Abstract
The innate immune system is important for the efficacy of vaccines, but excessive innate immune responses can cause adverse reactions after vaccination. Extracellular vesicles (EVs) are enriched in the blood and can deliver functional RNAs, such as microRNAs (miRNAs), to recipient cells, thereby mediating intercellular communication. However, the role of EVs in controlling the innate immune responses to vaccines has not been fully elucidated. Here, we found that miR-451a is abundant in human serum EVs and that its presence in blood-circulating EVs affects the innate immune responses of macrophages and dendritic cells to inactivated whole-virus vaccines (WV) against influenza. miR-451a in human serum EVs was stable for a week in healthy subjects, and its levels gradually fluctuated over several months. miR-451a within serum EVs was internalized into serum-cultured macrophages and dendritic cells and reduced endogenous 14-3-3ζ protein levels and decreased the expression of type I IFN and interleukin 6 in response to WV stimulation. miR-451a levels in blood-circulating EVs were positively correlated with intracellular miR-451a levels in mouse splenic CD11c+ cells and inversely correlated with the innate immune response to inactivated WV in vivo. These findings suggest that miR-451a in circulating EVs is internalized into recipient cells in vivo and that this internalization results in an attenuation of the innate immune response to WV. Moreover, a microarray analysis identified several other miRNAs that affect the macrophage response to inactivated WV. Our results reveal that miRNAs in circulating EVs significantly modify the responses of macrophages and dendritic cells to inactivated WV.
Keywords: innate immunity, microRNA (miRNA), extracellular vesicles, influenza virus, virus, vaccine, inflammation, antiviral response, macrophage, dendritic cell
Introduction
Cytokines and chemokines are important mediators for intercellular communication and modulating the innate and adaptive immune responses. In addition to these classical mediators, an accumulating body of evidence suggests that extracellular vesicles (EVs)2 deliver functional RNAs and proteins to immune cells and mediate intercellular communication (1–3). EVs are produced by various types of cells, and thus the vesicles are accumulated in body fluids, such as blood and saliva (4), and the blood contains high amounts of EVs (5). There are several types of EVs, including exosomes and microvesicles (6). Exosomes are small vesicles with a diameter of about 100 nm and are released from multivesicular bodies, whereas microvesicles are larger in diameter (50 nm to 1 μm) and are released from the plasma membrane (6, 7). Although initial studies reported that exosomes deliver functional RNAs, such as mRNA and microRNA (miRNA) (8), later studies reported that microvesicles also deliver miRNAs to recipient cells (9). miRNA is well-known to modulate gene function by targeting mRNAs and preventing their translation (10).
Recent studies have shown that miRNAs are specifically sorted into EVs. hnRNPA2B1 recognizes the EXO motif in miRNAs, which are selectively packaged into exosomes (11). In contrast, hnRNP-Q recognizes a different motif in miRNAs and sorts them into exosomes (12). There are several other pathways, in which miRNAs are selectively packaged into exosomes (7, 13–15). Although the underlying mechanism remains unclear, miRNAs are also sorted into microvesicles (16). EVs with miRNAs are transferred to specific types of cells. Dendritic cells (DCs) and macrophages efficiently uptake EVs via phagocytosis, and thus the destabilization of actin filaments inhibits the uptake of EVs (17).
The innate immune system recognizes pathogen-associated molecular patterns via pattern recognition receptors (PRRs) and triggers the signal to induce interferons (IFNs), pro-inflammatory cytokines, and chemokines. TLR3, TLR7, and RIG-I are PRRs that recognize viral RNA (18). TLR3 binds to endosomal viral double-stranded RNA (dsRNA) (19), and TLR7 recognizes viral single-stranded RNAs and a chemical, CL097, in the endosome (20, 21). dsRNA is also sensed by a cytoplasmic viral RNA sensor, RIG-I (22).
Recently, we found that EVs released from HBV-infected cells deliver host miRNAs to macrophages and control the innate immune response against HBV (7, 23). It has been reported that miR-146a and miR-155 in exosomes were internalized into dendritic cells and modulated the cytokine expression in response to LPS, a ligand for TLR4 in vivo (24). Moreover, Montecalvo et al. (25) have reported the underlying mechanisms of how miRNAs are transferred from donor to recipient dendritic cells via EVs. These observations show that miRNAs in EVs are internalized into dendritic cells and attenuate the target gene function in the recipient cells.
miR-451a is an miRNA that targets 14-3-3ζ, which is involved in pro-inflammatory cytokine expression pathway (26, 27). miR-451a reduces type I interferon (IFN) and IL-6 expression in response to influenza A virus infection via targeting 14-3-3ζ (26). Influenza A virus is known to be recognized by TLR3, TLR7, and RIG-I (20, 28, 29). Although formalin-inactivated whole-virus vaccine (WV) of influenza A virus induces type I IFN and pro-inflammatory cytokine expression (30), it remains unclear whether miR-451a in EVs modulates the innate immune response to inactivated WV.
Vaccination is the best prophylaxis for flu infection, and inactivated WV has superior immunogenicity (31–33). It has been shown that viral RNA within inactivated WV activates the innate immune response and is important for the efficacy of inactivated WV vaccine (30). In this study, we found that miR-451a in blood-circulating EVs affected intracellular miR-451a levels, resulting in modulation of the cytokine expression in response to inactivated WV. Moreover, our microarray study identified several other EV miRNAs that had a significant impact on the cytokine expression in response to inactivate WV. Our findings elucidated a role of blood-circulating EVs for controlling the responses of macrophages and dendritic cells in vivo.
Results
miR-451a levels in EVs increase after stimulation with inactivated WV
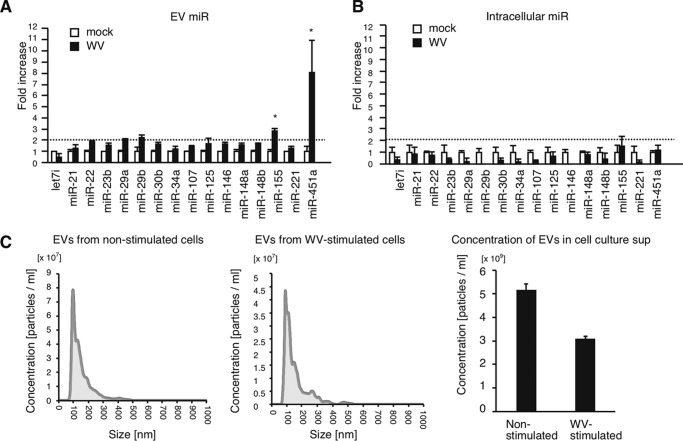
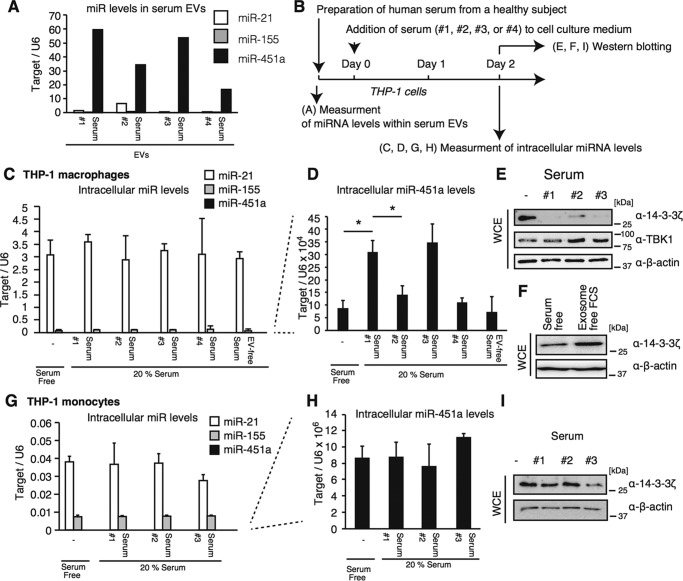
First, we focused on 20 immune regulatory miRNAs (34) and investigated their expression levels in EVs after stimulation with inactivated WV. THP-1 macrophages were treated with inactivated WV for 24 h, and the released EVs were subsequently collected. miRNA levels within EVs and THP-1 macrophages were determined by RT-qPCR. The expression levels were normalized to U6 RNA levels, because U6 is frequently used as a reference RNA to normalize miRNA levels within cells and EVs (35–37). Four of those miRNAs were not detectable in EVs, whereas the remaining 16 miRNAs could be quantified (Fig. 1, A and B). Interestingly, miR-155 and miR-451a levels in EVs were significantly increased by stimulation with inactivated WV (Fig. 1A). However, intracellular miR-155 and miR-451a levels were only slightly affected by stimulation (Fig. 1B), implying that sorting of miR-155 and miR-451a into EVs was increased by stimulation with inactivated WV, rather than transcription. Intracellular miR-155 and miR-451a are reported to control the innate immune response to LPS and influenza A virus infection, respectively (24, 26). The sizes of EVs were comparable before and after stimulation with inactivated WV (Fig. 1C), and the concentrations of EVs in cell culture supernatants were moderately reduced after stimulation (Fig. 1C, right panel). Because the role of exosomal miR-155 in EVs in the innate immune response has been reported previously (24), we focused on EV miR-451a in this study.
Figure 1.
miRNA levels in EVs released from THP-1 macrophages stimulated with WV. A and B, THP-1 macrophages were stimulated with 1 μg/ml inactivated WV for 6 h. Cells were washed with PBS and were further cultured in serum-free medium for 24 h. EVs were collected from culture supernatants. miRNA levels in EVs (A) and those in THP-1 macrophages (B) were determined by RT-qPCR and normalized to U6 expression levels (internal control). Data represent means ± S.D. (n = 3) (p < 0.05, two-way ANOVA). C, EVs were collected from cell culture medium of THP-1 macrophages stimulated with or without inactivated WV. Particle size and concentration of collected EVs were measured by NanoSight. The data are representative of three independent experiments. *, p < 0.05; NS, not significant. Data represent means ± S.D. (n = 3) (right panel).
