Abstract
Flavonoids are important polyphenolic natural products, ubiquitous in land plants, that play diverse functions in plants' survival in their ecological niches, including UV protection, pigmentation for attracting pollinators, symbiotic nitrogen fixation, and defense against herbivores. Chalcone synthase (CHS) catalyzes the first committed step in plant flavonoid biosynthesis and is highly conserved in all land plants. In several previously reported crystal structures of CHSs from flowering plants, the catalytic cysteine is oxidized to sulfinic acid, indicating enhanced nucleophilicity in this residue associated with its increased susceptibility to oxidation. In this study, we report a set of new crystal structures of CHSs representing all five major lineages of land plants (bryophytes, lycophytes, monilophytes, gymnosperms, and angiosperms), spanning 500 million years of evolution. We reveal that the structures of CHS from a lycophyte and a moss species preserve the catalytic cysteine in a reduced state, in contrast to the cysteine sulfinic acid seen in all euphyllophyte CHS structures. In vivo complementation, in vitro biochemical and mutagenesis analyses, and molecular dynamics simulations identified a set of residues that differ between basal-plant and euphyllophyte CHSs and modulate catalytic cysteine reactivity. We propose that the CHS active-site environment has evolved in euphyllophytes to further enhance the nucleophilicity of the catalytic cysteine since the divergence of euphyllophytes from other vascular plant lineages 400 million years ago. These changes in CHS could have contributed to the diversification of flavonoid biosynthesis in euphyllophytes, which in turn contributed to their dominance in terrestrial ecosystems.
Keywords: polyketide, flavonoid, redox regulation, enzyme catalysis, enzyme mechanism, chalcone synthase, cysteine, pKa, polyketide synthase, plant evolution
Introduction
In their transition from aquatic domains to terrestrial environments, early land plants faced several major challenges, including exposure to damaging UV-B radiation once screened by aquatic environments, lack of structural support once provided by buoyancy in water, drought, and novel pathogens and herbivores. To cope with many of these stresses, land plants have evolved a series of specialized metabolic pathways, among which phenylpropanoid metabolism was probably one of the most critical soon after the transition from water to land (1).
Flavonoids are a diverse class of plant phenolic compounds found in all extant land plants, with important roles in many aspects of plant life, including UV protection, pigmentation for attracting pollinators and seed dispersers, defense, and signaling between plants and microbes (2). Some flavonoids are also of great interest for their anti-cancer and antioxidant activities as well as other potential health benefits to humans (3). After the core flavonoid biosynthetic pathway was established in early land plants, new branches of the pathway continued to evolve over the history of plant evolution, producing structurally and functionally diverse flavonoids to cope with changing habitats, co-evolving pathogens and herbivores, and other aspects of plants' ecological niches. Basal bryophytes biosynthesize the three main classes of flavonoids, namely flavanones, flavones, and flavonols, which likely emerged as UV sunscreens (4). The lycophyte Selaginella biosynthesizes a rich diversity of biflavonoids, many of which were shown to be cytotoxic and may function as phytoalexins (5). The ability to synthesize the astringent, polyphenolic tannins, which defend against bacterial and fungal pathogens, seems to have evolved in euphyllophytes (4). Finally, seed plants, including gymnosperms and angiosperms, developed elaborate anthocyanin biosynthetic pathways to produce the vivid colors used to attract pollinators or ward off herbivores.
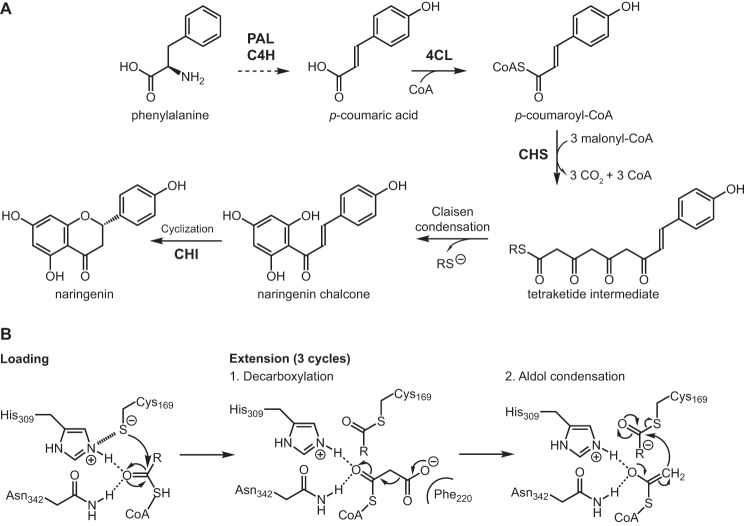
Chalcone synthase (CHS),2 a highly conserved plant type III polyketide synthase (PKS), is the first committed enzyme in the plant flavonoid biosynthetic pathway. CHS synthesizes naringenin chalcone from a molecule of p-coumaroyl-CoA and three molecules of malonyl-CoA (Fig. 1A) (6). The proposed catalytic mechanism of CHS involves loading of the starter molecule p-coumaroyl-CoA onto the catalytic cysteine, which also serves as the attachment site of the growing polyketide chain during the iterative elongation steps (7). This initial reaction step requires the cysteine to be present as a thiolate anion before loading of the starter molecule (Fig. 1B). Using thiol-specific inactivation and the pH dependence of the malonyl-CoA decarboxylation reaction, the pKa of the catalytic cysteine (Cys-164) of Medicago sativa CHS (MsCHS) was measured to be 5.5, a value significantly lower than 8.7 for free cysteine (8).
Figure 1.
A, phenylpropanoid and flavonoid metabolism. PAL, phenylalanine ammonia-lyase; C4H, trans-cinnamate 4-monooxygenase; 4CL, 4-coumarate-CoA ligase; CHI, chalcone isomerase; CoA, coenzyme A. Cyclization of naringenin chalcone to naringenin also proceeds spontaneously in aqueous solution. B, reaction mechanism of CHS. The extension step is performed three times to repeatedly extend the starter molecule malonyl-CoA to form a linear tetraketide intermediate, which then cyclizes to form naringenin chalcone.
Interestingly, we observed that the catalytic cysteine residues in the previously reported MsCHS crystal structures appear to be oxidized to sulfinic acid (PDB codes 1BI5 and 1BQ6) (11). Furthermore, the same phenomenon was observed in the crystal structures for several other plant type III PKSs evolutionarily derived from CHS, including Gerbera hybrida 2-pyrone synthase (PDB code 1QLV) (Fig. S1) (9). The other noncatalytic cysteines in these proteins do not appear to be oxidized. These findings suggest that the oxidation of the catalytic cysteine observed in several type III PKS crystal structures may not simply be an artifact of X-ray crystallography but rather reflects the intrinsic redox potential and reactivity of the catalytic cysteine evolved in this family of enzymes. Indeed, the propensity for a particular cysteine residue to undergo oxidation has been previously indicated to correlate with a low pKa (10).
Here, we present a set of new crystal structures of orthologous CHSs representing five major lineages of land plants, namely bryophytes, lycophytes, monilophytes, gymnosperms, and angiosperms, spanning 500 million years of land plant evolution. Through comparative structural analysis, in vivo complementation, in vitro biochemistry, mutagenesis studies, and molecular dynamics simulations, we reveal that CHSs of basal land plants, i.e. bryophytes and lycophytes, contain a catalytic cysteine less reactive than that of the CHSs from higher plants, i.e. euphyllophytes. We probe into the structure–function relationship of a set of residues that modulate the reactivity of the catalytic cysteine, which leads us to propose that euphyllophytes may have evolved a more catalytically efficient CHS to enhance flavonoid biosynthesis relative to their basal-plant relatives.
