Abstract
Purpose
This study aimed to associate DNA variants in promoter and exon flanking regions of the CYP19A1 gene with in vitro embryo production traits in cattle. The role of transcription factor binding sites created or lost due to DNA sequence variation and their possible effect on gene expression was also evaluated.
Methods
We collected date from Gyr dairy oocyte donor cows (Bos taurus indicus) at a commercial in vitro embryo production farm and analyzed the genotype–phenotype association with in vitro production traits. Using Sanger sequencing and web-based software, we assessed important CYP19A1 gene regions in oocyte donor cows and analyzed the effects of variants on the transcription factor binding sites.
Results
Two SNP mutations significantly associated with oocyte production, oocyte viability, embryo development, and pregnancies were found (T > C in the untranslated exon 1 flanking region ([GenBank: AJ250379.1]: rs718446508 T > C), and a T > C in the 5′-upstream region (1.1 promoter) ([GenBank: AC_000167.1]: rs41651668 T > C). Six new transcription factor binding sites were created. A binding site for transcription factors associated with the development of the placenta and embryo implantation was eliminated due to variations in the DNA sequence identified.
Conclusions
The CYP19A1 gene contributes to genetic variation of in vitro embryo production traits in cattle. The complexity of the physiological phenomena related to estrogen pathways and their influence on reproduction in cattle allow indication of the mutations evaluated here as possible genetic markers for embryo production traits, which should be validated in the next steps of marker-assisted selection.
Keywords: Embryo transfer, Estrogen, Genetic marker, Oocyte donor, Regulatory element, Transcription factor
Introduction
The features distinguishing phenotypes in complex traits in domestic animals are attributed to differences in gene expression [1]. The development of animals from zygotes to adults and the differentiation of cells into distinct tissues and organs require the expression of a specific set of genes at each developmental stage in each cell type [2].
The aromatase enzyme participates in follicular development, placenta function, and regulation of the female reproductive cycle, which makes it a potential candidate for fertility traits in cattle [3, 4]. This enzyme is encoded by the CYP19A1 gene (cytochrome P450, family 19, subfamily A, polypeptide 1). Located on chromosome 10q26 of the cow genome, it plays a key role in estrogen biosynthesis from androgens in ovarian granulosa cells [4, 5].
Aromatase transcription is driven by multiple tissue-specific promoters, resulting in the production of various mRNA transcripts. The CYP19A1 gene contains a noncoding alternative exon 1 followed by a common protein-coding region [1, 6, 7]. Expression of CYP19A1 in reproductive tissues (placenta, corpus luteum, and ovarian follicles) is mainly regulated by 1.1, 1.2, and 2 promoters [2, 6, 7].
During a follicular development wave, a specific number of follicles will be selected to become dominant, while the remaining follicles will be lost through a process known as atresia. The difference in diameter between the dominant and remaining follicles is known as the deviation [8]. Gene expression and transcriptome studies in the field of follicular development highlight the CYP19A1 gene in pre-deviation and onset of deviation stages of the follicular wave in cattle, as well as changes in the female reproductive cycle [9]. Reduced conversion of androgens to estrogens in the dominant follicles has been associated with low fertility in cattle due to altered follicular and oocyte development [10].
The epigenetic regulation mechanisms are related with the molecular control in CYP19A1 gene expression, so regulatory regions are important prerequisites for gene transcription initiation or silencing. [11]. The genetic variants that are significantly associated with complex traits are often not located in protein-coding regions, suggesting that these DNA sequences can affect the regulation of gene expression [12]. This study aimed to associate DNA variants in promoter and exon flanking regions of the CYP19A1 gene with in vitro embryo production traits in cattle.
Materials and methods
Data collection and OPU-IVFET traits
Data from 321 sessions of ultrasound-guided follicular puncture, or ovum pickup (OPU), followed by 304 in vitro fertilization sessions and 159 embryo transfer procedures (IVFET), were collected from 50 purebred Gyr dairy cows (Bos taurus indicus) at a commercial in vitro embryo production farm located in the state of Rio de Janeiro, Brazil. All cows were normally cycling and were kept only in pastures with shade. None of the cows were submitted to hormonal treatment before the OPU-IVFET procedure.
Prior to genotype–phenotype analysis, we analyzed the genetic diversity among the animals. Their origin was also investigated, and we determined that the population had three different origins. In this same evaluation, we observed a uniform distribution of the genotypic combinations evaluated, presenting the three genotypes in each subpopulation of origin.
The procedures of recovery and oocyte classification, in vitro maturation, fertilization, and embryo culture and transfer of in vitro-produced embryos followed the method described by Vega et al. [13]. Briefly, cumulus-oocyte complexes (COCs) were harvested by OPU. Imaging of ovaries was performed using a portable ultrasonic device with a 7.5-MHz intravaginal sector probe equipped with a needle guide (Scanner 100S, Pie Medical, Maastricht, Netherlands), and follicles between 2 and 8 mm were punctured using 19-G disposable hypodermic needles with an aspiration vacuum of 60–80 mmHg. Cumulus-oocyte complexes were ranked according to the number of layers of cumulus cells and cytoplasm: (I) viable (GI: more than 3 layers and homogeneous cytoplasm; GII: more than 3 layers and cytoplasm with granules, or fewer than 3 layers and homogeneous cytoplasm; GIII: fewer than 3 layers and cytoplasm with granules, or partially denuded and homogeneous or with mild cytoplasm granules) and (II) non-viable (GIV: partially denuded and cytoplasm with grooves, or naked, expanded or degenerate).
