Abstract
The natural amino acid hypusine (Nϵ-4-amino-2-hydroxybutyl(lysine)) is derived from the polyamine spermidine, and occurs only in a single family of cellular proteins, eukaryotic translation factor 5A (eIF5A) isoforms. Hypusine is formed by conjugation of the aminobutyl moiety of spermidine to a specific lysine residue of this protein. The posttranslational synthesis of hypusine involves two enzymatic steps, catalyzed by deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH). Hypusine is essential for eIF5A activity. Inactivation of either the eIF5A or the DHPS gene is lethal in yeast and mouse, underscoring the vital role of eIF5A hypusination in eukaryotic cell growth and animal development. The long and basic side chain of the hypusine residue promotes eIF5A-mediated translation elongation by facilitating peptide bond formation at polyproline stretches and at many other ribosome-pausing sites. It also enhances translation termination by stimulating peptide release. By promoting translation, the hypusine modification of eIF5A provides a key link between polyamines and cell growth regulation. eIF5A has been implicated in several human pathological conditions. Recent genetic data suggest that eIF5A haploinsufficiency or impaired deoxyhypusine synthase activity is associated with neurodevelopmental disorders in humans.
Keywords: spermidine, polyamine, cell growth, posttranslational modification (PTM), translation, eukaryotic translation factor, hypusine, neurodevelopment, eIF5A, neurodevelopment
Introduction
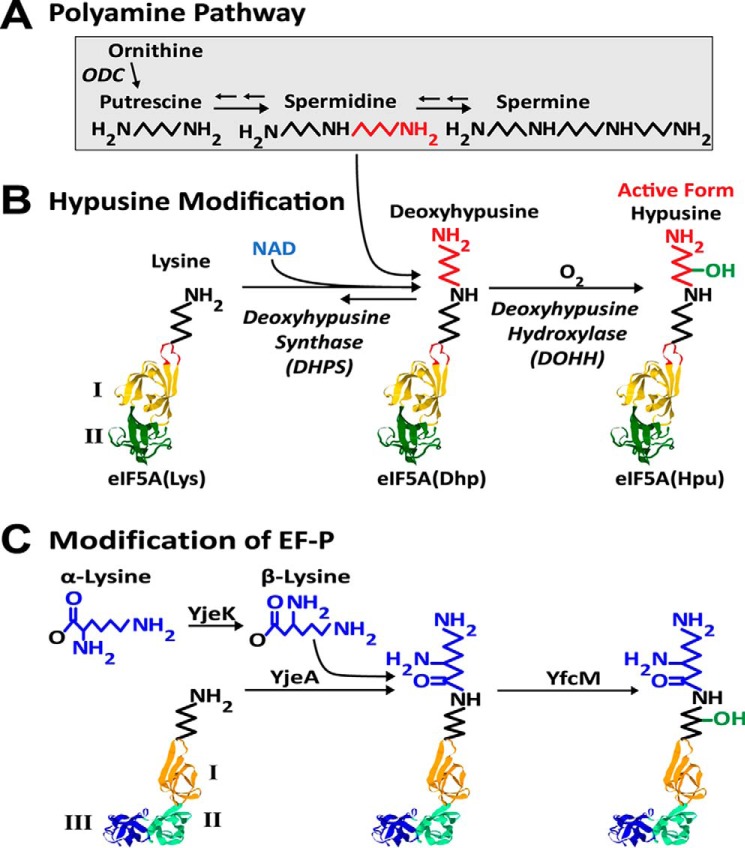
The polyamines, putrescine, spermidine, and spermine, exist as polycations under physiological environments and exert numerous effects on nucleic acids, proteins, phospholipids, and ion channels in living cells and organisms. They are vital for cell proliferation and regulate cellular activities at the transcriptional, translational, and posttranslational levels (for reviews, see Refs. 1–4). Thus, the polyamine pathways are intimately associated with cancer (5). In addition to their functions as polycations, a small portion (<2%) of the polyamine spermidine is metabolized to form an unusual amino acid, hypusine (Nϵ-4-amino-2-hydroxybutyl(lysine)), in a single family of cellular proteins, eukaryotic initiation factor 5A isoforms2 (eIF5A)3 (6) (Fig. 1, A and B). Hypusine synthesis occurs posttranslationally in two enzymatic steps (7). First, deoxyhypusine synthase (DHPS) catalyzes the cleavage of the spermidine and transfer of the 4-aminobutyl moiety to the ϵ-amino group of a specific lysine residue of eIF5A precursor to form a deoxyhypusine (Nϵ-4-aminobutyl(lysine)) residue (eIF5A(Dhp)) (8, 9). In the second step, this intermediate is hydroxylated by deoxyhypusine hydroxylase (DOHH) to form hypusine (10, 11) and thus to complete eIF5A maturation. The synthesis of hypusine exclusively in a single cellular protein represents one of the most specific posttranslational modifications known to date. Hypusine/deoxyhypusine is required for the activity of eIF5A and thereby for eukaryotic cell proliferation (12–15).