Results
Basal-plant CHSs contain reduced catalytic cysteine in their crystal structures
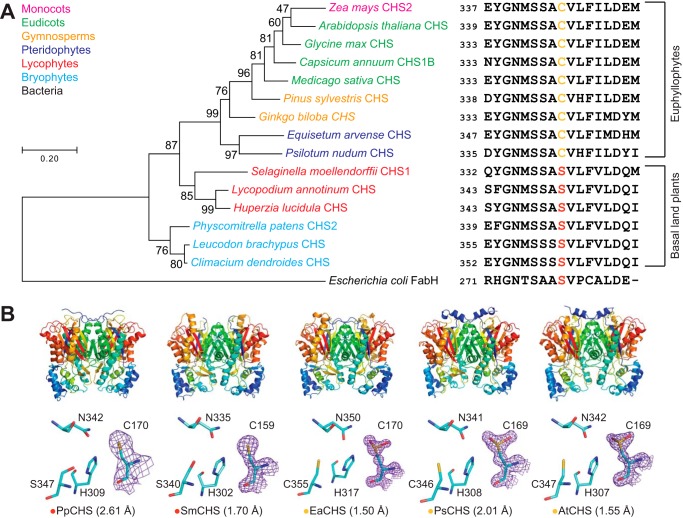
To examine the structural basis for the evolution of CHS across major land plant lineages, we cloned, expressed, and solved the crystal structures of the five CHS orthologs from the bryophyte Physcomitrella patens (PpCHS), the lycophyte Selaginella moellendorffii (SmCHS), the monilophyte Equisetum arvense (EaCHS), the gymnosperm Pinus sylvestris (PsCHS), and the angiosperm Arabidopsis thaliana (AtCHS) (Fig. 2 and Table 1). Like previously reported crystal structures of type III polyketide synthases, all five CHS orthologs form symmetric homodimers and share the same αβαβα thiolase fold, suggesting a common evolutionary origin (11). The catalytic triad of cysteine, histidine, and asparagine is found in a highly similar conformation to other PKS and related fatty acid biosynthetic β-ketoacyl-(acyl-carrier-protein) synthase III (KAS III) enzymes, suggesting that they share a similar general catalytic mechanism (Fig. 2B).
Figure 2.
Structural and in vivo functional characterization of diverse CHS orthologs. A, maximum-likelihood phylogenetic tree of CHSs from diverse land-plant species, with clades indicated by color. The tree is rooted on a bacterial KAS III enzyme (EcFabH). The scale bar indicates evolutionary distance in substitutions per amino acid. The sequence near the differentially conserved cysteine/serine (position 347 in AtCHS) is shown for each CHS. B, overall apo crystal structures and active-site structures of CHSs from diverse plant lineages. Top, the homodimeric form of CHS is shown with a color gradient from blue at the N terminus to red at the C terminus of each monomer. Bottom, backbone and side chains of the catalytic triad and the differentially conserved cysteine/serine are shown. The 2Fo − Fc electron density map contoured at 1.5σ is shown around the catalytic cysteine. CHSs from euphyllophytes show the catalytic cysteine oxidized to sulfinic acid, whereas CHSs from basal land plants have a reduced catalytic cysteine. The red or yellow dot next to the enzyme name indicates the presence of serine or cysteine, respectively, in position 347 (AtCHS numbering).
Table 1.
Crystallographic data collection and refinement statistics for the five wildtype CHSs
The highest-resolution shell values are given in parentheses.
| PpCHS | SmCHS | EaCHS | PsCHS | AtCHS | |
|---|---|---|---|---|---|
| PDB codes | 6DX7 | 6DX8 | 6DX9 | 6DXA | 6DXB |
| Data collection | |||||
| Total reflections | 161,385 (13,620) | 493,910 (50,217) | 1,164,176 (116,600) | 168,567 (13,442) | 430,437 (39,846) |
| Unique reflections | 82,406 (7763) | 81,737 (6434) | 125,911 (12,431) | 46,489 (4596) | 227,280 (21,992) |
| Multiplicity | 2.0 (1.8) | 6.0 (6.2) | 9.2 (9.4) | 3.6 (2.9) | 1.9 (1.8) |
| Completeness (%) | 98.95 (94.60) | 90.60 (79.18) | 99.97 (99.90) | 99.31 (98.75) | 98.48 (95.43) |
| Mean I/σ(I) | 14.31 (1.57) | 12.32 (1.62) | 13.26 (1.35) | 10.40 (2.62) | 10.17 (2.78) |
| R-merge | 0.03961 (0.4442) | 0.1427 (1.194) | 0.1013 (1.58) | 0.1424 (0.6168) | 0.04372 (0.2437) |
| CC1/2 | 0.998 (0.747) | 0.995 (0.51) | 0.998 (0.697) | 0.979 (0.406) | 0.996 (0.837) |
| Refinement | |||||
| Resolution range (Å) | 36.24–2.61 (2.703–2.61) | 39.01–1.7 (1.761–1.7) | 56.57–1.5 (1.554–1.5) | 52.45–2.01 (2.082–2.01) | 38.68–1.549 (1.604–1.549) |
| Space group | P 2 21 21 | P 1 21 1 | P 21 21 21 | P 1 21 1 | P 1 21 1 |
| Unit cell (Å) | 71.6 192.83 195.51 | 55.1702 66.6703 102.55 | 52.954 112.764 130.803 | 58.017 100.059 65.882 | 54.64 137.56 108.56 |
| Unit cell (°) | 90 90 90 | 90 91.35 90 | 90 90 90 | 90 110.807 90 | 90 95.59 90 |
| R-work | 0.1810 (0.3187) | 0.1927 (0.3111) | 0.1610 (0.4374) | 0.1591 (0.2512) | 0.1416 (0.1967) |
| R-free | 0.2627 (0.3951) | 0.2362 (0.3567) | 0.1796 (0.4500) | 0.2204 (0.3189) | 0.1640 (0.2151) |
| Non-hydrogen protein atoms | 17,592 | 5776 | 6054 | 6005 | 12,052 |
| Water molecules | 59 | 549 | 711 | 684 | 1680 |
| RMSD bonds (Å) | 0.014 | 0.012 | 0.009 | 0.012 | 0.009 |
| RMSD angles (°) | 1.5 | 1.32 | 1.26 | 1.31 | 1.27 |
| Ramachandran favored (%) | 94.07 | 97.74 | 97.8 | 96.75 | 97.91 |
| Ramachandran allowed (%) | 5.58 | 1.99 | 2.2 | 3.25 | 2.09 |
| Ramachandran outliers (%) | 0.35 | 0.27 | 0 | 0 | 0 |
| Average B-factor | 63.61 | 22.24 | 20.79 | 17.9 | 17.52 |
Based on the previously proposed reaction mechanism for MsCHS, the catalytic cysteine is Cys-169 in AtCHS and Cys-159 in SmCHS. This residue initiates the reaction mechanism by performing nucleophilic attack on p-coumaroyl-CoA (Fig. 1B). The other two members of the catalytic triad consist of His-309 and Asn-342 in AtCHS, and His-302 and Asn-335 in SmCHS. The catalytic histidine contributes to the lowered pKa of the catalytic cysteine by forming a stable imidazolium–thiolate ion pair (8). The histidine and asparagine also form the oxyanion hole that stabilizes the tetrahedral transition states formed during the initial nucleophilic attack by cysteine on p-coumaroyl-CoA and after malonyl-CoA decarboxylation (Fig. 1B).
