Abstract
The final steps of cell-wall biosynthesis in bacteria are carried out by penicillin-binding proteins (PBPs), whose transpeptidase domains form the cross-links in peptidoglycan chains that define the bacterial cell wall. These enzymes are the targets of β-lactam antibiotics, as their inhibition reduces the structural integrity of the cell wall. Bacterial resistance to antibiotics is a rapidly growing concern; however, the structural underpinnings of PBP-derived antibiotic resistance are poorly understood. PBP4 and PBP5 are low-affinity, class B transpeptidases that confer antibiotic resistance to Enterococcus faecalis and Enterococcus faecium, respectively. Here, we report the crystal structures of PBP4 (1.8 Å) and PBP5 (2.7 Å) in their apo and acyl-enzyme complexes with the β-lactams benzylpenicillin, imipenem, and ceftaroline. We found that, although these three β-lactams adopt geometries similar to those observed in other class B PBP structures, there are small, but significant, differences that likely decrease antibiotic efficacy. Further, we also discovered that the N-terminal domain extensions in this class of PBPs undergo large rigid-body rotations without impacting the structure of the catalytic transpeptidase domain. Together, our findings are defining the subtle functional and structural differences in the Enterococcus PBPs that allow them to support transpeptidase activity while also conferring bacterial resistance to antibiotics that function as substrate mimics.
Keywords: crystal structure, Enterococcus, antibiotic action, antibiotic resistance, enzyme structure, penicillin-binding proteins, transpeptidase, β-lactam, ESKAPE pathogen
Introduction
“ESKAPE” pathogens (1), including Staphylococcus aureus, Enterococcus faecalis, and Enterococcus faecium (the latter two collectively referred to as “enterococci”), are some of the leading causes of nosocomial (hospital-acquired) infections (2). In particular, E. faecium infections are highly prominent in hospital settings, causing bloodstream, soft-tissue, and urinary-tract infections in compromised patients with indwelling devices (implants), who have been exposed to multiple antibiotics. Unfortunately, treatment of enterococcal infections is significantly compromised by their increased resistance to most of the commonly employed antimicrobial agents, including aminoglycosides, clindamycin, and trimethoprim-sulfamethoxazole (3). Among β-lactam antibiotics, enterococci are resistant to the anti-staphylococcal penicillins and all but one cephalosporin. The three penicillins that possess any appreciable activity against enterococci (penicillin, ampicillin, and piperacillin) exhibit minimum inhibitory concentrations (MICs)3 that are significantly higher than those for streptococci. Further, rare strains of E. faecalis and most nosocomial strains of E. faecium exhibit even higher levels of resistance to these penicillins, effectively eliminating β-lactams as a treatment option (1, 2). Finally, enterococci also exhibit tolerance to the bactericidal activity of β-lactams (4), a phenomenon that compromises the use of β-lactam antibiotics as single agents in the treatment of enterococcal endocarditis (5). As a consequence, multiresistant E. faecium and E. faecalis represent one of the most dangerous threats in infectious disease therapeutics.
The bacterial cell wall, which is composed of layers of peptidoglycan modified with proteins and polymers, is essential for cell survival. In bacteria, this peptidoglycan layer is formed by the coordinated action of multiple proteins, including penicillin-binding proteins (PBPs). PBPs are transpeptidases, carboxypeptidases, and endopeptidases that synthesize new and remodel existing peptidoglycan. PBPs are classified by their enzymatic activity: 1) class A, bifunctional PBPs with both glycosyltransferase and transpeptidase activities; 2) class B, transpeptidases; and 3) class C, carboxypeptidases and endopeptidases. The focus of this study is a subgroup of the class B PBPs, which contain an N-terminal membrane-anchoring motif, an N-terminal extension of unknown function that is hypothesized to mediate protein interactions, and a C-terminal transpeptidase (TPase) domain.
Reduced susceptibility to β-lactam antibiotics in enterococci results from the expression of a single low-affinity class B PBP designated PBP4 in E. faecalis and PBP5 in E. faecium (6, 7). This low-affinity subgroup also includes PBP2a from S. aureus, an acquired PBP that confers resistance to many β-lactams (anti-staphylococcal penicillins, most cephalosporins, and carbapenems) (8). The PBP active site is located in the TPase domain and is defined by three conserved motifs: motif I, which includes the catalytic serine (SXXK; 424STFK427/422STFK425 for PBP4/PBP5); motif II, which is involved in the protonation of the β-lactam leaving group ((S/Y)XN; 482SDN484/480SDN482); and motif III, which facilitates substrate binding and defines the oxyanion hole (K(T/S)GT; 619KTGT622/617KTGT620) (Fig. 1) (9). The nucleophilic serine (Ser-424/Ser-422 for PBP4/PBP5) is located at the N terminus of helix α2, whereas the oxyanion hole is defined by the backbone nitrogen atoms of the nucleophilic serine and the motif III threonine (Thr-622/Thr-620). These motifs are bordered above by the “lid” (amino acids 445–473/443–471 for PBP4/PBP5) and below by the C-terminal helix (amino acids 657–680/655–678), which together enclose the active site in a deep cleft.
Figure 1.
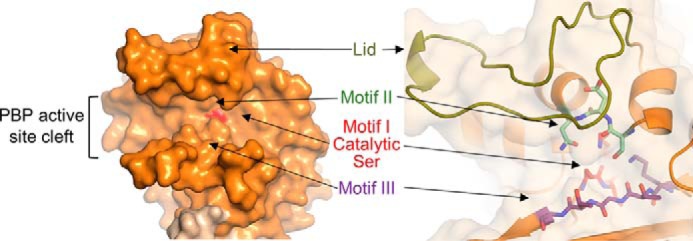
Key elements of PBP TPase domains. Left, surface representation of the PBP5 active-site cleft, with the location of the catalytic serine highlighted in red. Right, the key conserved motifs of the PBP TPase domains, including motif I, which includes the catalytic serine (SXXK, red), motif II (SXN, mint), motif III (KTG(T/S), purple), and the lid (yellow-green), which covers that active site to form the deep cleft seen at the left.
In class B PBP transpeptidases, the catalytic serine attacks the carbonyl of the penultimate d-Ala residue of a “donor” stem peptide, releasing the C-terminal d-Ala and forming a covalent acyl-enzyme adduct with the donor peptide. In a second step, the carbonyl of d-Ala adduct undergoes nucleophilic attack from a primary amine located at the extremity of a side chain of an acceptor stem peptide (9). This creates a bridge between the peptides and, in turn, links the glycan strands to one another. β-Lactams (penicillins, carbapenems, monobactams, and cephalosporins) mimic the d-Ala-d-Ala sequence in the donor substrate and function as suicide inhibitors. Since their discovery as the targets of β-lactam antibiotics, PBPs have been the subject of intense research, especially regarding their role in the resistance to β-lactams of both S. aureus and enterococci (6–8).
Because these PBPs have unusually low affinities for β-lactams, the β-lactam acylation rates are negligible compared with bacterial generation times, allowing the pathogens to survive antibiotic treatment. Of greater concern is the observation that prolonged β-lactam therapy can lead to the emergence of highly resistant strains. In E. faecium, high-level resistance correlates with mutations in the PBP5 catalytic domain that further weaken affinities for β-lactams (10). Whereas elevated resistance in E. faecalis strains is more rare, it can emerge after prolonged β-lactam treatments, often due to mutations in PBP4 (11).
