Abstract
Evolutionary mechanisms underlying innovation of cell types have remained largely unclear. In multicellular eukaryotes, the evolutionary molecular origin of sperm differentiation is unknown in most lineages. Here, we report that in algal ancestors of land plants, changes in the DNA-binding domain of the ancestor of the MYB transcription factor DUO1 enabled the recognition of a new cis-regulatory element. This event led to the differentiation of motile sperm. After neo-functionalization, DUO1 acquired sperm lineage-specific expression in the common ancestor of land plants. Subsequently the downstream network of DUO1 was rewired leading to sperm with distinct morphologies. Conjugating green algae, a sister group of land plants, accumulated mutations in the DNA-binding domain of DUO1 and lost sperm differentiation. Our findings suggest that the emergence of DUO1 was the defining event in the evolution of sperm differentiation and the varied modes of sexual reproduction in the land plant lineage.
The evolutionary mechanisms leading to the development of sperm are poorly understood. Here, the authors infer that neofunctionalisation and expression changes of the orthologue of DUO1 in algal ancestors of land plants were key events for sperm differentiation and sexual reproduction in this lineage.
Introduction
The development of differentiated cell types is related to multicellularity and the acquisition of novel functions. Morphological similarities between cell types across organisms might be indicative of their common evolutionary origin. The common origin of differentiated cell types and their underlying conserved gene regulatory networks illustrate the deep homology1 that exists amongst distant species2. However, while aspects of gene regulatory networks that cause differentiation of specific cell types might be conserved, cells still exhibit distinct traits that evolve independently. Such a scenario is illustrated by the evolution of male gametes. Across eukaryotes, small motile sperm that fertilize large immotile eggs evolved multiple times. This led to a mode of sexual reproduction defined as anisogamy, which is considered a key event in the evolution of sexual dimorphism3–6. Across kingdoms, spermatozoa share fusogenic properties7, yet show a widely varied morphology. In plants, motile sperm evolved first in Charophyceae, the freshwater algae sharing the last common ancestor with land plants, which are collectively referred to as Streptophyta (Fig. 1a). However, sperm motility is absent in conjugating green algae (Zygnematophyceae), the sister group of land plants8. Bryophytes, which include liverworts, mosses, and hornworts, are representatives of the first land plants and produce motile sperm that show characteristics identical to sperm from Charophyceae (Fig. 1a). Among vascular plants, while ferns and some gymnosperms produce motile flagellate sperm, flowering plants differentiate non-motile gametes, which are instead transported to the female gametes by the pollen tube. Such distinct modes of spermatogenesis in the land plant lineage suggest either the diversification of an ancestral gene regulatory network or that distinct types of spermatogenesis evolved independently.
Fig. 1.
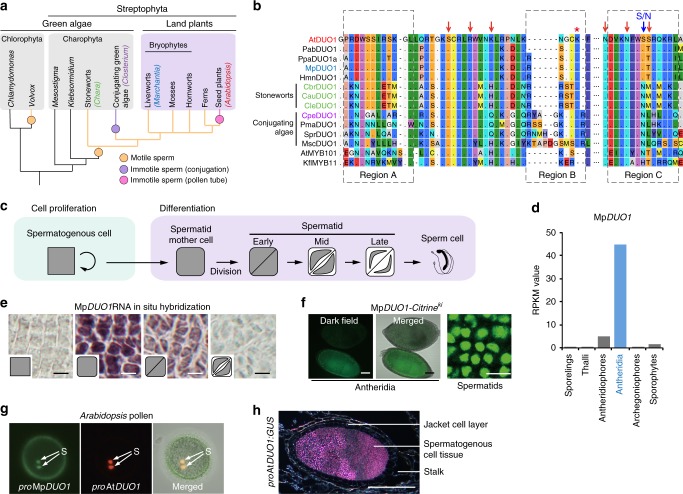
Characterization of MpDUO1. a A schematic phylogeny indicating the major groups discussed in this study. b Amino acid sequence alignment of the DUO1 MYB domains from streptophytes together with the non-DUO1 MYB domains from AtMYB101 and Klebsormidium KflMYB11. Regions A, B, and C are highlighted with a dashed box. Red arrows point to putative DNA-interacting amino acid residues. The blue arrow indicates the N to S change that occurred before land plant diversification. The red asterisk indicates the additional K residue typical of the DUO1 subfamily. c Schematic view of Marchantia male gamete development. d MpDUO1 expression profile during Marchantia sexual life cycle based on RNA-seq data of major tissues analyzed in ref. 14 Antheridiophores and archegoniophores are male and female reproductive branches, respectively. Thalli contain haploid somatic vegetative tissue. Sporophytes are diploid and develop from the embryo after fertilization. e RNA in situ hybridization of MpDUO1 during male gamete development (see Supplementary Figure 2 for the sense-probe control). Insets represent the developmental stages referred to in c. f The expression pattern of MpDUO1-Citrine fusion protein (green) in antheridia containing developing sperm (left two panels) and differentiating spermatids (right panel) from MpDUO1-Citrineki plants. g Z-projected confocal images of Arabidopsis pollen expressing proMpDUO1:H2B-Clover (green), and proAtDUO:H2B-mRuby2 (red). The pollen grain wall displays autofluorescence in the green channel. S, sperm cells. h Dark-field image of a section of an antheridium from a GUS-stained (pink) Marchantia plant expressing proAtDUO1:GUS. Scale bars, 5 μm (e), 100 µm (f, antheridia), 5 μm (f, spermatids), and 100 µm (h)
In Arabidopsis thaliana, DUO POLLEN 1 (DUO1) is a MYB transcription factor (TF) controlling male gamete development and differentiation9–11. The network controlled by DUO1 in Arabidopsis comprises the two closely related TFs, DUO1-ACTIVATED ZINC FINGER1 (DAZ1) and DAZ2 which, together with DUO1, regulate the expression of the sperm-specific histone variant H3.10 and the fusogenic factors GENERATIVE CELL-SPECIFIC 1 /HAPLESS 2 (GCS1/HAP2) and GAMETE EXPRESSED 2 (GEX2)12. Recent access to the transcriptome during male gametogenesis in the model bryophyte Marchantia polymorpha13,14 enabled us to address whether plant sperm share a common molecular origin.
Here we combine genetic and molecular analyses to show that DUO1-type MYB TF is present in land plants15. Using phylogenetic analyses we further identify DUO1 orthologs in major clades of bryophytes and in Charophyceae but not in Mesostigma and Klebsormidium representing earlier diverging groups of freshwater green algae. The ancient origin of DUO1 is supported by conservation of its essential function of DUO1 in spermatogenesis in the land plant lineage. Functional in vivo assays and DNA-binding analyses suggest that this strong conservation is explained by a change in the DNA-binding domain of an ancestral MYB TF in the common ancestor of Charophyceae and land plants. This change enabled DUO1 to bind a distinct motif in its target promoters and led to evolution of a sperm differentiation program. Our results support a single origin for spermatogenesis despite the remarkable diversity of sperm morphology amongst plant species.
Results and Discussion
Identification of DUO1 ortholog in Marchantia
DUO1 differs from all other R2R3 MYB TFs by a supernumerary lysine residue between the R2 and R3 repeats of its MYB domain11. Based on search for this signature amongst MYB TF families in land plants, DUO1 orthologs were identified in several species of bryophytes, including liverworts and mosses (Fig. 1a, b and Supplementary Figure 1a), indicating a common origin of DUO1 in the ancestor of land plants. In the liverwort Marchantia polymorpha, male reproductive branches (antheridiophores) differentiate from a vegetative thallus and host the antheridia where sperm cells are formed14,16. In the antheridia, proliferating sperm-cell precursors give rise to spermatid mother cells after a penultimate division. Sperm mother cells then undergo a diagonal cell division to produce spermatids, which differentiate into sperm (Fig. 1c)16,17. MpDUO1 is expressed in the sperm-cell lineage, exclusively in spermatid mother cells and spermatids (Fig. 1d, e and Supplementary Figs. 1b–d, 2). The MpDUO1 protein accumulates in the nucleus (Fig. 1f and Supplementary Fig. 1e). In Haplomitrium mnioides, a representative of the most basal liverworts, HmnDUO1 is also expressed in antheridia (Supplementary Figure 1f). The MpDUO1 promoter confers sperm-cell-lineage-specific expression in Arabidopsis mature pollen like the native AtDUO1 promoter (Fig. 1g). Conversely, the AtDUO1 promoter confers an expression pattern similar to that of MpDUO1 in developing antheridia (Fig. 1h and Supplementary Figure 1c). These results suggest that transcriptional control of DUO1 has remained conserved between liverworts and angiosperms, which diverged more than 450 MYA13.
In Arabidopsis, duo1 null mutants fail to undergo G2-to-M transition of the generative cell followed by differentiation of two sperm cells11. Marchantia polymorpha is dioecious with separate sexes determined by sex chromosomes. Marchantia male and female plants carrying a null Mpduo1-1ko allele (Supplementary Figs. 1b, 3a) show normal vegetative development. Female Mpduo1-1ko plants are fertile, but males Mpduo1-1ko are sterile (Fig. 2a, b and Supplementary Figure 3b), suggesting a specific role of MpDUO1 in male gametogenesis. The discharge of sperm masses as white aggregates from antheridiophores upon hydration is observed in wild-type (WT) but not in Mpduo1-1ko (Fig. 2b). In Mpduo1-1ko, the final diagonal division that produces spermatids takes place as in WT, but the subsequent sperm differentiation fails (Fig. 2c–e and Supplementary Figure 3c, d). In WT spermiogenesis, the sperm nucleus condenses and acquires a crescent shape that elongates as sperm mature. During formation of two flagella and nuclear elongation, microtubule arrays assemble the spline, which serves as a backbone structure for the elongated nucleus and motility apparatus located at the base of flagella. Eventually, reduction of cytoplasm occurs, and the mature sperm become almost devoid of cytoplasm. In Mpduo1-1ko, reduction of cytoplasm occurs as in WT, but neither nuclear condensation nor elongation takes place, producing round nuclei in sperm that eventually die (Fig. 2c, d and Supplementary Figures 3c, d). Neither the spline nor flagella are formed (Fig. 2d, e and Supplementary Figure 3d). All defects observed in Mpduo1-1ko sperm can be complemented by the expression of wild-type MpDUO1 (MpComp; Figs. 2c, e, 3a and Supplementary Figures 3a, c, e), showing that MpDUO1 is essential for male gamete development in Marchantia. The expression of AtDUO1 under control of the MpDUO1 promoter results in partial complementation of the Mpduo1-1ko phenotype (AtComp; Figs. 2c, 3a and Supplementary Figures 3a, c, e). We also tested whether MpDUO1 can substitute for AtDUO1 in Arabidopsis. In Arabidopsis duo1 mutants (Atduo1), the sperm precursor fails to divide, resulting in a single infertile cell that does not express the male germline-specific histone H3 variant (H3.10) encoded by HISTONE THREE RELATED 10 (HTR10)9–11,18. The expression of MpDUO1 under control of the AtDUO1 promoter almost completely rescues the division defect of Atduo1 pollen11 (MpComp; Fig. 3b). The seemingly rescued sperm do not express the AtDUO1 target gene HTR10 and are unable to transmit the Atduo1 allele to progeny. But sperm differentiation is fully restored when the MpDUO1 DNA-binding domain is fused to the AtDUO1 C-terminal activation domain and the resultant chimera is expressed under control of the AtDUO1 promoter (Chimera 1; Fig. 3b). Similarly, expression of the chimera combining AtDUO1 DNA-binding domain with the MpDUO1 C-terminal activation domain results in a better rescue of the Mpduo1 phenotype (AtDUO1/MpDUO1 chimera; Fig. 3a and Supplementary Figure 3e). Together these results show that DUO1 orthologs have controlled sperm-cell differentiation and morphogenesis since the evolution of land plants.