Notably, SmCHS and PpCHS are the first CHSs for which a reduced catalytic cysteine has been observed in the crystal structure (Fig. 2B). The catalytic cysteine in SmCHS can still become oxidized to sulfenic acid when the crystal is soaked in hydrogen peroxide, indicating that it is still susceptible to oxidation at a lower rate (Fig. S2 and Table S1). Like most other euphyllophyte type III PKS crystal structures solved to date, AtCHS, PsCHS, and EaCHS contain doubly oxidized catalytic cysteine sulfinic acid (Fig. 2B). This interesting observation suggests a functional divide between basal- plant and euphyllophyte CHSs. Despite shared orthology, the redox potential of the catalytic cysteine in PpCHS and SmCHS may differ from that of the euphyllophyte CHSs, resulting in different levels of sensitivity to oxidation under similar crystallization conditions. This could be due to the evolution of some novel molecular features in euphyllophyte CHSs not present in the lower-plant CHSs.
Basal-plant CHSs only partially complement the Arabidopsis CHS-null mutant
CHS orthologs have been identified in all land-plant species sequenced to date, suggesting a highly conserved biochemical function. To test whether the five CHSs from the five major plant lineages are functionally equivalent, we generated transgenic A. thaliana lines expressing each of the five different CHSs driven by the Arabidopsis CHS promoter in the CHS-null mutant transparent testa 4-2 (tt4-2) background (12) (Fig. S3).
Twenty independent T1 plants were selected for each construct. The phenotypes of the transgenic plants described below were represented by the majority of independent transgenic events for each unique construct. As the name indicates, the tt4-2 mutant is devoid of flavonoid biosynthesis and therefore lacks the accumulation of the brown condensed tannin pigments in seed coats, revealing the pale yellow color of the underlying cotyledons (12). Although AtCHS, PsCHS, and EaCHS fully complement the tt phenotype of tt4-2, PpCHS and SmCHS only partially rescue the seed tt phenotype of tt4-2 (Fig. S3), suggesting that PpCHS and SmCHS are likely less active than their higher-plant counterparts in vivo. This result also correlates with the crystallographic observation where the catalytic cysteine of basal-plant and euphyllophyte CHSs exhibits differential susceptibility to oxidation.
pKa of the catalytic cysteine is higher in basal-plant CHSs than in euphyllophyte CHSs
To perform nucleophilic attack on the p-coumaroyl-CoA substrate, the catalytic cysteine must be present in the thiolate anion form. As shown previously in MsCHS, the pKa of the catalytic cysteine is lowered to 5.5, well below physiological pH, to stabilize this deprotonated state (8). Two factors could contribute to the depressed pKa of Cys-164. First, His-303, one of the catalytic triad of CHS in the vicinity of Cys-164, provides an ionic interaction with Cys-164 that can further stabilize the cysteine thiolate anion. Second, Cys-164 is positioned at the N terminus of the MsCHS α-9 helix (11), which provides a stabilizing effect on the cysteine thiolate anion through the partial positive charge of the helix dipole (13). The acidic pKa of the catalytic cysteine in CHS ensures the presence of a cysteine thiolate anion in the enzyme active site at physiological pH to serve as the nucleophile for starter molecule loading.
To measure the pKa of the catalytic cysteine in the five land-plant CHS orthologs, we performed pH-dependent inactivation of CHS using iodoacetamide, a thiol-specific compound that reacts with sulfhydryl groups that are sufficiently nucleophilic, followed by a CHS activity assay at the usual reaction pH. At pH values above the pKa, the catalytic cysteine is deprotonated and able to react with iodoacetamide, thus inactivating CHS. At pH values below the pKa, the catalytic cysteine is protonated and protected from iodoacetamide modification, thus retaining CHS activity in the subsequent enzyme assay. The amount of CHS activity remaining after iodoacetamide treatment was expressed as a ratio compared with the CHS activity of a control treatment at the same pH but without iodoacetamide. The pKa value was calculated using nonlinear regression to fit a log(inhibitor) versus response equation, which gave the pH at which 50% of maximal inhibition was obtained.
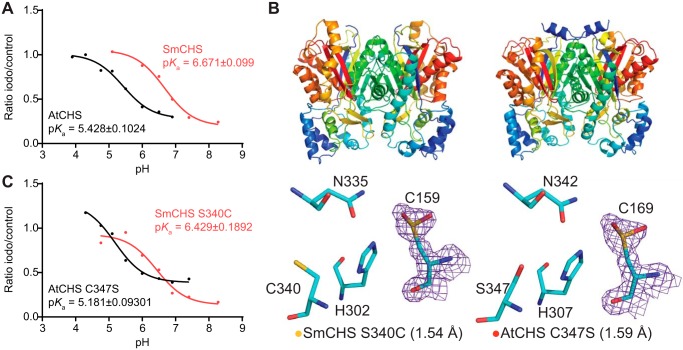
The pKa value for AtCHS was measured to be 5.428, which is close to the 5.5 measured for MsCHS (Fig. 3A). The pKa value for SmCHS was measured to be 6.468, ∼1 pH unit higher than that of the two angiosperm CHS orthologs. This elevated pKa value measured for SmCHS is consistent with the observation of a catalytic cysteine that is less reactive and less prone to oxidation. Also consistent with the crystallographic and plant complementation results, pKa values around 5.5 were measured for euphyllophyte orthologs PsCHS and EaCHS and around 6.5 for the basal-plant orthologs PpCHS (Fig. S4).
Figure 3.
pKa measurement of the catalytic cysteine and characterization of key residues that affect pKa. A, pKa measurement of AtCHS and SmCHS WT enzymes. CHS enzyme was pre-incubated at various pH values with or without the 25 μm iodoacetamide inhibitor for 30 s, and an aliquot was taken to run in a CHS activity assay. The ratio of naringenin product produced in the iodoacetamide treatment divided by the control treatment was calculated for each pH point. A nonlinear regression was performed to fit a log(inhibitor) versus response curve to determine the pH at which 50% of maximal inhibition was achieved, which was determined to be the pKa value of the catalytic cysteine residue. The pKa of AtCHS is close to the 5.5 determined for other euphyllophyte CHSs, whereas the pKa of SmCHS is over 1 pH unit higher. B, overall structures and active-site configurations of AtCHS C347S and SmCHS S340C single mutants. The 2Fo − Fc electron density map contoured at 1.5σ is shown around the catalytic cysteine. SmCHS S340C shows oxidation of Cys-159, unlike the SmCHS WT. AtCHS C347S has an oxidized Cys-169, like AtCHS WT. C, pKa measurements of AtCHS C347S and SmCHS S340C mutants.
Residues near the active-site cavity affect the pKa and reactivity of the catalytic cysteine
We next examined the sequence and structural differences between basal-plant and euphyllophyte CHSs that could play a role in modulating catalytic cysteine reactivity. This led us to first identifying a residue near the active site that is conserved as Cys-347 (AtCHS numbering) in AtCHS and other euphyllophyte sequences, and as Ser-340 (SmCHS numbering) in SmCHS and other lycophyte and bryophyte sequences (Fig. 2A).
To investigate the role of this residue in modulating catalytic cysteine reactivity, we generated the reciprocal mutations in SmCHS and AtCHS, respectively, and first characterized these mutant proteins using X-ray crystallography (Fig. 3B and Table 2). Under identical crystallization conditions as WT SmCHS, the SmCHS S340C mutant exhibits a partially oxidized catalytic cysteine in its crystal structure, suggesting that the residue does play some role in determining cysteine reactivity. The AtCHS C347S mutant, however, still retains an oxidized catalytic cysteine in its crystal structure.
Table 2.