Despite their clear biological and translational importance, only a handful of publications have reported fundamental molecular insights into E. faecalis PBP4 (11) or E. faecium PBP5 (12) activity, structure, and function, thereby limiting progress in the field for nearly 2 decades. Here, we determined the structures of PBP4 and PBP5 both alone and bound to multiple antibiotics. Our structures reveal that, like PBP2a from S. aureus (13), the catalytic serine changes conformation upon acylation for a subset of β-lactams. Moreover, unexpectedly, our structures reveal that N-terminal domains of these enzymes undergo unprecedented conformational changes without impacting the structure of the catalytic pocket. Together, our studies are beginning to define the subtle functional and structural differences in the enterococci PBPs that allow them to both support transpeptidase activity while also being resistant to antibiotics that function as substrate mimics.
Results
PBP4 is a critical locus responsible for reduced β-lactam susceptibility in E. faecalis
Prior studies describing deletion of pbp5 from the E. faecium genome and pbp4 from the E. faecalis genome have implicated PBP5 and PBP4 as important loci responsible for reduced penicillin susceptibility and cephalosporin resistance in clinical enterococcal strains (14, 15). Complementation studies in E. faecium have confirmed the involvement of PBP5 in resistance and detailed the role of different amino acid substitutions in higher levels of resistance (Table S1) (10). Here, we performed similar complementation experiments in E. faecalis with pbp4 expressed from the shuttle plasmid pBSU101 (16) in the pbp4-deletion mutant JH2-2Δpbp4 (Fig. 2) (14). Deletion of pbp4 from E. faecalis JH2-2 resulted in an increased sensitivity to five different β-lactam antibiotics (Table 1). The most striking increase in β-lactam sensitivity (measured as a reduction in MIC) was observed for ceftriaxone, whose MIC was reduced from >100 to 0.39 μg/ml. Ceftaroline also showed a substantial reduction, from 3.13 to <0.098 μg/ml. MICs for ampicillin, penicillin, and imipenem were less affected, reflecting the intrinsic activity of these agents against PBP4. Furthermore, reintroduction of pbp4 on a plasmid was associated with a return of resistance. Ampicillin, penicillin, and imipenem were somewhat less active than in JH2-2, presumably reflecting a 19.8-fold increase in expression from the plasmid compared with its chromosomal expression in JH2-2 (Fig. 2). Together, the data show that pbp4 is a critical locus responsible for reduced β-lactam susceptibility in E. faecalis.
Figure 2.
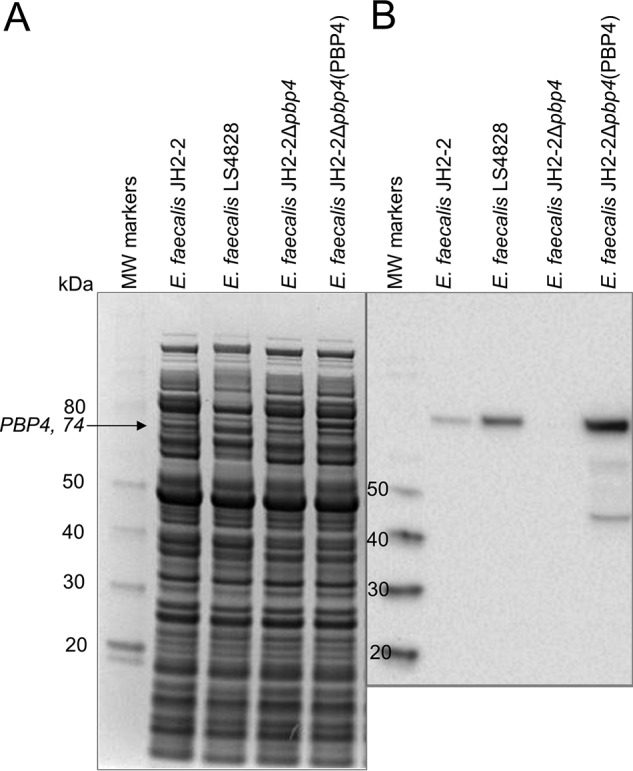
PBP4 expression from E. faecalis JH2-2, LS4828, a pbp4-deletion strain (JH2-2Δpbp4), and the deletion strain complemented with PBP4 in shuttle plasmid pBSU101 (pRIH304), JH2-2Δpbp4(PBP4). A, Coomassie-stained gel of proteins extracted for E. faecalis JH2-2, LS4828, E. faecalis JH2-2Δpbp4, and E. faecalis JH2-2Δpbp4(PBP4). B, Western blotting of the same gel using a polyclonal anti-PBP4 antibody, confirming expression of PBP4 from JH2-2 and LS4828; no band corresponding to PBP4 is observed from JH2-2Δpbp4. Expression of PBP4 in JH2-2Δpbp4(PBP4) is increased relative to JH2-2 due to the multicopy nature of the plasmid (19.8-fold compared with JH2-2).
Table 1.
MICs for different β-lactam antibiotics against E. faecalis strains
| Strain | MIC |
||||
|---|---|---|---|---|---|
| Ampicillin | Penicillin | Ceftriaxone | Imipenem | Ceftaroline | |
| μg/ml | |||||
| JH2–2 | 0.78 | 1.56 | >100 | 0.78 | 3.13 |
| JH2–2Δpbp4 | 0.39 | 0.78 | 0.39 | 0.39 | <0.098 |
| LS304Δpbp4(PBP4) | 1.56 | 6.25 | >100 | 3.13 | 6.25 |
PBP4 from E. faecalis adopts a structure similar to PBP5 from E. faecium
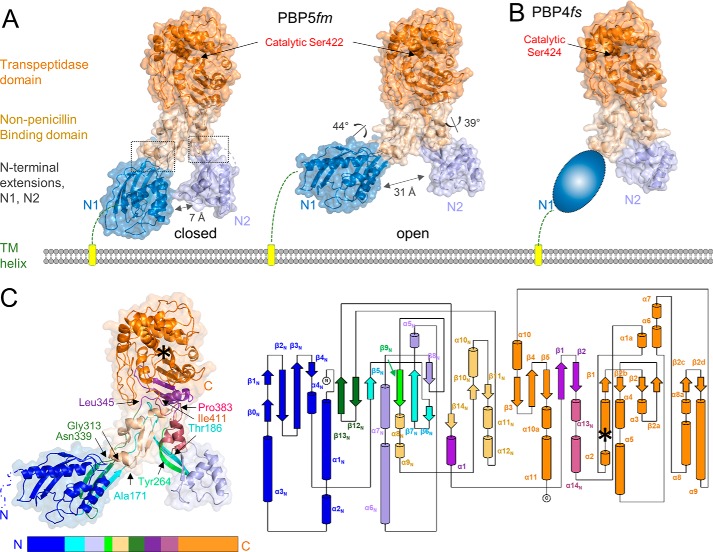
We used X-ray crystallography to determine the structures of PBP4 (E. faecalis PBP4(36–680), which lacks the N-terminal membrane anchor; hereafter referred to as PBP4) and PBP5 (E. faecium PBP5(37–678), which also lacks the N-terminal membrane anchor; hereafter referred to as PBP5) (Table 2). We obtained two different crystal forms of PBP5, which allowed us to determine the structures of PBP5 in two distinct conformations (open and closed) to resolutions of 2.7 and 2.9 Å, respectively (Fig. 3A). Strong electron density was observed for the entire sequence with the exception of three loops for the open conformation (140–144, 246–249, and 625–630). The structure of PBP4 was determined to 1.8 Å resolution. Strong electron density was observed for residues 172–680; interpretable electron density was not observed for the N1 domain and three short loops (Fig. 3B). The structures show that both PBPs are composed of four distinct structural domains: two N-terminal domains (N1 and N2), a nonpenicillin-binding domain (nPB), and a C-terminal catalytic TPase domain, which contains the nucleophilic serine (Fig. 3, A and B). With the exception of the N2 domain, all domains are composed of residues that are not linear in sequence, giving rise to an extensively interconnected topology (Fig. 3C; residue numbers for PBP5: N1, 43–171/314–339; N2, 192–258; nPB, 172–191/259–313/340–348/384–409; TPase, 349–383/410–678).