Fig. 2.
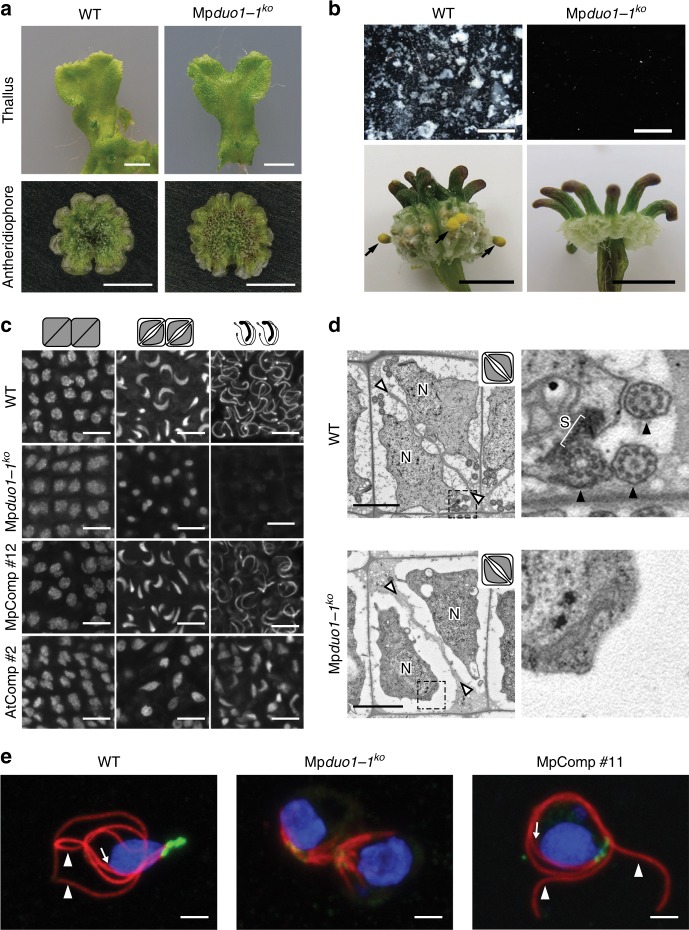
Mpduo1-1ko shows defects in sperm morphogenesis. a Images of thallus (top) and antheridiophore (bottom) of WT (left) and Mpduo1-1ko (right). b Top panels show sperm masses (white) discharged from mature antheridiophores into water in WT but not in Mpduo1-1ko. Bottom panels show the production of yellow sporangia (black arrows), which mark successful fertilization, on female WT about a month after crossing with male WT, but no sporangia production in crosses with male Mpduo1-1ko. c Feulgen staining of spermatids during spermiogenesis in WT, Mpduo1-1ko, and Mpduo1-1ko complemented by MpDUO1 genomic fragment (MpComp) and by proMpDUO1:AtDUO1 (AtComp). d Transmission electron micrographs of spermatids. White arrowheads indicate cell boundaries and N indicates nucleus. Areas in the dashed line boxes are enlarged in the right panels and show that the spline (S), a microtubular backbone-like structure, and flagella (black arrowheads) are present in WT but missing in Mpduo1-1ko. e Localization of centrins (green) and tubulins (red) in differentiating spermatids of WT, Mpduo1-1ko (the panel shows two sister cells), and MpComp. Centrin signals mark two basal bodies and an associated multi-layered structure. The blue signals indicate the DAPI stained nuclei. White arrowheads and arrows indicate flagella and spline, respectively. Scale bars, 5 mm (a), 400 μm (b, top), 5 mm (b, bottom), 5 μm (c), and 2 μm (d, e)
Fig. 3.
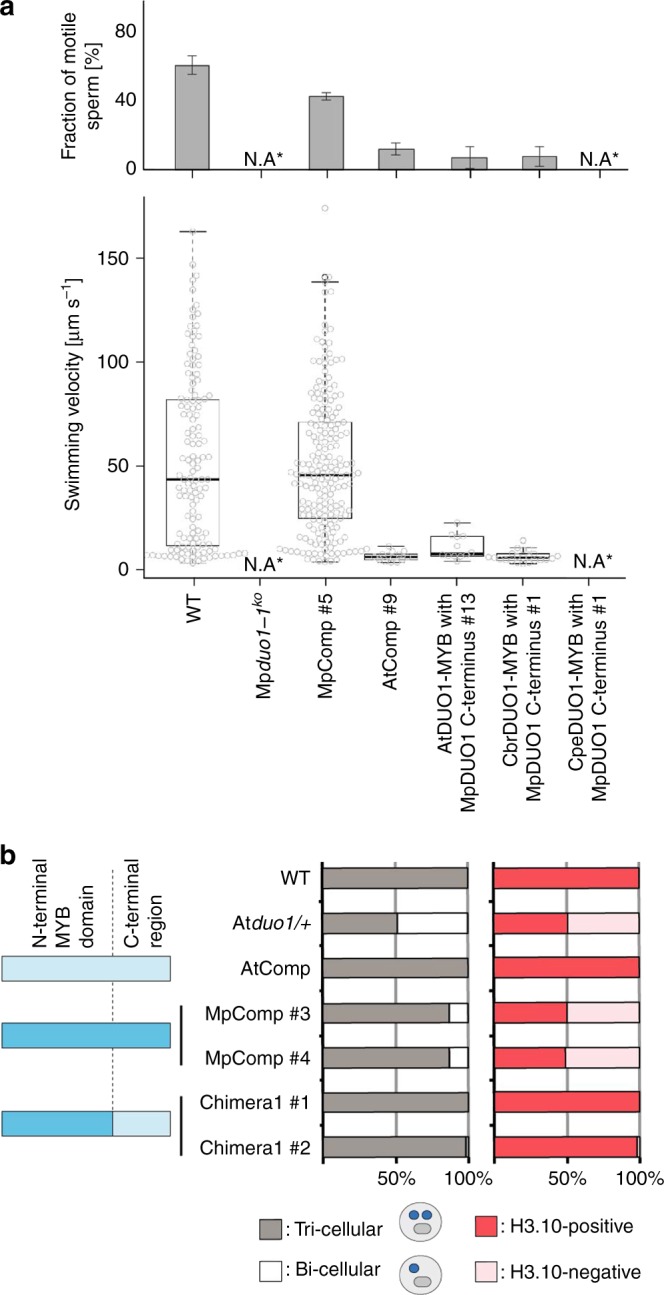
Functional conservation of DUO1 MYB domains. a Motility of mature sperm cells of WT and Mpduo1-1ko complemented with the MpDUO1 genomic fragment (MpComp #5), proMpDUO1:AtDUO1 (AtComp #9), proMpDUO1:AtChimera (AtDUO1-MYB with MpDUO1 C-terminus #13), proMpDUO1:CbrChimera (CbrDUO1-MYB with MpDUO1 C-terminus #1), and proMpDUO1:CpeChimera (CpeDUO1-MYB with MpDUO1 C-terminus #1). Percentage of motile sperm (upper) and average velocity (lower) of motile sperm based on analysis of three movies for each line are shown. N.A., not applicable, because spermatids abort in Mpduo1-1ko and Mpduo1-1ko complemented with proMpDUO1:CpeChimera (see Fig. 2b and Supplementary Figure 3e). The numbers of sperm observed are: n = 25, 23, 155 (WT), n = 46, 51, 58 (MpComp #5), n = 27, 32, 25 (AtComp #9), n = 7, 80, 94 (AtChimera #13), n = 44, 50, 59 (CbrChimera #1) for analysis of fraction of motile sperm; n = 117 (WT), n = 67 (MpComp #5), n = 10 (AtComp #9), n = 8 (AtChimera #13), n = 12 (CbrChimera #1) for analysis of swimming velocity. b Genetic complementation of Atduo1 by AtDUO1, MpDUO1, a chimera of the DNA-binding domain of MpDUO1 with the activation domain of AtDUO1. The degree of complementation is measured by rescue of pollen phenotype (left) and expression of H3.10 encoded by HTR10 (right). Schematic diagram of the constructs used for complementation are shown on the far left. Construct parts are color-coded light blue (AtDUO1) and dark blue (MpDUO1) (see Fig. 4b for details). n > 200 pollen
Molecular changes leading to DUO1 function
To dissect the distinct properties of the DUO1 DNA-binding domain, we tested the trans-activation potential of DUO1 TFs as well as chimeras thereof using an in vivo luciferase reporter assay with the DUO1-responsive HTR10 promoter9. In contrast to MpDUO1 (Chimera 1), the closely related MYB transcription factor MpR2R3-MYB21 (Fig. 4a and Supplementary Figure 4) does not trans-activate HTR10 (Chimera 2; Fig. 4b), so we used its R2R3 MYB domain as a negative control. Based on sequence alignment, we identified three specific conserved regions (A, B, and C) in the DUO1 DNA-binding domain (Figs. 1b, 4a). Region B includes the supernumerary lysine specific to DUO1 and region C contains several putative DNA-interacting residues, including a serine absent from MpR2R3-MYB21. We generated chimeric constructs swapping these three specific regions between MpDUO1 and MpR2R3-MYB21 and compared their trans-activation potential (Chimera 3 to 7; Fig. 4b). The exchange of region B or C significantly reduced activity, suggesting that these regions determine binding specificity for a distinct cis-regulatory element. Further, a screen for preferentially-bound DNA motifs using a protein-binding DNA microarray19,20 demonstrated the requirement of regions B and C for recognition of the conserved DUO1 consensus motif (Fig. 4c) distinct from that of other R2R3 MYB TFs19,20. Modeling the structure of MpDUO1 in complex with DNA shows that region B is located at the junction between two DNA-interacting helices (Fig. 4a), suggesting that variations at this position might alter DNA-binding specificity due to conformational changes. The region C contacts DNA and it is likely that discrete changes in this region consolidated the specific DNA-binding properties of DUO1. Thus, mutations in regions B and C conferred ancestral DUO1 with the potential to activate transcription of a new set of target genes.
