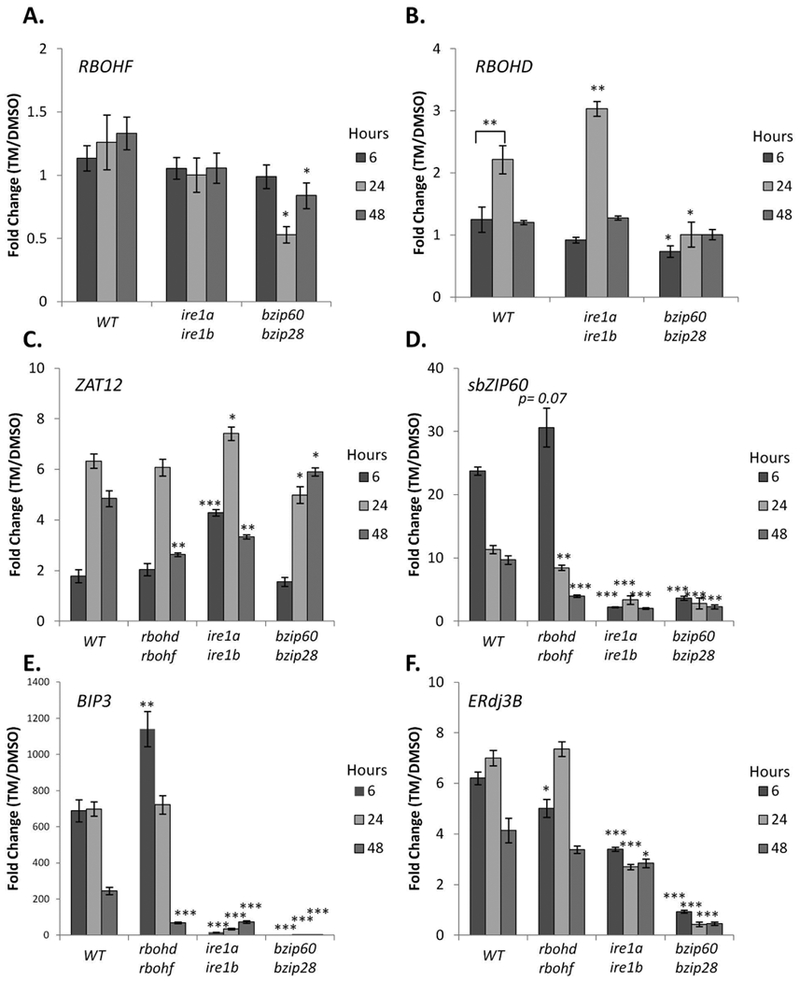
Time course gene expression analysis of WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 seedlings subjected to ER stress. qRT-PCR analyses were performed using primers specific for either RBOHF (A), RBOHD (B), ZAT12 (C), spliced bZIP60 (sbZIP60; D), BIP3 (E), or ERdj3B (E). Data represent the mean ratio ± SE (biological replicates=3); Statistical significance compared to equivalent WT value, unless a bracket is used to indicate comparison. Statistical significance determined using Student’s unpaired t-test and indicated by: *=p<0.05, **=p<0.005 ***=p<0.005, NS=not significant.
To provide further evidence that the observed increases in O2− (Figure 1) are dependent upon RBOHD and RBOHF activity during ER stress, we quantified the expression of ZAT12, an O2− responsive marker gene (Miller et al. 2009, Xu et al. 2017). Consistent with transiently increased levels of NBT staining at 24 hours in the root tip, which was largely independent of RBOHD and RBOHF activity (Figure 1A), we found a 6-fold upregulation in ZAT12 levels in the WT and rbohd rbohf lines (Figure 3C). Significantly at 48 hours, where we observed an increase in NBT staining of mature root tissues and upregulated H2O2 levels in WT but not rbohd rbohf seedlings (Figures 1, 2), we also found that rbohd rbohf seedlings had significantly lower levels of ZAT12 transcript (Figure 3C). These results demonstrate that 48 hours of ER stress induces O2− accumulation via RBOHD and RBOHF (Figure 1) to a level that elicits a transcriptional response.
We next aimed to test whether UPR regulation of RBOHD and RBOHF transcripts or H2O2 levels (Figures 2, 3A, 3B) was correlated with changes in superoxide-dependent signaling. We found that at 6 hours of Tm treatment in ire1a ire1b the levels of ZAT12 were significantly higher compared to WT. At 24 hours of treatment, ZAT12 levels in bzip60 bzip28 and ire1a ire1b mutants were slightly lower and higher than WT levels, respectively. Conversely, at 48 hours of treatment ZAT12 levels in bzip60 bzip28 and ire1a ire1b mutants were higher and lower than WT levels, respectively (Figure 3C). Although the increased ZAT12 levels in the ire1a ire1b mutant are consistent with the verified increase in transcription of RBOHD and H2O2 accumulation at 24 hours, the regulation of ZAT12 in both UPR mutant lines does not correlate with the RBOH transcription, or H2O2 accumulation at 48 hours. We therefore propose that an impaired UPR response likely has pleiotropic effects in the regulation of superoxide production, superoxide signaling, and H2O2 accumulation during adaptation to ER stress.
We next tested whether impaired RBOHD and RBOHF activity affected the canonical UPR at a transcriptional level. We tested the levels of ER stress-responsive genes such as spliced bZIP60 transcripts (sbZIP60) and BIP3, whose abundance is primarily regulated by IRE1-bZIP60, as well as ERDJ3B, whose expression is primarily regulated by bZIP28 (Ruberti et al., 2018). In WT, we found that the Tm treatment led to increased levels of sbZIP60 transcripts at 6 hrs, which were attenuated at 24 and 48 hours (Figure 3D). Compared to WT, in the rbohd robhf line, we observed no significant changes in the induction of sbZIP60 levels at 6 hours but a significant ~2 fold reduction of sbZIP60 levels at 48 hours (Figure 3D). An altered UPR signaling in rbohd rbohf was reflected in our analyses of BIP3 transcript levels, which were ~2-fold higher at 6 hours of treatment and ~4 fold lower at 48 hours in the rbohd rbohf line compared to WT (Figure 3E). Conversely, ERdj3B was found to be ~1.2-fold lower at 6 hours in the rbohd rbohf line compared to WT, but was otherwise insignificantly different. Taken together, these results indicate that RBOHD and RBOHF contribute to maintain UPR signaling homeostasis during ER stress.
