Abstract
Few data are available on the distribution and human infective potential of Cryptosporidium and Enterocytozoon bieneusi genotypes in bats. In this preliminary study, we collected 109 fecal specimens during April–July 2011 from a colony of straw-colored fruit bats (Eidolon helvum) in an urban park (Agodi Gardens) of Ibadan, Nigeria, and analyzed for Cryptosporidium spp., Giardia duodenalis and E. bieneusi using PCR targeting the small subunit rRNA gene, triosephosphate isomerase gene, and ribosomal internal transcribed spacer, respectively. Genotypes of these enteric parasites were determined by DNA sequencing of the PCR products. Altogether, 6 (5.5%), 0 and 16 (14.7%) specimens were positive for Cryptosporidium spp., G. duodenalis, and E. bieneusi, respectively. DNA sequence analysis of the PCR products indicated the presence of two novel Cryptosporidium genotypes named as bat genotype XIV (in 5 specimens) and bat genotype XV (in 1 specimen) and one known E. bieneusi genotype (Type IV in 1 specimen) and two novel E. bieneusi genotypes (Bat1 in 13 specimens and Bat2 in 2 specimens). In phylogenetic analysis of DNA sequences, the two novel Cryptosporidium genotypes were genetically related to Bat genotype II previously identified in fruit bats in China and Philippines, whereas the two novel E. bieneusi genotypes were genetically related to Group 5, which contains several known genotypes from primates. With the exception of Type IV, none of the Cryptosporidium and E. bieneusi genotypes found in bats in this study are known human pathogens. Thus, straw-colored fruit bats in Nigeria are mainly infected with host-adapted Cryptosporidium and E. bieneusi genotypes.
Keywords: Cryptosporidium, Enterocytozoon bieneusi, Fruit bat, Host-adapted genotypes
Graphical abstract
Highlights
-
•
Two novel Cryptosporidium genotypes were identified in fruit bats in Nigeria.
-
•
One known and two novel Enterocytozoon bieneusi genotypes were further detected.
-
•
Most Cryptosporidium and E. bieneusi genotypes in fruit bats are host adapted in nature.
1. Introduction
Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi are parasitic protists, causing diarrhea and other gastrointestinal symptoms in humans and animals (Wright, 2012). They are transmitted by direct contact with infected persons (anthroponotic transmission) or animals (zoonotic transmission) or through consumption of contaminated food or water (foodborne or waterborne transmission) (Checkley et al., 2015; Matos et al., 2012; Ryan et al., 2018).
Genetic diversity exists in each of the three groups of pathogens. Thus far, there are near 40 named Cryptosporidium species and as many unnamed species know as genotypes, each with some degree of host specificity (Feng et al., 2018). Similarly, there are at least eight genotypes of G. duodenalis known as assemblages A to H, which are likely cryptic species with different host ranges (Feng and Xiao, 2011). There are also over 250 E. bieneusi genotypes, forming at least 11 genotype groups with different host preferences (Santin et al., 2018; Zhang et al., 2018; Zhong et al., 2017). Only some of the species or genotypes are major human pathogens, such as C. parvum and C. hominis among Cryptosporidium spp., assemblages A and B in G. duodenalis, and Group 1 genotypes in E. bieneusi (Feng et al., 2018; Matos et al., 2012; Ryan et al., 2018). Molecular diagnostic tools are needed to differentiate the human-infective species and genotypes from animal-specific ones (Ghosh and Weiss, 2009; Xiao and Feng, 2017). As different human-pathogenic species and genotypes have distinct host range, the use of molecular diagnostic tools in epidemiologic investigations has significantly improved our understanding of the transmission of these pathogens in both industrialized and developing countries (Matos et al., 2012; Xiao and Feng, 2017).
Bats are known to play a major role in the transmission of emerging pathogens around the world (Han et al., 2015). This is especially the case with viruses such as coronavirus and rabies virus (Brook and Dobson, 2015). This is largely because of their large numbers, mobile nature, and tolerance to many of the pathogens (O'Connor, 2018; Serra-Cobo and Lopez-Roig, 2017). Their role in the transmission of Cryptosporidium spp., G. duodenalis, and E. bieneusi, however, remains unclear. There have been a few recent studies on the identity of Cryptosporidium spp. in bats in Asia, Australia and Europe, which have identified the occurrence of 12 Cryptosporidium genotypes in bats, all of which appear to be bat-specific (Kvac et al., 2015; Murakoshi et al., 2016, 2018; Schiller et al., 2016; Wang et al., 2013). Thus far, there has been no study on G. duodenalis in bats, but six E. bieneusi genotypes were identified in bats in South Korea recently, one of which belongs to Group 1, with the remaining ones belonging to Group 2, which contains E. bieneusi genotypes mostly found in ruminants (Lee et al., 2018).