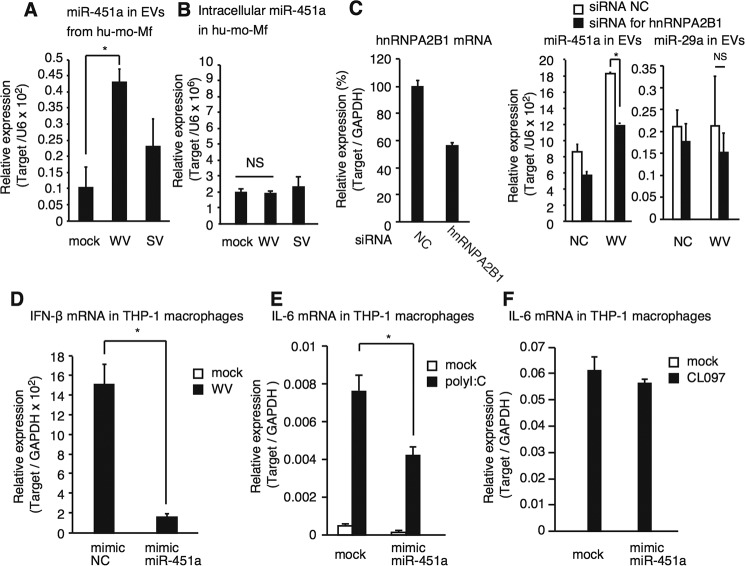
To investigate whether inactivated WV can increase miR-451a levels in EVs released from primary macrophages as well as immortalized macrophages, we used human monocyte-derived macrophages (mo-Mf) and found that the miR-451a level in mo-Mf–derived EVs increased after stimulation with inactivated WV as observed in THP-1 macrophages, although intracellular miR-451a levels did not increase (Fig. 2, A and B). We also used hemagglutinin (HA)-antigen–enriched split virus (SV) vaccine and found that a high concentration of SV could increase the miR-451a levels in EVs (Fig. 2A and Fig. S1A).
Figure 2.
Characterization of miR-451a. A and B, human blood monocytes were differentiated into macrophages in vitro and were stimulated with 1 μg/ml inactivated WV and SV for 24 h. Total RNA was isolated from EVs in cell culture supernatants (A) and from cells (B). miRNA expression levels were determined by RT-qPCR. Data represent means ± S.D. (n = 3) (p < 0.05, one-way ANOVA). C, siRNA for hnRNPA2B1 or negative control (NC), which contains a random sequence, was transfected into THP-1 macrophages using Lipofectamine RNAiMAX, according to the manufacturer's instruction, and then incubated for 2 days. Knockdown efficiency was determined by RT-qPCR (left panel). Cells were then stimulated with 20 μg/ml inactivated WV for 24 h. The expression levels of miR-451a and miR-29a in EVs were determined by RT-qPCR (middle and right panels). Data represent means ± S.D. (n = 3) (p < 0.05, t test). D–F, mimic miRNA of control (NC) and miR-451a were transfected into THP-1 macrophages for 48 h. Cells were then stimulated with mock, 20 μg/ml inactivated WV (D), 50 μg/ml poly(I-C) (E), or 5 μg/ml CL097 for 6 h, and the mRNA expressions of IFN-β (D) and IL-6 (E and F) were determined by RT-qPCR and normalized to GAPDH. Fold increase was calculated by dividing each value with that of control. Data represent means ± S.D. (n = 3) (p < 0.05, one-way ANOVA). *, p < 0.05.
miR-451a contains an EXO motif (Fig. S1C), and hnRNPA2B1 selectively sorts miRNA with the EXO motif into EVs, thus increasing the expression of miRNA in EVs (11). To test whether the increase of miR-451a levels in EVs depended on hnRNPA2B1, siRNA for hnRNPA2B1 was transfected into THP-1 macrophages, which were subsequently stimulated with inactivated WV and SV. Knockdown of hnRNPA2B1 markedly reduced the level of miR-451a in EVs released from cells stimulated with WV and SV (Fig. 2C and Fig. S1, A and B), whereas hnRNPA2B1 knockdown failed to reduce miR-29a levels in EVs (Fig. 2C, right panel), which does not contain the EXO motif (Fig. S1C). Collectively, these data suggest that miR-451a is sorted into EVs upon stimulation with influenza A virus vaccines.
miR-451a targets 14-3-3ζ and attenuates the expression of IL-6 and IFN-β in response to influenza A virus infection (26). Thus, we investigated whether miR-451a attenuates cytokine expression in response to inactivated WV as well as flu infection. We used a mimic miRNA for miR-451a (mimic miR-451a). Transfection of mimic miR-451a into THP-1 macrophages reduced IFN-β and IL-6 mRNA expression in response to inactivated WV (Fig. 2D and Fig. S1D). We confirmed that mimic negative control miRNA did not affect the cytokine expression in our experimental condition (Fig. S1E). Mimic miR-451a reduced the expression of endogenous 14-3-3ζ protein (Fig. S1F). Inactivated WV contains viral RNA, and thus we investigated whether mimic miR-451a affects TLR3- and TLR7-mediated signaling. Transfection with mimic miR-451a reduced IL-6 mRNA expression in response to poly(I-C), a TLR3 ligand, but not to CL097, a TLR7 ligand (Fig. 2, E and F). Because miR-451a modulated the innate immune response, we further focused on the function and role of EV miR-451a.
miR-451a levels in human serum EVs fluctuate severalfold during 1 year
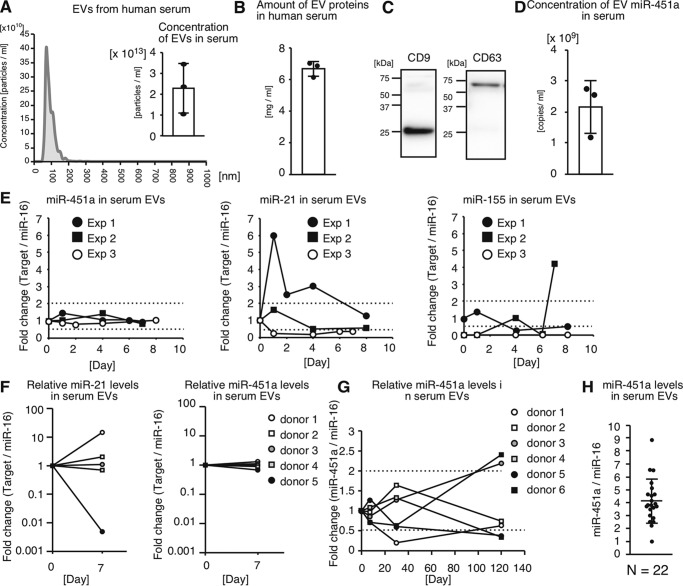
EVs are known to be abundant in human serum (5). Human sera contained 2.3 ± 1.2 × 1013 particles/ml of EVs (Fig. 3A), which were over 1000 times higher than those in serum-free cell culture supernatant (5.14 × 109 particles/ml and see Fig. 1C). The mean size of serum EVs was 93.2 nm (Fig. 3A), and the average concentration of EV proteins in human sera was 6.7 ± 0.48 mg/ml (Fig. 3B). Collected EVs expressed the exosome marker proteins, CD9 and CD63 (Fig. 3C). Because EVs with miR-451a were released from macrophages, we next investigated whether EVs in serum contain miR-451a. The concentration of EV miR-451a in serum was 2.2 ± 0.85 × 109 copies/ml (Fig. 3D), suggesting that human serum contains a high amount of EV miR-451a. We do not exclude the possibility that genetic backgrounds and environmental factors exhibit profound effects on the concentration of EVs and the miRNAs levels.
Figure 3.
miR-451a in human serum EVs. A, EVs were collected from sera of healthy human subjects. Concentration and size of EVs were determined by NanoSight. Histogram is a representative of three independent experiments. Average of EV concentrations in human sera (n = 3) is shown. B, EVs were collected from sera of healthy human subjects. Amounts of EV proteins were subsequently determined. The graph exhibits the concentrations of EV proteins in human serum (n = 3). C, collected EVs were subjected to SDS-PAGE, and the CD9 and CD63 proteins were detected by Western blotting with anti-CD9 and anti-CD63 antibodies. The data are representative of two independent experiments. D, RNA was extracted from collected EVs, and the concentration of EV miR-451a in serum was determined by RT-qPCR (n = 3). E, human sera were collected over the course of a week from healthy subjects. miRNA levels of miR-21, miR-155, and miR-451a in serum EVs were determined by RT-qPCR and normalized to that of miR-16. Experiments were repeated three times. F and G, six healthy subject sera were collected at day 0 and at day 7 (F), at day 30 and day 120 (G), and then miRNA levels of miR-21 and miR-451a in serum EVs were determined by RT-qPCR and normalized to that of miR-16. H, 22 healthy donor sera were collected. miR-451a levels in EVs within sera were determined by RT-qPCR and normalized to miR-16 levels. Healthy volunteers aged 22–56 years did not have any symptoms of disease and were not medicated when sera were collected. During the course of the experiments, those volunteers were not vaccinated with influenza A virus vaccine and had no symptom of influenza or any other disease.
To investigate the temporal changes of miR-451a levels in serum EVs, we collected sera from a healthy human subject during an 8-day period, and miR-451a levels in the EVs were determined. Because U6 RNA levels in human serum EVs were very low, we could not use U6 as a stable internal control. Therefore, we used miR-16 as an internal control to normalize miRNA levels in extracellular vesicles as described previously (38–40). miR-451a levels in sera were very high, and thus 5 μl of serum was sufficient to detect miR-451a expression in serum EVs. For comparison, we determined miR-21 and miR-155 levels in serum EVs, which are known to regulate inflammatory response. miR-21 and miR-155 levels in serum EVs fluctuated over a 4-fold range during 1 week; however, miR-451a levels were stable compared with those of miR-21 and miR-155, and its range of fluctuation was under 2-fold (Fig. 3, E and F). miR-451a levels in serum EVs were also stable during a single day (Fig. S2A). Next, we investigated them over several months. miR-451a levels in serum EVs gradually changed, and it took several weeks to change over 2-fold (Fig. 3G). It should be notable that miR-451a levels in serum EVs isolated from one donor changed over 10-fold during a 6-month period (Fig. S2B). We investigated the divergence of miR-451a expression levels in serum EVs of healthy human subjects and found a difference in miR-451a levels over a 10-fold range (Fig. 3H).