Fig. 4.
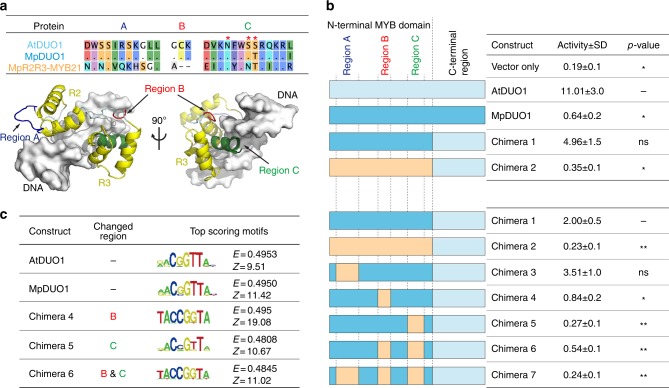
Characterization of DUO1 transcription factor activity. a Amino acid sequence alignment of regions A, B, and C among AtDUO1, MpDUO1, and MpR2R3-MYB21 (top). Dots indicate matching residues with AtDUO1. Asterisks indicate putative DNA-interacting residues in region C. Structural modeling of MpDUO1 in complex with DNA using SWISS-MODEL (bottom). The MpDUO1 MYB domain is overlaid onto the structure of the AMV v-MYB-DNA complex (PDB code: 1H8A). b in vivo transcriptional activation potentials of MpDUO1 and chimeras. Schematic diagram of constructs (left) are color-coded light blue (AtDUO1), dark blue (MpDUO1), and orange (MpR2R3-MYB21). DUO1 transcriptional activation potentials were measured by relative luciferase activity (right). n = 4 (upper), n = 8 (lower) (**p < 0.01; *p < 0.05; ns not significant; Student’s t-test). c Position weight matrix representations of the top-scoring 8-mer DNA sequences bound by different MYB domains on a protein-binding DNA microarray. Numbers denoted on the right side of each motif represent the motif E- and Z-scores19
Impact of MpDUO1 on gene expression in differentiating sperm
Next, we studied the impact of MpDUO1 on the expression of a set of genes associated with sperm differentiation (Supplementary Figure 5a). From the Marchantia antheridium transcriptome14, we selected genes encoding proteins likely to be important for flagella formation. These included the antheridium-specific tubulin genes MpTUA5 and MpTUB421, MpPACRG22, the ortholog of the gene PARKIN COREGULATED (PACRG) which is associated with the axoneme and is essential for normal sperm formation in mammals23–25, a homolog of the gene DYNEIN LIGHT CHAIN 7 (LC7)26 (MpLC714), and MpCEN1, a homolog of the gene encoding CENTRIN, which is essential for motility apparatus formation in Marsilea27. The gene PROTAMINE-LIKE (MpPRM)14 which encodes a protamine-like arginine-rich protein presumably involved in chromatin compaction and nuclear morphogenesis was also included. The expression of most of those genes followed MpDUO1 expression (Fig. 5a and Supplementary Figure 2; for MpLC7, MpPACRG, and MpPRM, see ref. 14) and depended on MpDUO1 (Fig. 5b and Supplementary Figure 5b), illustrating how MpDUO1 is essential for several key features of sperm differentiation. Moreover, MpDUO1 controls expression of MpDAZ1, the Marchantia ortholog of AtDAZ1 and AtDAZ2, two closely related downstream transcription factors targeted by DUO1 in Arabidopsis9,12 (Fig. 5, Supplementary Figs. 2, 5, and Supplementary Table 1), suggesting conservation of the DUO1-DAZ1 module among land plants. In contrast, MpDUO1 does not control expression of orthologs of AtDUO1 targets AtGCS1/HAP2 and AtGEX2, which are involved in gamete fusion and attachment, respectively9,28–30, and expressed in differentiating Marchantia sperm (Fig. 5b, Supplementary Figure 5, and Supplementary Table 1). This independence might be related to the fact that GCS1/HAP2 and GEX2 evolved prior to DUO1 in ancestors of plants, algae, and animals14,28,29,31. In Arabidopsis, HTR10 encoding a male germline-specific histone H3 variant (H3.10) is an important target of DUO19. But, no corresponding male germline-specific histone H3 variant was found in Marchantia14. Instead, a gene encoding a protamine-like sperm nuclear protein (MpPRM)14 is under the control of MpDUO1 (Fig. 5b and Supplementary Figure 5b). These data suggest that while the DUO1-DAZ1 regulatory module is conserved, loss and recruitment of new downstream target genes took place after the emergence of vascular plants.
Fig. 5.
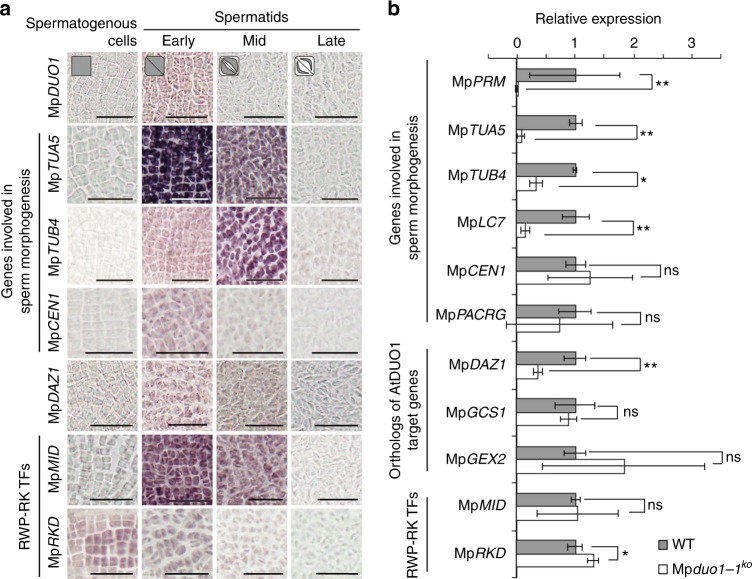
Characterization of potential DUO1 target genes in Marchantia. a RNA in situ hybridization of genes involved in sperm morphogenesis, the ortholog of AtDAZ1 and AtDAZ2 genes, and RWP-RK transcription factors during spermatogenesis (see Supplementary Figure 2 for sense-probe controls). Bars, 25 μm. Cell shape schematics (top) represent the developmental stages referred to in Fig. 1c. b Expression levels of genes involved in sperm morphogenesis, the ortholog of AtDUO1 target genes, and RWP-RK transcription factors in antheridiophores of WT (gray bars) and Mpduo1-1ko (white bars). The expression of each gene in WT is set to 1. Error bars indicate mean ± SD; n = 3 (*p < 0.05; **p < 0.01; ns, not significant; Student’s t-test)
Lack of impact of RWP-RK transcription factors in sperm
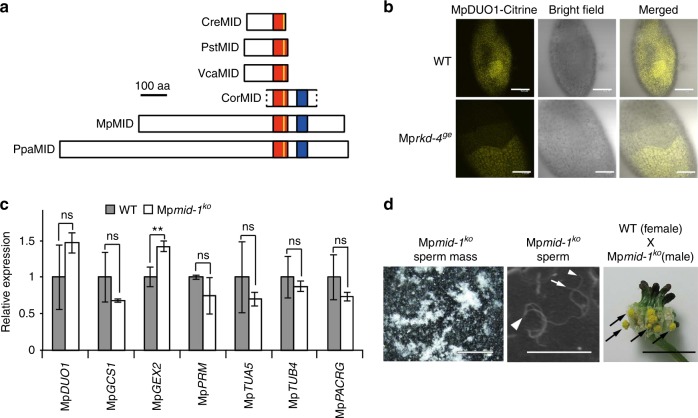
The RWP-RK domain containing transcription factors MpRKD and MpMID (also known as MpRWP2; Fig. 6a) are also expressed during male gametogenesis14,32,33. MpRKD expression is not confined to antheridium development14,32,33 (Supplementary Figure 5a). In antheridia, MpRKD expression occurs earlier than MpDUO1 expression and does not require MpDUO1 (Fig. 5 and Supplementary Figs. 2, 5b). In Mprkd-4ge mutant, some sperm progenitors express MpDUO1 (Fig. 6b and Supplementary Figure 6a) and generate fertile sperm32. Altogether, these results suggest that MpRKD plays a minor role in spermatogenesis compared with MpDUO1. MpMID is co-expressed with MpDUO1 (Fig. 5a and Supplementary Figs. 2, 5a), but MpDUO1 is not required for MpMID expression (Fig. 5b and Supplementary Figure 5b). Conversely, the loss of MpMID does not affect the expression of MpDUO1 or other sperm-cell-specific genes and, furthermore, male Mpmid-1ko mutants are fertile (Fig. 6c, d, Supplementary Figure 6b, c, and Supplementary Movies 1, 2). A MID ortholog is absent from Arabidopsis32–34. Taken together, these suggest that MID does not have a major role in male gametogenesis in land plants, in sharp contrast to its central role in sperm differentiation in the multicellular green alga Volvox carteri35,36, which belongs to distant relatives of land plants (Chlorophyta, Fig. 1a).
Fig. 6.