RBOHD and RBOHF are necessary for recovery from short-term and chronic ER stress
We next aimed to test whether RBOHD and RBOHF could contribute to ER stress resolution in situations subsequent to the adaptive phase. In this context, progression of the plant UPR has been studied in conditions of relief from ER stress (temporary ER stress; (Ruberti et al. 2018)) and in conditions of unresolved ER stress (chronic ER stress; (Chen et al. 2013)). Therefore, we first tested the role of RBOHD and RBOHF in recovery from temporary ER stress. In this assay, 7-day old-seedlings are transferred to liquid media containing Tm for 6 hrs, and then re-plated on growth medium without Tm. After 3 further days of growth, shoot fresh weight, root length and chlorophyll content are assayed to assess the ability of the various genetic backgrounds to overcome ER stress upon relief from an ER stress inducer. For the assay, we used WT, rbohd rbohf and ire1a ire1b. Consistent with previous findings (Ruberti et al. 2018), at the completion of the recovery phase, we found that the shoots of the ire1a ire1b mutant weighed significantly less that DMSO-treated controls and lost most of their chlorophyll; the root also ceased further growth (Figure 4A–D). We also found that although the shoots of the Tm-treated rbohd rbohf plants weighed similarly to the Tm-treated WT (Figure 4B), a significant reduction of chlorophyll content was evident (Figure 4C). The roots of Tm-treated rbohd rbohf also showed an overall reduction in length compared to WT (Figure 4D). These results indicate that RBOHD and RBOHF are necessary to successfully overcome temporary ER stress.
Figure 4. RBOHD and RBOHF are required in the recovery from temporary ER stress.
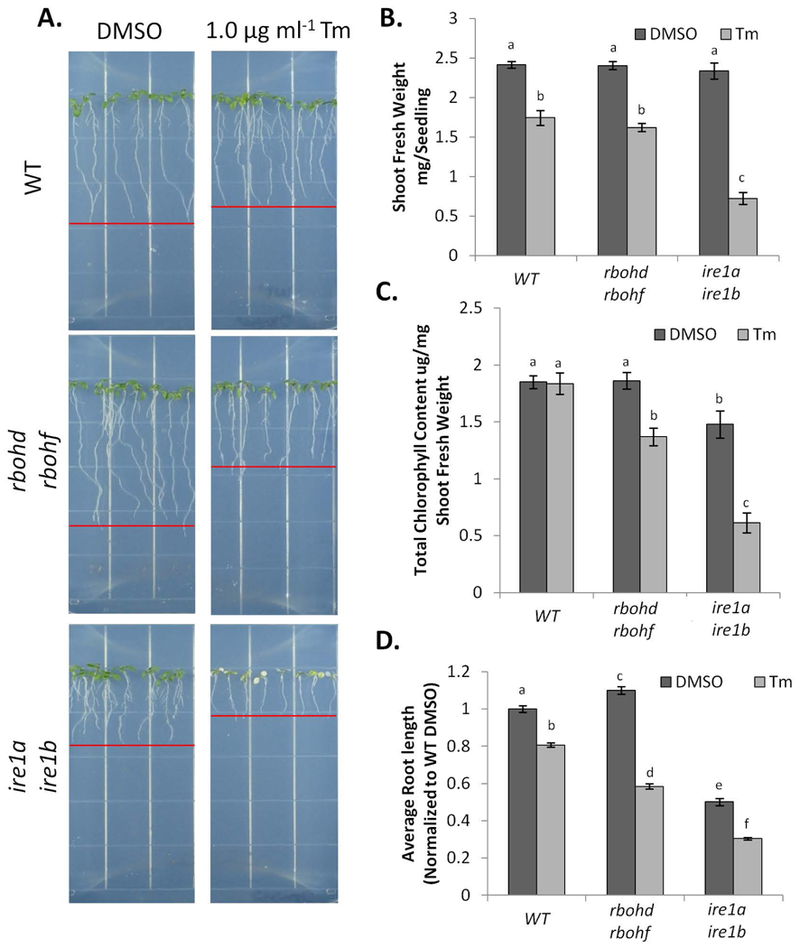
Phenotypic analyses of the various genetic backgrounds subjected to temporary ER stress. A. WT, rbohd rbohf, and ire1a ire1b seedlings were subjected to ER stress and were imaged after a 3 day recovery. From the seedlings grown in (A) shoot fresh weight (B), average total chlorophyll (C), and root length (D), were recorded as described in the materials and methods. Data represent the mean ± SE. (at least 10 biological replicates). Letters above each data point indicate statistically significant groups using Student’s unpaired t-test (p<0.05).
We then tested the requirement of RBOHD and RBOHF for overcoming chronic ER stress. To do so, we followed the customary approach to germinate seeds on growth medium containing Tm for comparison with seedlings germinated on control plates (Chen et al. 2012). As positive controls, we again used ire1a ire1b as well as bzip60 bzip28, which are hypersensitive to chronic ER stress (Liu et al. 2007a, Chen et al. 2012). After fourteen days of growth, we examined shoot fresh weight and chlorophyll content (Figure 5A–C). Consistent with previous findings (Chen et al. 2012, Ruberti et al. 2018), the ire1a ire1b and bzip60 bzip28 mutants showed a strong phenotype with marked reduction in shoot fresh weight and loss of chlorophyll content. When we analyzed rbohd rbohf, we found a significant reduction in the average shoot fresh weight and chlorophyll content compared to WT. These results indicate that, similar to temporary ER stress recovery, RBOHD and RBOHF are required to successfully overcome chronic ER stress.
Figure 5. RBOHD and RBOHF are required for adaptation to chronic ER stress.
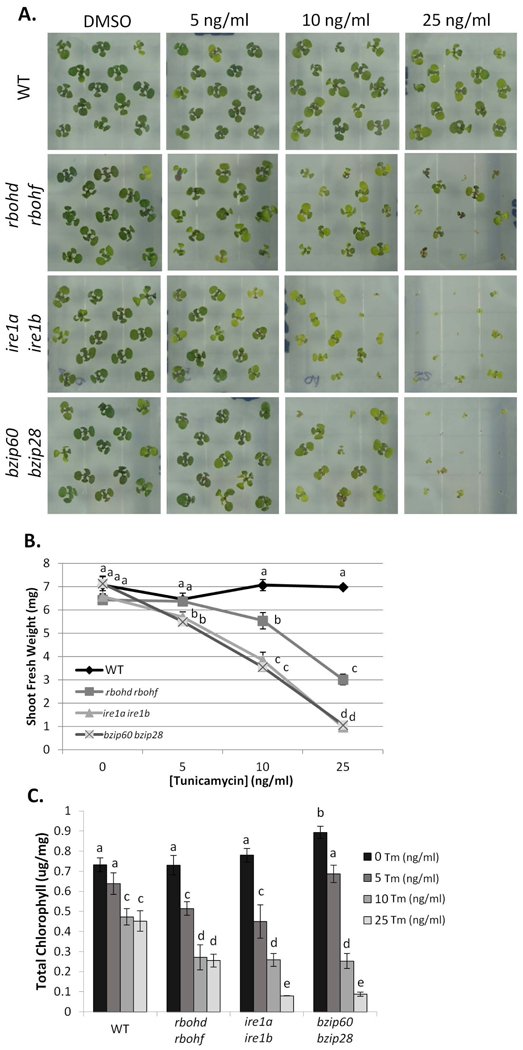
Phenotypic analysis of different seedling genetic backgrounds subjected to chronic ER stress. A. WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 were germinated on media containing the indicated concentration of Tm or DMSO grown for two weeks. Shoots fresh weight (B) and total chlorophyll content (C) were measured. Data represent the mean ± SE (at least 10 biological replicates). Letters above each data point indicate statistically significant groups using Student’s unpaired t-test (p<0.05).