We next investigated which types of cells release miR-451a–containing EVs, and we detected miR-451a in EVs released from HepG2, A549, and BJ cells (Fig. S2C). In mice, miR-451a levels were high in the lung, spleen, and heart compared with other tissues (Fig. S2D). These data suggest that miR-451a–containing EVs are released from various types of cells in vivo. Interestingly, our RNA-Seq analysis showed that miR-451a levels in mouse serum EVs were increased by intranasal infection with influenza A virus (Fig. S2E), suggesting that flu infection also affects miR-451a levels in serum EVs. It remains unclear whether other viral infections, other diseases, or physical conditions affect serum EV miR-451a levels (see “Discussion”).
Internalization of miR-451a in serum EVs into macrophages and dendritic cells
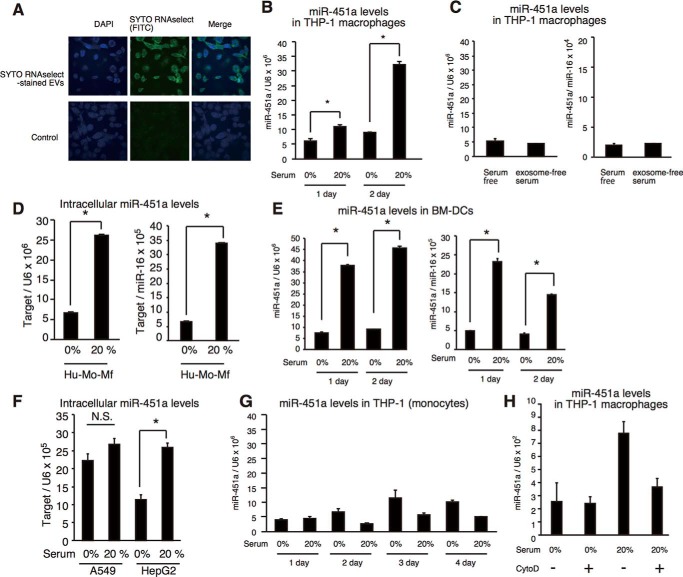
Previous studies reported that EVs deliver miRNAs into the cytoplasm of recipient cells (6, 7, 23). To observe the internalization of serum EV RNAs, EVs were collected from serum, and RNAs within EVs were stained with SYTO RNAselect dye, which is a membrane-permeable dye and binds to RNA, leading to emission of green fluorescence. 1 × 1012 particles of stained serum EVs were added to the cell culture of THP-1 macrophages (final concentration 2 × 1012 particles/ml) for 6 h, and we investigated whether stained RNAs within EVs were internalized into THP1 macrophages. We found that green fluorescence was detected in the cytoplasm of THP-1 macrophages, indicating that RNAs within serum EVs were internalized into THP-1 macrophages (Fig. 4A).
Figure 4.
Internalization of miR-451a into macrophages and dendritic cells. A, EVs were collected from human serum. EVs and mock were incubated with SYTO RNAselect dye to stain EV RNAs. 1 × 1012 of stained EVs and mock sample were added to THP-1 macrophages for 6 h. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and were observed by confocal microscopy. The data are representative of two independent experiments. B, THP-1 macrophages were cultured in medium containing 0 or 20% of human serum for the indicated time periods. Intracellular miR-451 levels were determined by RT-qPCR and were normalized to U6 RNA levels (internal control). Data represent means ± S.D. (n = 3) (p < 0.05, t test). C, THP-1 macrophages were cultured in serum-free medium or medium with exosome-free serum for 2 days. Intracellular miR-451 levels were determined by RT-qPCR and were normalized to U6 RNA levels (left panel) or miR-16 (right panel). Data represent means ± S.D. (n = 3) (p < 0.05, t test). D–F, human monocyte-derived macrophages (D), BM-DCs (E), A549 (F), and HepG2 cells (F) were cultured in medium containing 0 or 20% serum for 2 days. Intracellular miR-451a levels were then determined by RT-qPCR and normalized to U6 RNA (left panel) or miR-16 (right panel) levels. For BM-DCs, we used mouse sera. Human serum was used for other human cells. *, p < 0.05; NS, not significant. G, THP-1 monocytes were cultured in medium containing 0 or 20% of serum for 4 days. Intracellular miR-451a levels were determined by RT-qPCR. Data represent means ± S.D. (n = 3). H, THP-1 macrophages were treated with 250 nm cytochalasin D (CytoD) and cultured in medium containing 0 or 20% of human serum for 2 days. Intracellular miR-451a levels were determined by RT-qPCR. Data represent means ± S.D. (n = 3).
A previous in vitro study showed that exosomal miR-451a was internalized into a dendritic cell line, DC2.4, and repressed the expression of a target mRNA (25). The study reported that 12.5 μg/ml of the exosomes in cell culture was sufficient for DC2.4 cells to uptake exosomal miR-451a (25). Human sera contain much higher concentrations of EVs (6.7 ± 0.48 mg/ml) and a high amount of EV miR-451a (2.2 ± 0.85 × 109 copies/ml) as described above. In contrast, intracellular miR-451a levels were very low (0.48 copies/cell). Therefore, we reasoned that miR-451a in serum EVs would be internalized into macrophages and dendritic cells.
If serum EV miR-451a is internalized into macrophages, it is expected that intracellular miR-451a levels would increase after incubation of cells with human serum. We tested this hypothesis. First, THP-1 macrophages were cultured with human serum for several days, and their intracellular miR-451a levels in THP-1 macrophages were determined by RT-qPCR. We found that intracellular miR-451a levels increased after 1 and 2 days postincubation (Fig. 4B). We confirmed that serum-free conditions did not affect intracellular miR-451a levels (Fig. 4C), and normalization using either miR-16 or U6 did not affect the results (Fig. 4C, right and left panels). Increased miR-451a levels were also detected in human monocyte-derived macrophages and mouse bone marrow–derived dendritic cells (BM-DCs) after incubation with human or mouse serum, respectively (Fig. 4, D and E), suggesting that EVs can deliver miR-451a into not only immortalized macrophages but also primary macrophages and dendritic cells. We confirmed that normalization using either miR-16 or U6 did not affect the results (Fig. 4, D and E, right and left panels).
Second, we investigated the effect of serum EVs on other types of cells. Similar to THP-1 macrophages, the intracellular miR-451a level in HepG2 cells increased following incubation with human serum, but not in A549 cells or THP-1 monocytes (Fig. 4, F and G).
In general, internalization of EVs requires phagocytosis, and cytochalasin D treatment destabilizes actin filament formation and reduces the internalization of EV miRNAs (17, 25). Interestingly, treatment with cytochalasin D inhibited the increase of intracellular miR-451a levels after incubation with human serum (Fig. 4H). This observation weakened the possibility that an apparent increase of intracellular miR-451a was caused by attachment of EVs on the cell surface. Collectively, our data indicate that a physiological concentration of miR-451a in serum EVs is sufficient for the internalization of miR-451a from EVs into macrophages and dendritic cells.
Serum EV miR-451a levels affect its intracellular levels in macrophages
Next, we investigated whether the fluctuation of miR-451a levels in serum EVs, which was observed in Fig. 3, affects the intracellular miR-451a levels in recipient cells. Human sera (samples #1 to #4) with different miR-451a levels were collected from one donor (Fig. 5A). The expression levels of miR-21 and miR-155 were determined for comparison (Fig. 5A). THP-1 macrophages were incubated with the sera for 2 days, and then intracellular miR levels were determined by RT-qPCR as described in Fig. 5B. Intracellular levels of miR-21 and miR-155 were much higher than that of miR-451a and were not affected by incubation with any serum (Fig. 5C), whereas intracellular miR-451a levels increased by human serum (Fig. 5D) as observed in Fig. 4. The #2 and #4 sera contained lower amounts of EV miR-451a than #1 and #3 sera, and intracellular miR-451a levels in cells cultured with #2 or #4 serum were lower than those cultured with #1 or #3 sera (Fig. 5D). Next, we collected sera from other donors at different time points and cultured THP-1 macrophages with the collected sera. When cells were cultured in medium with human serum with a relatively high EV miR-451a level, their intracellular levels were also higher than those cultured with serum containing a low EV miR-451a level (Fig. S3, A and B). These data imply that the difference of EV miR-451a levels caused by fluctuation in single donor affects its intracellular levels of THP-1 macrophages.
Figure 5.
Macrophage-specific internalization of EV miR-451a. A, sera were collected four times from a single donor (sample #1 to #4). The expression levels of miR-21, miR-155, and miR-451a in EVs within the human serum were determined by RT-qPCR and were normalized to that of miR-16 in EVs. B, schematic of treatment schedule for following experiments (C–I). C–I, THP-1 macrophages (C–F) or THP-1 (monocyte) (G–I) were cultured in medium containing 20% of each human serum for 2 days. Intracellular miRNA expression levels of miR-21, miR-155, and miR-451a were determined by RT-qPCR and were normalized to U6 RNA levels (C, D, G, and H). Data represent means ± S.D. (n = 3) (p < 0.05, t test). Whole-cell extract of cells cultured with human serum (E and I) or exosome-free serum (F) for 2 days were prepared and were subjected to SDS-PAGE. Proteins were stained with indicated antibodies (E, F, and I) by Western blotting. *, p < 0.05.
miR-451a targets 14-3-3ζ and endogenous 14-3-3ζ protein levels decreased by incubation with human serum (Fig. 5E), suggesting that internalized miR-451a could target 14-3-3ζ and attenuate the expression. These data also weakened the possibility that the apparent increase of intracellular miR-451a was caused by attachment of EVs on the cell surface. We confirmed that the serum-free condition did not affect endogenous 14-3-3ζ protein levels in our experimental condition (Fig. 5F). EVs collected from human serum reduced endogenous 14-3-3ζ protein levels in THP-1 macrophages, but not TBK1 required for type I IFN production, suggesting the specific suppression of the 14-3-3ζ protein by miR-451a (Fig. S3C).
In contrast to THP-1 macrophages, intracellular miR-451a levels in nondifferentiated THP-1 cells (monocytes) did not increase by human serum (Fig. 5, G and H), and endogenous 14-3-3ζ protein levels did not change by incubation with human serum (Fig. 5I) as observed in Fig. 4G.