Table 2.
Data collection and refinement statistics
| PBP5 open | PBP5 closed | PBP5 + benzylpenicillin | PBP5 + imipenem | PBP4 apo | PBP4 + benzylpencillin | PBP4 + imipenem | PBP4 + ceftaroline | |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P6322 | P43212 | P6322 | P6322 | C2 | C2 | C2 | C2 |
| Cell dimensions | ||||||||
| a, b, c (Å) | 190.5, 190.5, 156.5 | 62.7, 62.7, 371.2 | 192.3, 192.3, 156.1 | 192.9, 192.9, 155.9 | 124.7, 84.6, 85.1 | 123.3, 85.5, 83.7 | 123.8, 85.4, 84.6 | 123.5, 85.5, 82.7 |
| α, β, γ (degrees) | 90, 90, 120 | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 | 90, 109.3, 90 | 90, 109.5, 90 | 90, 110.3, 90 | 90, 109.9, 90 |
| Resolution (Å) | 39.5–2.70 (2.79–2.70)a | 30.0–2.86 (2.94–2.86) | 39.0–2.93 (3.06–2.93) | 29.8–2.80 (2.91–2.80) | 17.5–1.80 (1.83–1.80) | 17.7–2.34 (2.38–2.34) | 37.9–2.62 (2.74–2.62) | 39.0–2.98 (3.17–2.98) |
| Rmerge | 0.059 (0.445) | 0.152 (0.575) | 0.255 (2.78) | 0.157 (1.66) | 0.072 (0.430) | 0.162 (0.606) | 0.058 (0.211) | 0.136 (0.396) |
| I/σ(I) | 13.0 (2.5) | 12.7 (3.0) | 9.2 (1.7) | 20.1 (1.8) | 16.5 (2.5) | 9.4 (2.1) | 12.5 (3.7) | 5.4 (2.3) |
| CC½ | 0.997 (0.707) | 0.997 (0.778) | 0.991 (0.379) | 0.999 (0.337) | 0.998 (0.685) | 0.991 (0.750) | 0.959 (0.321) | 0.890 (0.881) |
| Completeness | 0.99 (0.98) | 0.99 (0.91) | 0.99 (0.98) | 0.99 (0.99) | 0.99 (0.87) | 0.99 (1.00) | 0.96 (0.77) | 0.98 (0.92) |
| Redundancy | 3.1 (3.2) | 12.9 (9.9) | 7.5 (7.5) | 18.3 (12.9) | 6.7 (1.6) | 8.1 (6.0) | 4.6 (4.0) | 3.8 (3.8) |
| Refinement | ||||||||
| Resolution (Å) | 39.5–2.7 | 29.7–2.9 | 39.0–2.9 | 29.8–2.8 | 17.5–1.80 | 17.7–2.34 | 37.9–2.62 | 39.0–2.98 |
| Unique reflections | 45,727 | 18,016 | 36,572 | 42,395 | 76,417 | 34,537 | 23,930 | 16,139 |
| Rwork/Rfree | 0.18/0.21 | 0.22/0.28 | 0.19/0.21 | 0.17/0.20 | 0.19/0.21 | 0.20/0.22 | 0.20/0.23 | 0.23/0.26 |
| No. of atoms | 4,874 | 4,478 | 4,880 | 5,047 | 4,239 | 3,597 | 3,482 | 3,189 |
| Protein | 4,664 | 4,473 | 4,726 | 4,788 | 3,623 | 3,167 | 3,371 | 3,108 |
| Ligand/ion | 85 | 115 | 95 | 15 | 39 | 30 | 55 | |
| Water | 125 | 5 | 39 | 164 | 601 | 391 | 81 | 26 |
| B factors | ||||||||
| Protein | 56.8 | 57.6 | 63.5 | 76.8 | 20.7 | 28.3 | 65.2 | 56.8 |
| Ligand/ion | 106.5 | 101.8 | 100.7 | 41.0 | 46.4 | 91.7 | 83.9 | |
| Water | 50.0 | 38.3 | 45.8 | 67.7 | 33.3 | 37.8 | 62.0 | 46.8 |
| RMSDs | ||||||||
| Bond lengths (Å) | 0.006 | 0.002 | 0.002 | 0.002 | 0.015 | 0.002 | 0.002 | 0.002 |
| Bond angles (degrees) | 0.743 | 0.497 | 0.453 | 0.494 | 1.17 | 0.478 | 0.428 | 0.555 |
| PDB code | 6MKA | 6MKJ | 6MKG | 6MKF | 6BSQ | 6BSR | 6MKH | 6MKI |
a Values in parentheses are for the highest-resolution shell.
Figure 3.
PBP4 and PBP5 contain four distinct domains, whose topologies are highly interconnected. A, the closed (left) and open (right) conformations of E. faecium PBP5, colored by domain (N1 in dark blue, N2 in light blue, nPB in beige, and TPase in orange). The rotations of the N1 and N2 domains relative to the nBP/TPase domains, which result in a 24-Å opening of the cleft, are indicated, with the locations of the hinges shown as a dashed box. The location of catalytic Ser-422 is indicated by an arrow. The N-terminal TM helix is shown as a yellow cylinder. B, structure of E. faecalis PBP4 colored as in A. No interpretable electron density was observed for the N1 domain, presumably because it adopts multiple conformations; it is illustrated here as a cartoon. C, secondary structure (left) and topology map (right) of PBP5. Colors from the N-to-C terminus are indicated below the structure; a topology map of PBP5 is colored as in C. The location of the catalytic serine is indicated by an asterisk.
The PBP identified to be most similar to PBP4 is PBP5 (determined using the DALI structural homology server (17); Z-score of 62.5; sequence identity of 60%), with the individual domains of both proteins superimposing with root mean square deviations (RMSDs) of 0.75–1.1 Å (Table S2). The largest differences are observed in the TPase domain (structural elements that brace one side of the active site are shifted by 1.5–2.0 Å in PBP4 due to the presence of a tyrosine in PBP4, Tyr-605, which, in PBP5, is a much smaller threonine residue, Thr-603) (Fig. S1B). The next most similar PBP is PBP2a from S. aureus (hereafter referred to as PBP2a (13); Z-scores of 49.3 and 51.0 and sequence identities of 41 and 38% for PBP4 and PBP5 versus PBP2a, respectively). The PBP2a TPase domain aligns with those of PBP4 and PBP5 with >2.0-Å RMSD. Together, these proteins represent the defining members of the low-affinity, high-molecular-weight, class B PBPs.
The N1 and N2 domains are mobile, rotating as rigid bodies independently of the TPase domain
Overlaying the three members of this family (PBP4, PBP5 (open and closed), and PBP2a) on the TPase domain reveals that the N1 and N2 domains are mobile, adopting a wide range of conformations relative to the TPase domain. In particular, transforming the PBP5 closed to the PBP5 open state requires rotations of 44 and 39° by the N1 and N2 domains, respectively. This results in a widening of the N1–N2 cleft from 7 Å (closed) to 31 Å (open; Fig. 3A). The mobile nature of the N1 domain was confirmed by our structure of PBP4. Namely, no interpretable electron density was observed for the PBP4 N1 domain, suggesting that it adopts multiple conformations in the crystal. Finally, the N1 domain of PBP2a adopts a third conformation (13), with 75 and 88° rotations relative to those of PBP5 open and PBP5 closed, respectively. Similar observations were made for the N2 domain, in which all four N2 domains (PBP4, PBP5 open, PBP5 closed, and PBP2a) adopt distinct positions and are related to one another by rotations of 39–52° (Fig. 4A and Fig. S1A). How these domain rotations affect and/or direct transpeptidase activity is currently unknown.
