Abstract
Prebiotic chemistry, driven by changing environmental parameters provides canonical and a multitude of non-canonical nucleosides. This suggests that Watson-Crick base pairs were selected from a diverse pool of nucleosides in a pre-Darwinian chemical evolution process.
Life and LUCA
Life is a highly diverse phenomenon that occupies all conceivable geological niches on Earth. Its development is explained by Darwinian evolution, which must have begun with rudimentary “living” vesicles that at some point transitioned into what we call the last universal common ancestor (LUCA)1. LUCA is a hypothetical life form obtained from phylogenetic analysis from which all three kingdoms of life originated2. To our understanding, LUCA already possessed the capacity to synthesize specific building blocks such as amino acids, nucleotides and lipids1. While phylogenetics allow us to ascertain such information, any events that had occurred prior to LUCA’s emergence remain in the dark, leaving us with only the possibility of simulating plausible prebiotic scenarios in the laboratory.
Owing to the discovery of catalytic RNA, it is conceivable that life on Earth emerged from self-replicating RNA oligomers. A prerequisite for this RNA-world concept3 is that RNA was present on the early Earth. RNA, however, is a complex molecule (Fig. 1a) that consists of a sugar (ribose), heterocycles (A, C, G and U) for base pairing, and phosphodiesters to link the units. Since the formation of the ribose- and heterocyclic-portions of RNA required different chemistry, we must assume that the early Earth provided areas with different geological conditions to facilitate their syntheses. We can imagine dry desert-type mineralic surfaces that were only occasionally dampened by rain. These mineral fields may have experienced large temperature differences during day and night. Hot fields, close to active volcanos certainly existed with temperatures of above 200 °C, occasionally cooled by rain. We can imagine that much wetter climates existed as well, in which water could dissolve minerals that would later be (re)-precipitated in draughts. pH values may also have varied greatly. Acidic rain generated by NOx and SOx could have given rise to ponds with pH values below 3, while ponds filled with amines and amidines could have had pH values of above 10 or even 12. It seems plausible that the different RNA precursors formed separately in different geological settings and that they were incidentally washed together due to flooding or similar phenomena.
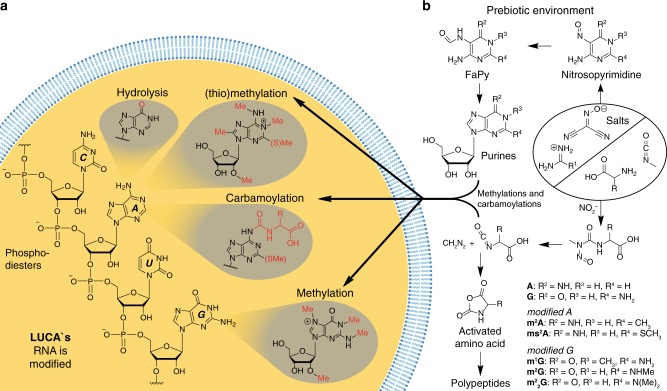
Fig. 1.
LUCA’s modified bases. a The chemical structure of ribonucleic acid (RNA) and of some modified bases. Different (thio)methylation sites and amino acid purine modifications in LUCA are marked in red. b Amidine salts are converted into nitrosopyrimidines, which then form formamidopyrimidines. Reaction with ribose provides a set of canonical and non-canonical nucleosides that are assumed to have been present in LUCA
Non-canonical bases and peptide-RNA hybrids
The chemistry of the early Earth must have been primitive and unspecific. It is hard to imagine that parameters such as temperatures or concentrations of reactants were so tightly controlled that single products formed selectively. Reactions likely took place in vessels such as aqueous ponds or on hot surfaces containing complex mixtures of different minerals. This leads us to believe that the conditions led to the formation of diverse pools of molecules (Fig. 1). Indeed, even today RNA is tremendously structurally diverse. Besides to the four canonical nucleobases (A, C, G and U) RNA contains a phlethora of additional modified nucleosides4. Some of these bases are found in all three kingdoms of life, and as such, can be considered to be molecular fossils of the original chemically diverse primordial soup. These non-canonical nucleosides are potential chemical ancestors; relics of early Earth’s chemistry1.