RBOHD and RBOHF contribute to preventing ER stress induced cell death
Previous reports have detailed the protective role that RBOHD and RBOHF play in preventing the spread of cell death during plant immune system response through yet undetermined mechanisms (Torres et al. 2005). Having established that RBOHD and RBOHF are required for successful recovery from temporary ER stress and survival from chronic ER stress, we postulated that these enzymes could be involved in mechanisms preventing cell death caused by ER stress. To test this hypothesis, we followed cell death by quantification of electrolyte leakage (Lee et al. 2010) in WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 seedlings exposed to Tm for two and six days (Figure 6A,B). We found that although the shoot and root of Tm-treated WT seedlings grew considerably less compared to DMSO control, no significant differences were detected in electrolyte leakage levels (Figure 6B). As expected, the bzip60 bzip28 and ire1a ire1b mutants showed considerable chlorophyll loss with electrolyte leakage reaching almost 50% at 6 days of Tm treatment. We found that rbohd rbohf plants also showed a significant chlorophyll loss and electrolyte leakage by 6 days of Tm-treatment (Figure 6B) compared to WT plants. These results indicate that RBOHD and RBOHF contribute in preventing cell death in ER stress conditions.
Figure 6. RBOHD and RBOHF act to prevent ER stress induced cell death.
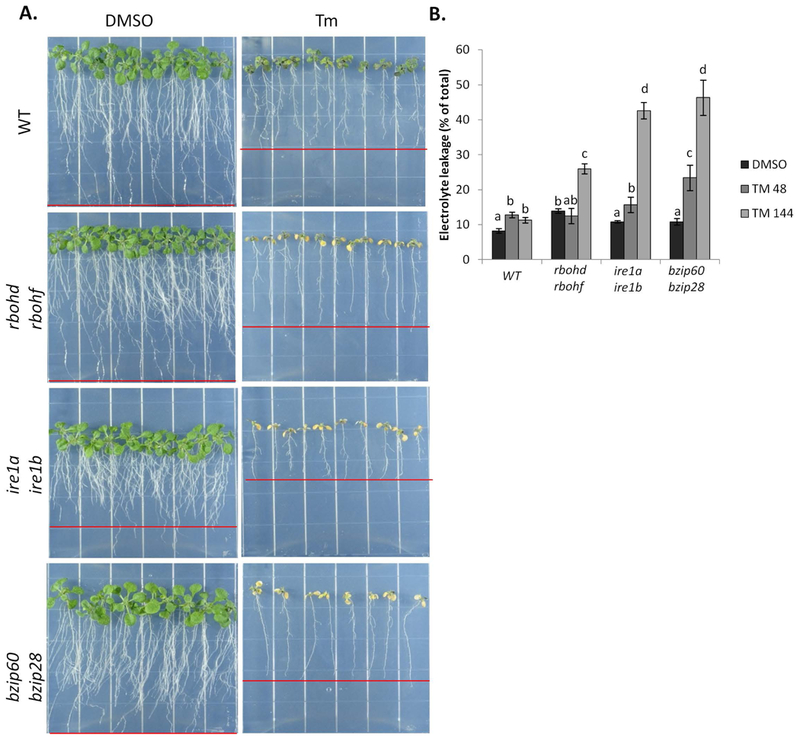
Determining the extent of cell death in seedlings of different genetic background subjected to prolonged ER stress. A. WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 seedlings treated with Tm in a plate system for 48 and 144 hours, and then photographed at 144 hours. B. The extent of cell death was determined by quantification of percent electrolyte leakage. Data represent the mean ± SE (4 biological replicates). Letters above each data point indicate statistically significant groups using Student’s unpaired t-test (p<0.05).
Discussion
In plants, the identity of several critical sensors and transducers of ER stress has been unraveled (Angelos et al. 2017), and the challenge ahead is to discover additional factors that participate in the UPR management. In this work, we have established that two NADPH Oxidases are necessary to overcome specific situations of ER stress, such as recovery from ER stress conditions and unresolved ER stress. These results enrich the general knowledge of the identity of the proteins and signaling pathways that contribute to ER stress survival in plants. The ability of mammalian cells to overcome ER stress is negatively influenced by NADPH Oxidases-produced ROS (Li et al. 2010). In plants, the relationship between ROS and the UPR was unknown. In this work, we showed that, in conditions of an intact UPR signaling, NADPH Oxidases-produced ROS exert a positive role on the ability of the plant to overcome ER stress. These findings support the conclusion that, despite the functional conservation of canonical ER stress sensors and transducers such as IRE1 and bZIP-transcription factors, plants have evolved unique strategies for ER stress survival.
NADPH Oxidases contribute to ROS production in conditions of ER stress
Several metabolic and signaling processes affect cellular concentrations of H2O2 in plants, including biotic and abiotic stress responses (Miller et al. 2009), nutrient availability (Contento et al. 2010), circadian rhythms (Zhong et al. 1994), and gravity (Hausmann et al. 2014). In Arabidopsis there are also studies indicating that ER stress affects cellular concentrations of H2O2, through both direct measurements (Ozgur et al. 2014, Ozgur et al. 2018) and analyses of redox-related biochemical activities that are intimately linked with cellular H2O2 levels (Ozgur et al. 2014). However, prior to our work the nature of the enzymes contributing to H2O2 levels in conditions of ER stress had yet to be clearly established in plants. In this work, we have set up a sensitive assay to measure H2O2 levels (Figure 2). Using this assay, superoxide staining and a genetic background lacking RBOHD and RBOHF, we showed that ER stress causes activation of NADPH Oxidase-coupled superoxide production, leading to the accumulation of H2O2 (Figures 1, 2). We further show that a superoxide-responsive signaling pathway (Xu et al. 2017) is also activated, as demonstrated by transcriptional induction of the marker gene ZAT12 (Figure 3C). The concomitant reduction of three different ROS reporters in the rbohd rbohf line compared to WT under conditions of ER stress (Figures 1, 2, 3; 48 hours) supports that the elevation of H2O2 in conditions of stress is contributed partially by RBOHD and RBOHF. Therefore, these findings identify two of the enzymes that operate in ROS management during ER stress potentially sharing redundant functions with other RBOH-enzymes.