In vitro maturation, fertilization, and embryo culture
Viable oocytes were matured in vitro (IVM) in TCM 199 medium (Invitrogen-Gibco BRL) with 10% inactivated estrous cow serum and 1.0 μg/mL of FSH (Pluset, Serono, Italy), 50 μg/mL of hCG (Profasi®, Serono, Italy), 1.0 μg/mL of estradiol (Sigma E-2758 St. Louis, MO, USA), 0.2 mM of sodium pyruvate (Biochemical 44,094), and 83.4 μg/mL of amikacin (Instituto Biochimico, Rio de Janeiro, Brazil). After IVM, viable sperm from frozen straws of sexed semen were separated using a Percoll discontinuous density gradient and in vitro fertilization (IVF) was performed in TALP-IVF, with 0.2 mM of pyruvate and 83.4 mg/mL of amikacin, supplemented with 6 mg/mL of bovine serum albumin. In vitro culture (IVC) was performed in synthetic oviduct fluid supplemented with 2.5% fetal bovine serum (Cripion, Industria Brasileira, Andradina, SP, Brazil) for 7 days. IVM, IVF, and IVC were carried out in an incubator at 38.5 °C in 5% CO2 in air with high relative humidity. The cleavage rate was assessed 96 h after IVF and blastocyst rate was assessed on day 7.
The selection of the in vitro production traits of embryos associated with commercial production was based on their wide use by companies engaged in commercial production of bovine embryos as well as research in the field of animal assisted reproduction technologies. The OPU-IVFET traits associated with the DNA variants were total oocyte number (Toc), ratio of viable cumulus-oophorus complexes (Rvcoc), ratio of cleaved embryos at day 4 of culture (Rced4), ratio of transferrable embryos at day 7 of culture (Rtembd7), and pregnancy rate at 30 days after transfer (PR30).
PCR and sequencing conditions
The genomic DNA was obtained from tail hair follicles. The extraction and purification steps were carried out using the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany), following the manufacturer’s instructions. The DNA concentrations were measured with a spectrophotometer (NanoDrop™ 2000, Thermo Science).
Specific primers were designed using the Primer Express® v2.0 program (Applied Biosystems). The features of primers and CYP19A1 gene regions selected are summarized in Table 1. The PCR was performed in a final volume of 20 μl, using 1× PCR “A” buffer [10 mM Tris-HCI (including Mg+2)], 0.5 mM dNTP mix (KAPA, Wilmington, Massachusetts, USA), 1 U of Taq DNA polymerase (KAPA), 10 pM of each primer (Invitrogen, Sao Paulo, SP, Brazil), 30 ng of extracted DNA, and deionized water. A negative control was included for each replicate of the PCR reaction. PCR purification was performed with Illustra™ ExoProStar™ (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The cycling conditions consisted of an initial stage of 95 °C for 1 min, followed by 35 amplification cycles with denaturation at 95 °C for 30 s, annealing of primers for 30 s (individual primer temperatures in Table 1), and extension at 72 °C for 1 min. After the last cycle, there was a final step of 7 min at 72 °C for the final extension of the strand. PCR was conducted using a thermocycler (Applied Veriti® 96-Well, Applied Biosystems, Foster City, CA, USA). The product was separated by polyacrylamide gel (8%) stained with silver nitrate (Sigma-Aldrich, Sao Paulo, SP, Brazil) to confirm the amplification.
Table 1.
Summary of features of primers and CYP19A1 gene regions analyzed
| Primer name and gene region | Primer sequence (5′-3′) | TA (°C) | Amplicon (bp) |
|---|---|---|---|
| UE1FR—untranslated exon 1 and flanking regions (1.2 promoter) | F-CTGAACGAGGTCCTGAAGAGAAG R-TAAGATACAACTATGCCACAAGCACT | 65.5 | 630 |
| 5-UTRPR 5′—upstream region (1.1 promoter) | F-GCTTGTCAACTGTTCATTCATTCCC R-CGTCTGAGCCTTGGTGTCCA | 65 | 275 |
| EX2COD—exon 2 protein-coding region (2 promoter) | F-CTCTCTTGGGCTTGCTTGTTTT R-ATTTTACTTTGCTGTCCCCATCTT | 65 | 665 |
| EX8SPL—exon 8 (splice region) | F-GCCACCTCCCTTTCTGTTCTG R-TCCCTTATTATTGCCTCTTCAACCT | 60 | 213 |
TA annealing temperature
After PCR, samples were sequenced using an ABI 3500 automatic sequencer equipped with 50-cm capillaries and POP7 polymer (Applied Biosystems). DNA templates (60 ng) were labeled with 2.5 pmol of each primer (UE1FR, 5-UTRPR, EX2COD, and EX8SPL, sequences in Table 1) and 0.5 μl of BigDye Terminator v3.1 Cycle Sequencing Standard (Applied Biosystems) in a final volume of 10 μl. Labeling reactions were performed in a LGC XP thermocycler with an initial denaturing step at 96 °C for 3 min followed by 25 cycles at 96 °C for 10 s, 55 °C for 5 s, and 60 °C for 4 min. Labeled samples were purified by 75% isopropanol precipitation followed by 60% ethanol rinsing. Precipitated products were suspended in 10 μl of Hi-Di formamide (Applied Biosystems), denatured at 95 °C for 5 min, ice-cooled for 5 min, and electro-injected in the automatic sequencer. Sequencing data were collected using the Data Collection 2 software (Applied Biosystems). The samples were sequenced in triplicate for genotype confirmation.