In this preliminary study, we have examined the occurrence and identity of Cryptosporidium spp., G. duodenalis, and E. bieneusi in straw-colored fruit bats in a popular public park in Ibadan, Nigeria.
2. Materials and methods
2.1. Specimens
The study was conducted with fecal specimens collected from straw-colored fruit bats (Eidolon helvum) living in the Agodi Gardens (N 07.40614; E 003.90073), a popular public park in central Ibadan, Nigeria (Fig. 1). It is located between a University College Hospital and a residential area and is 60.7 hectares in size. It had one single colony of bats with no interspecies co-roosting, with thousands of mixed ages of bats on trees with thick canopy. It was estimated that there were about 30,000 straw-colored fruit bats at the time of sampling, with no other species of bats in presence. Fecal droppings from bats hanging on tall forest trees were collected during April–July 2011 at 109 individual points. Only fresh droppings were collected from various locations on two separate occasions. They were stored at −20 °C prior to DNA extraction. There were no direct interactions between sampling personnel and animals at the time of sampling.
Fig. 1.
Location of the straw-colored fruit bats examined in the study.
2.2. Detection of Cryptosporidium spp., G. duodenalis and E. bieneusi
DNA was extracted from 200 μl of stored fecal specimens using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA). This technique was shown to be better in removing PCR inhibitors in environmental samples than other common commercial DNA extraction kits (Jiang et al., 2005). The extracted DNA was stored at −80 °C before analysis by PCR. To detect Cryptosporidium spp., a ∼830-bp fragment of the small subunit (SSU) rRNA gene was amplified by nested PCR, and Cryptosporidium genotypes were initially identified by restriction fragment length polymorphism (RFLP) analysis of the secondary PCR products using restriction enzymes SspI and VspI (New England BioLabs, Massachusetts, USA) (Xiao et al., 1999). To detect G. duodenalis, a ∼530-bp fragment of the triosephosphate isomerase (tpi) gene was amplified by nested PCR (Sulaiman et al., 2003a). To detect E. bieneusi, a 392-bp fragment of the rRNA unit containing the entire internal transcribed spacer (ITS) was amplified by nested PCR (Sulaiman et al., 2003b). Each specimen was analyzed by PCR twice using 1 μl of extracted DNA per PCR, with DNA from C. canis as the positive control for the SSU rRNA PCR, DNA from G. duodenalis assemblage C as the positive control for the tpi PCR, and DNA from E. bieneusi genotype PtEb IX as the positive control for the ITS PCR. Two negative controls (reagent-grade water) for primary PCR and secondary PCR were further used in each PCR run. Non-acetylated bovine serum albumin (Sigma-Aldrich, St, Louis, MO, USA) was used at the concentration of 400 ng/μl in the primary PCR to neutralize residual PCR inhibitors in DNA, as previously described (Jiang et al., 2005).
2.3. Genotyping pathogen by sequence analysis
All positive secondary PCR products in this study were sequenced in bi-directionally using the Big Dye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA) on an ABI 3130 Genetic Analyzer (Applied Biosystems). Sequences were assembled using ChromasPro 1.5 (http://technelysium.com.au/ChromasPro.html) and compared with reference sequences in NCBI database using ClustalX (http://clustal.org/) to determine the genotypes of pathogens of Cryptosporidium spp. and E. bieneusi in fecal specimens. Phylogenetic trees were constructed from the nucleotide sequence alignments using the Bayesian inference and Monte Carlo Markov Chain (MCMC) methods in MrBayes 3.2.6 (http://nbisweden.github.io/MrBayes), with the posterior probability (pp) values calculated by running 1,000,000 generations. Nucleotide sequences generated from the study were submitted to GenBank under accession numbers MK007969-MK007974.
3. Results
3.1. Occurrence of Cryptosporidium spp., G. duodenalis and E. bieneusi
Among the 109 specimens analyzed, 6 (5.5%) were positive for Cryptosporidium spp. by PCR analysis of the SSU rRNA gene and 16 (14.7%) were positive for E. bieneusi by PCR analysis of the ITS (Table 1). In contrast, none of them were positive for G. duodenalis by tpi-based PCR.
Table 1.