Figure 4.
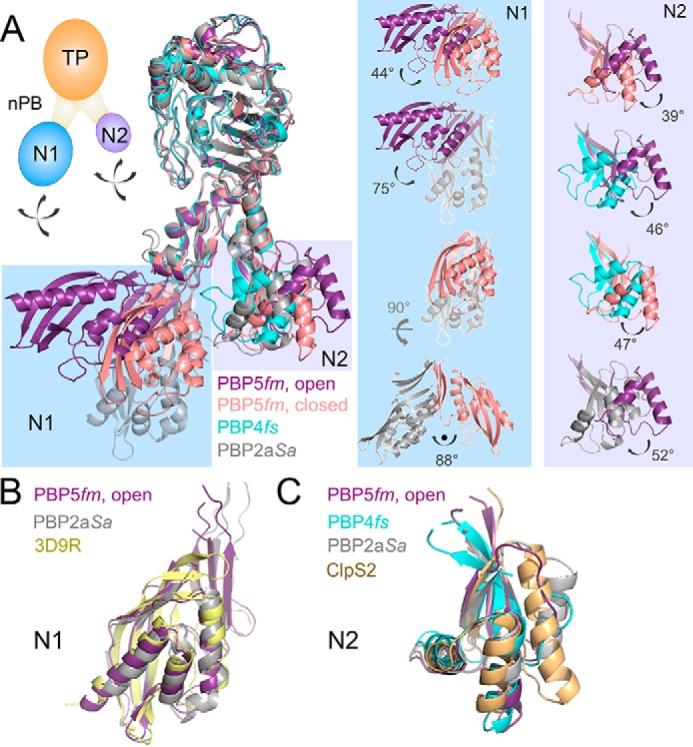
The N1 and N2 domains of the low-affinity subclass of class B PBPs are highly dynamic. A, left, superposition of PBP5fm (open, purple), PBP5fm (closed, pink), PBP4fs (cyan), and PBP2aSa (gray) using their respective TPase domains. The N1 and N2 domains of are highlighted in light blue and light purple, respectively. Right, the magnitudes of the rotations observed between the various N1 (light blue) and N2 (light purple) domains are indicated. Inset, cartoon of the PBP domains, with the N1 and N2 domains shown as dashed lines with arrows to highlight their ability to rotate freely of one another. Overlaid residues are defined in Tables S2 and S4. B, superposition of the N1 domains from PBP5 (purple), PBP2a (gray), and the ketosteroid isomerase-like protein from P. atrosepticum (PDB code 3D9R, yellow). C, superposition of the N2 domains from PBP5 (purple), PBP4 (cyan), PBP2a (gray), and ATP-dependent CLP protease adaptor protein CLPS2 (PDB code 4YJM, beige).
The function of the N-terminal domains (N1 and N2) of the high-molecular-weight class B PBP family is still under debate, with some suggesting that they function as protein–protein interaction domains (18) and others suggesting that they function as molecular “spacers” (19) (i.e. sterically constraining the catalytic site to remain a certain distance from the cell membrane). We used the DALI structural homology server to identify the proteins that are most similar to the N1 and N2 domains, to gain insights into the potential function of these domains. The data showed that the N-terminal domains are differentially conserved within the larger PBP family. Whereas the N1 domain (PBP5) is only found in PBP2a (S. aureus; Z-score, 17.7 (13)), the N2 domain (PBP5 and PBP4) is found in PBP2a (S. aureus; Z-score, 10.0 (13)), PBP3 (P. aeruinosa; Z-score, 7.3 (20)), and PBP1 (S. aureus; Z-score, 6.6). Unexpectedly, they were also shown to be structurally similar to proteins outside the PBP family. Namely, the N1 domain was identified as being similar to ketosteroid isomerase domains (Z-score, 12.9; Fig. 4B; PDB code 3D9R, yellow), whereas the N2 domain was identified as being similar to the ATP-dependent CLP protease adaptor protein CLPS2 (Z-score, 5.9; Fig. 4C; PDB code 4YJM, beige (21)). These latter observations are consistent with the hypothesis that the class B PBP N-terminal domains function as protein interaction domains. The proteins that interact directly with the N1 and N2 domains from PBP4, PBP5, and PBP2a, if they exist, are still unknown.
Conformational changes associated with β-lactam acylation are localized to the catalytic site
Previous studies of methicillin-resistant PBP2a showed that PBP2a motif III (strand β3) is twisted relative to the methicillin-susceptible PBP2. This twist causes the carbonyl oxygen of the motif III Thr (the last “T” in the K(T/S)GT motif) to point toward the oxyanion hole. However, PBP2a β-lactam acylation causes the carbonyl to rotate out of the oxyanion hole and adopt a position identical to that observed in PBP2 (13). These data suggested that the slow acylation rate of PBP2a may be due to this distorted conformation of β3.
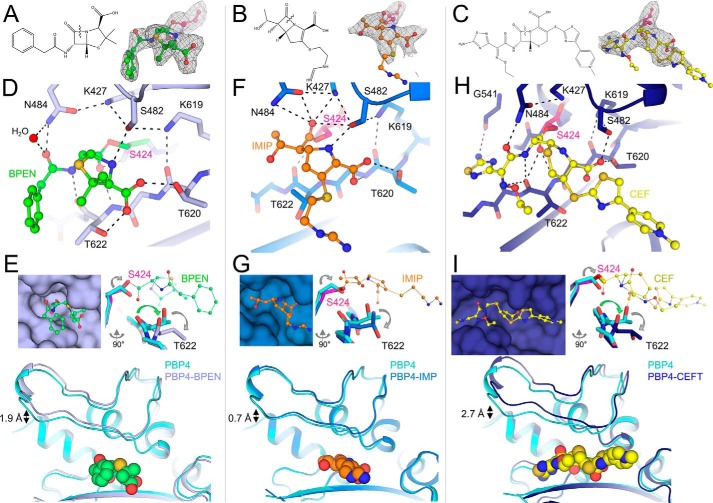
Like PBP2a, the β-lactam resistance of PBP5 and PBP4 has also been shown to be due to the inefficient formation of the acyl-PBP intermediate. To understand the molecular basis of PBP4 and PBP5 resistance to β-lactams, we determined the structures of five distinct acyl-enzyme complexes, PBP4:benzylpenicillin, PBP4:imipenem, PBP4:ceftaroline, PBP5:benzylpenicillin, and PBP5:imipenem, representing three different β-lactam classes (penicillins, benzylpenicillin; carbapenems, imipenem; cephalosporins, ceftaroline). Examination of the active sites revealed strong density for all covalent acyl-enzyme adducts (Fig. 5, A–C). As observed for PBP2a, superposition of the apo and β-lactam–bound structures revealed that acyl-adduct formation does not result in global conformational changes in either PBP5 or PBP4 (the RMSDs between the apo- and the drug-bound transpeptidase domains are between 0.31 and 0.53 Å (Table S3)). Rather, the changes are localized to domain movements about the active site and changes in the structures of the active-site motifs. However, as described below, the changes are not identical between the free and β-lactam–bound structures. Instead, they depend upon the particular acyl-adduct formed. Finally, the differences observed between apo-PBP4 and β-lactam-acyl-PBP4 and between apo-PBP5 and β-lactam-acyl-PBP5 are essentially identical for benzylpenicillin and imipenem.
Figure 5.