Variant analysis
Specific regions of the CYP19A1 gene including the alternative promoters of the first exon (P1.1 and P1.2), promoter and coding region of exon 2 (P2), and splice region of exon 8 were selected. The promoter regions were selected following sequences described by Fürbass et al. (1997), Vanselow et al. (2000), and Lenz et al. (2004) [3, 6, 7].
After sequencing, the identification of variants was performed with the aid of the Clustal Omega sequence alignment program [14] together with direct observation of genotypes in the electropherograms using the Sequence Scanner™ v2.0 software (Applied Biosystems).
Proportion of genetic variance explained by the SNPs
The individual variant effect was ascertained using a traditional SNP linear regression analysis (R2) with the REG routine of the SAS 9.2 program (2009).
Genotype–phenotype association analysis
The association between CYP19A1 genotypes and OPU-IVFET traits was determined using the GLM procedure of the SAS 9.2 program (SAS Institute, Cary, NC) (2009), with a model including genotype as fixed effect. The significance level was set at 0.05. The embryo production traits expressed as percentage were submitted to Box–Cox transformation before analysis using the TRANSREG procedure of SAS.
The model used in the study was:
where Yijk is the observation of the OPU-IVFET traits, μ is the mean of each trait, gi is the genotype effect, sj is the random effect of sire, yk is the effect of year, and εijk is the random error. The recipient effect was removed from the statistical model using backward elimination according to the Wald test result (when P > 0.20) [15]. The mean values of traits were compared using the Student–Newman–Keuls (SNK) method for unequal sample sizes.
None of the evaluated SNPs have been reported in association studies of OPU-IVFET traits. In the analysis, the markers were simultaneously considered because animals that had the two CYP19A1 mutations combined differed statistically from those that only had one mutation.
In silico analysis of transcription factor binding sites affected by SNPs in the bovine CYP19A1 gene
The prediction of transcription factor binding sites’ (TFBSs) affect by SNPs in the promoter and flanking regions analyzed was performed with the Genomatix SNPInspector software (http://www.genomatix.de/), using the standard configuration (selecting genome cattle, entering the SNP number, and the FASTA sequence of the SNP region of interest).
Results
Variant analysis
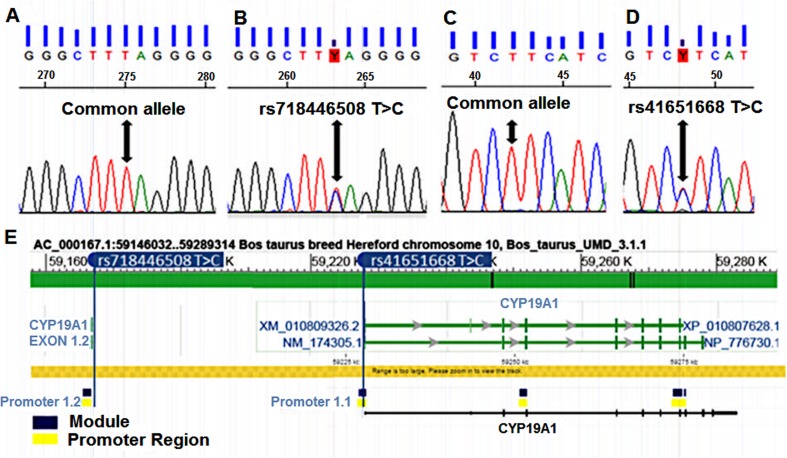
We sequenced ~ 1.8 kb of the CYP19A1 gene including the untranslated exon 1 and flanking regions, 5′-upstream region (1.1 promoter), exon 2 protein-coding region (2 promoter), and exon 8 (splice region) of 50 DNA samples from Gyr oocyte donor cows. Two SNPs were identified: a T > C in the untranslated exon 1 flanking region ([GenBank: AJ250379.1]: rs718446508 T > C), and a T > C in the 5′-upstream region (1.1 promoter) ([GenBank: AC_000167.1]: rs41651668 T > C) (Fig. 1).
Fig. 1.
Identification of DNA variants by Sanger sequencing in CYP19A1 gene of Gyr oocyte donor cows. a Common allele of the untranslated exon 1 flanking region (UE1FR). b Heterozygous mutation of the UE1FR. c Common allele of the 5′-upstream and promoter region (5-UTRPR). d Heterozygous mutation of the 5-UTRPR. e Schematic representation of the promoter regions and associated variants in the bovine CYP19A1 gene (modified output of Genomatix software)
Genotypic and allelic frequencies
The summary of genotypic and allelic frequencies is shown in Table 2. No homozygous animals were found for the mutant alleles. A total of seven mutant individuals were identified, four with heterozygous genotypes for both loci (UE1FR [C/T] and 5-UTRPR [T/C]), one UE1FR [C/T], and two 5-UTRPR [T/C]. These allelic combinations were considered in the phenotype–genotype association analysis irrespective of the number of phenotypic evaluations performed in UE1FR [C/T] and 5-UTRPR [T/C] genotypes (12 and 19 procedures of OPU-IVFET, respectively).
Table 2.
Genotypic and allelic frequency of two SNPs in the CYP19A1 gene in Gyr oocyte donor cows
| SNP | Genotypic frequency | Allele frequency | ||
|---|---|---|---|---|
| Genotype | % | n | ||
| rs718446508 (T/C) | TT | 0.92 | 46 | 0.96 T |
| CT | 0.08 | 4 | 0.04 C | |
| rs41651668 (T/C) | TT | 0.86 | 43 | 0.93 T |
| TC | 0.14 | 7 | 0.07 C | |
Genotype–phenotype association analysis
Means and standard deviations of OPU-IVFET traits in Gyr oocyte donor cows and their associations with CYP9A1 genotypes are shown in Table 3.