Although, the exact molecular mechanisms leading to activation of these enzymes in response to ER stress have yet to be elucidated, a regulation of RBOH enzymes by altered cytosolic calcium levels and association with G-protein signaling components may be postulated as contributing factors. In metazoans cellular calcium ion homeostasis is intimately linked with ER stress, as calcium is required for the proper functioning of ER luminal foldase activities (Krebs et al. 2015). Inhibition of sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA) transporters leads to the induction of ER stress and the UPR (Krebs et al. 2015). Upon prolonged UPR activation, a number of factors, including ER lumen hyper-oxidation by ER oxidase 1a (ERO1a), leads to calcium release from the ER through activation of inositol triphosphate receptors (IP3R) and apoptotic events (Sovolyova et al. 2014). Although IP3R-like ER calcium channels have yet to be identified in plants, SERCA-like transporters have been identified (Liang et al. 1997). Therefore it is possible that ER stress/UPR induction could alter ER and cytosolic calcium levels, which can promote RBOHD activation (Ogasawara et al. 2008). Furthermore, in plant responses to bacterial pathogen attack, it has also been demonstrated that a null mutation in the Arabidopsis Gβ subunit (agb1) of the heterotrimeric G-protein plasma membrane signaling complex was epistatic to the rbohd rbohf null mutation(Torres et al. 2013). The AGB1 gene acts in concert with receptor-like kinases to mediate oxidative burst response to pathogen-associated molecular pattern (PAMP) triggered immunity(Liu et al. 2013, Liu et al. 2016b). Although it is yet to be tested whether AGB1 interacts genetically with RBOHD and RBOHF in ER stress, similar to rbohd rbohf, agb1 is sensitive to chronic ER stress (Chen et al. 2012). It is therefore possible that AGB1 may contribute to RBOH signaling in conditions of ER stress.
Unlike in metazoans, NADPH Oxidase-produced ROS are beneficial to overcome ER stress in plants
Many aspects of the UPR regulators are conserved between plants and metazoans at a functional level even when compared to yeast wherein only IRE1 arm of the UPR is genetically encoded (Ron et al. 2007, Ruberti et al. 2015, Liu et al. 2016a). However, it is increasingly obvious that the different evolutionary contexts between plants and animals have forced the development of different survival strategies in overcoming ER stress. The lack of a plant homolog of the mammalian PERK UPR pathway is evidence to that effect. NADPH Oxidases are largely conserved between mammals and plants (Suzuki et al. 2011). However, the activities of mammalian NADPH Oxidase 2 (Nox2) and NADPH Oxidase 4 (Nox4), two homologs of RBOHD and RBOHF, promote apoptosis during ER stress (Pedruzzi et al. 2004, Li et al. 2010). Specifically, an intravenous tunicamycin challenge in Nox2−/− mice resulted in a dramatic reduction of renal cell apoptosis and increased protection against renal dysfunction when compared to WT mice (Li et al. 2010). In this work, we show that rbohd rbohf plants display a significant sensitivity to ER stress conditions (Figures 4, 5, 6). Specifically, in the recovery from acute Tm treatment, in rbohd rbohf, the resumption of plant growth is markedly delayed, and under chronic stress conditions (Figures 5, 6), rbohd rbohf plants exhibit enhanced progression of cell death (Figure 6). Therefore, differently from the mammalian system, during ER stress NADPH Oxidases RBOHD and RBOHF exert a pro-survival role in the ER stress response. As such, the results presented in this study identify RBOHD and RBOHF as important positive contributors to UPR adaptation in plants in a manner that runs differently from the mammalian cell system.
Homeostasis of NADPH Oxidase activity is required to maintain an effective UPR
Several studies, including studies of RBOH activity during heat stress response in plants, support a non-specific “transcriptional priming” by which fast acting ROS production by RBOHD amplifies initial stress-specific transcriptional responses in local and systemic tissues (Mittler et al. 2015). In our analyses, we established that the RBOHD and RBOHF are involved in the adaptive phase of the UPR (Figure 3D–F) as well as in recovery from temporary ER stress and in situations of chronic ER stress (Figures 4, 5). Consistent with a priming role of RBOH activity in stress responses, the verified misregulation of the adaptive phase of the UPR due to a compromised RBOH activity may lead to a defective actuation of proper cytoprotective responses necessary for surviving temporary and chronic ER stress. Interestingly, when compared to ERdj3B levels, we verified a stronger impact on the induction levels of sbZIP60 and BIP3 in rbohd rbohf compared to WT (Figure 3D–E). Based on the findings that BIP3 expression is principally controlled by the IRE1-bZIP60 arm and that the ERdj3B is principally controlled by bZIP28 arm, we propose that the activity of RBOHD and RBOHF positively influences the transcriptional role of bZIP60. This hypothesis does not exclude that other RBOH enzymes may affect the transcriptional role of bZIP60 as well as bZIP28. Indeed, in our work, while we demonstrate that RBOHD and RBOHF activity is required for successfully overcoming these specific conditions of ER stress, the activity of these enzymes is not strictly essential. This is supported by the relative survival of rbohd rbohf compared to ire1a ire1b and bzip60 bzip28 in temporary and chronic ER stress conditions (Figures 4, 5, 6). Given the relatively large size of RBOH family and the residual NADPH Oxidase marker activity observed in the rbohd rbohf line (Figure 1), it is possible that RBOHD and RBOHF share redundant activity with other RBOH enzymes in temporary and chronic ER stress conditions.