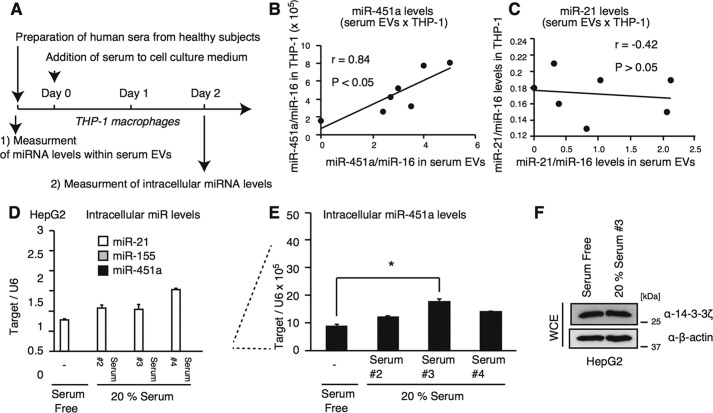
To perform a statistical analysis, we collected several human sera from healthy subjects and compared miR-451a levels in serum with intracellular miR-451a levels in cells cultured with serum (Fig. 6A). Interestingly, miR-451a levels in serum EVs were significantly correlated with intracellular miR-451a but not the miR-21 level in THP-1 macrophages (Fig. 6, B and C).
Figure 6.
A, schematic of preparation and treatment schedule. B and C, six healthy donor sera were collected. THP-1 macrophages were cultured in medium containing 0 or 20% of each human serum for 2 days. miR-451a (B) and miR-21 (C) levels in serum EVs and in THP-1 macrophages were determined by RT-qPCR. miR-21 levels in serum EVs were not correlated with intracellular miR-21 levels (Pearson correlation coefficient = −0.42, p > 0.05) (C), whereas miR-451a in serum EVs was significantly correlated with intracellular miR-451a levels (Pearson correlation coefficient = 0.84, p < 0.05, statistical power = 0.90) (B). The data of THP-1 cells cultured in serum-free medium was plotted as 0 on the x axis (miR-451a/miR-16 in serum EVs). D–F, HepG2 cells were cultured in medium containing 0 or 20% of human serum for 2 days. Intracellular miRNA levels of miR-21, miR-155, and miR-451a were determined by RT-qPCR (D and E). Whole-cell extract was prepared from cultured cells and subjected to SDS-PAGE. Proteins were detected by Western blotting with indicated antibodies (F). *, p < 0.05.
In contrast to THP-1 macrophages, among the three human serum samples (#2, #3, and #4), only serum #3 increased intracellular miR-451a levels in HepG2 cells (Fig. 6, D and E). However, serum #3 failed to reduce endogenous 14-3-3ζ protein levels in HepG2 cells (Fig. 6F). This suggests that internalized miR-451a is not functional in HepG2 cells under our experimental conditions. Collectively, our data suggest that the miR-451a in serum EVs is internalized into recipient cells in a cell-type–specific manner.
miR-451a levels in serum EVs affect the response of macrophages to inactivated WV
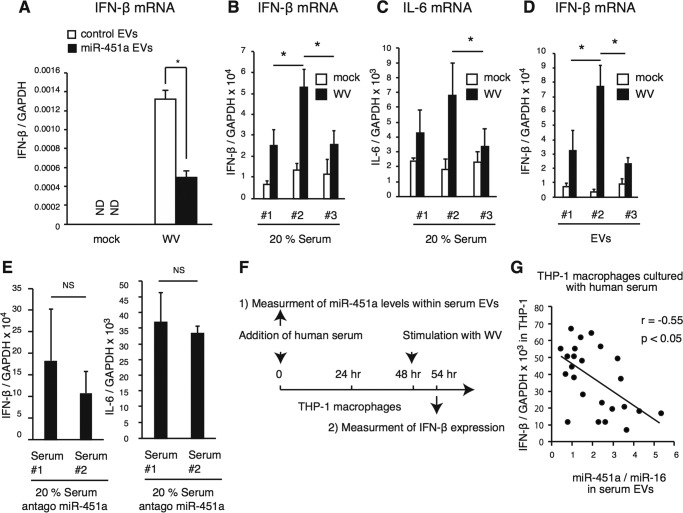
Because transfection of mimic miR-451a reduced IFN-β expression in macrophages in response to WV (Fig. 2D), we investigated whether EVs containing a high amount of miR-451a could reduce this response. First, mimic miR-451a was transfected into human cells, and EVs were collected from the cell culture medium of miR-451a–transfected (miR-451a EVs) or mock-transfected cells (mock EVs). Second, THP-1 macrophages were treated with collected EVs for 2 days and were subsequently stimulated with WV for 6 h, and IFN-β mRNA expression was determined. Treatment with miR-451a EVs significantly reduced IFN-β expression compared with that of mock EVs (Fig. 7A).
Figure 7.
Correlation of miR-451a levels in serum EVs with THP-1 macrophage response to inactivated WV. A, miR-451a mimic was transfected into THP-1 macrophages and was incubated for 2 days. EVs were collected from cell culture medium and then added to THP-1 macrophages. Cells were stimulated with inactivated WV for 6 h, and the expression of IFN-β mRNA was determined by RT-qPCR. Data represent means ± S.D. (n = 3) (p < 0.05, t test). B and C, THP-1 macrophages were cultured with each human serum (#1, #2, and #3), which was used in Fig. 5, for 2 days. Cells were stimulated with inactivated WV for 6 h, and the expression of IFN-β (B) and IL-6 (C) mRNA was determined by RT-qPCR. Data represent means ± S.D. (n = 3) (p < 0.05, t test). D, EVs were collected from 0.1 ml of human sera (#1, #2, and #3). Cells were cultured in serum-free medium with collected EVs in 24-well plates for 2 days and were subsequently stimulated with inactivated WV for 6 h. The expression of IFN-β mRNA was determined by RT-qPCR. Data represent means ± S.D. (n = 3) (p < 0.05, t test). E, antagomiR-451a was transfected into THP-1 macrophages, and cells were then cultured with or without human serum. IFN-β mRNA expressions in response to inactivated WV were determined by RT-qPCR. Data represent means ± S.D. (n = 3). “NS” represents “not significant” (p > 0.05, t test). F and G, schematic of treatment and preparation schedule (F). THP-1 macrophages were cultured with 20% of each human serum for 2 days. Cells were then stimulated with inactivated WV for 6 h, and IFN-β expression levels were determined by RT-qPCR. The expression levels of IFN-β mRNA were normalized to those of GAPDH. Correlation was investigated between miR-451a levels in serum EVs and type I IFN expression. Pearson correlation coefficient (r) was calculated, and a statistical analysis was performed (p < 0.05, n = 24, statistical power = 0.869) (G). The data are a representative of two independent experiments. *, p < 0.05.
Next, we investigated whether the fluctuations of miR-451a levels in serum EVs affects the response of macrophages to WV. First, we used sera collected from a single donor, which showed different EV miR-451a levels (Fig. 5A). THP-1 macrophages were incubated with human sera for 2 days and subsequently stimulated with inactivated WV for 6 h. IFN-β and IL-6 mRNA expression was determined by RT-qPCR. Although sera were collected from a single donor, each serum exhibited a different effect on the expression of IFN-β and IL-6 (Fig. 7, B and C). Interestingly, the cytokine expression appeared to be negatively correlated with miR-451a levels in serum EVs (Figs. 5A and 7, B and C, and Fig. S3D).
Second, EVs were collected from those sera. THP-1 macrophages were cultured with the collected EVs for 2 days, and IFN-β mRNA expression in response to inactivated WV was determined. As observed in a study using sera, EVs collected from #3 serum reduced the macrophage response more efficiently than EVs collected from other sera (#2 and #4) (Fig. 7D and Fig. S3E).
Third, inhibitory RNA for miR-451a (antagomiR-451a) transfection abrogated the difference of the IFN-β and IL-6 mRNA expression (Fig. 7E and Fig. S3F). AntagomiR-451a cancelled the reduction of 14-3-3ζ protein levels by serum EVs (Fig. S3G). These observations are consistent with our hypothesis that fluctuation of miR-451a levels in EVs affected the response of macrophages to WV. Because human serum could reduce the IFN-β mRNA expression even in the presence of antagomiR-451a (Fig. S3F), we did not exclude the possibility of the existence of other unknown factors in human serum that affect the response to inactivated WV (see below).
To perform a statistical analysis and to test whether the difference of EV miR-451a levels affects the macrophage response to WV, we used sera collected from several healthy subjects and investigated the correlation between miR-451a levels in serum EVs and IFN-β mRNA expression in THP-1 macrophages cultured with serum as shown in Fig. 7F. Interestingly, miR-451a levels in serum EVs were inversely correlated with IFN-β expression, and the correlation was statistically significant (Student's t test) (Fig. 7G). These data are consistent with our model that serum EVs deliver miR-451 into the cytoplasm of macrophages, thereby controlling cytokine expression.
miR-451a levels in blood-circulating EVs are correlated with its intracellular level of splenic CD11c+ cells
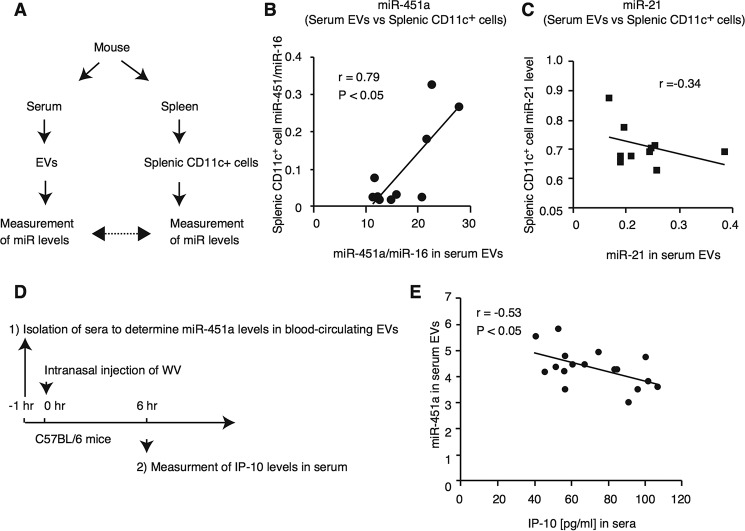
Our in vitro study suggested that serum EV miR-451a was internalized into dendritic cells and macrophages and increased its intracellular levels. Thus, we reasoned that miR-451a levels in blood-circulating EVs would be correlated with its intracellular levels in vivo. To test this hypothesis, we compared the miRNA levels of serum EVs with those of splenic CD11c+ cells that were collected from each C57BL/6 mouse as described in Fig. 8A. As expected, the miR-451a levels in mouse serum EVs were significantly correlated with intracellular miR-451a levels in splenic CD11c+ cells (Fig. 8B), although there was no correlation between miR-21 levels in EVs and in CD11c+ cells (Fig. 8C). These observations were consistent with our results obtained by in vitro analysis.