Table 3.
Means and standard deviations of OPU-IVFET traits of oocyte donor cows and their association with CYP19A1 genotypes
| OPU-IVFET traits | Genotypes (number of procedures per trait)1 | |||
|---|---|---|---|---|
| UE1FR [T/T] 5-UTRPR [T/T] | UE1FR [C/T] 5-UTRPR [T/T] | UE1FR [T/T] 5-UTRPR [T/C] | UE1FR [C/T] 5-UTRPR [T/C] | |
| Toc | 13.06 ± 4.4ª** (231) | 5.4 ± 3.3b** (12) | 12.82 ± 8.3a** (19) | 15.01 ± 3.8a** (59) |
| Rvcoc | 73.28 ± 8.8b** (231) | 72.09 ± 2.8b** (12) | 71.76 ± 11.2b** (19) | 78.90 ± 3.9a** (59) |
| Rced4 | 72.43 ± 10.5a (218) | 75.41 ± 7.3a (11) | 70.96 ± 12.1a (17) | 74.32 ± 6.8a (58) |
| Rtembd7 | 41.87 ± 14.4b** (200) | 20.16 ± 3.42c** (11) | 64.85 ± 19.89a** (14) | 41.93 ± 10.5 b** (58) |
| PR30 | 54.54 ± 22.8b* (108) | 33.33 ± 25.1c* (3) | 70.85 ± 25.2a* (7) | 44.51 ± 13.1b* (41) |
1Number of procedures per trait: obtained from the effective sessions in each variable of OPU-IVFET process in each analyzed genotype. UE1FR, untranslated exon 1 flanking region; 5UTRPR, 5′-upstream and promoter region; Toc, total oocyte number; Rvcoc, ratio of viable cumulus-oophorus complexes (COCs) (ratio between total COCs obtained and total of viable COCs); Rced4, ratio of cleaved embryos at day 4 of culture (ratio between total of COCs fertilized and total embryos cleaved); Rtembd7, ratio of transferable embryos at day 7 of culture (ratio between total embryos cleaved and total transferable blastocysts); and PR30, pregnancy rate at 30 days after transfer. a,bMeans between genotypes in the same line with different superscript letters are significantly different (*P < 0.05; **P < 0.001)
The traits associated with the commercial production of embryos in cattle evaluated were oocyte production (total oocyte number—Toc); oocyte viability (ratio of viable cumulus-oophorus complexes—Rvcoc); oocyte maturation and fertilization (ratio of cleaved embryos at day 4 of culture—Rced4); embryo development (ratio of transferable embryos at day 7 of culture—Rtembd7); and successful pregnancies (pregnancy rate at 30 days after transfer—PR30).
Numbers of procedures per trait were distributed in 321 sessions of ultrasound-guided follicular puncture, or ovum pickup (OPU); 304 in vitro fertilization sessions; and 159 embryo transfer procedures (IVFET).
Statistical differences in Toc, Rvcoc, Rtembd7 (P < 0.001), and PR30 (P < 0.05) were obtained.
Cows with the genotype combinations UE1FR [C/T] and 5UTRPR [T/C] showed similar performance to those of genotype combinations UE1FR [C/T]–5UTRPR [T/T] and UE1FR [T/T]–5UTRPR [T/C] for the Toc trait. Also, they proved to be superior to all other genotypic combinations in the Rvcoc trait. On the other hand, the genotype combination UE1FR [T/T]–5UTRPR [T/C] was superior to all other genotype combinations in the Rtembd7 and PR30 traits. The genotype combination UE1FR [C/T]–5UTRPR [T/T] presented the worst results for the Toc, Rtembd7, and PR30 traits.
For example, if cows with the genotype combination UE1FR [T/C]–5UTRPR [T/T] were the only ones responsible for the total oocyte production evaluated in this study (3936 oocytes), 163.6 more embryos and 408.8 more pregnancies would have been obtained than using cows whose genotype combination was UE1FR [T/C]–5UTRPR [C/T] and 401.1 more embryos and 426.8 more pregnancies than using homozygous cows for the two loci evaluated.
Proportion of genetic variance explain by the SNPs
The SNP rs41651668 (T/C) in cows with UE1FR [T/T] and 5-UTRPR [T/C] genotypes explained 0.1777 and 0.1402 of the genetic variance of the total oocyte number (Toc) and ratio of transferable embryos at day 7 of culture traits (Rtembd7). The SNP rs718446508 T > C explained 0.098 of the genetic variance of the ratio of transferable embryos at day 7 of culture. The proportion of genetic variance explained by the rs41651668 (T/C) SNP mentioned above in the other traits evaluated (Rvcoc, Rced4, and PR30) was less than 0.016, possibly due to the epistatic effect on these traits (Table 4).
Table 4.