Figure 1.
Pathways of polyamines (A), hypusine synthesis (B), and EF-P modification (C). A, simplified pathway of polyamine interconversion. Putrescine generated from ornithine is converted to spermidine and spermine. B, the posttranslational formation of hypusine in eIF5A by two enzymatic steps catalyzed by DHPS and DOHH. The aminobutyl moiety of spermidine (red) is used for hypusine synthesis at Lys50 of human eIF5A(Lys). The DHPS reaction is reversible (38). Model structure of eIF5A is based on the human eIF5A (PDB code 1FH4) (28) with the N-terminal domain I (yellow) containing the hypusine site loop (Ser45–Ala54) (orange) and the C-terminal domain II (green). C, EF-P (PDB code 1UEB) (30), the bacterial ortholog of eIF5A, contains domain III, which is similar to domain II and is modified by β-lysylation (31) followed by hydroxylation. eIF5A(Hpu), eIF5A final modified form containing hypusine.
Since the discovery of hypusine in 1971 (16), it has taken many years to reach the current status of knowledge on the hypusine-containing protein, eIF5A. In this review, we will discuss the history of hypusine research with emphasis on the specificity and mechanism of the hypusine modification enzymes, the role of eIF5A in cell growth and animal development, and its mode of action in translation. EF-P (elongation factor P), the bacterial ortholog of eIF5A, its modification by β-lysylation (Fig. 1C), and its function will also be discussed briefly, in light of the significant structural and functional similarities to eIF5A (17–19).
Discovery of hypusine and the hypusine-containing protein
Hypusine (Nϵ-4-amino-2-hydroxybutyl(lysine)) was first isolated from bovine brain extracts by Shiba et al. in 1971 (16) during their search for new amine components. Its chemical structure was determined, and it was named hypusine based on its relationship to hydroxyputrescine and lysine. Hypusine was found to occur in all animal tissues, as the free amino acid as well as a protein component (20, 21). Radioactivity was detected in the hypusine fraction isolated from rats injected with radioactive lysine, suggesting that hypusine was derived from lysine (21). However, no information was available on the nature of the protein(s) containing hypusine, and hypusine was buried in the literature merely as a new biochemical entity for a decade. The hypusine-containing protein was discovered in our laboratory in 1981 in an experiment designed to identify a cellular substrate protein into which polyamine is incorporated by the transglutaminase reaction (6). To this end, human lymphocytes were cultured in a medium containing radioactive spermidine, and the radiolabeled protein was analyzed by two-dimensional gel electrophoresis. To our surprise, only one specific protein (∼18 kDa, pI 5.2) was radiolabeled; the radioactive component of this protein turned out to be hypusine (6), and not a product of a transglutaminase reaction. This protein was later identified as eukaryotic translation initiation factor 4D (eIF-4D; current nomenclature, eIF5A) (22). We demonstrated that the direct polyamine precursor of hypusine is spermidine and that hypusine is formed posttranslationally, and not as a free amino acid. Thus, free hypusine initially isolated from soluble extracts of animal tissues must have originated from the proteolytic breakdown of eIF5A protein.
eIF5A, aIF5A, and their bacterial ortholog EF-P
Hypusine occurs in all eukaryotes and in certain archaea, but not in eubacteria (for reviews, see Refs. 7, 17, and 23). eIF5A, DHPS, and DOHH are all highly conserved in eukaryotes (23). eIF5A consists of two domains, shown in Fig. 1B: domain I (N-terminal domain in yellow) and domain II (C-terminal domain in green). The Lys residue (Lys50 in human eIF5A) that undergoes the hypusine modification is located at the tip of a flexible loop (orange) of domain I. The amino acid sequence of this loop (Ser45–Ala54) is strictly conserved in all eukaryotes, suggesting its importance in the hypusine modification as well as in eIF5A function (24, 25). eIF5A is one of the few translation factors universally conserved from bacteria to humans (25). It shares structural and functional analogy with its archaeal homolog, aIF5A (26) and the bacterial ortholog, EF-P (27). The crystal structures of eIF5A from humans (28) and yeast (PDB code 3ER0), aIF5A (29), and EF-P (30) show remarkable resemblance, except that EF-P contains an extra third domain similar to its second domain (Fig. 1C). eIF5A and aIF5A share the conserved lysine that undergoes deoxyhypusine/hypusine modification. EF-P does not undergo hypusine modification. Instead, some EF-P proteins that contain the lysine residue corresponding to the lysine modified to hypusine in eIF5A are modified by conjugation with β-lysine by YjeA (homolog of class II lysyl-tRNA synthetase) and are subsequently hydroxylated by YfcM (31, 32) (Fig. 1C). β-Lysine is produced from α-lysine by YjeK (lysine 2,3-aminomutase). Like the hypusine modification in eIF5A, β-lysylation is known to occur in EF-P only and generates a long basic side chain that is important for the factor's activity (33, 34).