In ER stress conditions, the production of ROS is antagonized by IRE1 and the bZIP60/bZIP28 transcription factors
The plant IRE1 as well as the availability of bZIP28 and bZIP60 together are strictly necessary to overcome temporary and prolonged stress, as demonstrated by the evidence that death is accelerated in the ire1a ire1b and bzip60 bzip28 mutants compared to WT in situations of recovery from temporary ER stress or in conditions of chronic ER stress ((Nagashima et al. 2011, Chen et al. 2012, Deng et al. 2013, Liu et al. 2016a); Figures 4, 5). An analysis of ER stress response in Chlamydomonas reinhardtii reported a significant transcriptional induction of ROS-dependent marker genes in IRE1 knockouts (Yamaoka et al. 2018). These findings led to the hypothesis that an increase in ROS may contribute to the ER stress sensitivity of ire1 knockout lines in this model systems (Yamaoka et al. 2018). Our results that in the Arabidopsis ire1a ire1b line, which is hypersensitive to ER stress ((Chen et al. 2012); Figures 4–6), the H2O2 levels are higher at 24 and 48 hours of Tm treatment compared to WT (Figure 2E) corroborate this hypothesis. Our results also provide direct evidence in support of the hypothesis that UPR regulators are required to manage cellular H2O2 concentrations under ER stress conditions in plants like they do in metazoans (Hourihan et al. 2016). However, the verified increases in H2O2 content in the ire1a ire1b and bzip60 bzip28 mutants were not directly correlated with the observed misregulation of ZAT12/RBOHD transcript levels in these lines compared to WT, which may indicate that an intact UPR signaling potentially coordinates ROS production/signaling through multiple mechanisms to promote proper ER stress management. Conversely, the verified differences in transcript induction of UPR marker genes (i.e., sbZIP60, BIP3 and ERdj3B) in rbohd rbohf compared to WT (Figure 3D–E) indicate that RBOH activity is required to maintain proper UPR signaling. How the observed increase in H2O2 levels may affect the UPR is yet undetermined at a mechanistic level. Recent findings have shown that ER stress alters the cytosolic redox potential, which in turn modulates the activity of the transcriptional coregulatory NPR1 (nonexpresser of PR genes), which binds and represses the transcriptional function of bZIP28 and bZIP60 (Lai et al. 2018). It is similarly possible that an excessive elevation of H2O2 in conditions of ER stress changes the activity of UPR and programmed cell death modulators. The ROS species produced by RBOHD and RBOHF suppress the spread of runaway cell death in a pathway parallel to the Lesion Stimulating Disease 1 (LSD1) zinc-finger protein under normal growth conditions, in response to salicylic acid, extracellular superoxide, and pathogen induced hypersensitive response (Torres et al. 2005). The results of our study indicate that RBOHD and RBOHF may also prevent the propagation of cell death in response to chronic ER stress (Figure 6). Under this light, the noted misregulation of RBOH activity and accumulation of H2O2 in ire1a ire1b and bzip60 bzip28 mutants during ER stress (Figures 2E, 3C) may represent convergent signals that initiate processes leading toward programmed cell death, and could at least partially explain the hypersensitivity of these lines to ER stress conditions (Figure 4–6) (Chen et al. 2012, Deng et al. 2013). Therefore, together our results support the hypothesis for a dual role of H2O2 in ER stress-induced programmed cell death, which is differently manifested in WT and mutants of the UPR signaling pathway. Specifically, in cells with an intact UPR, RBOH activity and H2O2 levels are maintained to levels that promote survival; however, a defective UPR likely causes misregulation of RBOH activity and excess H2O2 accumulation that may lead to cell death. The latter scenario may be beneficial for plants to favor the survival only of cells with an intact ability to overcome proteotoxic stress.
Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0), rbohd rbohf, (Torres et al. 2002) ire1a ire1b, and bzip60 bzip28 (Chen et al. 2012, Deng et al. 2013) plants were used in this study. For all experiments, surface-sterilized seeds were plated directly onto petri dishes containing half-strength Linsmaier and Skoog (½ LS) medium, 1.0% w/v sucrose, and 1.0% agar and then cold treated (4°C) in the dark for two days to synchronize germination. Plates were then transferred to a Percival growth chamber and incubated for the indicated time at 21°C under continuous light (130 μE).
Superoxide Histochemical Staining and Quantification
WT or rbohd rbohf seedlings were grown vertically on for 7 days, and then transferred to ½ LS media containing 1.0 μg/ml Tm, 1.0 μg/ml TM + 2.5 μM DPI, or DMSO and allowed to grow for a further 24 or 48 hours in the growth chamber. Whole seedlings were used for the subsequent Nitro Tetrazolium Blue (NBT) staining which took place at the same time on each day. NBT was dissolved to a concentration of 1 mg/ml in 20 mM HEPES pH 6.1, and protected from light until used. To reduce the inherent variability created by staggered staining of the seedlings for very short incubation periods, all experimental groups were treated with NBT solution at the same time. To do so, six 60mm Petri dishes containing 2 ml of ddH2O were prepared. Fifteen individuals from each genotype/treatment were then transferred to separate dishes with forceps. Once all seedlings were transferred, 15 ml of NBT solution was added to each dish and covered for 15 minutes. For each plate, the NBT solution was quickly removed with a seriological pipette and replaced with ~20 ml of ddH2O to remove excess NBT. Restarting at the first dish, the ddH2O was then removed, and seedlings were submerged in 95–100% ethanol for 10 minutes to fix the root tissues. Seedlings were then mounted on slides in a 1:1 ethanol:glycerol solution with a coverslip covering just the roots. Images were obtained using the microscope function of an Olympus Tough F2.0 camera outfitted with ring LED light guide. Over a white surface, slides were placed on a 60mm Petri dish marked with a reference distance. The slide and 60mm dish were then placed in the center of a 100mm square Petri dish and covered with the lid. The camera was placed directly on the square lid for imaging to ensure consistent distance from the subject. Three independent experiments were performed with similar results.
To quantify the stain intensity along the root, unedited images were converted to 32 bit black and white in ImageJ. Beginning at the root apex, pixel intensities were recorded along a traced line segment (1000 individual measurements). The same line traces were then moved immediately left or right of the root and background pixel intensities were recorded. For each root, relative pixel stain intensity was calculated by subtracting the background value from the original trace value. For each experimental group the relative stain intensity from 8 roots was averaged and standard error calculated. Three independent experiments were performed with similar results.
Extraction and Quantification of H2O2 in Seedling Tissues using Amplex Ultra Red
(Chakraborty et al. 2016)Two H2O2 extraction methods described previously (Chakraborty et al. 2016, Le et al. 2016) were tested: 1) phosphate buffer (K2HPO4, 20mM pH 6.5) and 2) neutralized Trichloroacetic Acid (TCA). For phosphate buffer extractions, 200 microliters was added to frozen tissue then briefly vortexed. Samples were then centrifuged at 21000 xg and 4°C for 20 minutes to ensure plant debris was pelleted then used immediately for H2O2 quantification. NTCA extractions were performed as follows: ten percent trichloroacetic acid (TCA), and 1M sodium bicarbonate solutions were prepared and kept on ice. Two hundred microliters of 10%TCA was added to still frozen samples on ice and briefly vortexed until mixture was homogenous and centrifuged at 21000 xg and 4°C for 20 minutes. In separate microcentrifuge tubes 86.6 microliters of sodium bicarbonate solution was aliquoted and 150 microliters of the centrifuged 10%TCA supernatant extract was added to bicarbonate containing tubes on ice. Any pink coloration due anthocyanin content in extracts should change to a dark blue hue after neutralization. One hundred microliters of the neutralized supernatants were arranged in 96 well plates on ice, to allow for sample transfer to assay with a multichannel pipette.