Percentage of genetic variance explained by significant SNPs associated with OPU-IVFET traits of oocyte donor cows
| Trait | UE1FR [C/T] (rs718446508 [C/T]) 5-UTRPR [T/T] | UE1FR [T/T] 5-UTRPR [T/C] (rs41651668 T/C) | UE1FR [C/T] (rs718446508 [C/T]) 5-UTRPR [T/C] (rs41651668 T/C) |
|---|---|---|---|
| Toc | 0.0026 | 0.1777 | 0.0357 |
| Rvcoc | 0.0022 | 0.0002 | 0.0610 |
| Rced4 | 0.0018 | 0.0054 | 0.0088 |
| Rtembd7 | 0.0980 | 0.1402 | 0.0013 |
| PR30 | 0.0167 | 0.0132 | 0.0095 |
UE1FR, untranslated exon 1 flanking region; 5UTRPR, 5′-upstream and promoter region; Toc, total oocyte number; Rvcoc, ratio of viable cumulus-oophorus complexes; Rced4, ratio of cleaved embryos at day 4 of culture; Rtembd7, ratio of transferable embryos at day 7 of culture, and PR30, pregnancy rate at 30 days after transfer
In silico analysis of transcription factor binding sites affected by SNPs in the CYP19A1 bovine gene
In this study, the substitution of a thymine (T) by a cytosine (C) in the SNP rs718446508 T > C resulted in the loss of one transcription factor binding site (glial cells missing homolog 1—GCM1). This SNP is located in an intergenic region (non-protein-coding), flanking the 1.2 promoter and the alternative exon 1 (to 23 bp from the end), and at ~ 6.0 kb from the 5′-UTR region of CYP19A1 gene.
On the other hand, six new transcription factor binding sites—for the androgen receptor family (GREF), the retinoid X receptor family (RXRF or nuclear hormone receptor), the downstream immunoglobulin control element (DICE), the ecotropic virus integration site-1(EVI1), the family of transcriptional regulatory proteins (GATA), and the prolactin regulatory element-binding protein (PCBE/PREB)—were generated due to the same nucleotide exchange in the SNP rs41651668 T > C. This SNP is within the promoter 1.1 (GeneBank accession number GXP_3838630 (+), 5′-UTR region) of the CYP19A1 gene. Other characteristics of the promoter 1.1 of the CYP19A1 bovine gene are shown in Table 5.
Table 5.
Characteristics of the promoter 1.1 of CYP19A1 bovine gene
| Promoter 1.1 | Coding transcript | Exons | TSS* |
|---|---|---|---|
| GXP_3838630 (+) | GXT_25140277 | 10 | 1038 |
| Length sequence (1138 bp) | GXT_25172617 | 10 | 1038 |
| TSS (1001–1038) | GXT_27476876 | 11 | 1001 |
*TSS, transcription starting site. Analysis performed with the Gene2Promoter tool of the Genomatix software
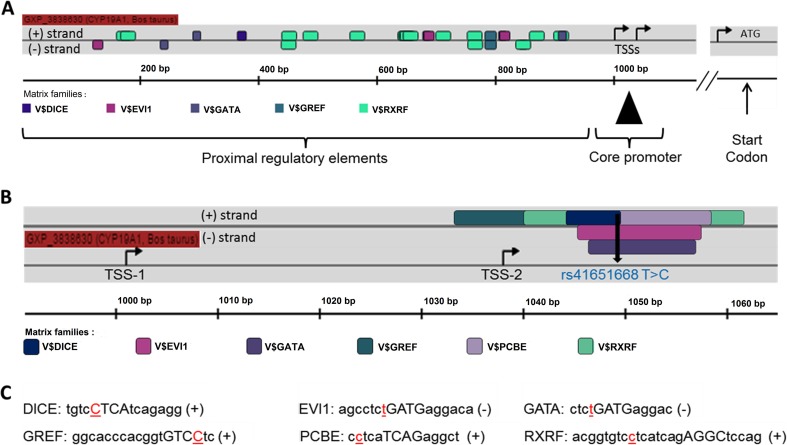
The SNP rs41651668 T > C found in animals with 5UTRPR [T/C] genotype is at position 1049 bp of the 1.1 promoter (Fig. 2b).
Fig. 2.
Schematic representation of promoter 1.1 of CYP19A1 bovine gene and associated TFBSs. DICE (downstream immunoglobulin control element), EVI1 (ecotropic virus integration site-1), GATA (family of transcriptional regulatory proteins), GREF (androgen receptor family), PCBE/PREB (prolactin regulatory element-binding protein), and RXRF (retinoid X receptor family). a Promoter structure without mutation effect. b Core promoter structure with new transcription binding sites created due to SNP rs41651668 T > C. c DNA sequences recognized by transcription factor families surrounding the second transcription start site (TSS-2). Nucleotides in red color denote the mutation site. Nucleotides in capitals denote the core sequence used by MatInspector (modified output of Genomatix-MatInspector software)
In silico identification of the transcription factor binding sites generated or deleted due to SNPs in the CYP19A1 cattle gene and other characteristics in TFBSs, such as starting and ending sites, core similarity, threshold value, and main effector tissues, are shown in Table 6. These features are related to the sequence of each SNP identified by the SNPInspector software for each SNP evaluated. The SNP rs718446508 sequence has 51 nucleotides and is at the 26-bp position. The SNP rs41651668 sequence has 501 bp and is at the 251-bp position. The generated and lost TFBSs by nucleotide exchange affect several tissues, but only the main effector tissues associated with the characteristics in this study were found to be related.
Table 6.