Characterization of two RWP-RK transcription factor genes, MpMID and MpRKD, in Marchantia. a Schematic diagrams of Volvocine MID proteins and proteins of MID/RKD(C) subfamily33 from Streptophytes. The red box represents the conserved RWP-RK domain while the yellow line indicates the RWP-RK motif. A blue box represents a conserved domain outside of the RWP-RK domain33, which corresponds to motif #12 of ref. 34 CreMID: Chlamydomonas reinhardtii (DQ355812), PstMID: Pleodorina starrii (BAF42661), VcaMID: Volvox carteri (ADI46915), CorMID: Coleochaete orbicularis (GBSL01000368 [partial sequence]), MpMID: Marchantia polymorpha (KU987912), PpaMID: Physcomitrella patens (XM_001779010). MID orthologs are not found in Arabidopsis thaliana and in Chara braunii. b The expression pattern of MpDUO1-Citrine fusion protein in developing antheridia of MpDUO1-Citrineki (WT) and MpDUO1-Citrineki; Mprkd-4ge (Mprkd-4ge) plants. Note that some sectors of spermatogenous cells differentiate to sperm while others fail in Mprkd mutant antheridia32. c Quantitative real-time PCR analysis of MpDUO1, MpGCS1, MpGEX2, and genes involved in sperm morphogenesis in stage 4 antheridiophores of WT (gray bars) and Mpmid-1ko (white bars). The relative expression of each gene in Mpmid-1ko was compared with WT. MpACT1 was used as an internal control. Error bars indicate mean ± SD; n = 3. (**p < 0.01; ns, not significant; Student’s t-test). d The sperm masses (white) discharged from mature antheridiophores of Mpmid-1ko into water (left) and the scanning electron micrograph of sperm of Mpmid-1ko (middle). White arrowhead and arrows indicate the nucleus and flagella, respectively. The right panel shows mature sporangia (black arrows) on a WT archegoniophore a month after crossing with sperm from Mpmid-1ko. Scale bars, 100 µm (b), 500 μm (d, left), 15 μm (d, middle), and 5 mm (d, right)
Origin of DUO1 in freshwater green algae
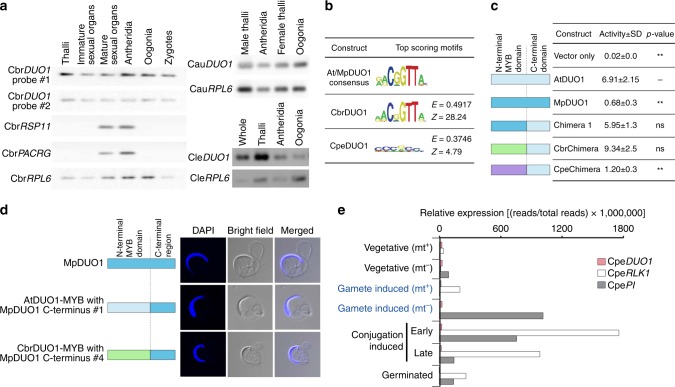
To further explore the origin of DUO1, we extended our search for DUO1 orthologs in freshwater green algae, the sister groups of land plants (Fig. 1a). We did not find DUO1 orthologs in the most basal algae Mesostigma and Klebsormidium, which do not differentiate sperm37. In contrast, stoneworts (Charophyceae) produce motile bi-flagellate sperm similar to those of liverworts37. In stoneworts, we did not find the ortholog of MID but discovered that they express the DUO1 ortholog (Fig. 1b) with a pattern broader than the sperm lineage-specific expression in land plants (Fig. 7a and Supplementary Figure 7). CbrDUO1 from the stonewort Chara braunii binds to the same motif as other DUO1 proteins, trans-activates the DUO1 target HTR10 in planta (CbrChimera; Fig. 7b, c), and complements the Marchantia Mpduo1-1ko mutant as efficiently as AtDUO1 (AtDUO1/MpDUO1 and CbrDUO1/MpDUO1 chimeras; Figs. 3a, 7d and Supplementary Figure 3e). These results suggest that an ancestral DUO1 gene first evolved in a stonewort ancestor and that DUO1 maintained its DNA-binding specificity and function during evolution leading to the land plants. It will be of particular interest to know whether an ortholog of DUO1 exists in Coleochate, which belongs to Charophyte and produces motile bi-flagellate sperm similar to those of liverworts37. But this lack of identification may be the result of limited availability of genome and transcriptome to vegetative cells only.
Fig. 7.
Characterization of green algal DUO1 orthologs. a Expression profiles of DUO1 orthologs from three Chara species, C. braunii (Cbr), C. australis (Cau), and C. leptospora (Cle) representing two distant clades (subgenera)41. Ribosomal protein L6 (RPL6) was used as the internal control. The expression of two flagella component genes (RSP11 and PACRG) was also analyzed in C. braunii. CbrDUO1 gene structure and amplified probed regions are shown in Supplementary Figure 7a. The whole untrimmed images are shown in Supplementary Figure 7b. Oogonia contain the egg. Antheridia contain developing sperm. Thalli are somatic vegetative tissue. b Position weight matrix representations of the top-scoring 8-mer DNA sequences bound by different MYB domains on a protein-binding DNA microarray. CpeDUO1 is DUO1 from Closterium peracerosum-strigosum-littorale complex (conjugating green alga). Numbers denoted on the right side of each motif represent the motif E- and Z-scores19. c In vivo transcriptional activation potentials of green algal DUO1 chimeras. DUO1 transcriptional activation potentials were measured by relative luciferase activity (right) (n = 4; **p < 0.01; ns, not significant; Student’s t-test). Schematic diagrams of constructs are color-coded light blue (AtDUO1), dark blue (MpDUO1), green (CbrDUO1), and purple (CpeDUO1). d The morphology of mature sperm of Mpduo1-1ko complemented by the expression of WT MpDUO1 (top), AtDUO1-MYB domain with MpDUO1 C-terminus chimera (middle), and CbrDUO1-MYB domain with MpDUO1 C-terminus chimera (bottom). The blue signals indicate the DAPI stained nuclei. Constructs are color-coded as in c. e Expression profiles of CpeDUO1 and sexual reproduction-specific CpeRLK1 and CpePI in Closterium peracerosum-strigosum-littorale complex. Stages equivalent to gametes in anisogamous plants are highlighted in blue. Relative expression ([reads per total sequenced reads] × 1,000,000) was used
Sperm and egg differentiation were lost and replaced by differentiation of mating types in conjugating green algae (Zygnematophyceae) (Fig. 1a). In contrast to stoneworts and land plants, DUO1 orthologs in conjugating green algae have accumulated insertions and substitutions in the DNA-binding domain (Fig. 1b, Supplementary Figure 4, and Supplementary Table 2). Consistent with the loss of sperm differentiation in Closterium peracerosum-strigosum-littorale complex, CpeDUO1 does not bind to the same motif as other DUO1 orthologs in vitro, does not efficiently trans-activate the DUO1 target HTR10 in planta (CpeChimera; Fig. 7b, c), and fails to complement the Marchantia Mpduo1-1ko mutant (CpeDUO1/MpDUO1 chimera; Fig. 3a and Supplementary Figure 3e). In addition, CpeDUO1 is barely expressed in gametes (Fig. 7e). Altogether, these data suggest that DUO1 function is not conserved in conjugating green algae, consistent with the notion that DUO1 was essential for sperm differentiation in algal ancestors of land plants.
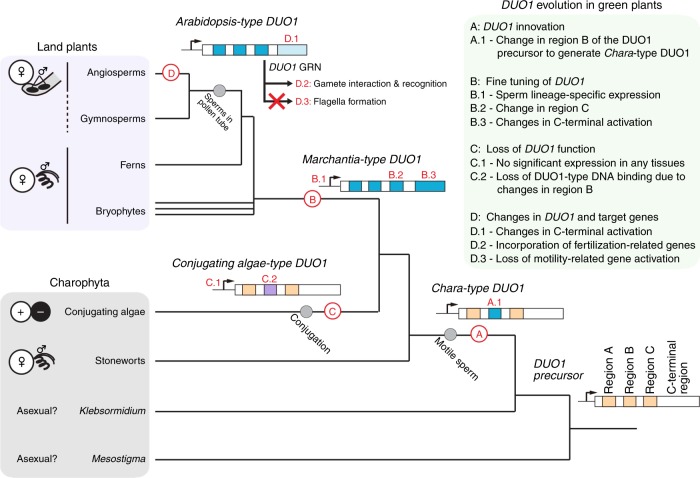
In conclusion, we show that DUO1 orthologs are essential for sperm differentiation in the land plant lineage. This supports the evolution of DUO1-type MYB TFs as a major event leading to the emergence and maintenance of sperm differentiation in the land plant lineage. We propose that a key change in region B took place in the common ancestor of stoneworts and land plants, defining the distinct DNA sequence-specificity and function of the ancestral DUO1 (Fig. 8). The change in region B event was followed by positive selection for cis-elements in the DUO1 promoter that led to sperm lineage-specific expression in the common ancestor of land plants. As land plants evolved, DUO1 retained a central control over this differentiation program, with gradual reconfiguration of downstream target genes, leading to sperm with distinct cytological features like bi-flagellate motile sperm in bryophytes and non-flagellate immotile sperm delivered by pollen tube in angiosperms. It will be of interest to determine if a comparable parsimonious mode of evolution took place in animals, which also show remarkable diversity of sperm morphologies.
Fig. 8.