A working solution of catalase was prepared by centrifuging ten microliters of ammonium sulfate catalase suspension (Sigma, C3515–10MG). The clear ammonium sulfate supernatant was then removed before catalase pellet was dissolved in 600 microliters of ddH2O. Five microliters of catalase working solution was then added to the remaining neutralized sample extracts in microcentrifuge tubes. These samples were incubated at room temperature for a minimum of 10 minutes. One hundred microliters of the catalase treated supernatants were then stored in the same 96 well plate on ice.
An H2O2 standard curve was prepared by serial dilution of 30% H2O2 to concentrations of 100 μM, 50 μM 25 μM 10 μM and 5 μM which were then diluted 10x (from 50 microliter aliquots) by sequential addition of 200 microliters of 10% TCA, 122 microliters of 1M sodium bicarbonate and 128 microliters of ddH2O to mimic extraction procedure. Final concentrations of H2O2 standards therefore ranged from 10 μM to 0.5 μM. NTCA extraction resulted in no net loss of H2O2. H2O2 standards were prepared fresh daily.
Amplex Ultra Red (AUR) working solution was prepared by dissolving 1 vial of AUR in 340 microliters of DMSO per manufacturer’s instructions to make a 10mM AUR stock. AUR assay solutions were prepared immediately before use. Fifty microliters of AUR stock was added to 10 ml of ddH2O containing 5 μg/ml commercial horseradish peroxidase (AUR-HRP) and to another 10 ml of ddH2O without HRP (AUR-NoHRP). Solutions were kept on ice and protected from light. All assays were performed in clear 96 well microplates and prepared on a cold block. Twenty five microliters of neutralized samples and catalase treated samples were always pipetted prior to addition of 75 microliters of the appropriate AUR assay solution by multichannel pipette. The plate was then briefly incubated (5 min) at room temperature in the dark. Longer incubation periods did not lead to greater differences between catalase treated samples and untreated samples, only increases in AUR chemical auto-oxidation. The microplates were read using a SpectramaxM2 (Molecular Devices) equipped with fluorescence detection capabilities. The excitation wavelength was set to 544 nm, and emission was recorded at 590 nm.
Experiments were performed to test the effectiveness of extraction procedures, dose response, and catalase treatment (Figure 2A,B,C). WT seedlings (~2g) were ground in a mortar with liquid nitrogen, to a fine powder and separated into triplicate 40, 30, 20, or 10 mg aliquots. Extraction procedures were performed as described above and assayed with AUR-HRP or AUR-NoHRP as indicated.
To assay H2O2 accumulation under ER stress conditions WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 were grown for 7 days and transferred to plates containing 1.0 μg/ml Tm or DMSO for 6, 24, or 48 hours as indicated, and harvested at the same time on consecutive days. Three biological replicates consisting of approximately 30–60 mg of seedling tissue each were briefly dried on a Kimwipe and exact fresh weight recorded before samples were placed in 1.7ml microcentrifuge tubes with two glass beads, and frozen in liquid nitrogen. Samples were ground to a fine powder in a Retch MM301 (Retch; Haan, Germany) by agitation at a frequency of 30/sec for two sets of 30 seconds which were separated by refreezing in liquid nitrogen and stored in liquid nitrogen. Samples were extracted with NTCA, and then treated with catalase as indicated. For each biological replicate the neutralized samples and catalase samples were assayed in two technical replicates in the same plate with a standard curve. Each genotype was assayed in a separate plate. The catalase labile signal was calculated by subtracting average fluorescence intensity of catalase treated replicates from the average fluorescence intensity of the neutralized sample replicates. The samples were compared to the individual H2O2 standard curves to derive an [H2O2] of the samples, and total micromoles of H2O2 in the extract. This was normalized to recorded sample fresh weights.
RNA extraction and Quantitative RT-PCR Analyses
The RNA measurement experiments were performed in parallel with the H2O2 quantification experiments. Seedlings germinated under normal growth conditions and grown for 7 days were transferred to plates containing ½ LS media with 1.0% sucrose and 1.0 μg/ml Tm or DMSO for 6, 24, or 48 hours as indicated, and harvested at the same time on two consecutive days. Groups of 5–10 seedlings were pooled placed in 1.7ml microcentrifuge tubes with two glass beads, and frozen in liquid nitrogen. Total RNA was extracted from whole seedlings using a NucleoSpin Plant RNA kit (Machery-Nagel) according to the manufacturer’s instructions, including on column DNase Digestion. All samples within the experiment were reverse-transcribed using iScript Reverse Transcriptase. RT-PCR with SYBR Green detection using a ΔΔct method was performed in technical triplicates using the Applied Biosystem 7500 Fast Real-Time 7500 PCR system, and data normalized to the expression of UBQ10 (AT4G05320). The values presented are the mean of three independent biological replicates ±SE.
Recovery from ER stress, and Chronic ER stress Phenotypic Analyses
For the recovery from ER stress experiments WT, rbohd rbohf, and ire1a ire1b were germinated on ½ LS plates grown vertically for 5 days, transferred to liquid ½ LS media containing 1.0 μg ml−1 Tm for 6 hours, then replated on ½ LS plates and grown vertically for a further 3 days. The plates were then photographed and the root lengths of at least 30 individuals from 4 separate plates were determined using ImageJ; the mean ±SE was then calculated. In groups of two shoots were excised, fresh weight recorded, placed in 1.7ml microcentrifuge tubes with two glass beads, and frozen in liquid nitrogen.
For the chronic ER stress experiments WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 were germinated on ½ LS plates containing DMSO, 5 ng/ml, 10 ng/ml, or 25 ng/ml Tm and grown for two weeks then photographed. In groups of five, shoots were excised, fresh weight recorded, placed in 1.7ml microcentrifuge tubes with two glass beads, and then frozen in liquid nitrogen.