Occurrence of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in straw-colored fruit bats in Ibadan, Nigeria.
| Pathogen | No. of specimens | No. positive (%) | Genotype |
|---|---|---|---|
| Cryptosporidium spp. | 109 | 6 (5.5%) | Bat genotype XIV (5), Bat genotype XV (1) |
| G. duodenalis | 109 | 0 | – |
| E. bieneusi | 109 | 16 (14.7%) | Type IV (1), Bat1 (13), Bat2 (2) |
3.2. Cryptosporidium genotypes in bats
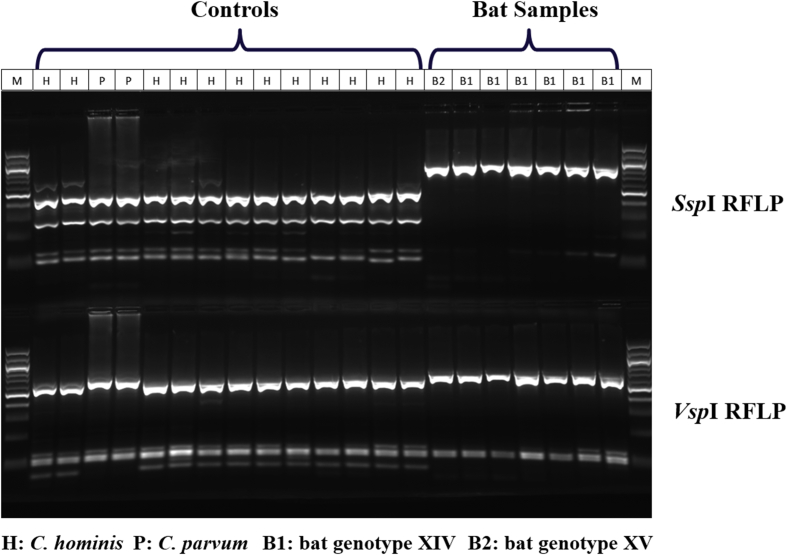
RFLP analysis of the SSU rRNA PCR products revealed a unique banding pattern for the Cryptosporidium spp. detected. All SSU rRNA products produced the same banding pattern, with the SspI restriction enzyme failing to digest the PCR product, while the VspI restriction enzyme producing a banding pattern that was similar to C. parvum (Fig. 2). DNA sequence analysis revealed the presence of two novel Cryptosporidium genotypes that were closely related to each other and to Cryptosporidium bat genotype II (Fig. 3). They were named as bat genotypes XIV and XV. The former was found in five specimens while the latter was found in one specimen. They differed from each other by about 28 single nucleotide polymorphisms (SNPs). Within Cryptosporidium bat genotype XIV, one specimen (No. 32604) produced a sequence that had one two-nucleotide insertion, one four-nucleotide deletion and one two-nucleotide deletion in two polymorphic areas of the SSU rRNA gene compared with other bat genotype XIV specimens.
Fig. 2.
Genotyping of Cryptosporidium spp. in straw-colored fruit bats by small subunit rRNA-based PCR-RFLP. Upper panel: SspI RFLP patterns; lower panel: VspI RFLP patterns; M: 100-bp molecular markers; H: C. hominis positive control; P: C. parvum positive control; B1: Cryptosporidium bat genotype XIV; B2: Cryptosporidium bat genotype XV.
Fig. 3.
Phylogeny of Cryptosporidium genotypes in bats based on Bayesian inference of sequences of the small subunit rRNA gene. The posterior probability values are indicated on the branches. Red ones are Cryptosporidium genotypes identified in straw-colored fruit bats in the present study, while blue ones are known Cryptosporidium genotypes previously reported in bats. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. E. bieneusi genotypes in bats
DNA sequence analysis showed the presence of three genotypes among the 16 E. bieneusi-positive specimens, including one known one and two novel ones. The former included Type IV, which was found in one specimens, while the latter were represented by two closely related genotypes named as Bat1 and Bat2, which differed from each other by 6 SNPs and were found in 13 and 2 specimens, respectively. Phylogenetically, the two new E. bieneusi genotypes belonged to Group 5, which contains several known genotypes from primates, such as CAF4, PtEb XII and KB-6 (Fig. 4).
Fig. 4.
Phylogeny of Enterocytozoon bieneusi genotypes in bats based on Bayesian inference analysis of sequences of the internal transcribed spacer of the rRNA gene. The posterior probability values are indicated on the branches. Red ones are E. bieneusi genotypes identified in straw-colored fruit bats in the present study. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Results of this preliminary study have shown the occurrence of Cryptosporidium spp. and E. bieneusi, but not G. duodenalis, in fruit bats living in an urban public park in Nigeria. The 5.5% infection rate for Cryptosporidium spp. is in line with the 2.1–8.9% infection rates of Cryptosporidium spp. reported in previous studies of several species of bats in Australia, USA, Czech Republic, China and Philippians (Kvac et al., 2015; Murakoshi et al., 2016; Schiller et al., 2016; Wang et al., 2013). The 14.7% detection rate of E. bieneusi in fruit bats examined in this study was significantly higher than the 1.9% detection rate of E. bieneusi in eight species of bats analyzed recently in South Korea (Lee et al., 2018). Giardia duodenalis was not examined in any of the previous studies of enteric protozoa in bats. Despite the use of a PCR assay that is designed to detect divergent Giardia species (Sulaiman et al., 2003a), we failed in obtaining any expected PCR products, indicating that G. duodenalis is not common in the bat species examined in Nigeria.