Crystallographic data collection and refinement statistics for the three mutant CHSs
The highest-resolution shell values are given in parentheses.
| SmCHS S340C | AtCHS C347S | AtCHS M7 | |
|---|---|---|---|
| PDB ID | 6DXC | 6DXD | 6DXE |
| Data collection | |||
| Total reflections | 185,024 (12,834) | 368,273 (17,370) | 293,345 (28,831) |
| Unique reflections | 105,252 (8805) | 201,521 (14,085) | 102,276 (7995) |
| Multiplicity | 1.8 (1.5) | 1.8 (1.2) | 2.9 (2.8) |
| Completeness (%) | 95.55 (80.52) | 93.32 (65.39) | 91.60 (76.72) |
| Mean I/σ(I) | 9.66 (3.06) | 8.04 (1.51) | 11.10 (1.18) |
| R-merge | 0.04892 (0.1642) | 0.05533 (0.3447) | 0.04526 (0.5779) |
| CC1/2 | 0.983 (0.891) | 0.994 (0.699) | 0.999 (0.778) |
| Refinement | |||
| Resolution range (Å) | 30.48–1.54 (1.595–1.54) | 32.92–1.59 (1.647–1.59) | 60.11–1.608 (1.665–1.608) |
| Space group | P 1 21 1 | P 1 21 1 | P 1 21 1 |
| Unit cell (Å) | 55.22 66.38 103 | 54.86 138.22 108.9 | 72.8 55.9 100.21 |
| Unit cell (°) | 90 91.73 90 | 90 95.73 90 | 90 92.51 90 |
| R-work | 0.1427 (0.1925) | 0.1455 (0.2608) | 0.1721 (0.2610) |
| R-free | 0.1725 (0.2471) | 0.1688 (0.2982) | 0.2023 (0.2675) |
| Nonhydrogen protein atoms | 5800 | 12,028 | 6058 |
| Water molecules | 881 | 1825 | 859 |
| RMSD bonds (Å) | 0.01 | 0.01 | 0.01 |
| RMSD angles (°) | 1.38 | 1.39 | 1.27 |
| Ramachandran favored (%) | 97.72 | 97.92 | 97.79 |
| Ramachandran allowed (%) | 2.28 | 2.02 | 2.08 |
| Ramachandran outliers (%) | 0 | 0.07 | 0.13 |
| Average B-factor | 16.52 | 20.78 | 21.36 |
We then measured the pKa value of the catalytic cysteine in both SmCHS S340C and AtCHS C347S mutants (Fig. 3C). The pKa value for SmCHS S340C decreases by about 0.25 pH units compared with WT SmCHS, consistent with the observation that the SmCHS S340C crystal structure contained a partially oxidized catalytic cysteine. The pKa value for AtCHS C347S decreases by about 0.25 pH units compared with WT AtCHS, also consistent with the observation that the AtCHS C347S crystal structure retained an oxidized catalytic cysteine. Taken together, the crystallographic and pKa measurement results suggest that the reciprocal mutation at this position is not sufficient to act as a simple switch between the active-site environments of euphyllophyte and basal-plant CHSs to modulate catalytic cysteine reactivity. Additional sequence and structural features likely contribute to an active-site environment that lowers the pKa of the catalytic cysteine in AtCHS.
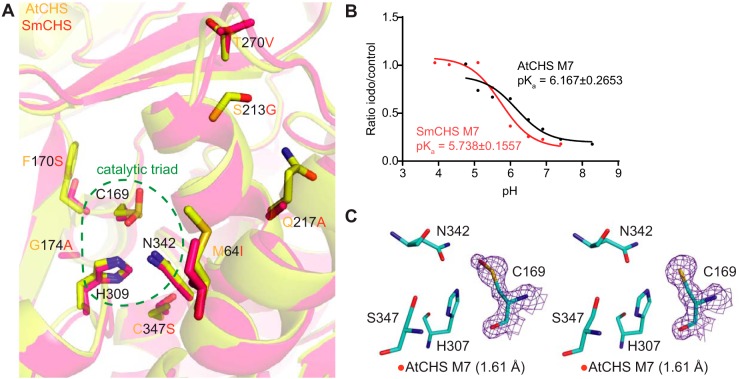
To identify these features, we examined a multiple sequence alignment of CHS orthologs from diverse plant species and identified residues that show conserved variations between euphyllophytes and basal-plant lineages (Fig. 4A and Fig. S5). Two residues, Phe-170 and Gly-173 in euphyllophyte CHSs, were found to be substituted as serine and alanine, respectively, in basal-plant lineages. Because of their positions in the α-helix immediately C-terminal to the catalytic Cys-169, we postulated that these two residues could play a role in determining the structure of the helix, which would have an effect on the electronic environment of the active site, due to the helix dipole's contribution to lowering the catalytic cysteine pKa value (11). Four additional residues near the active-site opening of CHS were also identified as differentially conserved between euphyllophytes and basal plants. We postulated that these positions might affect the dynamics of the active-site tunnel and solvent access to the active site. The six aforementioned residues were mutated in the SmCHS S340C background to their corresponding residues in AtCHS to generate the SmCHS I54M/S160F/A163G/G203S/A207Q/V258T/S340C septuple mutant, termed SmCHS M7. Likewise, the reciprocal mutations were also made in the AtCHS C347S background to generate AtCHS M7.
Figure 4.
Identification and characterization of additional key residues that affect CHS cysteine reactivity. A, overlaid crystal structures of AtCHS and SmCHS showing the seven conserved residue differences between euphyllophyte and basal-plant CHSs. B, pKa measurement of AtCHS M7 and SmCHS M7 mutants. The pKa of each M7 mutant is about 0.5 pH units higher or lower, respectively, than the corresponding WT CHS. C, active sites of the two monomers of the AtCHS M7 septuple mutant structure. The 2Fo − Fc electron density map contoured at 1.5σ is shown around the catalytic cysteine. The crystal structure shows oxidation to sulfenic acid in the catalytic cysteine of one chain (left) and a reduced cysteine in the other (right).
Compared with SmCHS S340C, the six additional mutations in SmCHS M7 lower the pKa by nearly 0.7 pH units from 6.429 to 5.738 (Fig. 4B). Similarly, the six mutations of AtCHS M7 raise the pKa by almost 1 pH unit from 5.181 to 6.167 compared with AtCHS C347S. Consistent with the pKa observation, the dimeric crystal structure of AtCHS M7 has one monomer with a catalytic cysteine singly oxidized to sulfenic acid and one monomer with a reduced cysteine (Fig. 4C). Sulfenic acid is more reduced than the doubly oxidized sulfinic acid seen in other euphyllophyte crystal structures, indicating that these six mutations decreased the reactivity of the catalytic cysteine. These mutations represent a part of a possible evolutionary path from ancestral basal-plant CHSs toward the stronger pKa-lowering properties of euphyllophyte CHSs. Any further attempts at engineering CHS to fully swap the pKa-lowering properties between AtCHS and SmCHS would likely require different methods of searching for conserved sequence differences, beyond visual observation of structural differences. An analysis of the CHS multiple sequence alignment using ancestral sequence reconstruction with FastML (14) identified eight additional positions that are differently conserved between euphyllophytes and basal plants and could affect CHS function based on their position in the CHS crystal structure (Fig. S6).
Molecular dynamics simulations reveal differences in active-site interactions between basal-plant and euphyllophyte CHSs
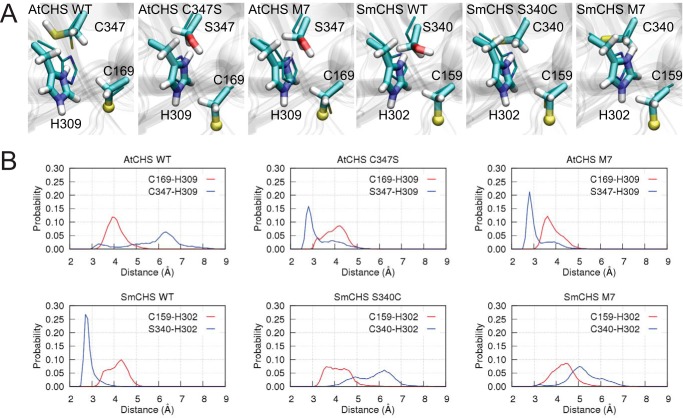
Our crystal structures revealed a correlation between the pKa value of the catalytic cysteine and a set of residues near the active site. To further investigate the mechanisms underlying these conserved differences between euphyllophyte and basal-plant CHSs, we employed molecular dynamics (MD) simulations to examine the interactions between these residues. We first surveyed the potential role of the C347S substitution (AtCHS numbering) in affecting the active-site environment in WT AtCHS and SmCHS (Fig. 5A). In WT AtCHS, where the largest cluster represents 70.3% of all structures sampled in this simulation, the thiol group of Cys-347 points away from the active site and cannot form any stable interaction with the catalytic His-309 (distance 6.5 Å). In contrast, the corresponding Ser-340 in SmCHS is 2.8 Å away from the histidine in the largest cluster, representing 98.7% of all structures sampled in the SmCHS simulation.