β-Lactam adduct formation results in similar, but distinct, sets of conformational changes at the PBP4 TPase catalytic cleft. A, left, the structure of benzylpenicillin. Right, Fo − Fc electron density (Polder map; contoured at 3.5σ to 2.4 Å resolution; top right) for the benzylpenicillin-acylated Ser-424. B, left, structure of imipenem. Right, Fo − Fc electron density (Polder map; contoured at 3.5σ to 2.6 Å resolution; top right) for the imipenem-acylated Ser-424. C, left, the structure of ceftaroline. Right, Fo − Fc electron density (contoured at 3.0σ to 3.0 Å resolution; top right) for the ceftaroline-acylated Ser-424. D, active site in the benzylpenicillin-acyl-PBP4 structure (benzylpenicillin in green, PBP4 in light blue, and Ser-424 in pink). Polar interactions are indicated by dashed lines. E, left, surface representation of PBP4 (light blue) with bound benzylpenicillin (green). Right, overlay of nonacylated PBP4 (cyan) with benzylpenicillin-acyl-PBP4 (light blue; acylated Ser-424 in pink; view rotated by 90° compared with that in D. Conformational changes in backbone and side-chain atoms are highlighted with arrows. Bottom, overlay orientation the same as that in D, highlighting opening of the lid to accommodate benzylpenicillin binding. F, active site in the imipenem-acyl-PBP4 structure (imipenem in orange, PBP4 in blue, and Ser-424 in pink). Polar interactions are indicated by dashed lines. G, left, surface representation of PBP4 (blue) with bound imipenem (orange). Right, overlay of nonacylated PBP4 (cyan) with imipenem-acyl-PBP4 (blue; acylated Ser-424 in pink; view rotated by 90° compared with that in D. Conformational changes in backbone and side-chain atoms are highlighted with arrows. Bottom, overlay orientation the same as that in F, highlighting opening of the lid to accommodate imipenem binding. H, active site in the ceftaroline-acyl-PBP4 structure (ceftaroline in yellow, PBP4 in dark blue, and Ser-424 in pink). Polar interactions are indicated by dashed lines. I, left, surface representation of PBP4 (dark blue) with bound ceftaroline (yellow). Right, overlay of nonacylated PBP4 (cyan) with ceftaroline-acyl-PBP4 (blue; acylated Ser-424 in pink; view rotated by 90° compared with that in H. Conformational changes in backbone and side chain atoms are highlighted with arrows. Bottom, overlay orientation the same as that in H, highlighting opening of the lid to accommodate ceftaroline binding.
Penicillins: Benzylpenicillin acylation induces a rotation of the nucleophilic serine and a twist of strand β3
As expected, the benzylpenicillin forms a covalent adduct with PBP4 via its catalytic serine, Ser-424. The electron density is well-defined for the entire molecule (Fig. 5A), with the benzylpenicillin carbonyl oxygen pointing toward the oxyanion hole defined by the backbone nitrogen atoms of Ser-424 and Thr-622 (Fig. 5D). The benzylpenicillin is further stabilized by 1) polar contacts with both PBP4 backbone atoms and the side chains of Ser-482, Asn-484, Lys-619, Thr-620, and Thr-622 and 2) the formation of intraprotein contacts between Lys-427 and Asn-484 and Ser-482.
The PBP4 apo and benzylpenicillin-acyl-PBP complex structures reveal that, like PBP2a, PBP4 has a distorted active site that undergoes both local and distributed conformational changes upon β-lactam acylation (Fig. 5E). First, in apo-PBP4, the nucleophilic Ser-424 hydroxyl points down toward the oxyanion catalytic pocket. However, upon acylation with benzylpenicillin, the Cβ–Oγ bond rotates by ∼150°. As a consequence, Oγ moves by 1.3 Å, and, in the acylated complex, is now oriented away from the oxyanion hole. Although this conformational change differs from that observed for Ser-403 in PBP2a, which results in a shift but not a rotation of the nucleophilic hydroxyl, it does suggest that the serine side chain is not ideally positioned for nucleophilic attack in the apo conformation. Because the β-lactam carbonyl now points toward the oxyanion hole, it displaces the carbonyl of Thr-622, which rotates out of the hole and upward toward the bound β-lactam moiety (Fig. 5E). This rotation of strand β3 results in the formation of two additional hydrogen bonds: 1) the first between the β-lactam carbonyl and the amide hydrogen of Thr-622 and 2) the second between the phenylacetamide nitrogen from benzylpenicillin and the Thr-620 side-chain hydroxyl. Finally, the Thr-618 side chain adopts a rotomer conformation that expands the active site to accommodate the β-lactam phenyl ring. The conformation of benzylpenicillin in the PBP4:benzylpenicillin complex is similar to other benzylpenicillin-bound PBP structures, with the exception that the phenylacetamidoyl group is observed in some structures to be rotated upward away from motif III. Finally, because the catalytic site of PBP4 is located in a deep, narrow cleft (Fig. 5E), the structural elements that enclose the catalytic site open in order to accommodate benzylpenicillin acylation. This results in a displacement of the “lid” moiety by 1.9 Å for benzylpenicillin-acyl-PBP4 compared with its apo conformation (Fig. 5E).
Carbapenems: Imipenem acylation does not alter the twist of strand β3
Imipenem also forms a covalent adduct with Ser-424, and its electron density is well-ordered for the entire molecule with the exception of its iminomethyl-amino tail (Fig. 5B). Overall, imipenem binds PBP4 in a manner similar to that observed in the benzylpenicillin-acyl-PBP complex; however, there are also distinct differences. As observed in the benzylpenicillin-acyl-PBP4 complex, the side chain of Ser-424 rotates out of the oxyanion hole to form the acyl-enzyme adduct (Fig. 5F). However, unlike in benzylpenicillin, where the β-lactam carbonyl points downward into the oxyanion hole, the imipenem carbonyl points upward, away from the oxyanion hole, where it hydrogen-bonds with Lys-427 and Asn-484 (Fig. 5F). As a result, the carbonyl of motif III Thr-622 does not rotate out of the oxyanion hole, but instead retains the twisted conformation of β3 observed in the apo-state (Fig. 5G). Additional polar contacts are observed between imipenem and the backbone and/or side-chain atoms of Ser-482, Thr-620, and Thr-622. The conformations of imipenem bound to PBP4 and PBP5 are identical to one another and other imipenem-transpeptidase complexes. The single exception is that the iminomethyl-amino-ethyl tail of imipenem adopts a wide range of conformations, an observation consistent with a lack of strong electron density for this element. Finally, as observed for benzylpenicillin, both the lid and central β-sheet of PBP4 open to accommodate imipenem acylation. However, they do so to a far lesser extent than for benzylpenicillin, with the lid moving by only 0.7 Å (Fig. 5G).
Cephalosporins: Ceftaroline acylation results in the widest opening of the catalytic cleft
We also determined the structure of the ceftaroline-acyl-PBP4 complex (Fig. 5C). Although the related PBP2a complex has been proposed to bind two molecules of ceftaroline (one forming the acyl-enzyme complex and a second molecule binding at a putative allosteric site located between the N1 and nBP domains), we observed density for only a single ceftaroline molecule in PBP4. The ceftaroline carbonyl oxygen points toward the oxyanion hole defined by the backbone nitrogen atoms of Ser-424 and Thr-622 (Fig. 5H). Ceftaroline is further stabilized by multiple polar interactions with the side chains of Ser-482, Asn-484, Lys-619, and Thr-620 and the backbone atoms of Gly-541 and Thr-622 and intraprotein polar interactions between Lys-427, Ser-482, Asn-484, and Lys-619.