Figure 8.
miR-451a levels in sera were correlated with splenic CD11c+ cells in vivo. A–C, schematic of preparation and measurement of samples (A). Mouse sera were collected from C57BL/6 mice. miR-21 and miR-451a levels in serum EVs were determined and were normalized to miR-16 levels. Splenic CD11c+ cells were isolated from C57BL/6 mice, and intracellular miR-21 and miR-451a levels were determined. The correlation between miRNA levels in serum EVs and in splenic CD11c+ cells was calculated, and statistical analyses were performed (p < 0.05, r represents Pearson correlation coefficient) (B and C). The data are representative of at least two independent experiments. D and E, schematic of treatment and preparation of the samples (D). Sera were collected from C57BL/6 mice, and miR-451a levels in serum EVs were determined. 25 μg of inactivated WV (50 μl volume) was intranasally administered into those mice. 6 h after administration, sera were collected. IP-10 levels in sera were determined by ELISA. The correlation between miRNA levels in serum EVs and in splenic CD11c+ cells was calculated, and statistical analyses were performed (p < 0.05, r represents Pearson correlation coefficient) (E). The data are a representative of two independent experiments.
Next, we investigated whether the difference of miR-451a levels in blood-circulating EVs affects the innate immune response to inactivated WV in vivo. Previously, Ishii and co-workers (30) reported that intranasal administration of inactivated WV increases the IP-10 levels in sera. IP-10 is known to be induced by type I IFNs (41). First, mouse sera were collected to determine miR-451a levels in blood-circulating EVs. Second, inactivated WV was then intranasally administered into mice, and sera were collected 6 h after administration to determine IP-10 levels in sera as described in Fig. 8D. Interestingly, a negative correlation was observed between miR-451a levels in serum EVs and produced IP-10 levels (Fig. 8E). These data are consistent with the in vitro data.
Several miRNA levels in serum EVs are correlated with macrophage responses to vaccine
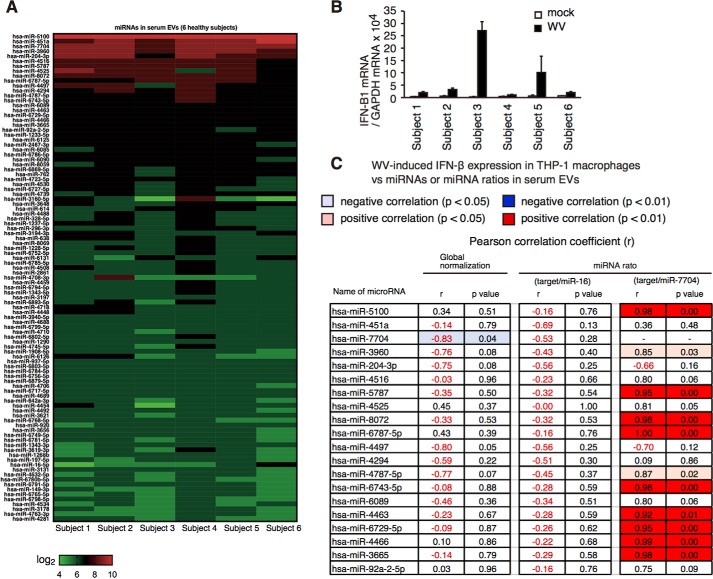
miR-451a was selected from only 20 immune regulatory miRNA. To investigate whether there are other EV miRNAs that affect the innate immune response to inactivated WV, we performed microarray analysis. Because over 5 ml of each serum was required to perform microarray analysis, we performed an in vitro study using human sera and THP-1 macrophages. First, sera were collected from six healthy subjects. EVs were isolated from each serum, and total RNAs were extracted from EVs and were subjected to microarray analysis. We have measured the expression of over 3000 miRNAs; however, most of the miRNAs were not detectable. This is because miRNAs were not amplified for microarray analysis, and the expression of most of miRNAs was very low. Therefore, the expression profiles of only highly expressed miRNAs in serum EVs of six healthy subjects are shown in Fig. 9A. Second, THP-1 macrophages were cultured for 2 days with each serum isolated from the healthy subjects, and they were then stimulated with inactivated WV for 6 h. The expression of IFN-β of macrophages was determined by RT-qPCR (Fig. 9B). Third, the correlation between IFN-β expression and serum EV miRNA expression was investigated (Fig. 9C and Table S1). When miR-451a levels were normalized to the total amount of RNAs (global normalization, Fig. 9C, 2nd column from left), the correlation coefficient between miR-451a levels and IFN-β expression was −0.14. But when miR-451a levels were normalized to miR-16, the correlation coefficient was −0.69 (Fig. 9C, 4th column from right), which was comparable with that of Fig. 7G. Considering that RNA is known to be unstable, normalization using an internal control may be better than that using the total RNA amount for normalization.
Figure 9.
Correlation between miRNA ratio and IFN-β expression in macrophages stimulated with vaccines. A, sera were collected from six healthy subjects, and EVs were collected from each serum. Total RNAs were isolated from EVs and subjected to microarray analysis. The expression profiles of highly expressed miRNAs (top 100 among 3160) of six healthy subjects were shown as a heat map. B, THP-1 macrophages were cultured with human serum used for the microarray analysis for 2 days and were stimulated with inactivated WV for 6 h. IFN-β expression was determined by RT-qPCR. C, Pearson correlation coefficients (r) between miRNA levels (or miRNA ratios) and IFN-β expression in macrophages were calculated, and p values were determined. 2nd column from left, correlation coefficients were calculated using the values of global normalization of miRNA levels. 2nd column from right, correlation coefficients were calculated using the ratio of target miRNA/miR-7704. 4th column from right, correlation coefficients were calculated using the ratio of target miRNA/miR-16. The values that are statistically significant are highlighted in blue (negative correlation) or red (positive correlation).
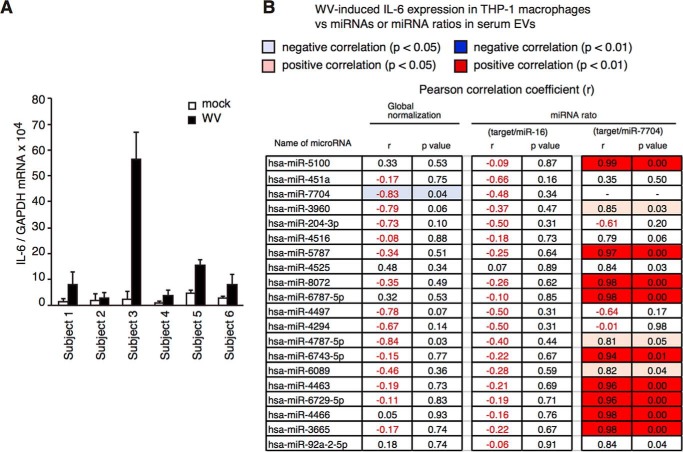
When we looked for other internal control miRNAs, we found that ratios of miR-7704 and other miRNAs gave rise to a statistically significant correlation between miRNA levels and IFN response (Fig. 9C, right two columns, and Fig. S4, A–D). For instance, the correlation coefficient between miR-5100/miR-7704 ratio and the type I IFN expression was statistically significant (p = 0.008) (Fig. 9C). The ratio was also significantly correlated with IL-6 levels (Fig. 10, A and B).
Figure 10.
Correlation between miRNA ratio and IL-6 expression in macrophages stimulated with vaccines. A, THP-1 macrophages were cultured with human serum used for the microarray analysis in Fig. 9 for 2 days and were stimulated with inactivated WV for 6 h. IL-6 expression was determined by RT-qPCR. B, Pearson correlation coefficients (r) between miRNA levels (or miRNA ratios) and IL-6 expression in macrophages were calculated, and p values were determined. 2nd column from left, correlation coefficients were calculated using the values of global normalization of miRNA levels. 2nd column from right, correlation coefficients were calculated using the ratio of target miRNA/miR-7704. 4th column from right, correlation coefficients were calculated using the ratio of target miRNA/miR-16. The values that are statistically significant are highlighted in blue (negative correlation) or red (positive correlation).
Because miR-451a regulated the cytokine expression, we reasoned that identified miRNAs would be involved in the cytokine expression pathway. As expected, miR-7704 mimic RNA reduced the expression of IFN-β by inactivated WV in THP-1 macrophages (Fig. S5A). Conversely, miR-5100 mimic RNA increased the expression of IFN-β in THP-1 macrophages in response to inactivated WV or influenza A virus infection (Fig. S5B). These data suggest that miR-5100 and miR-7704 are positive and negative regulators for inactivated WV-induced IFN-β expression in THP-1 macrophages, respectively.
Although SV hardly induced IFN-β expression in THP-1 macrophages compared with inactivated WV (Fig. S6A), IFN-γ-treated THP-1 expressed IFN-β and IL-6 in response to SV (Fig. S6, B and C). When SV instead of inactivated WV was used for the experiments, we found that the ratios of miR-3160-5p and miR-7704 were significantly correlated with SV-induced IFN-β expression in THP-1 macrophages (Fig. S6C) and that the ratios of miR-6085 and miR-2467-3p were significantly correlated with SV-induced IL-6 expression in THP-1 macrophages (Fig. S6D). Collectively, our data indicate that not only miR-451a but also other miRNAs in serum EVs affect the macrophage response.