Figure 5.
Molecular dynamics simulations of CHS orthologs and mutants. A, centroid structure of the largest cluster of the catalytic pair Cys-169–His-309. For visualization purposes, the sulfur atom in the ionic cysteine Cys-169 is shown in a ball representation, and crystal structures are depicted as thin sticks. B, distributions of inter-residue distances obtained from simulations.
Next, we determined the inter-residue distances between the ionic pair Cys-169–His-309 as in AtCHS or Cys-159–His-302 as in SmCHS and between residue Cys-347 (AtCHS)/Ser-340 (SmCHS) and the catalytic histidine (Fig. 5B). For the WT SmCHS simulation, we observed a sharp peak at around 2.8 Å between Ser-340 and His-302, reflecting a stable hydrogen bond between the two residues. On the contrary, no such short-distance peak was observed for WT AtCHS. These results suggest that the catalytic histidine is stabilized upon forming a hydrogen bond with Ser-340 in SmCHS, but such an interaction is relatively loose in AtCHS. Similar differences between the other euphyllophyte and basal-plant CHSs are also seen for PsCHS, EaCHS, and PpCHS (Fig. S7).
To further investigate the motion of the catalytic histidine in various mutant enzyme active-site environments, we also performed MD simulations of AtCHS C347S, SmCHS S340C, AtCHS M7, and SmCHS M7 (Fig. 5A). The largest cluster sizes were 86.0, 66.6, 96.7, and 71.0%, respectively. Ser-347 and His-309 in AtCHS mutants adopt a similar conformation to the corresponding residues in WT SmCHS. In contrast, no stable hydrogen bond between Cys-340 and His-302 is formed in the largest cluster of the SmCHS mutant simulations. Introducing point mutations dramatically changes the distributions of those key inter-residue distances. In the AtCHS C347S mutant, the Ser-347–His-309 distance dramatically shortens to a peak around 2.8 Å, and introducing the six additional mutations in AtCHS M7 further increases the height of the peak. This suggests that mutating these seven positions in AtCHS to the corresponding residues in SmCHS can allow the active-site residues to approximate the interactions of WT SmCHS. The opposite effect is seen in SmCHS S340C and SmCHS M7 mutants, which recapitulate the weak interaction between Cys-347 and His-309 seen in WT AtCHS.
Based on these results, we hypothesize that the strong Ser-340–His-302 interaction facilitated by the SmCHS active-site environment may weaken the stabilizing effect of His-302 on the catalytic cysteine thiolate compared with that in AtCHS, thus contributing to the higher pKa value. Meanwhile, the inter-residue distance of the catalytic cysteine–histidine ionic pair is rather stable in all CHS simulations, ranging from 3 to 5 Å and centered around 4.1 Å. This suggests that the C347S substitution (AtCHS numbering) does not directly break this ionic interaction but may subtly influence the charge distribution on the histidine imidazole ring to perturb the catalytic cysteine pKa value (Fig. 6). In addition, the presence of a cysteine appears to decrease solvent content in the active site compared with serine, which would increase the pKa-lowering effect of the ionic interaction between histidine and the catalytic cysteine (Fig. S8 and supporting Note). Taken together, our results suggest that euphyllophyte CHSs have evolved to enhance the reactivity of the catalytic cysteine through the modification of specific interactions between active-site residues to allow for stronger stabilization of the thiolate.
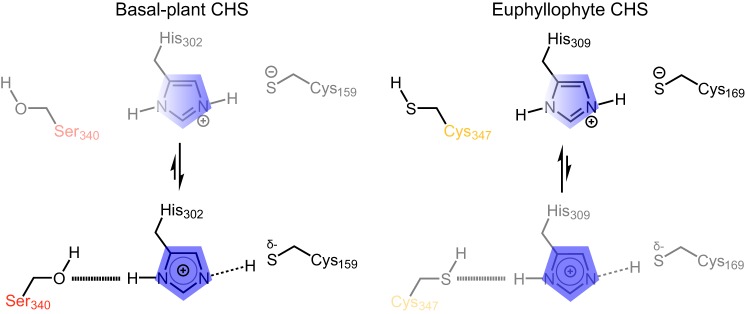
Figure 6.
Proposed model for differential modulation of catalytic cysteine nucleophilicity in basal-plant (left) and euphyllophyte (right) CHSs. In basal-plant CHSs (left), the serine (Ser-340 in SmCHS) interacts more strongly with the histidine of the catalytic triad, weakening the ionic interaction that stabilizes the thiolate form of the catalytic cysteine. This is depicted as a shift of the equilibrium toward a state in which the positive charge on the histidine (blue) is shifted away from the catalytic cysteine (Cys-159 in SmCHS), and the shared proton interacts more closely with cysteine. In euphyllophyte CHSs (right), this position mutated to a cysteine (Cya-347 in AtCHS), which interacts relatively loosely with the catalytic histidine, in turn strengthening the ionic interaction between the catalytic histidine and the activated thiolate of the catalytic cysteine. This is depicted as a shift of the equilibrium toward a state in which the positive charge on the histidine (blue) is shifted toward the catalytic cysteine (Cys-169 in AtCHS).
Discussion
As early plants initially migrated from water to land and further radiated to occupy diverse terrestrial environmental niches, they continuously encountered new challenges from biotic and abiotic stresses. The greatly expanded diversity and increased abundance of flavonoids in certain plant lineages could have increased the demand for metabolic flux into flavonoid biosynthesis. One adaptive strategy to meet this demand, among many others, is to increase the enzymatic efficiency of chalcone synthase, the first committed enzyme of flavonoid biosynthesis that gates flux from general phenylpropanoid metabolism. One property of CHS that affects its enzymatic efficiency is the reactivity of the first step of nucleophilic attack on p-coumaroyl-CoA. To investigate this, we performed structural, biochemical, mutagenesis, and molecular dynamics experiments on CHS orthologs from five major plant lineages. Our results suggest that euphyllophyte CHSs have indeed evolved new structural features to increase the reactivity of their catalytic cysteine compared with basal-plant CHSs.
To identify sequence and structural features between euphyllophyte and basal-plant CHSs that lead to this difference in enzymatic properties, we generated mutants in the background of AtCHS and SmCHS at various positions with conserved sequence differences segregating euphyllophyte and basal-plant CHSs. AtCHS M7 and SmCHS M7 had pKa values raised by about 0.7 pH units and lowered by about 1 pH unit from the WT enzymes, respectively. Furthermore, AtCHS M7 also exhibits a less oxidized catalytic cysteine in its crystal structure than in WT AtCHS. These results indicate that we were able to identify residue changes that partially traced the evolutionary path from SmCHS to AtCHS that increased the reactivity of the catalytic cysteine. In the type III PKS family, the introduction of a large number of mutations to yield subtle changes in enzyme activity is not unprecedented. Stilbene synthase produces resveratrol, a tetraketide product whose biosynthetic mechanism differs from that of naringenin chalcone in only the final cyclization step. In a previous study, a total of 18 point mutations were required to convert CHS activity to stilbene synthase activity, through small changes in the hydrogen-bonding network in the active site (15).