A model for DUO1 evolution during green plant evolution. DUO1 arose from a pre-existing MYB TF in an ancestor of stoneworts by acquiring a DUO1-type feature in region B of its MYB domain (A.1). DUO1 innovation in ancestral plants promoted a specific program of differentiation of male gametes. Before the emergence of land plants, the sperm lineage-specific expression of DUO1 was acquired (B.1) and a change in region C occurred to refine DNA-binding specificity (B.2), which has been conserved among land plants. The DUO1 C-terminal domain also underwent large changes, presumably adapting its transcription activation function for individual species during evolution (B.3). In conjugating green algae, the DUO1 ortholog is no longer expressed in the sperm lineage (C.1) and has accumulated mutations in the MYB domain (C.2), resulting in a loss of DUO1 function. During land plant evolution, the DUO1 C-terminal activation domain has been modified, establishing lineage-specific activity (D.1). In seed plants, DUO1 retained its DNA-binding specificity and other male gamete-related genes, such as those important for fertilization, were recruited to (D.2) and motility-related genes were removed from (D.3) the set of genes controlled by DUO1, leading to significant rewiring of the DUO1 regulatory network. Modes of sexual reproduction are shown on the left37. Sexual reproduction is unknown in Mesostigma, and Klebsormidium. Note that some gymnosperms (Ginkgo and cycads) have flagellate sperm cells delivered by a pollen tube
Methods
Plant materials and growth conditions
Male and female accessions of Marchantia polymorpha L., Takaragaike (Tak)-1 and Tak-2, respectively, were used as wild-type plants38. Plants were cultured on half-strength Gamborg’s B5 medium containing 1% sucrose and 1.3% agar under continuous light condition from white fluorescent tubes (50 to 60 µmol m−2 s−1) at 22 °C. To induce the sexual reproduction, thalli developed from gemmae on half-strength Gamborg’s B5 medium were transferred to soil under continuous white light supplemented with far-red light irradiation39. Arabidopsis thaliana plants were grown on soil in growth chambers at 24 °C under continuous illumination. duo1-4/+ has been described previously12. Male plants of Haplomitrium minioides were collected in Higashi-Hiroshima City (Hiroshima Prefecture, Japan) on 24 May, 2012. Plants of Chara braunii, C. australis, and C. leptospora (=C. globularis40,41) that have been maintained in culture in Sakayama lab (Kobe University, Japan)41 were used. The strains of heterothallic Closterium peracerosum-strigosum-littorale complex used in this work were NIES-67 (mt+) and NIES-68 (mt-), which were obtained from the National Institute for Environmental Studies, Ibaraki, Japan. The respective vegetative cells were cultured in nitrogen-supplemented medium (C medium; http://mcc.nies.go.jp/02medium-e.html#c) under a 16 h light/8 h dark cycle.
Plasmid constructions and plant transformation
The genome regions of MpDUO1 and MpMID used for plasmid constructions in this study are shown in Supplementary Figs. 1b and 6b, respectively. To generate the Mpduo1-1ko and Mpmid-1ko, the upper and lower arms were amplified from Tak-1 genomic DNA and were cloned into the PacI and AscI sites, respectively, of the pJHY-TMp1 vector42. To generate the MpDUO1-Citrineki, the upper and lower arms were amplified from Tak-1 genomic DNA and were cloned into the AscI and PacI sites, respectively, of pJHY-TMp1-Cit in which a Citrine ORF was cloned into the HindIII site of pJHY-TMp1 vector. The resultant plasmids were introduced into sporelings derived from crosses between Tak-1 and Tak-2. Screening for gene-targeted lines was performed by genomic PCR42. A single gemma from each T1 line was isolated to establish the line.
To generate Mprkd mutants in MpDUO1-Citrineki background, RKD locus was edited using the CRISPR-Cas9 system43. Candidates were selected on the basis of gemma cup phenotype32,33 and PCR products with genomic DNA as the template were analyzed for each candidate to confirm mutations. Several male mutant lines with phenotypes in antheridia32 were chosen for analysis of MpDUO1-Citrine expression.
To construct proMpDUO1:GUS, the genomic fragment of the upstream region of MpDUO1 (proMpDUO1-1) was amplified from Tak-1 genomic DNA and cloned into the EcoRI site of pENTR1A vector (Life Technologies) and then transferred to pMpGWB30444 through the Gateway technology (Life Technologies). The resultant plasmid was introduced into regenerated thalli of Tak-1 and Tak-2. To construct proMpDUO1:AtDUO1, the 5′-upstream region (proMpDUO1-2) and 3′-downstream region (MpDUO1 3′ region) of MpDUO1 were amplified from Tak-1 genomic DNA. The proMpDUO1-2 was cloned between SalI and BamHI sites and MpDUO1 3′ region was cloned in EcoRV site, respectively, of pENTR1A to generate pENTR1A_proMpDUO1_3′ MpDUO1. AtDUO1 ORF was inserted between BamHI and NotI sites of pENTR1A_proMpDUO1_3′ MpDUO1 and the proMpDUO1:AtDUO1:3′MpDUO1 fragment was transferred into pMpGWB30144 through the Gateway technology. To construct proMpDUO1:MpDUO1 for MpDUO1-1ko complementation, the genomic fragment containing the 5′-upstream region and coding region of MpDUO1 was amplified from Tak-1 genomic DNA. The fragment was cloned into pDONR221 (Life Technologies), then into pMpGWB10144 through the Gateway technology (Life Technologies). The resultant plasmids were introduced into regenerated thalli of Mpduo1-1ko. T1 lines were selected on the half-strength Gamborg’s B5 medium containing 0.5 µM chlorsulfuron and a single gemma from each T1 line was isolated to establish the line.
For Arabidopsis complementation assay in Atduo1-4/+, proAtDUO1:AtDUO1-Clover, proAtDUO1:MpDUO1-Clover and proAtDUO1:Chimera1 were inserted into the multisite Gateway T-DNA destination vector pAlligatorR4345 through the Gateway technology (Life Technologies). AtDUO1 cDNA synthesized from Col-0 pollen RNA as well as MpDUO1 cDNA synthesized from Tak-1 young antheridiophore RNA were cloned into pDONR221 with attB1 and attB2 sites. The additional Gateway technology vectors used in this study are pENTR_2-r3_Clover45, pENTR_4-1r_proAtDUO19,10, and pAlligatorG43_proHTR10:HTR10-mCherry45.
For promoter conservation assay, proMpDUO1:H2B-Clover and proAtDUO1:H2B-Clover were inserted into the multisite Gateway T-DNA destination vector pAlligatorR43 and pAlligatorG4345, respectively. Promoters of MpDUO1 (proMpDUO1-2) and AtDUO1 were cloned into pDONR-P4P1r (Life Technologies). pENTR221-H2B vector was previously described45. The mRuby2 clone (Addgene) was introduced into pDONR-P2rP3 (Life Technologies).
For transient luciferase assay, the firefly and Renilla luciferase constructs built with pK7m24GW346 and pB2GW747, respectively, were used9. Chimera 1 to 7 DNA fragments with attB1 and attB2 sites were generated by fusion PCR method48, and then cloned into pDONR221 vector (Life Technologies) through the Gateway technology. These entry clones as well as At DUO1 and Mp DUO1 cDNA entry clones were then recombined into pB2GW7 to generate Pro35S constructs for the transient luciferase assay.
For the protein-binding DNA microarray assay, synthesized Escherichia coli-codon-optimized fragments (ThermoFisher Scientific) encoding MYB domains of AtDUO1, MpDUO1, Chimera 4, 5, 6 as well as MpR2R3-MYB21 and KflMYB were cloned into pDONR221 and then transferred to the destination vector pMAL-C2 vector (New England Biolabs) through Gateway technology, generating the in-frame fusion of Maltose Binding Protein (MBP) and MYB DNA-binding domain.
Transgenic Arabidopsis plants were generated using the floral dip method49 and T1 transgenic plants were screened based on each selection marker of the construct. All oligonucleotides and synthesized fragment sequences used for this study are listed in Supplementary Table 3.
Histochemical GUS staining
Histochemical staining for GUS activity was performed by a common procedure50 with some modifications. The thalli, antheridiophores, archegoniophores, and sporophytes of proMpDUO1:GUS-expressing plants were fixed in 90% (v/v) acetone, vacuum-infiltrated and incubated at 37 °C overnight in the GUS assay solution containing 100 mM sodium phosphate buffer (pH 7.2), 5 mM potassium-ferrocyanide, 5 mM potassium-ferricyanide, 0.1% (v/v) Triton X-100 and 0.5 mg ml−1 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-Gluc). Chlorophyll in the tissue were removed by incubation in 70% (v/v) ethanol.
RNA in situ hybridization
Probe and tissue preparation and hybridization were performed as described14. A probe fragment for each gene was amplified from Tak-1 cDNA with a set of gene-specific primers (Supplementary Table 3) and was cloned into the pCR-BluntII-TOPO vector (Life Technologies). DIG-labeled anti-sense and sense RNA probes were synthesized with a DIG RNA Labeling kit (SP6/T7) (Roche) according to the manufacturer’s instructions. Antheridiophore receptacles of Tak-1 at stage 3 to 414 were fixed in a solution containing 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M phosphate buffer, pH 7.0, and 0.05% Triton X-100, dehydrated and embedded in paraffin. Eight-µm sections were made with a microtome, applied to an APS-coated glass slide, and then deparaffinized and rehydrated. They were treated with 1 µg ml−1 of proteinase K (ThermoFisher Scientific) in 100 mM Tris-HCl, pH 8.0, 50 mM EDTA, pH 8.0 at 37 °C for 30 min, subsequently fixed in 4% (w/v) paraformaldehyde in PBS (7 mM Na2HPO4, 3 mM NaH2PO4, 130 mM NaCl) for 10 min, and treated with 0.5% (v/v) acetic anhydride in 100 mM triethanolamine for 10 min. Sections were incubated at 55 °C for 2 h in pre-hybridization buffer [50% formamide (w/v), 5 × SSC, 40 µg ml−1 salmon sperm DNA] and then in the same buffer with 100 ng of probes for more than 16 h. After treatment with 50 µg ml−1 of RNase A (SIGMA) in RNase buffer (10 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM EDTA) at 37 °C for 30 min, sections were washed in 0.2 × SSC (30 mM NaCl, 3 mM sodium citrate, pH 7.0) at 55 °C for 1 h, incubated with 1% (w/v) Blocking Reagent (Roche) in buffer 1 (100 mM Tris-HCl, pH 7.5, 150 mM NaCl) for 1 h and then incubated with 1/1000 diluted Anti-Digoxigenin-AP, Fab fragments (Roche) with buffer 1 containing 1% (w/v) blocking reagent for 1 h. The slides were subsequently washed three times with buffer 1 for 10 min each, rinsed with buffer 2 (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2) for 5 min and then covered with NBT/BCIP (Roche) diluted 1/125 in buffer 2. After incubation at 22 °C for more than 16 h in the dark, the reaction was stopped by immersing the slides in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
Confocal microscopy
The accumulation of MpDUO1-Citrine was observed in isolated antheridia and sperm of MpDUO1-Citrineki plant. The Citrine fluorescence was detected in a range from 525 to 565 nm with confocal laser scanning microscopy (FV-1000; Olympus and LSM780; Zeiss) after excitation at 515 nm. An antheridiophore of the MpDUO1-Citrineki plant was hand-sectioned with a blade and immersed in 1 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI) solution. Antheridia were isolated from the sections and observed immediately under a confocal microscope (FV-1000, Olympus) with the following setting: MpDUO1-Citrine was excited by 488-nm laser and detected by a GaAsP detector with 535−565-nm window; DAPI was excited by 405-nm laser and detected by a photomultiplier tube with 425−475-nm window.