Discussion
Cytokines and chemokines mediate intercellular communication. An accumulating body of evidence indicates that EVs also mediate intercellular communication, and our data indicate that EVs affect the innate immune response of macrophages and dendritic cells to inactivated WV. miR-451a levels in serum EVs gradually fluctuated over several weeks. Because miR-451a is abundant in serum EVs and are internalized into recipient cells, fluctuations of miR-451a concentration in serum EVs affected intracellular miR-451a levels in macrophages and dendritic cells, thereby modulating the innate immune response to inactivated WV. In addition, our microarray analysis identified several miRNAs that affected IFN-β expression in THP-1 macrophages in response to inactivated WV or SV. These data elucidate a role of blood-circulating EVs with miRNAs in the response of macrophage to influenza A virus vaccines.
Unlike miR-451a, intracellular miR-21 or miR-155 levels in macrophages did not increase following incubation with human serum. Considering that intracellular miR-21 and miR-155 levels were high compared with that of miR-451a, we suspect that the amounts of internalized miRNAs might be not sufficient to increase the basal intracellular miR-21 and miR-155 levels. This hypothesis can explain why specific miRNAs within human serum EVs can affect miRNA levels in recipient cells. Both of these conditions, a high level of miRNA in EVs and a low level in recipient cells, appear to be important for the miRNAs in blood EVs to influence intracellular miRNA levels in macrophages and DCs in vivo.
Although EVs delivered miRNAs to macrophages, our study showed that EVs barely delivered miR-451a into the cytoplasm of monocytes. This suggests that blood-circulating monocytes are not affected by EVs with miR-451a. In contrast, macrophages reside in tissues. Thus, EVs have to cross blood vessels to deliver miR-451a into macrophages in each tissue. It was shown that miR-155 and miR-146 in exosomes pass immune cells in vivo (24). In addition, adipose-derived EVs deliver miRNAs into other tissues (42). Moreover, EVs can even cross the blood–brain barrier (43). These observations are consistent with our model that circulating EVs deliver miR-451a to macrophages and dendritic cells in each tissue.
It is possible that the apparent increase of intracellular miR-451a levels is caused by attachment of EV on cell surface. However, our data weakened the possibility. First, cytochalasin D treatment, which destabilizes intracellular actin filaments, suppressed the increase of intracellular miR-451a levels after incubation of cells with serum. Second, the increase of intracellular miR-451a was correlated with the decrease of 14-3-3ζ protein levels, which is a target of miR-451a.
It has been shown that the SIDT2 protein transports internalized dsRNA from the endosome to the cytoplasm (44). However, siRNA for SIDT2 failed to inhibit EV-mediated reductions of 14-3-3ζ protein levels (Fig. S3C). It is possible that other pathways, such as SIDT1, compensate the defect of SIDT2. There is another possibility that miRNAs are released into the cytoplasm by fusion of the EV membrane with the endocytic vesicle membrane (1). Further studies are required to fully reveal the underlying mechanism.
The blood contains sufficient amounts of miRNAs. Previous studies reported that there are many free miRNAs that make a complex with the Ago2 protein and thus are stable in the blood (45, 46). Therefore, it is possible that serum miRNAs that are not included in EVs affected macrophage response. However, this possibility is weakened by the observation that EV miRNA levels do not correlate with the blood miRNA levels (47). In addition, we showed that the cytokine expression in macrophages correlated with EV miR-451a levels (Fig. 7G). These observations support our model that EV miR-451a, but not free miR-451a, affects macrophage response.
miR-451a targets 14-3-3ζ and reduces 14-3-3ζ protein levels (26). Incubation of macrophages with human serum containing miR-451a EVs markedly reduced endogenous 14-3-3ζ protein levels. The 14-3-3ζ protein binds FOXO3 and ZFP36 and inhibits these proteins by sequestering them from nucleic acids (48). FOXO3 and ZFP36 are known to be negative regulators for the expression of type I IFN and pro-inflammatory cytokine (49, 50). Therefore, it is expected that the action of miR-451a on 14-3-3ζ leads to the release of sequestrated FOXO3 and ZFP36, and these negatively regulate cytokine expression. This is consistent with our observation that the expression of IFN-β and IL-6 in response to inactivated WV was inversely correlated with miR-451a levels in serum EVs.
The miR-451a level in serum EVs gradually fluctuated over several months, suggesting that usual daily life events, such as eating and moderate exercise, do not affect its level in serum EVs. Considering that miR-451a in serum EVs affects the macrophage response, the stability of miR-451a expression during daily life events seems to be important to avoid excessive inflammation or attenuated immune response to pathogens. In contrast, we detected viral infection-induced change of miR-451a levels in serum EVs. In a mouse animal model, we found that the miR-451a level in serum EVs increased 5 days after influenza A virus infection. miR-451a levels in EVs depended on hnRNPA2B1 (Fig. 2), and hnRNPA2B1 is known to bind viral proteins of influenza A virus (51, 52). Thus, it is possible that viral proteins affect hnRNPA2B1 function, resulting in the change of miR-451a levels in EVs. Alternatively, it is also possible that influenza A virus affects sumoylation of hnRNPA2B1 required for the binding of hnRNPA2B1 to miRNAs (11), because it has been reported that influenza A virus interacts with the cellular sumoylation system (53). A previous study reported that intracellular miR-451a expression in the liver was reduced in high-fat diet-induced nonalcoholic steatohepatitis mice (54). Considering that donors who provided sera were healthy without symptoms of diseases, physical conditions might affect miR-451a levels in blood-circulating EVs. miR-451a levels were relatively high in the lung and spleen compared with other tissues, and thus, these tissues seem to be major sources of miR-451a–containing EVs in the blood. This means that the function of macrophages and DCs is regulated by other tissues. Recently, it was shown that adipose-derived exosomes regulate the gene expression in other tissues (42). These observations imply the inter-tissue communication via blood-circulating EVs.
Because there are various miRNAs in serum EVs, it is not surprising that not only miR-451a but also other miRNAs affect in vivo DC and macrophage functions. Because high expression levels in EVs seem to be important for miRNA to increase the intracellular miRNA levels, we expected that microarray analysis would uncover candidate miRNAs. We performed a microarray assay and focused on miRNAs that were highly expressed in blood-circulating EVs. At the same time, we investigated the response of macrophages cultured with serum against influenza A virus vaccine, and we unexpectedly found high correlation coefficients between miRNA ratios, such as miR-5100 and miR-7704, and macrophage response. This is consistent with our hypothesis that miRNAs packaged into EVs have significant impact on the response of macrophages to vaccine. miR-7704 and miR-5100 were negative and positive factors for type I IFN expression, respectively. Thus, we prefer the interpretation that the ratio of positive and negative factor levels is more efficiently correlated with type I IFN expression than positive or negative factor expression level alone.
miR-7704 was first identified by a microarray analysis to isolate miRNA expressed in macrophages after stimulation with IL-27 (55). Our study showed that mimic miR-7704 reduced type I IFN expression in response to inactivated WV, suggesting that miR-7704 is a negative regulator for the innate immune response. The miRNA target prediction software, TargetScanHuman, predicted that IRF-1 is a candidate of miR-7704–targeted genes. IRF-1 is well-known to regulate the innate immune response (56). Further studies are required to reveal the underlying mechanism of how miR-7704 attenuates the IFN response. Nevertheless, those miRNAs as well as miR-451a are expected to concomitantly modulate the innate immune response.
Vaccines are important for preventing infection. However, adverse reactions are sometimes caused by an excessive innate immune response to vaccines. Although we could not detect systemic IL-1β, IL-6, and TNFα production in mice injected with inactivated WV, at least under our experimental conditions, IFN-β and IL-6 expression was induced by stimulation of macrophages with inactivated WV. It was reported that IL-1β, IL-6, and TNFα produced by macrophages and other types of cells are known to be endogenous pyrogens, which are causes of fever (57). TNFα, IL-6, and other cytokines induce vascular permeability, a cause of swelling and redness (58, 59). Inflammation caused by innate immune responses is a cause of pain (60). Because the miRNA ratios were highly correlated with the innate immune response, it is expected that these miRNA ratios could be used for biomarkers to predict the adverse reaction before vaccination. Further studies would reveal the usefulness of EV miRNAs to predict the adverse reaction before vaccination.
Experimental procedures
Ethics statement
All of the animal studies were conducted in accordance with the Guidelines for Animal Experimentation of the Japanese Associations for Laboratory Animal Science and were approved by the Animal Care and Use Committee of Kumamoto University (A29-027, J27-232). All human samples were obtained and used in accordance with the Declaration of Helsinki and with the procedures approved by Ethics Committee of the Faculty of Life Sciences at Kumamoto University (RINRI 1203). All adult subjects provided informed consent, which was written in a document. We did not use any human samples derived from children.
Animals, cells, and reagents
THP-1 (JCRB Cell Bank) monocytes and THP-1 macrophages were cultured in RPMI 1640 medium with 5% FCS. THP-1 monocytes were differentiated into macrophages with phorbol 12-myristate 13-acetate (60 ng/ml) for 16 h, as described previously (23, 61). Human monocyte-derived macrophages were generated from CD14+ monocytes isolated from human blood and cultured with human GM-CSF for 6 days (62). HepG2 and A549 cells (provided by T. Seya at Hokkaido University) were cultured in DMEM (high Glc) and DMEM (low Glc) with 10% FCS, respectively. To obtain BM-DCs, BM cells were prepared from the femur and tibia. Isolated BM cells were cultured in RPMI 1640 medium with 10% FCS, 0.1 mm 2-mercaptoethanol, and 10 ng/ml murine GM-CSF for 6 days. Formalin-inactivated WV and SV were derived from the influenza A virus strain, A/California/7/2009[H1N1], and were provided by KAKETSUKEN (Kumamoto) (63). Mimic miRNAs, siRNAs for hnRNPA2B1, and negative control siRNA (scrambled siRNA, which has no significant sequence similarity to mouse or human gene sequences) were purchased from Bioneer Corp. and ThermoFisher Scientific. C57BL/6 mice were purchased from KYUDO Co. (Tosu). Poly(I-C), CL097, and cytochalasin D were purchased from GE Healthcare, Invivogen, and Wako, respectively. Anti-14-3-3ζ antibody and anti-β-actin mAb (AC-15) were purchased from Cell Signaling Technology and Sigma, respectively.