The two Cryptosporidium genotypes found in bats in this study are not known human pathogens. This agrees with previous characterizations of Cryptosporidium spp. in bats in several countries, where mainly novel Cryptosporidium genotypes were detected in both fruit bats and insectivorous bats (Kvac et al., 2015; Murakoshi et al., 2016, 2018; Schiller et al., 2016; Wang et al., 2013). Human-pathogenic C. hominis and C. parvum, however, were identified in four bats in Australia, Czech Republic, and Australia (Kvac et al., 2015; Schiller et al., 2016). Similarly, the two common E. bieneusi genotypes found in fruit bats in this study are not known human pathogens, only with one fruit bat positive for Type IV, a common zoonotic E. bieneusi subtype in humans (Matos et al., 2012). Thus, in contrast to their role in the transmission of emerging pathogens, bats can be only minor reservoirs of human-pathogenic Cryptosporidium spp. and E. bieneusi.
The two novel Cryptosporidium genotypes identified in straw-colored fruit bats in Nigeria appear to be host-adapted Cryptosporidium spp. Phylogenetically, the two Cryptosporidium genotypes in this study formed the basal branch of the SSU rRNA-based Bayesian inference tree, together with Cryptosporidium bat genotypes II and XIII previously identified in fruit bats in China and Philippines (Murakoshi et al., 2016; Wang et al., 2013). Similarly, bat genotypes V and XI from fruit bats in Australia and Philippines formed another cluster (Murakoshi et al., 2016; Schiller et al., 2016), while bat genotypes VIII, IX, and X from fruit bats in Australia formed a third cluster (Schiller et al., 2016). In contrast, other six Cryptosporidium genotypes identified in insectivorous bats are mostly dispersed over the SSU rRNA-based tree, probably as a results of the more diverse nature of their hosts (Fig. 3). As only a small number of bat species have been examined in a few countries, more novel Cryptosporidium genotypes are likely to be identified in future.
The two novel E. bieneusi genotypes identified are very divergent from common human-pathogenic E. bieneusi genotypes, which are mostly belong to Group 1. In phylogenetic analysis of the ITS sequences, they formed a cluster with several E. bieneusi genotypes from primates. They, however, have 10–13 SNPs compared to other genotypes in the group, thus could be bat-adapted E. bieneusi genotypes rather than related genotypes from primates. The host specificity of the various E. bieneusi genotype groups described thus far appears to be less strict than previously believed (Guo et al., 2014).
In conclusion, this preliminary study has shown the occurrence of Cryptosporidium spp. and E. bieneusi, but not G. duodenalis, in straw-colored fruit bats in an urban public park in Nigeria, mostly with the host-adapted Cryptosporidium and E. bieneusi genotypes. More genetic characterization of the pathogens in divergent host species, geographic areas, and environmental settings are needed to have better understanding of the prevalence, host specificity and evolution of Cryptosporidium spp., Giardia spp., and E. bieneusi in bats.
Declaration of interest
We have no conflict of interest to declare with this work.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31425025).
Contributor Information
Na Li, Email: nli@scau.edu.cn.
Adekunle B. Ayinmode, Email: ayins2000@yahoo.com.
Hongwei Zhang, Email: Zhwei69@163.com.
Yaoyu Feng, Email: yyfeng@scau.edu.cn.
Lihua Xiao, Email: lxiao1961@gmail.com.