Prebiotic routes to purines and amino acid modified purines
Prebiotic chemical processes likely occurred under conditions far away from defined laboratory chemistry. Today, Chemists use special glassware for their reactions and reactants are added step-wise in a tightly controlled manner, often in a protecting atmosphere and regularly in inert solvents. Concentration and temperature are controlled, and importantly, the reaction product is carefully isolated and purified before it is introduced into the next reaction. All of this was impossible on the early Earth. Prebiotic chemistry forces chemists therefore to think about robust reactions that proceed under “dirty” conditions. Reactions that are general enough that they tolerate different temperatures and concentrations, and that are selective enough to proceed even in mixtures are privileged in a prebiotic setting. They are driven by fluctuations of outside physico-chemical parameters such as day–night or seasonal cycles, which provide intermittent wet and dry conditions along with changing temperatures. Drying out could have triggered selective precipitation and crystallization, which leads to purification and concentration to enable successive reactions5. Prebiotic reactions have to work in water, in dry-state conditions, or otherwise in higher-boiling solvents such as formamide. Reductions and oxidations may have occurred in the presence of iron- and sulphur-containing compounds, for example, by the conversion of iron sulphide (FeS) to pyrite (FeS2)6. High-energy irradiation could also have initiated chemical reactions, particularly when we assume the absence of a shielding ozone layer. Since DNA and RNA are not stable under UV irradiation (λ < 300 nm), photocatalysis was probably more important for the formation of small reactive organic molecules. Without sophisticated DNA/RNA repair mechanisms, oligonucleotides could survive only in niches devoid of UV light7. It is safe to assume that UV light was a thread to early life.
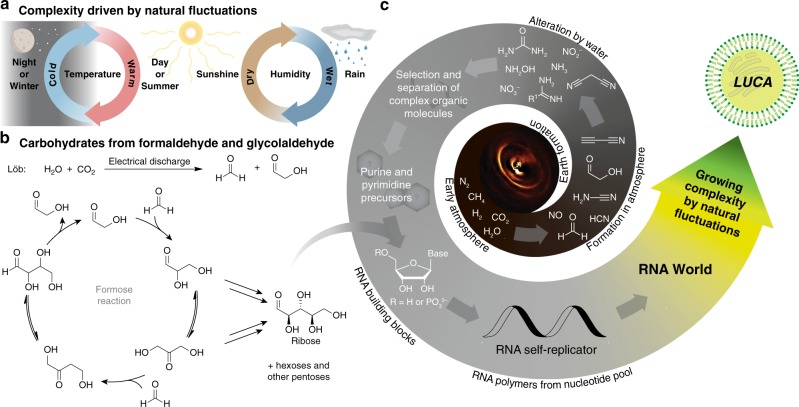
When we think about the formation of small prebiotic starting molecules, electrical discharge needs to be considered (Fig. 2a). In a nearly neutral N2 atmosphere composed also of H2O, CO2, H2 and CH4, electrical discharge converts N2 into NO8 which can be reduced to NH2OH and then into NH3. In addition, N2 reacts with CH4 under discharge conditions to give products including HCN, cyanamide and cyanoacetylene9. Under such electrical discharge conditions (Fig. 2a) humid CO2 produces the starting materials for sugars such as formaldehyde and glycolaldehyde (Fig. 2b). This all together provides a rather large set of small prebiotic organic molecules that can act as starting materials for the formation or RNA nucleosides (Figs. 1b and 2b).
Fig. 2.
Natural fluctuations steer chemical transformations towards greater complexity. a Molecular complexity driven by fluctuations of physico-chemical conditions, such as day–night or seasonal changes, leading to wet–dry cycles. b Since the early Earth was not uniform, carbohydrates could have emerged separately from nucleobases or their precursors e.g. via the formose reaction. c When washed into the same environment, different nucleosides/nucleotides could have formed
Cyanamide, for example, can give different amidinium compounds by nucleophilic addition, which form low-melting organic salts with nitrosated malononitrile (Fig. 1b)5. We found that that these salts form formamidopyrimidines (FaPys, Fig. 1b), which react efficiently with ribose to give purine nucleosides10. This new FaPy-pathway generates not only the two canonical purine bases (A and G), but also a variety of RNA modifications5. Under special conditions, even amino acid modified purine nucleosides are generated. Most importantly all these modified purine bases are found today in contemporary RNA, thus strengthening the idea that non-canonical RNA bases are vestiges of our primordial anscestor11.
Ribose and the oligomerization problem
The use of ribose as a prebiotic starting material for nucleoside formation is sometimes criticised because no clear prebiotically plausible route to ribose has been found. We still believe that ribose was present on the early Earth and that we have simply to discover the right conditions. It is well known that formaldehyde and glycolaldehyde produce ribose in the formose reaction (Fig. 2b), but admittedly, the yields are low (<1%)12. It is known that borates increase the yield of ribose13 and we are sure that with more time and research even better and more selective conditions will be found. We should also not forget that life could have begun with oligonucleotides composed of sugars other than ribose14. Even ribose can exist in a 5- or a 6-membered ring form. The 5-membered arrangement (furanosides) are what today constitute our RNA, but we know from A. Eschenmosers’ seminal work that the 6-membered pyranosidic RNA also produces wonderful double strands with selective base pairing15. It is a riddle, why the thermodynamically less favoured 5-membered furanosides were chosen to create life. One argument is that the 5-membered furanosides are more easily phosphorylated because they possess a very reactive primary hydroxyl group16. Such phosphorylation is needed to stitch the nucleosides together to enable strand formation17. Given that prebiotic RNA nucleosides and consequently RNA strands may have been structurally diverse, containing for example, amino acid modifications, we can envision that some had physico-chemical properties or catalytic properties that offered a survival advantage. Pre-Darwinian evolution may have acted first on molecules rather than living species to select the fittest, initially maybe just the most stable molecules or RNA strands (Fig. 2c). At some point nature must have discovered that the ultimate solution to molecular survival is reproduction via self-replication and catalysis. This is best achieved in a shielded environment within which the molecules needed for replication can be autonomously generated (metabolism). This unit is called a cell.