In silico identification of the TFBSs generated or deleted due to SNPs in CYP19A1 cattle gene
| SNP allele number | New/ lost | TFBSs family/matrix | Start pos. | End pos. | Core sim. | Threshold | Main effector tissues |
|---|---|---|---|---|---|---|---|
| rs41651668 T > C | New | V$GREF/ARE.03 | 236 | 254 | 0.87 | 0.87 | Endocrine system, urogenital system |
| rs41651668 T > C | New | V$RXRF/THRB.03 | 243 | 267 | 0.79 | 0.74 | Endocrine system, pituitary gland |
| rs41651668 T > C | New | V$DICE/DICE.01 | 247 | 261 | 0.81 | 0.8 | Embryonic structures, immune system |
| rs41651668 T > C | New | V$EVI1/MEL1.02 | 247 | 263 | 1 | 0.99 | Hematopoietic system, immune system |
| rs41651668 T > C | New | V$GATA/GATA5.01 | 248 | 260 | 0.75 | 0.83 | Embryonic structures, hematopoietic system |
| rs41651668 T > C | New | V$PCBE/PREB.01 | 250 | 264 | 1 | 0.86 | Endocrine system, pituitary gland |
| rs718446508 T > C | Lost | V$GCMF/GCM1.03 | 22 | 36 | 1 | 0.83 | Embryonic structures, endocrine system |
TFBSs, transcription factor binding sites
Discussion
The genetic architecture of complex traits is controlled by many genes and environmental factors [16]. The search for biomarkers associated with complex phenotypes such as fertility has been reported in several works. Despite the importance of CYP19A1 in reproductive systems, little is known about its regulation.
This study is the first to associate SNP mutations in the CYP19A1 gene regions with in vitro embryo production traits in cattle. The method applied in this study is based on the targeted search for markers in potentially important genomic regions for expression of traits (prior knowledge method).
This paper reports OPU-IVFET results in Bos taurus indicus cattle harboring two significant and different T > C substitutions in the CYP19A1 gene, one at the promoter region (UE1FR) and another at the 5′-untranslated region (5-UTRPR). The two “C” alleles were found to be present in 8 and 14%, respectively, of the studied population and always in heterozygosity, so we analyzed four possible combinations of alleles: UE1FR (T/T) or (T/C) and 5-UTRPR (T/T) or (T/C).
Recent studies evaluating quantitative traits in cattle have ranked the proportion of SNP effects based on the total variance explained by them, concluding that SNPs in or within 50 kb of candidate genes explained 15.75% of the total variance of the trait while non-synonymous SNPs within these genes explained 8.6% of the total variance [16].
In this study, the proportions of genetic variance explained by significant SNPs (R2) were 9.8% and 14.02% (in Rtembd7 trait) and 17.7% (in Toc). The R2 value tends to be higher when the number of markers with estimated effects and the sample size of the population under analysis are larger [17]. It is necessary to clarify that our population was small (n = 50), as was the number of markers analyzed. Although the R2 values obtained indicate the genetic participation of the variants in the significant traits, they have limited value in terms of accuracy of genetic variance explained by the SNPs (values can be underestimated).
The results obtained for oocyte production (Toc), embryo development (Rtembd7) (P < 0.001), and pregnancy success (PR30) (P < 0.05) were negatively affected by SNP rs718446508 T > C. This SNP flanks the alternative exon 1 of CYP19A1 gene, an important reproductive control region mainly active in the placenta, although also in the ovary and brain [3, 7].
The nucleotide substitution due to SNP rs718446508 T > C leads to loss of a binding site for GCM1 (glial cells missing homolog 1) transcription factor. This TF is essential for placental development, promoting syncytiotrophoblast formation and placental vasculogenesis by activating fusogenic and proangiogenic gene expression in the placenta [18]. Besides the embryonic structures, GCM1 also affects the endocrine system, immune system, parathyroid glands, and thymus gland [19].
The effects of the immune system are present in reproductive organs and are related with normal reproductive functions like inflammation accompanying ovulation and follicular atresia. Lower apoptosis in fertile follicles is logical, since apoptosis occurs in atretic follicles and is a normal process of follicle selection [20].
We hypothesize that the proportion of genetic variance explained by this SNP (0.098 for the Rtembd7), together with the physiological mechanisms derived from the in silico analysis of the mutation mentioned, may be associated with the poor results obtained in this study in cows with UE1FR [C/T]–5UTRPR [T/T] genotype for the Rtembd7 and PR30 traits, while low oocyte production might be associated with abnormal follicular atresia of immunological origin.
The second mutation evaluated in this study (SNP rs41651668 T > C) is found in the core promoter gene region of CYP19A1 bovine at positions + 11 and + 48 from the TSSs and created a new site for the families of transcription factors GREF, RXRF, DICE, EVI1, GATA, and PCBE (Fig. 2b).
The results showed there are many potential transcription factors in the DNA region affected by SNP rs41651668 T > C. Several binding sites for transcription factors were predicted for the region containing the second mutation, thus forming a cluster of TFBSs.
The transcription factor family GREF/ARE.03 (androgen receptor binding site-AR) is associated with follicular development in females. In ovarian follicles of humans and other mammals, androgens are synthesized in the theca cells and act as a substrate for estradiol synthesis in granulosa cells (GCs), so expressed androgen receptors (ARs) and intrafollicular androgens are central to fertility [21]. Abnormal androgen levels or deficiency in androgen/AR signaling in the ovary can affect critical events in oogenesis, such as the first meiotic division and epigenetic reprogramming [22]. The ARs are expressed in the ovarian follicles throughout folliculogenesis. In the pre-antral and antral follicles, ARs are found in both theca cells and GC but become primarily expressed in GC as the follicle develops into the antral and preovulatory stages [23].