To examine in detail the intramolecular interactions that lead to enhanced cysteine reactivity, we performed molecular dynamics simulations on CHS. In comparing different CHS orthologs and point mutants, we observed that the presence of a cysteine in position 347 (AtCHS numbering) leads to a weak interaction between that cysteine and histidine, as indicated by the broad distribution of inter-residue distances centered at a distance greater than 5 Å, too long for a stable hydrogen bond. In contrast, when a serine is present, the sharp peak of serine–histidine inter-residue distance around 2.75 Å suggests the presence of a strong hydrogen bond. This hydrogen bonding likely shifts the electron density of the histidine away from the catalytic cysteine, weakening the imidazoline–thiolate ion pair. This weakened ionic interaction would lead to less pKa depression compared with CHS orthologs and mutants containing a cysteine in the nearby position, where the histidine is able to maintain a stronger ion pair with the catalytic cysteine and lower the pKa to a greater degree. This is reminiscent of the role of aspartate 158 in papain, a cysteine protease that also uses a cysteine–histidine–asparagine catalytic triad for nucleophilic attack on its substrates (16). Although Asp-158 is not essential for papain activity, its side chain affects the pH-activity profile by forming a hydrogen bond with the backbone amide of the catalytic histidine. This interaction stabilizes the catalytic ionic pair and maintains an optimal orientation of active-site residues. A D158E mutant papain had a pH-activity profile shifted by 0.3 pH units, about the same magnitude of the effect we observed on pKa for CHS cysteine/serine mutants.
We propose a model of the role of position 347 in enhancing CHS reactivity (Fig. 6). In the basal example of SmCHS, the serine interacts more strongly with the histidine of the catalytic triad, weakening the ionic interaction that stabilizes the thiolate form of the catalytic cysteine. In euphyllophyte CHSs, this position mutated to a cysteine, which interacts more poorly with the histidine, strengthening the ionic interaction and stabilizing the activated thiolate of the catalytic cysteine.
Although the mechanism of how the other six mutations in the M7 mutants affect the catalytic cysteine is not entirely clear, we noticed that, possibly due to the smaller side chains of the S213G and Q217A mutations, AtCHS M7 has a surface helix in a slightly different conformation than WT AtCHS, leading to a slightly wider active-site opening (Fig. S9). There is also a newly solvent-accessible cavity as determined by a computational cavity-finding software (Fig. S8). These structural differences could lead to subtle changes in the amino acid backbone dynamics near the active site and thus alter the active-site volume or electronic environment, which could alter the pKa value of the catalytic cysteine (8).
Although cysteine sulfenic and sulfinic acid have been thought of as crystallographic artifacts, an increasing number of studies have shown that this type of cysteine oxidation can play an important functional role. In particular, cysteine sulfinic acid has been shown to play a regulatory role in reversible inhibition of the activity of enzymes such as protein-tyrosine phosphatase 1B and glyceraldehyde-3-phosphate dehydrogenase, suggesting that cysteine redox potential can be an evolved trait (17, 18).
Our results demonstrate that euphyllophytes could have evolved a CHS enzyme that is intrinsically more active, with increased cysteine reactivity as a component, as one adaptation to produce the larger suite of flavonoids needed to counter the various environmental stresses they face. Although it may seem counterintuitive for euphyllophytes, which encounter more oxidative environments than do basal plants, to rely on a CHS enzyme that is more susceptible to oxidation, this susceptibility may be an unavoidable trade-off resulting from the chemical nature of a more nucleophilic cysteine: a catalytic cysteine more reactive toward substrate is also more reactive toward oxidants like hydrogen peroxide. To compensate for this increased susceptibility to oxidation, euphyllophytes may have evolved other systems to better maintain the redox environment inside the cell, one of those systems being the antioxidant flavonoids themselves.
Materials and methods
Cloning and site-directed mutagenesis of CHSs
Total RNA was obtained from A. thaliana, P. sylvestris, E. arvense, S. moellendorffii, and P. patens. Reverse transcription was performed to obtain cDNA. The open reading frames (ORFs) of five CHS orthologs were amplified via PCR from cDNA, digested with NcoI and XhoI, and ligated into NcoI- and XhoI-digested pHis8-3 or pHis8-4B Escherichia coli expression vectors. Site-directed mutagenesis was performed according to the QuikChange II site-directed mutagenesis protocol (Agilent Technologies).
Transgenic Arabidopsis
The AtCHS promoter (defined as 1328 bp of sequence upstream of the CHS transcription start site) was amplified via PCR from Arabidopsis genomic DNA, digested with HindIII and XhoI, and ligated into HindIII- and XhoI-digested pCC 1136, a promoterless Gateway cloning binary vector containing a BAR resistance gene marker, to generate pJKW 0152. The five CHS ORFs described above were then PCR-amplified from cDNA and cloned into pCC 1155, an ampicillin-resistant version of the pDONR221 Gateway cloning vector, with BP clonase in the Gateway cloning method (ThermoFisher Scientific). The resulting vectors were recombined with pJKW 0152 using LR clonase in the Gateway cloning method to generate the final binary constructs. Agrobacterium tumefaciens-mediated transformation of Arabidopsis was performed using the floral dipping method (44).
Recombinant protein expression and purification
CHS genes were cloned into pHis8-3 or pHis8-4B, bacterial expression vectors containing an N-terminal His8 tag followed by a thrombin or tobacco etch virus cleavage site, respectively, for recombinant protein production in E. coli. Proteins were expressed in the BL21(DE3) E. coli strain cultivated in terrific broth and induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside overnight at 18 °C. E. coli cells were harvested by centrifugation, resuspended in 150 ml of lysis buffer (50 mm Tris, pH 8.0, 500 mm NaCl, 30 mm imidazole, 5 mm DTT), and lysed with five passes through an M-110L microfluidizer (Microfluidics). The resulting crude protein lysate was clarified by centrifugation (19,000 × g, 1 h) prior to Qiagen nickel-nitrilotriacetic acid (Ni-NTA) gravity flow chromatographic purification. After loading the clarified lysate, the Ni-NTA resin was washed with 20 column volumes of lysis buffer and eluted with 1 column volume of elution buffer (50 mm Tris, pH 8.0, 500 mm NaCl, 300 mm imidazole, 5 mm DTT). 1 mg of His-tagged thrombin or tobacco etch virus protease was added to the eluted protein, followed by dialysis at 4 °C for 16 h in dialysis buffer (50 mm Tris, pH 8.0, 500 mm NaCl, 5 mm DTT). After dialysis, the protein solution was passed through Ni-NTA resin to remove uncleaved protein and His-tagged tobacco etch virus. The recombinant proteins were further purified by gel filtration on an ÄKTA Pure fast protein liquid chromatography (FPLC) system (GE Healthcare). The principal peaks were collected, verified by SDS-PAGE, and dialyzed into a storage buffer (12.5 mm Tris, pH 8.0, 50 mm NaCl, 5 mm DTT). Finally, proteins were concentrated to >10 mg/ml using Amicon Ultra-15 centrifugal filters (Millipore).