Analysis of EVs
To culture human and mouse cells, FCS was heat-inactivated at 56 °C for 30 min to culture human and mouse cells. Human serum without heat inactivation was used for culture of THP-1 cells or human mo-Mf. Mouse serum without heat inactivation was used for culture of BM-DCs when we observed miRNA internalization in recipient cells.
To determine miRNA levels in EVs released from culture cells, cells were seeded onto 24-well plates. The next day, cells were washed four times with PBS and subsequently cultured in serum-free medium for 1 or 2 days. EVs were isolated with total exosome isolation reagent (from cell culture media and from serum) (ThermoFisher Scientific), according to the manufacturer's instructions. Briefly, sera were prepared from healthy human subjects. Serum was mixed with 0.2 volume of the Total Exosome Isolation (from serum) reagents and incubated at 4 °C for 30 min. After incubation, samples were centrifuged at 10,000 × g for 10 min. Supernatants were discarded, and the pellet was resuspended with 1× PBS. To isolate EVs from cell culture medium, cell culture supernatants were mixed with 0.5 volume of the Total Exosome Isolation (from culture media) and were incubated at 4 °C overnight. Samples were centrifuged at 10,000 × g for 1 h. Supernatants were discarded, and pellets were suspended with 1× PBS.
To transfer EVs to cells, collected EVs was suspended with PBS and was added to the cell culture medium. To observe miRNA levels in serum EVs sequentially over 1 day, 1 week, or several months, about 10 μl of human blood were collected from the first finger of the left hand of healthy donors using Medisafe Finetouch Dispo (Terumo), and sera were prepared. Five microliters of sera were used for isolating EVs. To obtain human sera for cell culture and for measurement of miRNA levels in serum EVs, blood was collected from the vein of healthy donors, and sera were prepared.
Microarray
To perform the microarray analysis, 10 ml of each serum were collected from six healthy subjects. EVs were isolated from 5 ml of each serum. Total RNA was isolated from EVs using the mRNeasy mini kit (Qiagen) and was labeled with Cy5 using the 3D-gene miRNA labeling kit (TORAY). RNAs were hybridized for 16 h at 37 °C with a rotary shaker (250 rpm). Hybridization buffer and washing protocols were followed by the protocol supplied by TORAY. 3D-gene scanner (TORAY) was used for scanning. Images were quantified using Extraction software (TORAY). The raw data of each spot was normalized by substitution with a mean intensity of the background signal determined by all blank spot signal intensities of 95% confidence intervals. The microarray data were deposited to GEO. The accession number is GSE100128. The heatmap was drawn using Prism software.
miRNA expression
To determine the miRNA expression levels, total RNA was isolated with TRIzol (ThermoFisher Scientific). The reverse transcription reaction for the miRNA generation was performed using miR-X miRNA first-strand synthesis kit (Clontech). The SYBR Green real-time PCR master mix (ABI) was used for qPCR using the StepOne real time PCR system (ABI). When EVs were isolated from human serum, miRNA levels were normalized to miR-16 expression levels. Otherwise, the miRNA levels were normalized to U6 RNA expression levels. Copy numbers of miR-451a in human sera were also determined using the StepOne real time PCR system with standard synthesized RNA, according to the manufacturer's instructions.
Measurement of EVs
EV size and concentration were determined using NanoSight LM10V-HS. Each sample was diluted 10,000× and was measured five times to determine the average. The data were analyzed with NTA3.2 software.
Statistical analysis
All statistical analyses were carried out using Prism 7 (version 7) (GraphPad Software, Inc.), MS-Excel, or G*Power software. One-way ANOVA was performed to investigate the significance of differences between the means of several independent groups. To examine correlation, Pearson correlation coefficient (r) was calculated. To perform power analysis, we used G*Power software (version 3.1.9.3) and performed Student's t test to determine statistical power values (two-tails, effect size = Pearson correlation coefficient, α error probability = 0.05) (64).
Author contributions
M. O. and Y. F. data curation; M. O. and Y.F. formal analysis; M. O. and Y. F. investigation; T. K., T. D., and M. K. methodology; H. K. and H. O. supervision; H. O. writing-original draft; H. O. project administration; H. O. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Drs. T. Seya and M. Matsumoto at Hokkaido University for helpful discussion and K. Yoshinaga at Kumamoto University for technical assistance. We also thank Global Institute for Collaborative Research and Education (GI-CoRE) program at Hokkaido University.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, and Technology (MEXT), a grant-in-aid from the Japan Agency for Medical Research and Development (AMED), JST PREST, Tokyo Metropolitan Government, the Takeda Science Foundation, Kao Research Council for the Study of Healthcare Science, The NOVARTIS Foundation (Japan) for the Promotion of Science, The Uehara Memorial Foundation, the Japanese Association for Complement Research, the Waksman Foundation of Japan, and the Daiichi Sankyo Foundation of Life Science. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Table S1.
The microarray data were deposited to the Gene Expression Omnibus (GEO) database under GEO accession number GSE100128.
- EV
- extracellular vesicle
- miRNA
- microRNA
- WV
- whole-virus vaccine
- IFN
- interferon
- IL
- interleukin
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- FCS
- fetal calf serum
- DMEM
- Dulbecco's modified Eagle's medium
- PRR
- pattern recognition receptor
- DC
- dendritic cell
- mo-Mf
- monocyte-derived macrophage
- SV
- split virus
- BM
- bone marrow
- EXO
- exosome
- HBV
- hepatitis B virus.
References
- 1. Robbins P. D., and Morelli A. E. (2014) Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simons M., and Raposo G. (2009) Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 3. Grasedieck S., Sorrentino A., Langer C., Buske C., Döhner H., Mertens D., and Kuchenbauer F. (2013) Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood 121, 4977–4984 10.1182/blood-2013-01-480079 [DOI] [PubMed] [Google Scholar]
- 4. Gallo A., Tandon M., Alevizos I., and Illei G. G. (2012) The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 7, e30679 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caby M. P., Lankar D., Vincendeau-Scherrer C., Raposo G., and Bonnerot C. (2005) Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 17, 879–887 10.1093/intimm/dxh267 [DOI] [PubMed] [Google Scholar]
- 6. Colombo M., Raposo G., and Théry C. (2014) Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 7. Kouwaki T., Okamoto M., Tsukamoto H., Fukushima Y., and Oshiumi H. (2017) Extracellular vesicles deliver host and virus RNA and regulate innate immune response. Int. J. Mol. Sci. 18, 666 10.3390/ijms18030666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 9. Muralidharan-Chari V., Clancy J. W., Sedgwick A., and D'Souza-Schorey C. (2010) Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 123, 1603–1611 10.1242/jcs.064386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filipowicz W., Bhattacharyya S. N., and Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 11. Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D. J., Pascual-Montano A., Mittelbrunn M., and Sánchez-Madrid F. (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santangelo L., Giurato G., Cicchini C., Montaldo C., Mancone C., Tarallo R., Battistelli C., Alonzi T., Weisz A., and Tripodi M. (2016) The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 17, 799–808 10.1016/j.celrep.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 13. Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., and Ochiya T. (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 10.1074/jbc.M110.107821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shurtleff M. J., Temoche-Diaz M. M., Karfilis K. V., Ri S., and Schekman R. (2016) Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, e19276 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koppers-Lalic D., Hackenberg M., Bijnsdorp I. V., van Eijndhoven M. A. J., Sadek P., Sie D., Zini N., Middeldorp J. M., Ylstra B., de Menezes R. X., Würdinger T., Meijer G. A., and Pegtel D. M. (2014) Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 8, 1649–1658 10.1016/j.celrep.2014.08.027 [DOI] [PubMed] [Google Scholar]
- 16. Moldovan L., Batte K., Wang Y., Wisler J., and Piper M. (2013) Analyzing the circulating microRNAs in exosomes/extracellular vesicles from serum or plasma by qRT-PCR. Methods Mol. Biol. 1024, 129–145 10.1007/978-1-62703-453-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morelli A. E., Larregina A. T., Shufesky W. J., Sullivan M. L., Stolz D. B., Papworth G. D., Zahorchak A. F., Logar A. J., Wang Z., Watkins S. C., Falo L. D. Jr., and Thomson A. W. (2004) Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 104, 3257–3266 10.1182/blood-2004-03-0824 [DOI] [PubMed] [Google Scholar]
- 18. Loo Y. M., and Gale M. Jr. (2011) Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alexopoulou L., Holt A. C., Medzhitov R., and Flavell R. A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- 20. Diebold S. S., Kaisho T., Hemmi H., Akira S., and Reis e Sousa C. (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- 21. Hemmi H., Kaisho T., Takeuchi O., Sato S., Sanjo H., Hoshino K., Horiuchi T., Tomizawa H., Takeda K., and Akira S. (2002) Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3, 196–200 10.1038/ni758 [DOI] [PubMed] [Google Scholar]