Interestingly, the family matrix of transcription factors V$PCBE/PREB.01 was the only one of the families affected by the second mutation that did not present another site within the evaluated promoter (Fig. 2a). The prolactin regulatory element-binding protein (PREB) is a transcription factor that specifically binds to a Pit-1 binding element in the prolactin (PRL) promoter, to regulate PRL gene expression in mammals [24]. In the reproductive system, PRL induces transcription of the estrogen receptor (ER). Female mice in the knockout model (PRL−/−genotype) are completely infertile, with only a few oocytes reaching the stage of blastocysts.
The matrix transcription factor V$THRB.03 (thyroid hormone receptor, beta) is directly involved with folliculogenesis, ovulation, embryonic structure development, and pregnancy maintenance in vertebrates. The majority of reproductive effects are due to changes in thyroid hormone levels [25, 26].
The high number of binding sites for this transcription factor in the CYP19A1 promoter 1.1 (14 in the proximal promoter and one in core promoter, due to the mutation) may demonstrate the importance of regulating the thyroid hormone gene expression in the different physiological conditions mentioned above.
The other families of transcription factors (V$DICE/DICE.01, V$EVI1/MEL1.02, and V$GATA/GATA5.01) participate as co-regulators in functions associated with angiogenic development of the hematopoietic and immune system, development, differentiation, and control of cell proliferation, i.e., acting on other genes or transcription factors. The downstream immunoglobulin control element (DICE) regulates the vascular endothelial growth factor (VEGF), an angiogenic factor that has been suggested as playing a physiological role in growth and atresia of the bovine dominant follicle [27] and implantation process and pregnancy maintenance [28].
Genomic regions linked to a target gene that can direct spatio-temporal-specific gene expression are commonly known as Cis-regulatory modules (CRMs) [29].
We believe that the cluster formation of TBFSs in the core promoter region suggests the formation of a CRM that can direct specific expression of the CYP19A1 gene and this mechanism is associated with the positive results obtained by the UE1FR [T/T]–5UTRPR [C/T] genotype. The percentage of genetic variance explained by the SNP rs41651668 T > C in the genotype above mentioned in Toc and Pemb7d traits (0.140 and 0.177) showed their important participation in genetic variance of these traits in in vitro embryo production in cattle. Future studies to evaluate the effect of the mutations presented here on the expression of regulated, co-regulated, or CYP19A1-associated genes should be performed.
On the other hand, the similar results presented by the homozygous and heterozygous genotypes in both loci in most of the evaluated traits can be explained by the compensatory effect of the mutation rs41651668 T > C relative to the deleterious effect of the mutation rs718446508 T > C, besides the mode of genetic action of the trait, commonly epistatic or overdominant, in which the genotypic value of the heterozygote is outside the amplitude of the genotypic values of the two homozygotes.
Conclusions
The mutations with significant associations found in this study in the CYP19A1 bovine gene participate in the genetic variance of in vitro embryo production traits, due to their possible role as regulatory regions of gene expression; so, they can be useful to increase OPU-IVFET process efficiency in commercial populations. It should be noted that these markers need to be validated in additional populations before they are used for marker-assisted selection in a commercial setting. We believe that information brought by reproductive physiology will provide new markers and new improved phenotypes, which will increase the efficiency of selection schemes for embryo production traits.
Acknowledgements
We are particularly grateful to the Embrapa Dairy Cattle research unit for providing real-world data and expert support for this study.
Funding
We thank the Office to Coordinate Improvement of University Personnel (CAPES) for the grant awarded to the first author and the National Council for Technological and Scientific Development (CNPq) for financial support.
Ethical approval
This study was approved by the Ethics Committee on Animal Experimentation of Norte Fluminense State University (UENF Protocol no. 243, March 11, 2014).
Informed consent
Does not apply
Conflict of interest
The authors declare they have no competing interests.
Contributor Information
Wilder Hernando Ortiz Vega, Email: wilortvet@yahoo.es.
Celia Raquel Quirino, Email: crqster@gmail.com.
Aylton Bartholazzi-Junior, Email: junior_barth@hotmail.com.
Miguel Alejandro Silva Rua, Email: miguelvet-rua@gmail.com.
Raquel Varella Serapião, Email: raquel@pesagro.rj.gov.br.
Clara Slade Oliveira, Email: clara.oliveira@embrapa.br.
References
- 1.Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- 2.Davidson EH, Erwin DH. REVIEW gene regulatory networks and the evolution of animal body plans. Science (80- ) 2006;311:796–801. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 3.Fürbass R, Kalbe C, Vanselow J. Tissue-specific expression of the bovine aromatase- encoding gene uses multiple transcriptional start sites and alternative first exons*. Endocrinology. 1997;138:2813–2819. doi: 10.1210/endo.138.7.5257. [DOI] [PubMed] [Google Scholar]
- 4.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, et al. The Ensembl gene annotation system. Database. 2016;2016:1–19. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Li R, Hu Y. The alternative noncoding exons 1 of aromatase (Cyp19) gene modulate gene expression in a posttranscriptional manner. Endocrinology. 2009;150:3301–3307. doi: 10.1210/en.2008-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz S, Pöhland R, Becker F, Vanselow J. Expression of the bovine aromatase cytochrome P450 gene (Cyp19) is primarily regulated by promoter 2 in bovine follicles and by promoter 1.1 in Corpora Lutea. Mol Reprod Dev. 2004;67:406–413. doi: 10.1002/mrd.20000. [DOI] [PubMed] [Google Scholar]
- 7.Vanselow J, Kuhn C, Furbass R, Schwerin M. Isolation of the bovine CYP19 promoter 1.2 and identification of genetic variants. Anim Genet. 2000;31:337–338. doi: 10.1046/j.1365-2052.2000.00668.x. [DOI] [PubMed] [Google Scholar]
- 8.Beg MA, Ginther OJ. Follicle selection in cattle and horses: role of intrafollicular factors. Reproduction. 2006;132:365–377. doi: 10.1530/rep.1.01233. [DOI] [PubMed] [Google Scholar]
- 9.Li P, Meng J, Liu W, Smith GW, Yao J, Lyu L. Transcriptome analysis of bovine ovarian follicles at predeviation and onset of deviation stages of a follicular wave. Int J Genomics. 2016;2016:1–9. doi: 10.1155/2016/3472748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summers AF, Pohlmeier WE, Sargent KM, Cole BD, Vinton RJ, Kurz SG, et al. Altered theca and cumulus oocyte complex gene expression, follicular arrest and reduced fertility in cows with dominant follicle follicular fluid androgen excess. PLoS One. 2014;9:1–13. doi: 10.1371/journal.pone.0110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanselow1 J. RF. Epigenetic control of folliculogenesis and luteinization. Anim Reprod. 2010;49:134–9.