Protein crystallization
All protein crystals were grown by hanging drop vapor diffusion at 4 °C, except for EaCHS at 20 °C. For AtCHS WT and C347S crystals, 1 μl of 10 mg/ml protein was mixed with 1 μl of reservoir solution containing 0.1 m HEPES, pH 7.5, 0.3 m ammonium acetate, 14% (v/v) PEG 8000, and 5 mm DTT. For AtCHS M7 crystals, 1 μl of 16.33 mg/ml protein was mixed with 1 μl of reservoir solution containing 0.125 NaSCN, 20% (v/v) PEG 3350, and 5 mm DTT; 0.2 μl of a crystal seed stock from previous rounds of crystal optimization was also added. For SmCHS WT and S340C crystals, 1 μl of 10 mg/ml protein was mixed with 1 μl of reservoir solution containing 0.1 m MOPSO, pH 6.6, 0.3 m Mg(NO3)2, 19% (v/v) PEG 4000, and 5 mm DTT. For EaCHS, 1 μl of 16.92 mg/ml protein was mixed with 1 μl of reservoir solution containing 0.15 m LiCl, 8% PEG 6000, and 5 mm DTT. For PsCHS, 1.66 μl of 14.65 mg/ml protein was mixed with 0.67 μl of reservoir solution containing 0.14 m NH4Cl, 20% (v/v) PEG 3350, and 5 mm DTT; 0.2 μl of a crystal seed stock from previous rounds of crystal optimization was also added. For PpCHS, 1 μl of 10 mg/ml protein was mixed with 1 μl of reservoir solution containing 0.1 m MES, pH 6.9, 18% (v/v) PEG 20000, and 5 mm DTT. Crystals were harvested within 1 week and transferred to a cryoprotection solution of 17% glycerol and 83% reservoir solution. H2O2 soaking of SmCHS crystals was performed by adding H2O2 to 1 mm to the cryoprotection solution and incubating at 4 °C for 75 min. Single crystals were mounted in a cryoloop and flash-frozen in liquid nitrogen.
X-ray diffraction and structure determination
X-ray diffraction data were collected at beamlines 8.2.1 and 8.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory on ADSC Quantum 315 CCD detectors for AtCHS WT, AtCHS C347S, and SmCHS S340C crystals. X-ray diffraction data were collected at beamlines 24-ID-C and 24-ID-E of the Advanced Photon Source at Argonne National Laboratory on an ADSC Quantum 315 CCD detector, Eiger 16M detector, or Pilatus 6M detector for SmCHS WT, EaCHS, PsCHS, and AtCHS M7 crystals. Diffraction intensities were indexed and integrated with iMosflm (19) and scaled with Scala under CCP4 (20, 21). The phases were determined with molecular replacement using Phaser under Phenix (22). Further structural refinement utilized Phenix programs. Coot was used for manual map inspection and model rebuilding (23). Crystallographic calculations were performed using Phenix.
Comparative sequence and structure analyses
CHS protein sequences were derived from NCBI and the 1000 Plants (1KP) Project (24, 25). In all cases, AtCHS was used as the search query. Amino acid alignment of CHS orthologs was created using MUSCLE with default settings (26). UCSF Chimera and ESPript were used to display the multiple-sequence alignments shown in Fig. 2 and Figs. S5 and S6 (27, 28). Phylogenetic analysis was performed using MEGA7 (29). All structural figures were created with the PyMOL Molecular Graphics System, version 1.3 (Schrödinger, LLC) (30). Active-site cavity measurements for the AtCHS and AtCHS M7 structures were determined using KVFinder (31).
Enzyme assays and pKa measurement
A 4CL-CHS-coupled assay was used for kinetic analysis. A 4CL reaction master mix was made by incubating 917 nm A. thaliana 4CL1 (NCBI accession number NP_175579.1) in 100 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 5 mm ATP, 100 μm p-coumaric acid, 100 μm CoA, and 10 or 50 μm malonyl-CoA for 30 min at room temperature to generate p-coumaroyl-CoA at a final concentration of 70 μm. This 4CL was divided into individual aliquots of 196 μl in Eppendorf tubes. CHS enzyme was incubated for 30 or 60 s in 16-μl volumes using a triple buffer system (50 mm AMPSO, 50 mm sodium phosphate, 50 mm sodium pyrophosphate, various pH values) (32, 33) at room temperature in the presence of 25 μm iodoacetamide for the inactivation sample or water for the control sample. Aliquots (4 μl) were withdrawn from the incubation mixture and added to the standard coupled CHS assay system. The CHS reaction was run for 10 min at room temperature and stopped by addition of 200 μl of methanol.
The assay samples were centrifuged and analyzed directly by LC-MS. LC was conducted on a Dionex UltiMate 3000 UHPLC system (ThermoFisher Scientific), using water with 0.1% formic acid as solvent A and acetonitrile with 0.1% formic acid as solvent B. Reverse-phase separation of analytes was performed on a Kinetex C18 column, 150 × 3 mm, 2.6-μm particle size (Phenomenex). The column oven was held at 30 °C. Samples were eluted with a gradient of 5–60% B for 9 min, 95% B for 3 min, and 5% B for 3 min, with a flow rate of 0.7 ml/min. MS analysis was performed on a TSQ Quantum Access Max mass spectrometer (ThermoFisher Scientific) operated in negative ionization mode with a SIM scan centered at 271.78 m/z to detect naringenin chalcone.
The pH profiles (pH on the x axis, ratio of naringenin chalcone produced with iodoacetamide-treatment to control on the y axis) were determined by fitting raw data to the log(inhibitor) versus response equation using nonlinear regression in Prism, version 6.0f (GraphPad software).
Molecular dynamics
All MD simulations were performed using the GROMACS 5.1.4 package (34) and CHARMM force field (35). The catalytic residues were modeled as protonated histidine (His-309 in AtCHS number) and deprotonated cysteine (Cys-169 in AtCHS numbering). All CHSs were constructed as dimers and were pre-aligned to the WT AtCHS crystal structure using the Multiseq plugin of VMD (36). All CHS dimers were solvated with 0.1 m NaCl in a dodecahedron box. Before the production runs, all systems were submitted to a minimization, followed by a 500-ps NVT and a 500-ps NPT run with heavy atoms constrained. This was followed by another 5-ns NPT simulation with protein backbone constrained. In all simulations, an integration time step of 2 fs was used, with bonds involving hydrogens constrained using LINCS (37, 38). The van der Waals interaction was smoothly switched off starting from 10 Å, with a cutoff distance of 12 Å. The neighboring list was updated every 10 steps with Verlet cutoff scheme. The electrostatic interaction was evaluated using Particle-Mesh-Ewald (PME) summation (39) with a grid spacing of 1.5 Å to account for the long-range interaction, whereas its short-range interaction in real space had a cutoff distance of 12 Å. The velocity-rescaling thermostat (40) and Parrinello-Rahman barostat (41, 42) were employed to maintain the temperature at 300 K and the pressure at 1 bar.
For each CHS, three copies of 200-ns production runs were performed. The aggregated simulation time of all CHS WT and mutant systems is 5.4 μs. The two monomers of a given CHS were treated equivalently in the analysis, i.e. the three copies of trajectories of each monomer were combined after they were aligned to chain A of the associated crystal structure, resulting in a total of 1.2-μs trajectory for analysis of a given CHS system. Clustering analysis was carried out with GROMACS gmx cluster with RMSD cutoff of 0.1 nm. The inter-residue distance was measured using the tcl scripting abilities provided by VMD (43). The minimum distance between the two nitrogen atoms of the catalytic histidine and the associated hydroxyl, thiol, or thiolate group of its serine or cysteine partner was taken as the inter-residue distance. Water occupancy calculation was performed using the volmap plugin of VMD (43).
Author contributions
G. L. and J.-K. W. conceptualization; G. L., Y. W., and J.-K. W. data curation; G. L., Y. W., and J.-K. W. formal analysis; G. L., Y. W., and J.-K. W. validation; G. L., Y.-C. C., Y. W., and J.-K. W. investigation; G. L., Y.-C. C., and Y. W. visualization; G. L., Y.-C. C., Y. W., and J.-K. W. methodology; G. L., Y.-C. C., Y. W., and J.-K. W. writing-original draft; G. L., Y.-C. C., Y. W., and J.-K. W. writing-review and editing; Y.-C. C. software; Y. W. and J.-K. W. supervision; Y. W. and J.-K. W. project administration; J.-K. W. funding acquisition.