- 22. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., and Fujita T. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 23. Kouwaki T., Fukushima Y., Daito T., Sanada T., Yamamoto N., Mifsud E. J., Leong C. R., Tsukiyama-Kohara K., Kohara M., Matsumoto M., Seya T., and Oshiumi H. (2016) Extracellular vesicles including exosomes regulate innate immune responses to hepatitis B virus infection. Front. Immunol. 7, 335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexander M., Hu R., Runtsch M. C., Kagele D. A., Mosbruger T. L., Tolmachova T., Seabra M. C., Round J. L., Ward D. M., and O'Connell R. M. (2015) Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 6, 7321 10.1038/ncomms8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montecalvo A., Larregina A. T., Shufesky W. J., Stolz D. B., Sullivan M. L., Karlsson J. M., Baty C. J., Gibson G. A., Erdos G., Wang Z., Milosevic J., Tkacheva O. A., Divito S. J., Jordan R., Lyons-Weiler J., et al. (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 10.1182/blood-2011-02-338004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenberger C. M., Podyminogin R. L., Navarro G., Zhao G. W., Askovich P. S., Weiss M. J., and Aderem A. (2012) miR-451 regulates dendritic cell cytokine responses to influenza infection. J. Immunol. 189, 5965–5975 10.4049/jimmunol.1201437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patrick D. M., Zhang C. C., Tao Y., Yao H., Qi X., Schwartz R. J., Jun-Shen Huang L., and Olson E. N. (2010) Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3ζ. Genes Dev. 24, 1614–1619 10.1101/gad.1942810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., and Gale M. Jr. (2008) Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82, 335–345 10.1128/JVI.01080-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Le Goffic R., Pothlichet J., Vitour D., Fujita T., Meurs E., Chignard M., and Si-Tahar M. (2007) Cutting edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J. Immunol. 178, 3368–3372 10.4049/jimmunol.178.6.3368 [DOI] [PubMed] [Google Scholar]
- 30. Koyama S., Aoshi T., Tanimoto T., Kumagai Y., Kobiyama K., Tougan T., Sakurai K., Coban C., Horii T., Akira S., and Ishii K. J. (2010) Plasmacytoid dendritic cells delineate immunogenicity of influenza vaccine subtypes. Sci. Transl. Med. 2, 25ra24 [DOI] [PubMed] [Google Scholar]
- 31. Gross P. A., Ennis F. A., Gaerlan P. F., Denson L. J., Denning C. R., and Schiffman D. (1977) A controlled double-blind comparison of reactogenicity, immunogenicity, and protective efficacy of whole-virus and split-product influenza vaccines in children. J. Infect. Dis. 136, 623–632 10.1093/infdis/136.5.623 [DOI] [PubMed] [Google Scholar]
- 32. Gross P. A., and Ennis F. A. (1977) Influenza vaccine: split-product versus whole-virus types–how do they differ. N. Engl. J. Med. 296, 567–568 10.1056/NEJM197703102961012 [DOI] [PubMed] [Google Scholar]
- 33. Rhorer J., Ambrose C. S., Dickinson S., Hamilton H., Oleka N. A., Malinoski F. J., and Wittes J. (2009) Efficacy of live attenuated influenza vaccine in children: a meta-analysis of nine randomized clinical trials. Vaccine 27, 1101–1110 10.1016/j.vaccine.2008.11.093 [DOI] [PubMed] [Google Scholar]
- 34. Smyth L. A., Boardman D. A., Tung S. L., Lechler R., and Lombardi G. (2015) MicroRNAs affect dendritic cell function and phenotype. Immunology 144, 197–205 10.1111/imm.12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang Z. P., Chen J., Seok H. Y., Zhang Z., Kataoka M., Hu X., and Wang D. Z. (2013) MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 112, 1234–1243 10.1161/CIRCRESAHA.112.300682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao C., and Rajewsky K. (2009) MicroRNA control in the immune system: basic principles. Cell 136, 26–36 10.1016/j.cell.2008.12.027 [DOI] [PubMed] [Google Scholar]
- 37. Xiao C., Calado D. P., Galler G., Thai T. H., Patterson H. C., Wang J., Rajewsky N., Bender T. P., and Rajewsky K. (2007) miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 38. Xiang M., Zeng Y., Yang R., Xu H., Chen Z., Zhong J., Xie H., Xu Y., and Zeng X. (2014) U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem. Biophys. Res. Commun. 454, 210–214 10.1016/j.bbrc.2014.10.064 [DOI] [PubMed] [Google Scholar]
- 39. Song J., Bai Z., Han W., Zhang J., Meng H., Bi J., Ma X., Han S., and Zhang Z. (2012) Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 57, 897–904 10.1007/s10620-011-1981-7 [DOI] [PubMed] [Google Scholar]
- 40. Zhou W., Fong M. Y., Min Y., Somlo G., Liu L., Palomares M. R., Yu Y., Chow A., O'Connor S. T., Chin A. R., Yen Y., Wang Y., Marcusson E. G., Chu P., Wu J., et al. (2014) Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Padovan E., Spagnoli G. C., Ferrantini M., and Heberer M. (2002) IFN-α2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. J. Leukoc. Biol. 71, 669–676 [PubMed] [Google Scholar]
- 42. Thomou T., Mori M. A., Dreyfuss J. M., Konishi M., Sakaguchi M., Wolfrum C., Rao T. N., Winnay J. N., Garcia-Martin R., Grinspoon S. K., Gorden P., and Kahn C. R. (2017) Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455 10.1038/nature21365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Balusu S., Van Wonterghem E., De Rycke R., Raemdonck K., Stremersch S., Gevaert K., Brkic M., Demeestere D., Vanhooren V., Hendrix A., Libert C., and Vandenbroucke R. E. (2016) Identification of a novel mechanism of blood-brain communication during peripheral inflammation via choroid plexus-derived extracellular vesicles. EMBO Mol. Med. 8, 1162–1183 10.15252/emmm.201606271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen T. A., Smith B. R. C., Tate M. D., Belz G. T., Barrios M. H., Elgass K. D., Weisman A. S., Baker P. J., Preston S. P., Whitehead L., Garnham A., Lundie R. J., Smyth G. K., Pellegrini M., O'Keeffe M., et al. (2017) SIDT2 transports extracellular dsRNA into the cytoplasm for innate immune recognition. Immunity 47, 498–509.e6 10.1016/j.immuni.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., Mitchell P. S., Bennett C. F., Pogosova-Agadjanyan E. L., Stirewalt D. L., Tait J. F., and Tewari M. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andreu Z., Rivas E., Sanguino-Pascual A., Lamana A., Marazuela M., González-Alvaro I., Sánchez-Madrid F., de la Fuente H., and Yáñez-Mó M. (2016) Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles 5, 31655 10.3402/jev.v5.31655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xie J. X., Fan X., Drummond C. A., Majumder R., Xie Y., Chen T., Liu L., Haller S. T., Brewster P. S., Dworkin L. D., Cooper C. J., and Tian J. (2017) MicroRNA profiling in kidney disease: plasma versus plasma-derived exosomes. Gene 627, 1–8 10.1016/j.gene.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fu H., Subramanian R. R., and Masters S. C. (2000) 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 10.1146/annurev.pharmtox.40.1.617 [DOI] [PubMed] [Google Scholar]
- 49. Luron L., Saliba D., Blazek K., Lanfrancotti A., and Udalova I. A. (2012) FOXO3 as a new IKK-ϵ-controlled check-point of regulation of IFN-β expression. Eur. J. Immunol. 42, 1030–1037 10.1002/eji.201141969 [DOI] [PubMed] [Google Scholar]
- 50. Schichl Y. M., Resch U., Hofer-Warbinek R., and de Martin R. (2009) Tristetraprolin impairs NF-κB/p65 nuclear translocation. J. Biol. Chem. 284, 29571–29581 10.1074/jbc.M109.031237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang C. K., Chen C. J., Wu C. C., Chen S. W., Shih S. R., and Kuo R. L. (2017) Cellular hnRNP A2/B1 interacts with the NP of influenza A virus and impacts viral replication. PLoS ONE 12, e0188214 10.1371/journal.pone.0188214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y., Zhou J., and Du Y. (2014) hnRNP A2/B1 interacts with influenza A viral protein NS1 and inhibits virus replication potentially through suppressing NS1 RNA/protein levels and NS1 mRNA nuclear export. Virology 449, 53–61 10.1016/j.virol.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pal S., Santos A., Rosas J. M., Ortiz-Guzman J., and Rosas-Acosta G. (2011) Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158, 12–27 10.1016/j.virusres.2011.02.017 [DOI] [PubMed] [Google Scholar]
- 54. Hur W., Lee J. H., Kim S. W., Kim J. H., Bae S. H., Kim M., Hwang D., Kim Y. S., Park T., Um S. J., Song B. J., and Yoon S. K. (2015) Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 64, 265–276 10.1016/j.biocel.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 55. Swaminathan S., Hu X., Zheng X., Kriga Y., Shetty J., Zhao Y., Stephens R., Tran B., Baseler M. W., Yang J., Lempicki R. A., Huang D., Lane H. C., and Imamichi T. (2013) Interleukin-27 treated human macrophages induce the expression of novel microRNAs which may mediate anti-viral properties. Biochem. Biophys. Res. Commun. 434, 228–234 10.1016/j.bbrc.2013.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sato M., Taniguchi T., and Tanaka N. (2001) The interferon system and interferon regulatory factor transcription factors–studies from gene knockout mice. Cytokine Growth Factor Rev. 12, 133–142 10.1016/S1359-6101(00)00032-0 [DOI] [PubMed] [Google Scholar]
- 57. Dinarello C. A. (2004) Infection, fever, and exogenous and endogenous pyrogens: some concepts have changed. J. Endotoxin Res. 10, 201–222 10.1179/096805104225006129 [DOI] [PubMed] [Google Scholar]
- 58. Pober J. S., and Sessa W. C. (2007) Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815 10.1038/nri2171 [DOI] [PubMed] [Google Scholar]
- 59. Paul R., Koedel U., Winkler F., Kieseier B. C., Fontana A., Kopf M., Hartung H. P., and Pfister H. W. (2003) Lack of IL-6 augments inflammatory response but decreases vascular permeability in bacterial meningitis. Brain 126, 1873–1882 10.1093/brain/awg171 [DOI] [PubMed] [Google Scholar]
- 60. Scholz J., and Woolf C. J. (2007) The neuropathic pain triad: neurons, immune cells and glia. Nat. Neurosci. 10, 1361–1368 10.1038/nn1992 [DOI] [PubMed] [Google Scholar]
- 61. Atay S., Gercel-Taylor C., Suttles J., Mor G., and Taylor D. D. (2011) Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 65, 65–77 10.1111/j.1600-0897.2010.00880.x [DOI] [PubMed] [Google Scholar]
- 62. Lacey D. C., Achuthan A., Fleetwood A. J., Dinh H., Roiniotis J., Scholz G. M., Chang M. W., Beckman S. K., Cook A. D., and Hamilton J. A. (2012) Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J. Immunol. 188, 5752–5765 10.4049/jimmunol.1103426 [DOI] [PubMed] [Google Scholar]
- 63. Uno S., Kimachi K., Kei J., Miyazaki K., Oohama A., Nishimura T., Ibaragi K., Odoh K., Kudo Y., and Kino Y. (2011) Effect of prior vaccination with a seasonal trivalent influenza vaccine on the antibody response to the influenza pandemic H1N1 2009 vaccine: a randomized controlled trial. Microbiol. Immunol. 55, 783–789 10.1111/j.1348-0421.2011.00381.x [DOI] [PubMed] [Google Scholar]
- 64. Faul F., Erdfelder E., Lang A. G., and Buchner A. (2007) G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 10.3758/BF03193146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.