DHPS and DOHH: Mechanism, inhibitors, and specificity
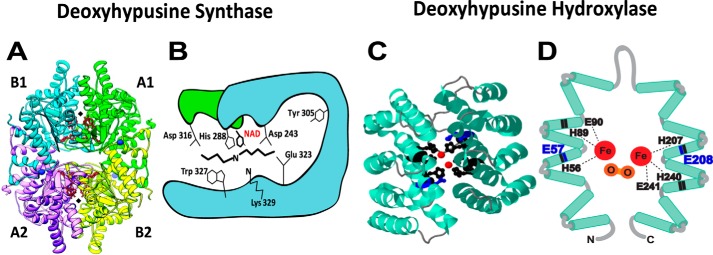
The first evidence that hypusine is formed in two independent enzymatic steps was obtained by the use of iron chelators (35). In the presence of a metal chelator, α,α-dipyridyl, the unhydroxylated intermediate, deoxyhypusine (Nϵ-4-aminobutyl(lysine)) was formed in eIF5A. The DHPS reaction is proposed to occur in four steps (Fig. 2) (36): step I, NAD-dependent dehydrogenation of spermidine with generation of enzyme-bound NADH; step II, cleavage of dehydrospermidine to produce an enzyme-butylimine intermediate and 1,3-diaminopropane; step III, transfer of the butylimine moiety from the enzyme intermediate to the ϵ-amino group of a specific lysine of the eIF5A precursor; and step IV, reduction of the eIF5A imine intermediate by the enzyme-bound NADH to form deoxyhypusine. The evidence that the NADH generated in step I remains bound to the enzyme until step IV to reduce the eIF5A imine intermediate was obtained by NADH fluorescence measurement (37). The involvement of the enzyme imine intermediate was validated by trapping it into a stable adduct using NaBH3CN reduction; this active site was identified as Lys329 (in human DHPS) (36). If the eIF5A precursor is omitted, DHPS can catalyze an abortive reaction (i.e. spermidine cleavage) to produce 1,3-diaminopropane, Δ1-pyrroline, and NADH (Fig. 2, partial reaction, dotted arrows). Apparently, the DHPS reaction is reversible, as radioactive spermidine can be generated upon incubation of the radiolabeled deoxyhypusine protein with NAD and DHPS (38). However, there is no evidence of back-conversion of the hypusine-containing eIF5A protein to eIF5A(Lys). DHPS can also catalyze the synthesis of homospermidine from spermidine, using putrescine as an acceptor of butylimine moiety (38), instead of eIF5A(Lys), although the Km of the human enzyme for putrescine is much higher (∼700-fold) than that for eIF5A(Lys). Interestingly, a plant enzyme, homospermidine synthase, catalyzes synthesis of homospermidine by the same mechanism, but does not catalyze synthesis of deoxyhypusine (39), suggesting its evolution by DHPS gene duplication and selection of the minor function of this enzyme for plant secondary metabolism. DHPS is a tetrameric enzyme composed of four identical subunits of ∼40 kDa, and its active sites are located at the interface of dimers (Fig. 3A) (40, 41). The crystal structure of the enzyme–NAD complex showed a restricted spermidine binding pocket, contributing to its narrow specificity. The role of each of the conserved amino acid residues of the predicted spermidine-binding pocket was confirmed by alanine substitution (42). The proposed model of spermidine binding at the active site shows the two terminal amino groups of spermidine, anchored by acidic residues Asp243, Asp316, and Glu323 (Fig. 3B) (23, 42). Only those compounds with two terminal basic groups spaced by a distance of 7–8 methylenes are effective inhibitors of DHPS. Of many analogs of spermidine tested, N1-monoguanyl 1,7-diaminoheptane (GC7) is the most potent inhibitor (43). It is actively taken up by cells and effectively inhibits eIF5A hypusination intracellularly. Either alone or in combination with other agents, GC7 caused inhibition of growth in various mammalian cells (44–46) and in an animal tumor model (47). High-throughput screening is in progress to identify new, specific inhibitors or enhancers of DHPS (48).