Supplementary Material
Acknowledgments
We thank Joseph Noel and members of the Noel lab of the Salk Institute for early discussion regarding catalytic cysteine oxidation that the Noel lab first observed in various plant type III PKSs, which motivated this work. This work is based on research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by National Institutes of Health Grant P41 GM103403 from NIGMS. The Pilatus 6M detector on the NE-CAT 24-ID-C beam line is funded by an National Institutes of Health-ORIP HEI Grant S10 RR029205. This research used resources of the Advanced Photon Source, a United States Department. of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This work was supported by the Howard Hughes Medical Institute, National Science Foundation Grant CHE-1709616 (to J. K. W.), Pew Scholars Program in the Biomedical Sciences Grant 27345 (to J. K. W.), Searle Scholars Program of the Kinship Foundation Grant 15-SSP-162 (to J. K. W.), and direct grants from the Chinese University of Hong Kong (to Y. W.). J. K. W. is a co-founder, a member of the Scientific Advisory Board, and a shareholder of DoubleRainbow Biosciences, which develops biotechnologies related to natural products. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S9, Table S1, supporting Note, and supporting Ref. 1.
- CHS
- chalcone synthase
- PKS
- polyketide synthase
- MD
- molecular dynamics
- Ni-NTA
- nickel–nitrilotriacetic acid
- RMSD
- root mean square deviation
- MOPSO
- 2-hydroxy-3-morpholinopropanesulfonic acid
- PDB
- Protein Data Bank.
References
- 1. Weng J.-K., and Chapple C. (2010) The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285 10.1111/j.1469-8137.2010.03327.x [DOI] [PubMed] [Google Scholar]
- 2. Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yao L. H., Jiang Y. M., Shi J., Tomás-Barberán F. A., Datta N., Singanusong R., and Chen S. S. (2004) Flavonoids in food and their health benefits. Plant Foods Hum. Nutr. 59, 113–122 10.1007/s11130-004-0049-7 [DOI] [PubMed] [Google Scholar]
- 4. Rausher M. D. (2006) in The Science of Flavonoids (Grotewold E., ed) pp. 175–211, Springer, New York [Google Scholar]
- 5. Weng J.-K., and Noel J. P. (2013) Chemodiversity in Selaginella: a reference system for parallel and convergent metabolic evolution in terrestrial plants. Front. Plant Sci. 4, 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weng J.-K., and Noel J. P. (2012) Structure–function analyses of plant type III polyketide synthases. Methods Enzymol. 515, 317–335 10.1016/B978-0-12-394290-6.00014-8 [DOI] [PubMed] [Google Scholar]
- 7. Austin M. B., and Noel J. P. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 10.1039/b100917f [DOI] [PubMed] [Google Scholar]
- 8. Jez J. M., and Noel J. P. (2000) Mechanism of chalcone synthase: pKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. J. Biol. Chem. 275, 39640–39646 10.1074/jbc.M008569200 [DOI] [PubMed] [Google Scholar]
- 9. Jez J. M., Austin M. B., Ferrer J., Bowman M. E., Schröder J., and Noel J. P. (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem. Biol. 7, 919–930 10.1016/S1074-5521(00)00041-7 [DOI] [PubMed] [Google Scholar]
- 10. Reddie K. G., and Carroll K. S. (2008) Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 12, 746–754 10.1016/j.cbpa.2008.07.028 [DOI] [PubMed] [Google Scholar]
- 11. Ferrer J. L., Jez J. M., Bowman M. E., Dixon R. A., and Noel J. P. (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784 10.1038/11553 [DOI] [PubMed] [Google Scholar]
- 12. Shirley B. W., Kubasek W. L., Storz G., Bruggemann E., Koornneef M., Ausubel F. M., and Goodman H. M. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671 10.1046/j.1365-313X.1995.08050659.x [DOI] [PubMed] [Google Scholar]
- 13. Kortemme T., and Creighton T. E. (1995) Ionisation of cysteine residues at the termini of model α-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 253, 799–812 10.1006/jmbi.1995.0592 [DOI] [PubMed] [Google Scholar]
- 14. Ashkenazy H., Penn O., Doron-Faigenboim A., Cohen O., Cannarozzi G., Zomer O., and Pupko T. (2012) FastML: a web server for probabilistic reconstruction of ancestral sequences. Nucleic Acids Res. 40, W580–W584 10.1093/nar/gks498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Austin M. B., Bowman M. E., Ferrer J.-L., Schröder J., and Noel J. P. (2004) An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11, 1179–1194 10.1016/j.chembiol.2004.05.024 [DOI] [PubMed] [Google Scholar]
- 16. Storer A. C., and Ménard R. (1994) Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 244, 486–500 10.1016/0076-6879(94)44035-2 [DOI] [PubMed] [Google Scholar]
- 17. van Montfort R. L., Congreve M., Tisi D., Carr R., and Jhoti H. (2003) Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 423, 773–777 10.1038/nature01681 [DOI] [PubMed] [Google Scholar]
- 18. Peralta D., Bronowska A. K., Morgan B., Dóka É., Van Laer K., Nagy P., Gräter F., and Dick T. P. (2015) A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 11, 156–163 10.1038/nchembio.1720 [DOI] [PubMed] [Google Scholar]
- 19. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 24. NCBI Resource Coordinators (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–D19 10.1093/nar/gkv1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matasci N., Hung L.-H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., Nguyen N., Warnow T., Ayyampalayam S., Barker M., Burleigh J. G., Gitzendanner M. A., Wafula E., Der J. P., dePamphilis C. W., et al. (2014) Data access for the 1,000 plants (1KP) project. Gigascience 3, 17 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 28. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeLano W. L. (2016) The PyMOL Molecular Graphics System, Version 1.3, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 31. Oliveira S. H., Ferraz F. A., Honorato R. V., Xavier-Neto J., Sobreira T. J., and de Oliveira P. S. (2014) KVFinder: steered identification of protein cavities as a PyMOL plugin. BMC Bioinformatics 15, 197 10.1186/1471-2105-15-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ellis K. J., and Morrison J. F. (1982) Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87, 405–426 10.1016/S0076-6879(82)87025-0 [DOI] [PubMed] [Google Scholar]
- 33. Schlegel B. P., Jez J. M., and Penning T. M. (1998) Mutagenesis of 3α-hydroxysteroid dehydrogenase reveals a “Push-Pull” mechanism for proton transfer in aldo-keto reductases. Biochemistry 37, 3538–3548 10.1021/bi9723055 [DOI] [PubMed] [Google Scholar]
- 34. Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindahl E. (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 [Google Scholar]
- 35. Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr. (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 10.1021/ct300400x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts E., Eargle J., Wright D., and Luthey-Schulten Z. (2006) MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7, 382 10.1186/1471-2105-7-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hess B., Bekker H., Berendsen H. J., Fraaije J. G., and Others (1997) LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 10.1002/(SICI)1096-987X(199709)18:12%3C1463::AID-JCC4%3E3.0.CO%3B2-H [DOI] [Google Scholar]
- 38. Hess B. (2008) P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 10.1021/ct700200b [DOI] [PubMed] [Google Scholar]
- 39. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 10.1063/1.464397 [DOI] [Google Scholar]
- 40. Bussi G., Donadio D., and Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 41. Parrinello M., and Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 10.1063/1.328693 [DOI] [Google Scholar]
- 42. Nosé S., and Klein M. L. (1983) Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50, 1055–1076 10.1080/00268978300102851 [DOI] [Google Scholar]
- 43. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–8, 27–8 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 44. Weigel D., and Glazebrook J. (2002) Arabidopsis: A Laboratory Manual, CSHL Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.