Clover and mRuby2 in the transgenic Arabidopsis pollens were excited at 488 and 561 nm, respectively and detected in a range from 495 to 540 and from 566 to 600 nm, respectively, with confocal laser scanning microscopy (LSM780; Zeiss). In brief, 3-4 open flowers were collected in a microfuge tube containing 300 μl of the solution [0.1 M sodium phosphate (pH 7.0), 1 mM EDTA, 0.1% Triton X-100]. After brief vortexing and centrifugation, 15 μl of the pollen pellet was transferred to a microscope slide and imaged12.
Feulgen staining of antheridia
Feulgen staining51 was performed with some modifications. The antheridiophores receptacles of Tak-1 at stage 514 were fixed overnight in 3:1 mixture of ethanol and glacial acetic acid at 4 °C overnight. After fixation, the equal volume of ethanol was added and the samples were incubated for 1 h. Then the samples were hydrated in graded ethanol solutions. The samples were rinsed three times with distilled water for 15 min each and hydrolyzed in 5 N HCl for 1 h. After hydrolysis, the samples were rinsed three times with distilled water for 5 min each and stained with Schiff’s reagent (Sigma-Aldrich) for 2 h. The samples were rinsed twice for 15 min each with distilled water and were dehydrated in graded ethanol solutions and 100% ethanol. The 100% ethanol was exchanged with the fresh one at hourly intervals until it remained colorless after the exchange. Then LR White resin (Sigma-Aldrich) was added to make a 1:1 mixture of 100% ethanol and LR White and the samples were left for 1 h at room temperature. Then the mixture was replaced with pure LR White and left at room temperature overnight. The antheridia were manually dissected from antheridiophores and placed in fresh LR White on a standard glass microscope slide and a cover glass was gently lowered over the antheridia in LR White. The samples were incubated at 60 °C overnight. The cover glass can be carefully removed from the polymerized LR White. The fluorescence was detected at 535 nm and longer with LSM510 META Confocal Imaging System (Zeiss) after excitation at 488 nm by an argon laser.
Nuclear shape quantification
Fiji package52 was used to quantify shapes [circularity (4π*area/perimeter2), aspect ratio (major_axis/minor_axis), and solidity (area/convex_area)] of Feulgen-stained Marchantia antheridia nuclei obtained from Z-projected confocal images. In brief, nuclear images were smoothened through Gaussian blur function, followed by threshold adjustment to capture the shape of nuclei. Nuclear images which are either partial or overlapping with others were excluded from quantification.
TEM analysis of antheridia
The antheridiophore receptacles of Tak-1 at stage 414 were fixed with 2% each of paraformaldehyde and glutaraldehyde in 0.05 M cacodylate buffer pH 7.4 at 4 °C overnight. After fixation, the samples were washed three times with 0.05 M cacodylate buffer for 30 min each, and were post-fixed with 2% osmium tetroxide in 0.05 M cacodylate buffer at 4 °C for 3 h. The samples were dehydrated in graded ethanol solutions then in 100% ethanol. The samples were infiltrated with propylene oxide twice for 30 min each and were put into a 7:3 mixture of propylene oxide and resin, Quetol-651 (Nissin EM Co.) for 1 h, then the mixture was kept in a tube without cap overnight to volatilize propylene oxide. The samples were transferred to a fresh resin and were polymerized at 60 °C for 48 h. The polymerized resins were ultra-thin sectioned at 80 nm with a diamond knife using Ultracut UCT (Leica) and the sections were mounted on copper grids. They were stained with 2% uranyl acetate at room temperature for 15 min, and then they were washed with distilled water followed by being secondary-stained with Lead stain solution (Sigma-Aldrich) at room temperature for 3 min. The grids were observed by JEM-1400Plus (JEOL Ltd.) at an acceleration voltage of 80 kV. Digital images were taken with a CCD camera, VELETA (Olympus).
Immunofluorescence staining
The fixation and permeabilization of antheridia were carried out essentially according to previous report53. Isolated spermatid cells were incubated with a polyclonal anti-centrin antibody against a recombinant protein corresponding to a full-length centrin from a brown alga Scytosiphon lomentaria (a gift from Dr. Taizo Motomura, Hokkaido University, Japan) diluted 1:500 with PBS [137 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, and 1.47 mM KH2PO4, pH 7.4] in a moist chamber for 90 min at 37 °C. After a PBS wash, incubation with a monoclonal anti α-tubulin (T5168, Sigma) diluted 1:500 with PBS was performed for 60 min at 37 °C. After a wash again with PBS, the samples were incubated for 60 min at 37 °C with an equal mixture of Alexa 488-conjugated goat anti-rabbit IgG (H + L) (A11034, Invitrogen) diluted 1:200 with PBS and Alexa 568-conjugated goat anti-mouse IgG (H + L) (A11031, Invitrogen) diluted 1:200 with PBS. After washing with PBS for 10 min, the nuclei were stained with 1 μg ml−1 DAPI in PBS. Slides were mounted using Vectashield (Vector Laboratory) and observed using a confocal laser scanning microscope (FV-1000, Olympus).
Quantitative RT-PCR analysis
Total RNA was extracted with an RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s protocol and the quality and quantity of the resultant RNA were evaluated using a NanoDrop 2000c spectrophotometer (ThermoFisher Scientific). One μg of total RNA was reverse-transcribed in a 20 μl reaction mixture using Transcriptor (Roche). After the reaction, the mixture was diluted with 180 μl of distilled water and 2 μl aliquots were used for PCR in a 10 μl PCR reaction mixture containing 1 μl of 10× Ex Taq buffer, 1 μl of 2 mM dNTPs, 0.4 μl of 10 μM each of primers, and 0.05 μl of Ex Taq DNA polymerase (Takara) for semi-quantitative RT-PCR analysis. The PCR products were separated on a 2.5% (w/v) agarose gel, stained with ethidium bromide, and visualized under UV light. The primers used in these experiments are listed in Supplementary Table 3. For quantitative RT-PCR analysis, the cDNA samples were diluted with 220 µl of distilled water and 2 μl aliquots were amplified with the CFX96 Real-time PCR Detection System (Bio-Rad) using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara). The two-step PCR cycling program was performed according to manufacturer’s protocol. The primers used in these experiments are listed in Supplementary Table 3. MpACT1 was used as an internal control.
Genetic complementation assay on Atduo1 and Mpduo1-1ko
For Arabidopsis complementation assay, mature pollen grains from WT and T3 stable homozygous complemented lines (AtComp, MpComp #3 and #4, and Chimera 1 #1 and #2) were examined by fluorescence microscopy. The frequency of bicellular and tricellular pollen grains was determined by scoring the number of respective pollen grains by DAPI staining. The ability of H3.10 activation was assessed by counting the frequency of pollen grains expressing mRuby2 fluorescence marker in sperm (proAtHTR10:HTR10-mRuby2).
For Marchantia complementation assay, the discharged sperm was directly observed without fixation on a Miniscope TM3000 (HITACHI, Japan) to obtain scanning electron microscopic images. To observe the nuclear morphology of sperm, the discharged sperm were stained with DAPI and observed under a confocal microscope (FV-1000, Olympus) with the following setting: DAPI was excited by 405-nm laser and detected by a photomultiplier tube with 425−475-nm window. Movies of discharged sperm were taken under a microscope BX43 (Olympus) with a dry dark-field condenser U-DCD (Olympus). For quantitative motility analysis, sperm discharged in water was observed under a phase-contrast/DIC microscope and video-recorded at the resolution of 1216 × 960 pixels and at the rate of 7 frames per second (fps) for 10 s by a microscope camera DP26 (Olympus). To track individual sperm, an open-source software ImageJ (ver. 1.51n, the Fiji distribution)52 was used. Movies of sperm were first converted into a sequence of individual frames. Their green-channel images were extracted and converted into 8-bit gray-scale, and their black and white values were inverted. To reduce stationary objects in the background, each image was subtracted with a Z projection of the entire frames. Moving paths of sperm were detected by TrackMate ver. 3.4.254, a plugin bundled with the Fiji distribution of ImageJ, with the following modified parameters: initial threshold, auto; linking max distance, 50 pixel; gap-closing max distance, 50 pixel. Tracks with duration of at least 3 s, or 21 frames, were selected and the length of each track was measured to calculate average speed in µm s-1. Three movies were analyzed to obtain sperm swimming velocity for each genotype, and the total numbers of sperm observed are: n = 132 (WT), n = 193 (MpComp #5), n = 14 (AtComp #9), n = 12 (AtChimera #13), n = 22 (CbrChimera #1) for analysis of fraction of motile sperm; n = 117 (WT), n = 67 (MpComp #5), n = 10 (AtComp #9), n = 8 (AtChimera #13), n = 12 (CbrChimera #1) for analysis of swimming velocity.
To test the fertilization ability, mature antheridiophores were immersed in water and aliquots of sperm suspension were observed for discharged sperm and deposited onto archegoniophore receptacles of Tak-2. Sporophyte and sporangium development was observed about a month after crossing.
Protein-binding DNA microarray assay
Recombinant plasmids harboring MBP-MYB fusions were introduced into the BL21 strain of E. coli, and the expression of recombinant proteins was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 6 h at 25 °C. Pellets corresponding to 25 ml of induced E. coli culture for each construct were stored at −80 °C and resuspended in 1 ml 1× binding buffer prior to DNA-binding assay20. Bacterial lysates were sonicated twice for 30 s, and centrifuged twice at 20,000×g to obtain cleared extracts of soluble proteins.