Acknowledgements
We thank the Deutsche Forschungsgemeinschaft (DFG) for financial support via the programs SFB1032 (TP-A5), SFB749 (TP-A4), SPP-1784 and CA275/11-1. Additional funding from the Excellence Cluster EXC114 CiPSM is acknowledged. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement n° EPiR 741912).
Author contributions
T.C. conceived, developed and wrote this manuscript. S.B., C.S. and A.C. helped writing this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sidney Becker, Christina Schneider.
Change history
1/15/2019
The original version of this Article contained errors in the citations in the second, third and fourth sentences of the first paragraph of the ‘Life and LUCA’ section, which incorrectly read ‘Its development is explained by Darwinian evolution, which must have begun with rudimentary “living” vesicles that at some point transitioned into what we call the last universal common ancestor (LUCA)2. LUCA is a hypothetical life form obtained from phylogenetic analysis from which all three kingdoms of life originated3. To our understanding, LUCA already possessed the capacity to synthesize specific building blocks such as amino acids, nucleotides and lipids2.’ The correct version states ‘(LUCA)1’ in place of ‘(LUCA)2’, ‘originated2’ instead of ‘originated3’ and ‘lipids1’ rather than ‘lipids2’. This has been corrected in both the PDF and HTML versions of the Article.
References
- 1.Weiss MC, et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- 4.Carell T, et al. Structure and function of noncanonical nucleobases. Angew. Chem. Int. Ed. 2012;51:7110–7131. doi: 10.1002/anie.201201193. [DOI] [PubMed] [Google Scholar]
- 5.Becker S, et al. Wet-dry cycles enable the parallel origin of canonical and non-canonical nucleosides by continuous synthesis. Nat. Commun. 2018;9:163. doi: 10.1038/s41467-017-02639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol. Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mees A, et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 8.Airapetian VS, Glocer A, Gronoff G, Hébrard E, Danchi W. Prebiotic chemistry and atmospheric warming of early Earth by an active young Sun. Nat. Geosci. 2016;9:452. doi: 10.1038/ngeo2719. [DOI] [Google Scholar]
- 9.Benner, S. A., Kim, H.-J. & Biondi, E. in Prebiotic Chemistry and Chemical Evolution of Nucleic Acids 31–83 (Springer, Berlin, 2018).
- 10.Benner SA, Kim HJ, Carrigan MA. Asphalt, water, and the prebiotic synthesis of ribose, ribonucleosides, and RNA. Acc. Chem. Res. 2012;45:2025–2034. doi: 10.1021/ar200332w. [DOI] [PubMed] [Google Scholar]
- 11.Schneider C, et al. Noncanonical RNA nucleosides as molecular fossils of an early Earth—Generation by prebiotic methylations and carbamoylations. Angew. Chem. Int. Ed. 2018;57:5943–5946. doi: 10.1002/anie.201801919. [DOI] [PubMed] [Google Scholar]
- 12.Butlerow A. Formation synthétique d’une substance sucrée. Comptes Rendus Acad. Sci. 1861;53:145–147. [Google Scholar]
- 13.Ricardo A, Carrigan MA, Olcott AN, Benner SA. Borate minerals stabilize ribose. Science. 2004;303:196–196. doi: 10.1126/science.1092464. [DOI] [PubMed] [Google Scholar]
- 14.Fialho DM, et al. Glycosylation of a model proto-RNA nucleobase with non-ribose sugars: implications for the prebiotic synthesis of nucleosides. Org. Biomol. Chem. 2018;16:1263–1271. doi: 10.1039/C7OB03017G. [DOI] [PubMed] [Google Scholar]
- 15.Krishnamurthy R, et al. Pyranosyl‐RNA: base pairing between homochiral oligonucleotide strands of opposite sense of chirality. Angew. Chem. Int. Ed. 1996;35:1537–1541. doi: 10.1002/anie.199615371. [DOI] [Google Scholar]
- 16.Kim HJ, et al. Evaporite borate‐containing mineral ensembles make phosphate available and regiospecifically phosphorylate ribonucleosides: borate as a multifaceted problem solver in prebiotic chemistry. Angew. Chem. Int. Ed. 2016;55:15816–15820. doi: 10.1002/anie.201608001. [DOI] [PubMed] [Google Scholar]
- 17.Li L, et al. Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J. Am. Chem. Soc. 2017;139:1810–1813. doi: 10.1021/jacs.6b13148. [DOI] [PMC free article] [PubMed] [Google Scholar]