- 12.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet. 2012;13:469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega WHO, Quirino CR, Serapião RV, Oliveira CS, Pacheco A. Phenotypic correlations between ovum pick-up in vitro production traits and pregnancy rates in Zebu cows. FunpecrpComBr Genet Mol Res Genet Mol Res. 2015;14:7335–7343. doi: 10.4238/2015.July.3.9. [DOI] [PubMed] [Google Scholar]
- 14.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bursac Z, Gauss CH, Williams DK, Hosmer DW. Purposeful selection of variables in logistic regression. Source Code Biol Med. 2008;3:1–8. doi: 10.1186/1751-0473-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goddard ME, Kemper KE, MacLeod IM, Chamberlain AJ, Hayes BJ. Genetics of complex traits: prediction of phenotype, identification of causal polymorphisms and genetic architecture. Proc R Soc B. 2016;283(20160):1–9. doi: 10.1098/rspb.2016.0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin FY, Chang CW, Cheong ML, Chen HC, Lee DY, Chang GD, Chen H. Dual-specificity phosphatase 23 mediates GCM1 dephosphorylation and activation. Nucleic Acids Res. 2011;39:848–861. doi: 10.1093/nar/gkq838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashemolhosseini S, Wegner M. Mini-review impacts of a new transcription factor family: mammalian GCM proteins in health and disease. J Cell Biol JCB J Cell Biol. 2004;166:765–768. doi: 10.1083/jcb.200406097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YM, Rao CV, Lei ZM. Macrophages in human reproductive tissues contain luteinizing hormone/chorionic gonadotropin receptors. Am J Reprod Immunol. 2003;49:93–100. doi: 10.1034/j.1600-0897.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- 21.Borgbo T, Macek Sr M, Chrudimska J, Jeppesen J, Hansen L, Andersen Cy. Size matters: associations between the androgen receptor CAG repeat length and the intrafollicular hormone milieu. Mol Cell Endocrinol 2016;419:12–17. [DOI] [PubMed]
- 22.Pan J-X, Zhang J-Y, Ke Z-H, Wang F-F, Barry JA, Hardiman PJ, Qu F. Androgens as double-edged swords: induction and suppression of follicular development. Hormones. 2015;14:190–200. doi: 10.14310/horm.2002.1580. [DOI] [PubMed] [Google Scholar]
- 23.Kristensen SG, Ebbesen P, Andersen CY. Transcriptional profiling of five isolated size-matched stages of human preantral follicles. Mol Cell Endocrinol. 2015;401:189–201. doi: 10.1016/j.mce.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Hiyama G, Kansaku N, Tanaka T, Wakui S, Zadworny D. Characterization of chicken prolactin regulatory element binding protein and its expression in the anterior pituitary gland during embryogenesis and different reproductive stages. Japan Poult Sci Assoc. 2015;52(1):42–51. doi: 10.2141/jpsa.0140036. [DOI] [Google Scholar]
- 25.Habibi HR, Nelson ER, Allan ERO. New insights into thyroid hormone function and modulation of reproduction in goldfish. Gen Comp Endocrinol. 2012;175:19–26. doi: 10.1016/j.ygcen.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Krassas GE, Poppe K, Glinoer D. Thyroid function and human reproductive health. Endocr Rev. 2010;31:702–755. doi: 10.1210/er.2009-0041. [DOI] [PubMed] [Google Scholar]
- 27.Tasaki Y, Nishimura R, Shibaya M, Lee H-Y, Acosta TJ, Okuda K. Expression of VEGF and its receptors in the bovine endometrium throughout the estrous cycle: effects of VEGF on prostaglandin production in endometrial cells. J Reprod Dev Reprod Dev. 2010;56:9–139. doi: 10.1262/jrd.09-206E. [DOI] [PubMed] [Google Scholar]
- 28.Pfarrer C, Ruziwa S, Winther H, Callesen H, Leiser R, Schams D, et al. Localization of vascular endothelial growth factor (VEGF) and its receptors VEGFR-1 and VEGFR-2 in bovine Placentomes from implantation until term. Placenta. 2006;27:889–898. doi: 10.1016/j.placenta.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Nelson AC, Wardle FC. Conserved non-coding elements and cis regulation: actions speak louder than words. Development. 2013;140:1385–1395. doi: 10.1242/dev.084459. [DOI] [PubMed] [Google Scholar]