The changes in PBP4-associated ceftaroline-acyl-PBP4 adduct formation are more similar to those observed with benzylpenicillin than with imipenem. First, as observed for both benzylpenicillin and imipenem, ceftaroline acylation results in a rotation of the nucleophilic serine upward away from the oxyanion hole (Fig. 5I). However, as observed for benzylpenicillin, acylation of PBP4 by ceftaroline displaces the Thr-622 carbonyl out of the oxyanion hole, causing strand β3 to twist outward. This new orientation of the Thr-622 carbonyl is stabilized by a hydrogen bond with ceftaroline. An identical polar interaction is present in the penG-acyl-PBP4 complex between the phenylacetamide nitrogen from benzylpenicillin and the Thr-622 carbonyl. In contrast, imipenem lacks a nitrogen in the corresponding position, explaining why the imipenem-acyl-PBP complex does not result in a rotation of strand β3. Finally, adduct formation with ceftaroline results in the greatest opening of the catalytic cleft, with the lid and central β-sheet both moving by ∼2.7 Å to accommodate ceftaroline binding (Fig. 5I).
β-Lactam acylation does not increase PBP4 and PBP5 thermal stability
To determine whether β-lactam acylation increases PBP4 and PBP5 stability, we used differential scanning fluorimetry (DSF). We first compared the melting temperature (Tm) values of free PBP4 and PBP5. The DSF data show that they unfold at similar temperatures, with Tm of 54.4 and 53.2 °C for PBP4 and PBP5, respectively (Table 3). We then incubated both PBPs with either benzylpenicillin, imipenem, or ceftaroline for either 1 or 12 h and repeated the DSF measurements. The data show that all three β-lactams alter the PBP Tm values and do so in a manner that depends on both the β-lactam and the PBP protein. First, imipenem modestly increases the Tm values of both PBP4 and PBP5, with the largest increase observed for PBP4 incubated for 12 h (ΔTm +1.8 °C), suggesting that it slightly stabilizes the conformation of both PBPs. In contrast, whereas benzylpenicillin results in an increase in the PBP5 Tm (up to Tm +1.8 °C at a 12-h incubation), it decreases the Tm for PBP4 (−0.8 °C for the 1-h incubation). This suggests that acylation by benzylpenicillin slightly increases the stability of PBP5 while having the opposite effect on PBP4. The largest differences in Tm values between the apo and acylated PBPs were observed with ceftaroline (between −2.0 and 4.2 °C). Further, whereas ceftaroline destabilized PBP4 (ΔTm −3.7 °C for a 1-h incubation), it stabilized PBP5 (ΔTm +4.2 °C for a 12-h incubation). Taken together, the data show that both benzylpenicillin and imipenem have measurable but modest effects on the Tm values of both PBP4 and PBP5, whereas ceftaroline has much larger effects, consistent with the larger conformational changes observed between PBP4 and the ceftaroline-acyl-PBP4 adduct.
Table 3.
DSF Tm values for free and acylated PBP4 and PBP5
Tm values reported are the average and S.D. of four independent measurements.
| PBP | 1-h incubation |
12-h incubation |
||
|---|---|---|---|---|
| Tm | ΔTm* | Tm | ΔTm* | |
| °C | °C | |||
| PBP4 | ||||
| Apo | 54.4 ± 0.1 | 53.4 ± 0.2 | ||
| Benzylpenicillin | 53.6 ± 0.1 | −0.8 | 53.0 ± 0.1 | −0.4 |
| Imipenem | 54.9 ± 0.2 | +0.5 | 55.2 ± 0.1 | +1.8 |
| Ceftaroline | 50.8 ± 0.1 | −3.6 | 51.4 ± 0.1 | −2.0 |
| PBP5 | ||||
| Apo | 53.2 ± 0.1 | 53.0 ± 0.1 | − | |
| Benzylpenicillin | 53.9 ± 0.3 | +0.7 | 54.8 ± 0.2 | +1.8 |
| Imipenem | 53.4 ± 0.2 | +0.1 | 53.2 ± 0.3 | +0.2 |
| Ceftaroline | 56.4 ± 0.1 | +3.2 | 57.2 ± 0.3 | +4.2 |
aΔTm is the difference in Tm relative to that of the corresponding apo-PBP.
Discussion
PBP4, PBP5, and PBP2a are the defining members of the low-affinity high-molecular-weight class B PBPs (22). The availability of the three-dimensional structures of all three PBPs in multiple states provides a unique opportunity to define the structural and functional similarities and differences within this family and how this impacts their ability to be inhibited by β-lactams. As observed for other PBPs (13, 23, 24), β-lactam binding to PBP4 and PBP5 does not lead to extensive conformational changes. Rather, the changes are exclusively localized to and around the catalytic pocket. The conformations observed both in the apo- and β-lactam–bound states are similar for all members of the low-affinity class B high-molecular-weight PBPs (i.e. PBP4, PBP5, and PBP2a) (13) (this work). Namely, as observed for apo-PBP2a (13), the catalytic serines of PBP4 and PBP5 are not ideally positioned for nucleophilic attack. Further, strand β3 is twisted toward the catalytic pocket, with the PBP4 Thr-622 carbonyl (PBP5, Thr-620) pointing toward the oxyanion hole. As observed for PBP2a upon acylation with nitrocefin, the catalytic serines of both PBP4 and PBP5 rotate upward upon acylation with benzylpenicillin (PBP4/PBP5) or ceftaroline (PBP4), whereas strand β3 rotates upward and displaces the PBP4 Thr-622 (PBP5 Thr-620) carbonyl out of the oxyanion hole. However, this is only observed for a subset of β-lactams, as it is not observed when either PBP4 or PBP5 are acylated with imipenem. Although the unusual twisted conformation of strand β3 (and its rotation upon acylation with a subset of β-lactams) was originally identified in PBP2a, and now in PBP4 and PBP5, it has also been identified in other PBPs, including PBPa from Mycobacterium tuberculosis (23) and PBP2x from Streptococcus pneumoniae (24). Further, the observation in PBP4 and PBP5 that the rotation of strand β3 upon β-lactam acylation depends on the nature of the β-lactam also holds true for PBPa (23). Namely, in PBPa, strand β3 also rotates upon acylation with benzylpenicillin but remains in a twisted conformation upon acylation with imipenem (i.e. identical to what we observed for PBP4 and PBP5).
The second major conformational change associated with β-lactam acylation is the widening of the catalytic cleft that accommodates β-lactams and substrates. PBP4 and PBP5, in particular, are characterized by an unusually deep catalytic pocket that is bounded by multiple structural elements, which includes a lid that braces the top of the catalytic pocket. This lid must open in order for β-lactams (and substrates) to access the catalytic serine. Once acylated, these elements maintain a more open conformation (between 0.7 and 2.7 Å) due to the presence of the β-lactam. This widening of the catalytic cleft is routinely observed in PBPs whose structures have been determined with and without β-lactam acylation.
Notably, kinetic and structural studies have suggested that one member of this family, PBP2a, contains an allosteric pocket at the N1-nBP junction that, when occupied, biases the conformation of the PBP2a active site toward a state that is more susceptible to ceftaroline acylation (25), a model that is still controversial (26). Our discovery that the N1 and N2 domains rotate independently of the TPase domain and do not alter the conformation of the catalytic site suggests that PBP4 and PBP5 may not be regulated by allostery. This is supported by our structures, which show that all β-lactams tested bind PBP4 and PBP5 with 1:1 stoichiometries, forming only acyl-enzyme complexes with the catalytic serine. Overlaying our PBP5 structures with that of PBP2a bound to the putative allosteric ceftaroline demonstrates that, both in the open and closed PBP5 conformations, the corresponding pocket is not present in these PBPs. This is because, in PBP5, the short nBP α-helix clashes with the PBP2a-bound allosteric ceftaroline molecule (the corresponding helix in PBP2a is in a different position). Further, the very few PBP2a residues that contact the noncovalently bound ceftaroline are not conserved in either PBP4 or PBP5. Together, these data suggest that PBP4 and PBP5 do not contain an allosteric binding pocket and instead bind ceftaroline in a 1:1 ratio.