Second strand of DNA was synthesized in a primer extension reaction with 32 U Thermo Sequenase Polymerase (USB), 163 μM dNTPs, 1.63 μM Cy5-dUTP (GE Healthcare) and 1.17 μM oligonucleotide primer (5′-CAGCACGGACAACGGAACACAGAC-3′) in a 900 μl total volume reaction. DNA microarray was incubated with the mixture in a hybridization oven for 10 min at 85 °C, the temperature gradually reduced up to 60 °C during 30 min and hold at this temperature for 90 min. The slide was then rapidly transferred to wash solution (1× PBS, 0.01% Triton X-100), incubated at 37 °C for 10 min with agitation and rinsed in 1× Phosphate Buffered Saline (PBS) for 3 min at room temperature. The slide was spun dry by centrifugation (1 min at 500 rpm) and scanned at 2 μm resolution in a Agilent’s DNA Microarray Scanner for monitoring the amount of dsDNA. The binding mixture obtained from cleared bacterial lysates was adjusted to 175 μl and to contain 2% milk and 0.89 μg of denatured salmon sperm DNA (ssDNA). Double stranded DNA microarray was incubated with the binding mixture in a humid chamber for 2.5 h at room temperature. Slides were then washed three times PBS-1% Tween 20 (5 min), three times in PBS-0.01% Triton X-100 (5 min) and spun dry by centrifugation. DNA–protein complexes were incubated with 16 μg of Rabbit polyclonal to Maltose Binding Protein (Abcam) in PBS-2% milk for 16 h at room temperature. Slides were washed 3× in PBS-0.05% Tween 20, 3× in PBS-0.01% Triton X-100 (5 min each wash) and dried. Labeling of DNA–protein complexes were performed by incubating the microarrays with 0.4 μg of goat anti-rabbit IgG DyLight 549 conjugated (Pierce) in PBS-2% milk for 3 h at room temperature, followed by the same washes as before and the slides dried for scanning.
We used the nPBM11 design containing 167,773 different oligonucleotide probes19 synthesized in an Agilent’s SurePrint G3 4 × 180k format (Agilent Technologies). DNA microarrays were scanned in a DNA Microarray Scanner at 2-μm resolution and quantified with Feature Extraction 9.0 software (Agilent Technologies). Normalization of probe intensities and calculation of E- and Z-scores of all the possible 8-mers were carried out with the PBM Analysis Suite55. Perl scripts were modified to adapt them to nPBM11 microarray dimensions and to input files generated by Feature Extraction software.
Transient luciferase assay
Agrobacterium-mediated transient transformation of Nicotiana tabacum leaf was carried out as described by Sparkes et al.56 modified as detailed below. Agrobacterium strains were combined as required at an OD600 of 0.1 for reporter and effector vectors and an OD600 of 0.02 for the Renilla luciferase control vector in infiltration media (280 mM d-glucose, 50 mM MES, 2 mM Na3PO4·12H2O, 0.1 mM acetosyringone). 4–6-week-old Nicotiana tabacum plants were grown in greenhouse conditions. Agrobacterium suspensions were taken up in 1 ml syringes and the underside of leaves gently rubbed to remove a small region of the cuticle. The syringe tip was placed at these regions and Agrobacterium suspensions were gently infiltrated. Plants were placed in a growth chamber under normal growth conditions and left for 2 days. Each leaf disc from an infiltrated region was excised using a 9 mm cork-borer and ground to homogeneity in 300 μl of 1× Passive Lysis Buffer (Promega) in a chilled mortar and pestle. Leaf extracts were centrifuged at 16,000×g for 5 minutes at 4 °C to pellet cell debris. Two separate 25 μl aliquots were assayed separately for firefly and Renilla luciferase activities in 100 μl of the respective assay buffer. The firefly luciferase assay buffer (25 mM glycylglycine, 15 mM KPO4 pH 8.0, 4 mM EGTA, 2 mM ATP, 1 mM DTT, 15 mM MgSO4, 0.1 mM CoA, 75 μM luciferin with final pH adjusted to 8.0) and Renilla luciferase assay buffer (1.1 M NaCl, 2.2 mM Na2EDTA, 0.22 M KPO4 pH 5.1, 0.44 mg/ml BSA, 1.43 μM coelenterazine with final pH adjusted to 5.0) were prepared immediately before measurement57. Luminescence was measured in white 96-well plates with a FLUOstar Omega (BMG LABTECH Ltd) microplate reader as relative luminescence units (RLUs) integrated over 10 seconds. Extracts of discs taken from non-infiltrated leaves were assayed to determine mean background RLU values, which were subtracted from those for extracts of infiltrated leaves. Normalized luciferase activity (FLuc/RLuc) was calculated for each extract from the ratio of background subtracted RLUs obtained in firefly luciferase (FLuc) and Renilla (RLuc) luciferase assays.
Sequences, alignment, and phylogenetic tree construction
Sequences of Klebsormidium MYB TFs and DUO1 MYB TFs of conjugating green algae were obtained from published data58–60. The 5′ and 3′ parts of CpeDUO1 cDNA fragment were amplified separately by PCR from cDNA from conjugation induced early stage. Sequences of land plant DUO1, DAZ1, GCS1/HAP2, and GEX2 were obtained from www.phytozome.jgi.doe.gov. Amino acid sequence alignment illustrated in Supplementary Figure 1a was generated by CLC Workbench 7 package (QIAGEN).
RNA preparation and RNA-seq analysis
For the preparation of total RNA from vegetative cultures of Closterium peracerosum-strigosum-littorale complex, mating-type (mt)+ and mt− cells were harvested at 0 (start time of light illumination), 6, 12, and 18 h, respectively. For the preparation of total RNA from mating cultures, vegetative growing cells of the mt+ and mt− were collected, washed three times with nitrogen-depleted medium (MI medium)61, and incubated separately in MI medium (3.6 × 105 cells/72 ml in 300-ml Erlenmeyer flasks) under continuous light for 2, 8, and 24 h (gamete induced mt+ and mt-, respectively). The cells of both mating types, which had been separately cultured in MI medium (at 3.6×105/72 ml in 300-ml Erlenmeyer flasks) for 24 h, were mixed and co-incubated (at 3.6 × 105 each/72 ml in 300-ml Erlenmeyer flasks) for 1, 2, 4, 6, 8, 12, 16, 20, and 24 h (conjugation induced early) and for 48, 72, 96 h (conjugation induced late). For the preparation of total RNA from germinated zygote, zygotes were dried once and incubated in C medium for 12, 24, 48, 72 h under a 16 h light/8 h dark cycle (germinated). The harvested cells were frozen in liquid nitrogen and total RNA was isolated using TRIZOL Plus Kit (Invitrogen, Carlsbad, CA, USA), in accordance with supplier instructions. Paired-end libraries were generated with TruSeq RNA Sample Preparation Kit (Illumina, www.illumina.com), according to manufacturer’s instructions. Sequencing was carried out 76 bps with a Genome Analyzer IIx using standard reagents. All high-quality sequences were de novo assembled with Trinity62. Expression frequency of the contigs was calculated by RSEM63, using all high-quality sequences.
DUO1 DNA-binding motif search
The DUO1 consensus DNA-binding motif (5′-RRCSGTT-3′) generated in this study was used to search the upstream regions of AtDAZ1, AtGCS1/HAP2, and AtGEX2 homologs of some angiosperms, a lycophyte, and some bryophytes by using the dna-pattern tool from Regulatory Sequence Analysis Tools (RSAT) web server (http://www.rsat.eu/)64 with default parameters. Up to 2-kb upstream regions between the putative transcriptional start site and the upstream neighboring gene were collected from Phytozome ver.11 (https://phytozome.jgi.doe.gov/pz/portal.html). The genes used in this experiment are listed in Supplementary Table 4.
Selection pressure analyses
Branch model tests were performed using codeML in PAML65 on DUO1 and S18 MYB domain sequences from genes in Supplementary Table 415,58,59. Default parameters were used, to the exception of cleandata = 0 to prevent removal of codon information around region B. Likelihood ratio tests were performed between models and chi-squared tests were performed to assess statistical significance. Maximum-likelihood tree shown in Supplementary Figure 4 was generated in PhyML 3.166 using the LG + I + G protein substitution model, selected according to ProtTest 367. Full results are listed in Supplementary Table 2.
RT-PCR and Southern hybridization analysis
Gene expression analysis of three Chara species was performed using standard procedures40 with the following modifications. A probe fragment for each gene was amplified from cDNA of each species with a set of gene-specific primers (Supplementary Table 3) and was cloned into the pCR-BluntII-TOPO vector (Life Technologies), which contains SP6 and T7 polymerase-binding sites. RT-PCR was performed with a set of gene-specific primers (Supplementary Table 3). Probe synthesis, hybridization, washing, and detection were performed by using DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) according to the manufacturer’s protocol. Hybridization was performed at 55 °C.
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
We thank M. Endo, S. Akimcheva, B. Jamge, K. Nagao, and L. Collins for technical assistance and suggestions and Dominique Bergmann for critical reading of the manuscript. We thank T. Motomura, C. Nagasato, K. Ura (Hokkaido University, Japan), and K. Kimura (Saga University, Japan) for the anti-centrin antibody, and the Vienna Biocenter Core Facilities for technical help. This work was supported by FWF (I2163-B16 to F.B. and W1238-B20 to S.A.M.) and the Biotechnology and Biological Research Council (BB/N005090 to D.T.); ERA-CAPS EVOREPRO project to DT and FB; Spanish MINECO grant BIO2017-86651-P (AEI/FEDER) to J.F.-Z.; JSPS KAKENHI grant 15K07185 to H. Sa, MEXT grants (25113005, 23370022, and 24657031 to T.A.; 25113001 and 15K21758 to T.Ko. and T.A.; and 221S0002 to H. Sa., H. Se., and T.N.; and 26291081 to T.N., KS, and M.S.); a Grant-in-Aid for the Japan Society for the Promotion of Science Fellows (to A.H. and A.O.); the Kyoto University BRIDGE program (to A.H.); Lise-Meitner fellowship (M1818-B21 to M.B.); and GMI (T.Ka., M.B., O.A., and F.B.).
Author contributions
A.H., T.Ka., F.B., T.A., and D.T. conceived the study; A.H., T.Ka., M.B., M.Z., D.H., M.S., T.A., Y.T., T.T., K.K., and A.O. performed molecular and cellular analyses; I.L.-V. and J.F.-Z carried out DNA microarray experiments; S.M. performed molecular evolution analysis; H.Se., H.Sa., T.N., and Y.S. performed algal genome and expression analyses; K.Y., K.I., R.N., and T.Ko. generated Marchantia genome resources and vectors; T.N., KS, and M.S. identified HmnDUO1; A.H., T.Ka., J.F.-Z., D.T., F.B., and T.A. analyzed the data; T.Ka., F.B., and A.H. wrote the manuscript with critical reviewing and editing by T.A., D.T., M.B., and J.F.-Z.