For both ER stress recovery and chronic stress experiments, total chlorophyll content (chlorophyll a+ chlorophyll b) per mg fresh weight was determined as described previously (Tait et al. 2003). Samples were ground to a fine powder in a Retch MM301 (Retch; Haan, Germany) by agitation at a frequency of 30/sec for two sets of 30 seconds which were separated by refreezing in liquid nitrogen. From the liquid nitrogen 1 ml of DMSO was added to the sample tubes, inverted until mixed then incubated at room temperature for 20 minutes in the dark. Samples were then centrifuged at 21000xg and 200ul aliquots added to a clear 96 well plate for spectrophotometric quantification of chlorophyll content using a SpectramaxM2 (Molecular Devices). For all experiments at least 10 biological replicates for each experimental group were recorded, presented data represents the mean ±SE.
Electrolyte Leakage Measurements
WT, rbohd rbohf, ire1a ire1b, and bzip60 bzip28 seedlings were grown for 7 days then transferred to plates containing 1.0 μg/ml Tm, or DMSO. Seedlings were imaged after six days on treatment plates. After 48 and 144 hours the extent of cell death was determined by quantification of percent electrolyte leakage as described previously with minor modifications (Dong et al. 2006, Lee et al. 2010). Groups of five seedlings were briefly washed in ddH2O, and then incubated in 4 ml of ddH2O for 3 hours in glass culture tubes with gentle agitation. Liquid conductivity was measured (Measurement 1). The tubes were autoclaved with caps and allowed to cool under gentle agitation for 3 hours. Total conductivity was measured (measurement 2) and percentage of the total was calculated as (%=measurement1/measurement2*100).
Acknowledgements
We acknowledge support by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (award number DE-FG02–91ER20021) for infrastructure, NASA (award NNX12AN71G), NIH R01-GM101038, and a fellowship from Michigan State University under the Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523).
Footnotes
The authors declare no conflicts of interest.
Literature Cited
- Angelos E, Ruberti C, Kim SJ and Brandizzi F (2017). Maintaining the factory: the roles of the unfolded protein response in cellular homeostasis in plants. Plant J 90(4): 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Scheuner D, Han J, Song B, Ribick M, Wang J, Gildersleeve RD, Pennathur S and Kaufman RJ (2009). Translation Attenuation through eIF2alpha Phosphorylation Prevents Oxidative Stress and Maintains the Differentiated State in Beta Cells. Cell Metabolism 10(1): 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Møller ALB, Kristiansen KA, Schulz A, Møller IM, Schjoerring JK and Jahn TP (2007). Specific Aquaporins Facilitate the Diffusion of Hydrogen Peroxide across Membranes. Journal of Biological Chemistry 282(2): 1183–1192. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Hill AL, Shirsekar G, Afzal AJ, Wang GL, Mackey D and Bonello P (2016). Quantification of hydrogen peroxide in plant tissues using Amplex Red. Methods 109: 105–113. [DOI] [PubMed] [Google Scholar]
- Chen Y and Brandizzi F (2012). AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69(2): 266–277. [DOI] [PubMed] [Google Scholar]
- Chen Y and Brandizzi F (2013). Analysis of Unfolded Protein Response in Arabidopsis. G Protein-Coupled Receptor Signaling in Plants: Methods and Protocols M. P. Running. Totowa, NJ, Humana Press, 10.1007/978-1-62703-532-3_8: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL and Bassham DC (2010). Increase in catalase-3 activity as a response to use of alternative catabolic substrates during sucrose starvation. Plant Physiology and Biochemistry 48(4): 232–238. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ and Diehl JA (2003). Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23(20): 7198–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ and Howell SH (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci 108(17): 7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R and Howell SH (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci 110(48): 19633–19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Hu X, Tang W, Zheng X, Kim YS, Lee BH and Zhu JK (2006). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 26(24): 9533–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M and Penel C (2007). Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytologist 174(2): 332–341. [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C and Larkin RM (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105(42): 16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper C and Dolan L (2006). Control of Plant Development by Reactive Oxygen Species. Plant Physiology 141(2): 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething MJ and Sambrook J (1992). Protein folding in the cell. Nature 355(6355): 33–45. [DOI] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin Y-H, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA and Kaufman RJ (2013). ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature Cell Biology 15: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M and Ron D (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6(5): 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y and Ron D (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397(6716): 271–274. [DOI] [PubMed] [Google Scholar]
- Hausmann N, Fengler S, Hennig A, Franz-Wachtel M, Hampp R and Neef M (2014). Cytosolic calcium, hydrogen peroxide and related gene expression and protein modulation in Arabidopsis thaliana cell cultures respond immediately to altered gravitation: parabolic flight data. Plant Biology 16: 120–128. [DOI] [PubMed] [Google Scholar]
- Hourihan John M., Moronetti Mazzeo, Lorenza E, Fernández-Cárdenas LP and Blackwell TK (2016). Cysteine Sulfenylation Directs IRE-1 to Activate the SKN-1/Nrf2 Antioxidant Response. Molecular Cell 63(4): 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Van Aken O, Schwarzländer M, Belt K and Millar AH (2016). The Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiology 171(3): 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y and Koizumi N (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci 102(14): 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs J, Agellon LB and Michalak M (2015). Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochemical and Biophysical Research Communications 460(1): 114–121. [DOI] [PubMed] [Google Scholar]
- Lai Y-S, Renna L, Yarema J, Ruberti C, He SY and Brandizzi F (2018). Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc Natl Acad Sci, 10.1073/pnas.1802254115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le CTT, Brumbarova T, Ivanov R, Stoof C, Weber E, Mohrbacher J, Fink-Straube C and Bauer P (2016). ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) Interacts with FER-LIKE IRON DEFICIENCY- INDUCED TRANSCRIPTION FACTOR (FIT) Linking Iron Deficiency and Oxidative Stress Responses. Plant Physiology 170(1): 540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.-h. and Zhu J-K (2010). Phenotypic Analysis of Arabidopsis Mutants: Electrolyte Leakage after Freezing Stress. Cold Spring Harbor Protocols 2010(1): pdb.prot4970. [DOI] [PubMed] [Google Scholar]