Finally, although modest conformational changes in the N-terminal domains have been observed in other PBPs, our structures demonstrate that the N1 and N2 domains in this family of proteins are highly mobile (Fig. 4), despite their highly interconnected nature (Fig. 3C). As an example, our PBP5 structures show that rotations of only 45° result in an opening of the N1–N2 cleft by more than 24 Å. Importantly, these structures also show that these rotations occur without altering the conformation the domains themselves (i.e. the domains rotate as rigid bodies (the nBP and TPase domains of the open and closed states superimpose on one another with RMSD values < 0.25 Å)). As a consequence, the structure of the catalytic site is unchanged between the two conformations. To our knowledge, this is the first time such large conformational changes have been observed in this family of PBPs. One consequence of this interdomain mobility is that the distance between the catalytic site and the cell membrane can change. For example, overlaying PBP2a onto PBP5 via their N1 domains shows that whereas the PBP5 active site is 65 Å away from the cell membrane, the PBP2a active site is 30 Å further, 95 Å away from the cell membrane (Fig. S2). Thus, although the function of PBP domain mobility is currently unknown, it is possible that these motions, via a mechanism yet to be discovered, facilitate the building of the cell wall.
Experimental procedures
Complementation of pbp4 in E. faecalis pbp4-deletion mutant JH2-2Δpbp4
pbp4 was amplified from E. faecalis JH2-2 (including the 200 bp upstream of the structural gene; native promoter) and ligated to pBSU101 after removal of the pBSU101 native promoter and gfp gene. This plasmid was then introduced by electroporation into Escherichia coli DH10B. Successful transformation was confirmed by plasmid extraction and restriction digestion, along with PCR amplification and sequencing of pbp4 within the plasmid. The final plasmid (pRIH304) was then transformed into E. faecalis JH2-2Δpbp4 by electroporation (spectinomycin selection; 125 μg/ml). Expression of PBP4 was confirmed by Western blotting using a polyclonal anti-PBP4 antibody as described previously (Fig. 2) (11).
Cloning and expression of PBP4 and PBP5
E. faecalis PBP4(36–680) and E. faecium PBP5(37–678), which lack the N-terminal transmembrane domains, were subcloned into the pRP1b expression vector (27) encoding an N-terminal His6 tag and a tobacco etch virus cleavage site and expressed in E. coli BL21 (DE3) cells. Cells were grown in Luria broth in the presence of selective antibiotics at 37 °C up to A600 of 0.8–1.0, and expression was induced by the addition of isopropyl-β-d-1-thiogalactopyranoside (1.0 and 0.5 mm for PBP4 and PBP5, respectively). Proteins were expressed for ∼18 h at 18 °C prior to harvesting by centrifugation at 8,000 × g for 15 min at 4 °C. Bacterial pellets were used immediately or stored at −80 °C. For purification, cell pellets were resuspended in lysis buffer (50 mm Tris, pH 8.5, 500 mm NaCl, 5 mm imidazole, 0.1% Triton X-100) and lysed using high-pressure homogenization (Avestin EmulsiFlex C3). The cell lysate was centrifuged at 42,500 × g for 55 min at 4 °C. The clarified supernatant was loaded onto a pre-equilibrated HisTrap HP column (GE Healthcare) and eluted using an imidazole gradient. The fractions containing the PBP (PBP4 or PBP5) were pooled and dialyzed for 48 h at 4 °C with tobacco etch virus protease to remove the N-terminal His6 tag. A second nickel-nitrilotriacetic acid purification step was used to remove the cleaved His6 tag, any remaining uncleaved protein, and the His6-tagged protease. The cleaved PBP4 and PBP5 were then dialyzed for 3 h at 20 °C against 10 mm Tris, pH 8.0, 1.5 m (NH4)2SO4, after which they were loaded onto a pre-equilibrated HiTrap Phenyl HP hydrophobic interaction column (GE Healthcare). Fractions containing the PBP were pooled, dialyzed against size-exclusion chromatography (SEC) buffer (10 mm Tris, pH 8.5, and 300 mm NaCl (PBP4) or 800 mm NaCl (PBP5)) for 3 h at 20 °C and purified by SEC (Superdex 200 26/60; GE Healthcare). The resulting monomeric peaks were pooled and concentrated to 12 mg/ml (PBP4) or 15 mg/ml (PBP5).
Crystallization
PBP4
Purified PBP4 crystallized in 40 mm KH2PO4, pH 3.25, 16% PEG 8000, 20% glycerol, 10 mm TCEP (vapor diffusion). PBP4-acylated complexes with benzylpenicillin, imipenem, and ceftaroline were obtained by soaking PBP4 crystals with a 40-fold molar excess of the β-lactam for 45 min (benzylpenicillin) or 1 h (imipenem/ceftaroline). Crystals used for soaking were grown in the same conditions used for apo-PBP4 with the exception that the TCEP was eliminated, and, for the benzylpenicillin-acyl-PBP4 complex, the crystals were grown in 24% glycerol. PBP4 apo and complex crystals were harvested directly from the well and immediately flash-frozen in liquid nitrogen.
PBP5, open state
The open state of PBP5 crystallized in 0.1 m trisodium citrate, pH 5.5, 2.0 m ammonium sulfate (vapor diffusion). The penicillin-acyl-PBP5 complex was obtained using co-crystallization using the same conditions, but also including a 20-fold molar excess of benzylpenicillin. The impenem-acyl-PBP5 complex was obtained by soaking PBP5 crystals in crystallization buffer supplemented with a 20-fold molar excess of imipenem for 12 h. PBP5 apo and β-lactam–acylated complexes were cryoprotected using 5.0 m ammonium sulfate, after which they were immediately flash-frozen in liquid nitrogen.
PBP5, closed state
Crystals were obtained using the acoustic droplet ejection robot (28) at National Synchrotron Light Source II in 100 mm BisTris, pH 6.5, 7.5% (w/v) PEG 3350, 7.5% (v/v) PEG 400, 25 mm sucrose, 25 mm trehalose, 25 mm glucose, and 25 mm galactose.
Data collection, processing, and solution
Data were collected at the Stanford Synchrotron Radiation Lightsource (beamline 12-2; PBP5, open state, benzylpenicillin-acyl-PBP5, imipenem-acyl-PBP4 and ceftaroline-acyl-PBP4), National Synchrotron Light Source II (PBP5, closed state), Advanced Photon Source (beamline 23ID; impenem-acyl-PBP5), or the University of Arizona (Bruker liquid Gallium MetalJet with a Photon II CPAD detector; PBP4, benzylpenecillin-acyl-PBP4). Data were processed using either AutoXDS, XDS, or SAINT/XPREP (29, 30). The open state of PBP5 and PBP4 were both phased using molecular replacement (PHASER as implemented in Phenix (31)), using PDB entry 5DVY as a search model. The closed state of PBP5 was phased using MR (same programs) but using the PBP5 open state as a search model. The β-lactam-acyl-PBP complexes were phased using either MR or difference Fourier methods. Clear electron density was visible for all acylated β-lactams. All structures were completed using iterative rounds of manual building (Coot (32)) and refinement (Phenix). Molecular figures were generated using PyMOL (33).
Differential scanning fluorimetry
Purified PBP4 and PBP5 either alone or in the presence of either a 40-fold (PBP4) or 20-fold (PBP5) molar excess of benzylpenicillin, imipenem, and ceftaroline were incubated for 1 h or overnight. Forty-five microliters of each protein at a concentration of 1.1 mg/ml were added to 5 μl of 20× SYPRO (5000X, Invitrogen) for a final reaction volume of 50 μl. DSF experiments were performed on a CFX96 touch RT-PCR System (Bio-Rad); temperature was ramped from 4 to 80 °C in 0.2 °C increments (5 s for each increment). Data were analyzed with the Bio-Rad CFX manager version 3.1 software and SigmaPlot.