References
- Brook C.E., Dobson A.P. Bats as 'special' reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., Huston C.D., Kotloff K.L., Kang G., Mead J.R., Miller M., Petri W.A., Jr., Priest J.W., Roos D.S., Striepen B., Thompson R.C., Ward H.D., Van Voorhis W.A., Xiao L., Zhu G., Houpt E.R. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Feng Y., Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K., Weiss L.M. Molecular diagnostic tests for microsporidia. Interdiscip. Perspect. Infect. Dis. 2009;2009:926521. doi: 10.1155/2009/926521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Alderisio K.A., Yang W., Cama V., Feng Y., Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl. Environ. Microbiol. 2014;80:218–225. doi: 10.1128/AEM.02997-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Wen H.L., Zhou C.M., Chen F.F., Luo L.M., Liu J.W., Yu X.J. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Alderisio K.A., Singh A., Xiao L. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 2005;71:1135–1141. doi: 10.1128/AEM.71.3.1135-1141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvac M., Horicka A., Sak B., Prediger J., Salat J., Sirmarova J., Bartonicka T., Clark M., Chelladurai J.R., Gillam E., McEvoy J. Novel Cryptosporidium bat genotypes III and IV in bats from the USA and Czech Republic. Parasitol. Res. 2015;114:3917–3921. doi: 10.1007/s00436-015-4654-1. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Oem J.K., Lee S.M., Son K., Jo S.D., Kwak D. Molecular detection of Enterocytozoon bieneusi from bats in South Korea. Med. Mycol. 2018;56:1033–1037. doi: 10.1093/mmy/myx136. [DOI] [PubMed] [Google Scholar]
- Matos O., Lobo M.L., Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012;2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi F., Koyama K., Akasaka T., Horiuchi N., Kato K. Molecular and histopathological characterization of Cryptosporidium and Eimeria species in bats in Japan. J. Vet. Med. Sci. 2018;80:1395–1399. doi: 10.1292/jvms.18-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi F., Recuenco F.C., Omatsu T., Sano K., Taniguchi S., Masangkay J.S., Alviola P., Eres E., Cosico E., Alvarez J., Une Y., Kyuwa S., Sugiura Y., Kato K. Detection and molecular characterization of Cryptosporidium and Eimeria species in Philippine bats. Parasitol. Res. 2016;115:1863–1869. doi: 10.1007/s00436-016-4926-4. [DOI] [PubMed] [Google Scholar]
- O'Connor K.C. Bats are "blind" to the deadly effects of viruses. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aau2259. [DOI] [PubMed] [Google Scholar]
- Ryan U., Hijjawi N., Feng Y., Xiao L. Giardia: an under-reported foodborne parasite. Int. J. Parasitol. 2018:30246–30247. doi: 10.1016/j.ijpara.2018.07.003. S0020-7519. [DOI] [PubMed] [Google Scholar]
- Santin M., Calero-Bernal R., Carmena D., Mateo M., Balseiro A., Barral M., Lima Barbero J.F., Habela M.A. Molecular characterization of Enterocytozoon bieneusi in wild carnivores in Spain. J. Eukaryot. Microbiol. 2018;65:468–474. doi: 10.1111/jeu.12492. [DOI] [PubMed] [Google Scholar]
- Schiller S.E., Webster K.N., Power M. Detection of Cryptosporidium hominis and novel Cryptosporidium bat genotypes in wild and captive Pteropus hosts in Australia. Infect. Genet. Evol. 2016;44:254–260. doi: 10.1016/j.meegid.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Serra-Cobo J., Lopez-Roig M. Bats and emerging infections: an ecological and virological puzzle. Adv. Exp. Med. Biol. 2017;972:35–48. doi: 10.1007/5584_2016_131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Fayer R., Bern C., Gilman R.H., Trout J.M., Schantz P.M., Das P., Lal A.A., Xiao L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Fayer R., Lal A.A., Trout J.M., Schaefer F.W., 3rd, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003;69:4495–4501. doi: 10.1128/AEM.69.8.4495-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cao L., He B., Li J., Hu T., Zhang F., Fan Q., Tu C., Liu Q. Molecular characterization of Cryptosporidium in bats from Yunnan province, southwestern China. J. Parasitol. 2013;99:1148–1150. doi: 10.1645/13-322.1. [DOI] [PubMed] [Google Scholar]
- Wright S.G. Protozoan infections of the gastrointestinal tract. Infect. Dis. Clin. North Am. 2012;26:323–339. doi: 10.1016/j.idc.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Xiao L., Escalante L., Yang C., Sulaiman I., Escalante A.A., Montali R.J., Fayer R., Lal A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8-9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Koehler A.V., Wang T., Haydon S.R., Gasser R.B. New operational taxonomic units of Enterocytozoon in three marsupial species. Parasites Vectors. 2018;11:371. doi: 10.1186/s13071-018-2954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Tian Y., Song Y., Deng L., Li J., Ren Z., Ma X., Gu X., He C., Geng Y., Peng G. Molecular characterization and multi-locus genotypes of Enterocytozoon bieneusi from captive red kangaroos (Macropus Rufus) in Jiangsu province, China. PLoS One. 2017;12:e0183249. doi: 10.1371/journal.pone.0183249. [DOI] [PMC free article] [PubMed] [Google Scholar]