Data availability
Novel data generated in this study have been deposited at GenBank under the accessions: MpDUO1 (LC172177), MpR2R3-MYB07 (KX683859), MpR2R3-MYB21 (KX683860) MpACT1 (LC172182) MpCEN1 (LC379265) MpTUA5 (LC172181) MpDAZ1 (LC172178) MpGCS1 (LC172179) MpGEX2 (LC172180) HmnDUO1 (LC379264) HmnACT1 (LC379378) CbrDUO1 (LC199499), CauDUO1 (LC221833), CleDUO1 (LC221832), CbrRSP11 (LC382020) CbrPACRG (LC382019) CbrRPL6 (LC382018) CauRPL6 (LC382017) CpeDUO1 (LC176570). Previously reported sequences used in this study are available at GenBank under the accession: MpPACRG (LC102460), MpPRM (LC102462) MpTUB4 (KM096548) MpLC7 (LC102461), MpMID/RWP2 (KU987912) CleRPL6 (AB035569), CpeRLK1 (AB920609) CpePI (AB012698). All other data are available from the authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Asuka Higo, Tomokazu Kawashima.
Contributor Information
Frédéric Berger, Email: Frederic.Berger@gmi.oeaw.ac.at.
Takashi Araki, Email: taraqui@lif.kyoto-u.ac.jp.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-07728-3.
References
- 1.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 2.Tschopp P, Tabin CJ. Deep homology in the age of next-generation sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20150475. doi: 10.1098/rstb.2015.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell G. The evolution of anisogamy. J. Theor. Biol. 1978;73:247–270. doi: 10.1016/0022-5193(78)90189-3. [DOI] [PubMed] [Google Scholar]
- 4.Charlesworth B. The population genetics of anisogamy. J. Theor. Biol. 1978;73:347–357. doi: 10.1016/0022-5193(78)90195-9. [DOI] [PubMed] [Google Scholar]
- 5.Parker GA. The sexual cascade and the rise of pre-ejaculatory (Darwinian) sexual selection, sex roles, and sexual conflict. Cold Spring Harb. Perspect. Biol. 2014;6:a017509. doi: 10.1101/cshperspect.a017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker GA, Baker RR, Smith VG. The origin and evolution of gamete dimorphism and the male-female phenomenon. J. Theor. Biol. 1972;36:529–553. doi: 10.1016/0022-5193(72)90007-0. [DOI] [PubMed] [Google Scholar]
- 7.Wong JL, Johnson MA. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 2010;20:134–141. doi: 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. U. S. A. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg M, et al. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell. 2011;23:534–549. doi: 10.1105/tpc.110.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownfield L, et al. A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 2009;5:e1000430. doi: 10.1371/journal.pgen.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotman N, et al. A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 2005;15:244–248. doi: 10.1016/j.cub.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Borg M, et al. An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell. 2014;26:2098–2113. doi: 10.1105/tpc.114.124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman JL, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304 e215. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Higo A, et al. Transcriptional framework of male gametogenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2016;57:325–338. doi: 10.1093/pcp/pcw005. [DOI] [PubMed] [Google Scholar]
- 15.Du H, et al. The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci. Rep. 2015;5:11037. doi: 10.1038/srep11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimamura M. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 2016;57:230–256. doi: 10.1093/pcp/pcv192. [DOI] [PubMed] [Google Scholar]
- 17.Durand EJ. The development of the sexual organs and sporogonium of Marchantia polymorpha. Bull. Torre. Bot. Club. 1908;35:16. doi: 10.2307/2485335. [DOI] [Google Scholar]
- 18.Durbarry A, Vizir I, Twell D. Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 2005;137:297–307. doi: 10.1104/pp.104.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl Acad. Sci. USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godoy M, et al. Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J. 2011;66:700–711. doi: 10.1111/j.1365-313X.2011.04519.x. [DOI] [PubMed] [Google Scholar]
- 21.Buschmann H, Holtmannspotter M, Borchers A, O’Donoghue MT, Zachgo S. Microtubule dynamics of the centrosome-like polar organizers from the basal land plant Marchantia polymorpha. New Phytol. 2016;209:999–1013. doi: 10.1111/nph.13691. [DOI] [PubMed] [Google Scholar]
- 22.Yamato KT, et al. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl Acad. Sci. USA. 2007;104:6472–6477. doi: 10.1073/pnas.0609054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda K, Ikeda T, Morikawa K, Kamiya R. Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil. Cytoskelet. 2007;64:814–821. doi: 10.1002/cm.20225. [DOI] [PubMed] [Google Scholar]
- 24.Lorenzetti D, Bishop CE, Justice MJ. Deletion of the Parkin coregulated gene causes male sterility in the quaking(viable) mouse mutant. Proc. Natl Acad. Sci. USA. 2004;101:8402–8407. doi: 10.1073/pnas.0401832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagimachi R, et al. Production of fertile offspring from genetically infertile male mice. Proc. Natl Acad. Sci. USA. 2004;101:1691–1695. doi: 10.1073/pnas.0307832100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiBella LM, Sakato M, Patel-King RS, Pazour GJ, King SM. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell. 2004;15:4633–4646. doi: 10.1091/mbc.e04-06-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klink VP, Wolniak SM. Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell. 2001;12:761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 2006;8:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- 29.von Besser K, Frank AC, Johnson MA, Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. doi: 10.1242/dev.02683. [DOI] [PubMed] [Google Scholar]
- 30.Engel ML, Holmes-Davis R, McCormick S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005;138:2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedry J, et al. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 2017;168:904–915 e910. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koi S, et al. An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr. Biol. 2016;26:1775–1781. doi: 10.1016/j.cub.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Rovekamp M, Bowman JL, Grossniklaus U. Marchantia MpRKD Regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 2016;26:1782–1789. doi: 10.1016/j.cub.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 34.Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014;65:5577–5587. doi: 10.1093/jxb/eru261. [DOI] [PubMed] [Google Scholar]
- 35.Geng S, De Hoff P, Umen JG. Evolution of sexes from an ancestral mating-type specification pathway. PLoS Biol. 2014;12:e1001904. doi: 10.1371/journal.pbio.1001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T. Males evolved from the dominant isogametic mating type. Curr. Biol. 2006;16:R1018–R1020. doi: 10.1016/j.cub.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 37.McCourt RM, Delwiche CF, Karol KG. Charophyte algae and land plant origins. Trends Ecol. Evol. 2004;19:661–666. doi: 10.1016/j.tree.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 2008;49:1084–1091. doi: 10.1093/pcp/pcn085. [DOI] [PubMed] [Google Scholar]
- 39.Chiyoda S, Ishizaki K, Kataoka H, Yamato KT, Kohchi T. Direct transformation of the liverwort Marchantia polymorpha L. by particle bombardment using immature thalli developing from spores. Plant Cell Rep. 2008;27:1467–1473. doi: 10.1007/s00299-008-0570-5. [DOI] [PubMed] [Google Scholar]
- 40.Tanabe Y, et al. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl Acad. Sci. USA. 2005;102:2436–2441. doi: 10.1073/pnas.0409860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakayama H, et al. Taxonomic reexamination of Chara globularis (Charales, Charophyceae) from Japan based on oospore morphology and rbcL gene sequences, and the description of C. leptospora sp. nov. J. Phycol. 2009;45:917–927. doi: 10.1111/j.1529-8817.2009.00700.x. [DOI] [PubMed] [Google Scholar]
- 42.Ishizaki K, Johzuka-Hisatomi Y, Ishida S, Iida S, Kohchi T. Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci. Rep. 2013;3:1532. doi: 10.1038/srep01532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugano SS, et al. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55:475–481. doi: 10.1093/pcp/pcu014. [DOI] [PubMed] [Google Scholar]
- 44.Ishizaki K, et al. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS ONE. 2015;10:e0138876. doi: 10.1371/journal.pone.0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawashima T, et al. Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. eLife. 2014;3:e04501. doi: 10.7554/eLife.04501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant. Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Karimi M, Inze D, Depicker A. GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant. Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 48.Horton RM. In vitro recombination and mutagenesis of DNA: SOEing together tailor-made genes. Methods Mol. Biol. 1993;15:251–261. doi: 10.1385/0-89603-244-2:251. [DOI] [PubMed] [Google Scholar]
- 49.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 50.Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T. Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J. Plant. Res. 2012;125:643–651. doi: 10.1007/s10265-012-0477-7. [DOI] [PubMed] [Google Scholar]
- 51.Braselton JP, Wilkinson MJ, Clulow SA. Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem. 1996;71:84–87. doi: 10.3109/10520299609117139. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimamura Masaki. Plant Microtechniques and Protocols. Cham: Springer International Publishing; 2015. Whole-Mount Immunofluorescence Staining of Plant Cells and Tissues; pp. 181–196. [Google Scholar]
- 54.Tinevez JY, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 55.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 57.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
- 58.Cooper, E. & Delwiche, C. Green algal transcriptomics for phylogenetics and comparative genomics. figshare, 10.6084/m6089.figshare.1604778 (2016).
- 59.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ju C, et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants. 2015;1:14004. doi: 10.1038/nplants.2014.4. [DOI] [PubMed] [Google Scholar]
- 61.Ichimura T. Sexual cell division and conjugation-papilla formation in sexual reproduction of Closterium strigosum. In Proceedings of the Seventh International SeaweedSymposium (ed. Nishizawa, K) 208–214 (University of Tokyo Press, Tokyo, 1971).
- 62.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Medina-Rivera A, et al. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015;43:W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 66.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 67.Darriba Diego, Taboada Guillermo L., Doallo Ramón, Posada David. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Novel data generated in this study have been deposited at GenBank under the accessions: MpDUO1 (LC172177), MpR2R3-MYB07 (KX683859), MpR2R3-MYB21 (KX683860) MpACT1 (LC172182) MpCEN1 (LC379265) MpTUA5 (LC172181) MpDAZ1 (LC172178) MpGCS1 (LC172179) MpGEX2 (LC172180) HmnDUO1 (LC379264) HmnACT1 (LC379378) CbrDUO1 (LC199499), CauDUO1 (LC221833), CleDUO1 (LC221832), CbrRSP11 (LC382020) CbrPACRG (LC382019) CbrRPL6 (LC382018) CauRPL6 (LC382017) CpeDUO1 (LC176570). Previously reported sequences used in this study are available at GenBank under the accession: MpPACRG (LC102460), MpPRM (LC102462) MpTUB4 (KM096548) MpLC7 (LC102461), MpMID/RWP2 (KU987912) CleRPL6 (AB035569), CpeRLK1 (AB920609) CpePI (AB012698). All other data are available from the authors on reasonable request.