Author contributions
T. M. M., É. D. D'A., and C. L. expressed and purified proteins. T. M. M. and É. D. D'A. performed the crystallization experiments. T. M. M., É. D. D'A., A. S., and J. J. performed crystallographic data collections. T. M. M., É. D. D'A. and R. P. analyzed the crystallographic data. C. D. and M. G. S. performed PBP4 complementation experiments. L. B. R., R. P., and W. P. conceived the experiments. T. M. M., É. D. D'A., R. P., and W. P. analyzed and interpreted the data and with L. B. R. wrote the manuscript.
Supplementary Material
Acknowledgments
This research used the AMX 17-ID-1 beamline of the National Synchrotron Light Source II, a United States Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Brookhaven National Laboratory under Contract DE-SC0012704. This research also used beamline 12.2 at the Stanford Synchrotron Radiation Lightsource. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the United States Department of Energy, Office of Science, and Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by NIGMS, National Institutes of Health (including Grant P41GM103393).
This work was supported by National Institutes of Health Grant R56AI045626 (to L. B. R. and W. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Figs. S1 and S2.
- MIC
- minimum inhibitory concentration
- PBP
- penicillin-binding protein
- TPase
- transpeptidase
- nPB
- nonpenicillin-binding domain
- RMSD
- root mean square deviation
- DSF
- differential scanning fluorimetry
- SEC
- size-exclusion chromatography
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Rice L. B. (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- 2. Weiner L. M., Webb A. K., Limbago B., Dudeck M. A., Patel J., Kallen A. J., Edwards J. R., and Sievert D. M. (2016) Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control. Hosp. Epidemiol. 37, 1288–1301 10.1017/ice.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arias C. A., and Murray B. E. (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moellering R. C. Jr., and Weinberg A. N. (1971) Studies on antibiotic syngerism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labeled streptomycin by enterococci. J. Clin. Invest. 50, 2580–2584 10.1172/JCI106758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jawetz E., and Sonne M. (1966) Penicillin-streptomycin treatment of enterococcal endocarditis: a re-evaluation. N. Engl. J. Med. 274, 710–715 10.1056/NEJM196603312741304 [DOI] [PubMed] [Google Scholar]
- 6. Williamson R., Calderwood S. B., Moellering R. C. Jr., and Tomasz A. (1983) Studies on the mechanism of intrinsic resistance to β-lactam antibiotics in group D streptococci. J. Gen. Microbiol. 129, 813–822 [DOI] [PubMed] [Google Scholar]
- 7. Williamson R., le Bouguénec C., Gutmann L., and Horaud T. (1985) One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 131, 1933–1940 [DOI] [PubMed] [Google Scholar]
- 8. Chambers H. F. (1988) Methicillin-resistant staphylococci. Clin. Microbiol. Rev. 1, 173–186 10.1128/CMR.1.2.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghuysen J. M. (1991) Serine β-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45, 37–67 10.1146/annurev.mi.45.100191.000345 [DOI] [PubMed] [Google Scholar]
- 10. Rice L. B., Bellais S., Carias L. L., Hutton-Thomas R., Bonomo R. A., Caspers P., Page M. G. P., and Gutmann L. (2004) Impact of specific pbp5 mutations on expression of β-lactam resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 48, 3028–3032 10.1128/AAC.48.8.3028-3032.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rice L. B., Desbonnet C., Tait-Kamradt A., Garcia-Solache M., Lonks J., Moon T. M., D'Andréa É. D., Page R., and Peti W. (2018) Structural and regulatory changes in PBP4 trigger decreased β-lactam susceptibility in Enterococcus faecalis. MBio 9, e00361–18 10.1128/mBio.00361-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sauvage E., Kerff F., Fonzé E., Herman R., Schoot B., Marquette J. P., Taburet Y., Prevost D., Dumas J., Leonard G., Stefanic P., Coyette J., and Charlier P. (2002) The 2.4-Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell Mol. Life Sci. 59, 1223–1232 10.1007/s00018-002-8500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim D., and Strynadka N. C. J. (2002) Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9, 870–876 10.1038/nsb858 [DOI] [PubMed] [Google Scholar]
- 14. Arbeloa A., Hugonnet J.-E., Sentilhes A.-C., Josseaume N., Dubost L., Monsempes C., Blanot D., Brouard J.-P., and Arthur M. (2004) Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in Gram-positive bacteria. J. Biol. Chem. 279, 41546–41556 10.1074/jbc.M407149200 [DOI] [PubMed] [Google Scholar]
- 15. Eliopoulos G. M., Wennersten C., and Moellering R. C. (1982) Resistance to β-lactam antibiotics in Streptococcus faecium. Antimicrob. Agents Chemother. 22, 295–301 10.1128/AAC.22.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aymanns S., Mauerer S., van Zandbergen G., Wolz C., and Spellerberg B. (2011) High-level fluorescence labeling of Gram-positive pathogens. PLoS One 6, e19822 10.1371/journal.pone.0019822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Höltje J. V. (1998) Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Macheboeuf P., Contreras-Martel C., Job V., Dideberg O., and Dessen A. (2006) Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30, 673–691 10.1111/j.1574-6976.2006.00024.x [DOI] [PubMed] [Google Scholar]
- 20. Ren J., Nettleship J. E., Males A., Stuart D. I., and Owens R. J. (2016) Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms. FEBS Lett. 590, 288–297 10.1002/1873-3468.12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stein B. J., Grant R. A., Sauer R. T., and Baker T. A. (2016) Structural basis of an N-degron adaptor with more stringent specificity. Structure 24, 232–242 10.1016/j.str.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zapun A., Contreras-Martel C., and Vernet T. (2008) Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 32, 361–385 10.1111/j.1574-6976.2007.00095.x [DOI] [PubMed] [Google Scholar]
- 23. Fedarovich A., Nicholas R. A., and Davies C. (2012) The role of the β5-α11 loop in the active-site dynamics of acylated penicillin-binding protein A from Mycobacterium tuberculosis. J. Mol. Biol. 418, 316–330 10.1016/j.jmb.2012.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamada M., Watanabe T., Miyara T., Baba N., Saito J., Takeuchi Y., and Ohsawa F. (2007) Crystal structure of cefditoren complexed with Streptococcus pneumoniae penicillin-binding protein 2X: structural basis for its high antimicrobial activity. Antimicrob. Agents Chemother. 51, 3902–3907 10.1128/AAC.00743-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otero L. H., Rojas-Altuve A., Llarrull L. I., Carrasco-López C., Kumarasiri M., Lastochkin E., Fishovitz J., Dawley M., Hesek D., Lee M., Johnson J. W., Fisher J. F., Chang M., Mobashery S., and Hermoso J. A. (2013) How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc. Natl. Acad. Sci. U.S.A. 110, 16808–16813 10.1073/pnas.1300118110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frère J.-M., and Page M. G. P. (2014) Penicillin-binding proteins: evergreen drug targets. Curr. Opin. Pharmacol. 18, 112–119 10.1016/j.coph.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 27. Peti W., and Page R. (2007) Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 51, 1–10 10.1016/j.pep.2006.06.024 [DOI] [PubMed] [Google Scholar]
- 28. Teplitsky E., Joshi K., Ericson D. L., Scalia A., Mullen J. D., Sweet R. M., and Soares A. S. (2015) High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density. J. Struct. Biol. 191, 49–58 10.1016/j.jsb.2015.05.006 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonzolez A., and Tsai Y. (2010) AutoXDS, Stanford University, Stanford, CA [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.