- Li G, Scull C, Ozcan L and Tabas I (2010). NADPH oxidase links endoplasmic reticulum stress, oxidative stress, and PKR activation to induce apoptosis. J Cell Biol 191(6): 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Cunningham KW, Harper JF and Sze H (1997). ECA1 complements yeast mutants defective in Ca2+ pumps and encodes an endoplasmic reticulum-type Ca2+-ATPase in Arabidopsis thaliana. Proc Natl Acad Sci 94(16): 8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-X, Srivastava R, Che P and Howell SH (2007a). An Endoplasmic Reticulum Stress Response in Arabidopsis Is Mediated by Proteolytic Processing and Nuclear Relocation of a Membrane-Associated Transcription Factor, bZIP28. The Plant Cell 19(12): 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR and Zhang Y (2013). Heterotrimeric G Proteins Serve as a Converging Point in Plant Defense Signaling Activated by Multiple Receptor-Like Kinases. Plant Physiology 161(4): 2146–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX and Howell SH (2016a). Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol 211(2): 418–428. [DOI] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P and Howell SH (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal 51(5): 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y and He C (2016b). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Reports 35(5): 995–1007. [DOI] [PubMed] [Google Scholar]
- Maity S, Rajkumar A, Matai L, Bhat A, Ghosh A, Agam G, Kaur S, Bhatt NR, Mukhopadhyay A, Sengupta S and Chakraborty K (2016). Oxidative Homeostasis Regulates the Response to Reductive Endoplasmic Reticulum Stress through Translation Control. Cell Rep 16(3): 851–865. [DOI] [PubMed] [Google Scholar]
- Malhotra JD and Kaufman RJ (2007). Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal 9(12): 2277–2293. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP and Ron D (2004). CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes & Development 18(24): 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL and Mittler R (2009). The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Science Signaling 2(84): ra45-ra45. [DOI] [PubMed] [Google Scholar]
- Mittler R and Blumwald E (2015). The Roles of ROS and ABA in Systemic Acquired Acclimation. The Plant Cell 27(1): 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Kadota Y, Zipfel C, Molina A and Torres M-A (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. Journal of Experimental Botany 67(6): 1663–1676. [DOI] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X and Orellana A (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7(2): e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC and Schroeder JI (2004). Reactive Oxygen Species Activation of Plant Ca2+ Channels. A Signaling Mechanism in Polar Growth, Hormone Transduction, Stress Signaling, and Hypothetically Mechanotransduction. Plant Physiology 135(2): 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y and Koizumi N (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, Nara M, Suzuki K, Tanokura M and Kuchitsu K (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. Journal of Biological Chemistry 283(14): 8885–8892. [DOI] [PubMed] [Google Scholar]
- Ozgur R, Turkan I, Uzilday B and Sekmen AH (2014). Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J Exp Bot 65(5): 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur R, Uzilday B, Iwata Y, Koizumi N and Turkan I (2018). Interplay between unfolded protein response and reactive oxygen species: a dynamic duo. J Exp Bot, 10.1093/jxb/ery040. [DOI] [PubMed] [Google Scholar]
- Ozgur R, Uzilday B, Sekmen AH and Turkan I (2015). The effects of induced production of reactive oxygen species in organelles on endoplasmic reticulum stress and on the unfolded protein response in arabidopsis. Ann Bot 116(4): 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi E, Guichard C, Ollivier V, Driss F, Fay M, Prunet C, Marie J-C, Pouzet C, Samadi M, Elbim C, O’Dowd Y, Bens M, Vandewalle A, Gougerot-Pocidalo M-A, Lizard G and Ogier-Denis E (2004). NAD(P)H Oxidase Nox-4 Mediates 7-Ketocholesterol-Induced Endoplasmic Reticulum Stress and Apoptosis in Human Aortic Smooth Muscle Cells. Molecular and Cellular Biology 24(24): 10703–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Hager J, Gakière B and Noctor G (2008). Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. Journal of Experimental Botany 59(2): 135–146. [DOI] [PubMed] [Google Scholar]
- Ron D and Walter P (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews Molecular cell biology 8(7): 519–529. [DOI] [PubMed] [Google Scholar]
- Ruberti C, Kim S-J, Stefano G and Brandizzi F (2015). Unfolded protein response in plants: One master, many questions. Current opinion in plant biology 27: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti C, Lai YS and Brandizzi F (2018). Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor1 upon the pro-adaptive signaling mediated by bZIP28. Plant Journal 93(1): 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan A, Kobayashi Y, Nakano Y, Iuchi S, Kobayashi M, Sahoo L and Koyama H (2017). Genome-wide Association Study Reveals that the Aquaporin NIP1;1 Contributes to Variation in Hydrogen Peroxide Sensitivity in Arabidopsis thaliana. Molecular Plant 10(8): 1082–1094. [DOI] [PubMed] [Google Scholar]
- Sevier CS and Kaiser CA (2008). Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783(4): 549–556. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L and Prywes R (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental cell 3(1): 99–111. [DOI] [PubMed] [Google Scholar]
- Sovolyova N, Healy S, Samali A and Logue Susan E (2014). Stressed to death – mechanisms of ER stress-induced cell death. Biological Chemistry. 395: 1. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG and Howell SH (2013). BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. The Plant Cell 25(4): 1416–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lu S-J, Zhang S-S, Zhou S-F, Sun L and Liu J-X (2013). The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Molecular plant 6(5): 1605–1615. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA and Mittler R (2011). Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14(6): 691–699. [DOI] [PubMed] [Google Scholar]
- Tait MA and Hik DS (2003). Is dimethylsulfoxide a reliable solvent for extracting chlorophyll under field conditions? Photosynthesis Research 78(1): 87–91. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL and Jones JD (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A 99(1): 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD and Dangl JL (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37(10): 1130–1134. [DOI] [PubMed] [Google Scholar]
- Torres MA, Morales J, Sánchez-Rodríguez C, Molina A and Dangl JL (2013). Functional Interplay Between Arabidopsis NADPH Oxidases and Heterotrimeric G Protein. Molecular Plant-Microbe Interactions 26(6): 686–694. [DOI] [PubMed] [Google Scholar]
- Tripathy BC and Oelmüller R (2012). Reactive oxygen species generation and signaling in plants. Plant Signaling & Behavior 7(12): 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP and Weissman JS (2004). Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164(3): 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tran T, Padilla Marcia CS, Braun DM and Goggin FL (2017). Superoxide-responsive gene expression in Arabidopsis thaliana and Zea mays. Plant Physiology and Biochemistry 117: 51–60. [DOI] [PubMed] [Google Scholar]
- Yamaoka Y, Choi BY, Kim H, Shin S, Kim Y, Jang S, Song W-Y, Cho CH, Yoon HS, Kohno K and Lee Y (2018). Identification and functional study of the ER stress sensor IRE1 in Chlamydomonas reinhardtii. The Plant Journal, 10.1111/tpj.13844. [DOI] [PubMed] [Google Scholar]
- Zhong HH, Young JC, Pease EA, Hangarter RP and McClung CR (1994). Interactions between Light and the Circadian Clock in the Regulation of CAT2 Expression in Arabidopsis. Plant Physiology 104(3): 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Romero R and Petty HR (2010). Amplex UltraRed enhances the sensitivity of fluorimetric pyruvate detection. Anal Biochem 403(1–2): 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E (2015). ERO1: A protein disulfide oxidase and H2O2 producer. Free Radic Biol Med 83: 299–304. [DOI] [PubMed] [Google Scholar]