Figure 2.
Mechanism of the DHPS reaction. The reaction occurs in four steps (I–IV) to form deoxyhypusine in eIF5A (36). However, if eIF5A(Lys) is omitted, DHPS catalyzes the cleavage of spermidine and generates 1,3-diaminopropane, Δ1-pyrroline, and NADH (broken arrows). The transient enzyme–imine intermediate at Lys329 was confirmed by trapping it into a stable form by reduction with NaBH3CN (36).
Figure 3.
Crystal structures of DHPS and DOHH and their active sites. A, human DHPS homotetramer (PDB codes 1ROZ and 1RLZ) complex with NAD (41). The two active sites at the dimer interfaces are indicated by black diamonds. B, active site residues of DHPS critical for catalysis (Lys329 and His288) and binding of spermidine (Asp243, Asp316, Glu323, and Trp327) (23, 42). C, crystal structure of DOHH peroxo–diiron intermediate (PBD code 4D4Z) (50) consisting of eight HEAT repeats (helical hairpins), diiron center (red), and critical active-site residues (black and blue). D, a diagram of DOHH (11) active site showing peroxo (orange)–diiron (red) center and the four conserved His-Glu motifs critical for catalysis. His56, His89, Glu90, His207, His240, and Glu241 (black) are required for the binding of iron (49), and Glu57 and Glu208 (blue) are required for binding of the terminal amino group of deoxyhypusine side chain (blue) (53).
DOHH catalyzes a stereospecific hydroxylation at C2 of the deoxyhypusine side chain. It is a unique nonheme diiron monooxygenase (49) with a superhelical HEAT-repeat structure (Fig. 3C) (11, 50). The structure (50) and mechanism of DOHH (51, 52) are distinct from other known protein hydroxylases. DOHH consists of eight tandem repeats of α-helical pairs with the diiron center anchored by four strictly conserved His-Glu motifs that are critical for the enzyme activity (11, 49). Of these, His56, His89, Glu90, His207, His240, and Glu241 are involved in the binding of the two irons, whereas Glu57 and Glu208 are responsible for the binding of the deoxyhypusine side chain of the protein substrate eIF5A(Dhp) (Fig. 3D) (49, 53). The enzyme is inhibited by iron chelators, such as ciclopirox olamine or deferiprone, two Food and Drug Administration–approved antifungal agents. These compounds are also effective against retroviral infections (54), including HIV-1 (55), and in the inhibition of cancer cell growth (56). It was suggested that they confer these effects at least partially through inhibition of DOHH (55, 56).
Both DHPS and DOHH are totally specific for eIF5A and do not modify any other cellular proteins. DHPS and Lia1 (ligand of eIF5A, later identified as DOHH) were identified as two strong eIF5A-binding partners from yeast two-hybrid screening (57). Formation of high-affinity complexes, between eIF5A and either enzyme, was demonstrated by pulldown assays, native gel electrophoresis, or equilibrium ultracentrifugation. eIF5A and DHPS form a stable complex with a Kd < 0.5 nm (58). However, no crystal structure of the eIF5A–DHPS complex is yet available. To determine the minimum structure of eIF5A required for modification, truncated eIF5A polypeptides were tested as substrates for DHPS and DOHH. When truncated peptides of human eIF5A(Lys) (aa 1–90, 1–80, 1–70, 1–60, 10–90, 20–90, 30–90, 20–80, and 30–80) were used as substrates for DHPS, deoxyhypusine was formed in all of the peptides, except aa 1–70, 1–60, and 30–80, indicating that aa 20–80 is the minimum size required (59). When the peptides were used in the combined DHPS and DOHH assay, hypusine formation was detected with aa 1–90, 10–90, and 20–90 peptides, but not with aa 1–80, 1–70, and 30–90, indicating that aa 20–90 is minimally required to be substrates for both DHPS and DOHH (23, 53). These findings suggest that the nearly intact domain I of eIF5A is required for its modification by DHPS or DOHH and thereby uncover the structural basis of the extraordinary selectivity of hypusine synthesis in a single protein.
Role of polyamines and eIF5A in eukaryotic translation, cell proliferation, and animal development
A critical role of eIF5A in cell growth regulation was first implied by a drastic increase in the hypusine-containing protein in lymphocytes upon activation with a mitogen (6, 22, 60). A correlation between hypusine formation and growth was also observed in other mammalian cells, including rat hepatoma tissue culture cells (61), and in Ras oncogene–transfected NIH3T3 cells (62).
Gene inactivation studies in the yeast Saccharomyces cerevisiae and in mice provide firm evidence for the essential nature of eIF5A and its deoxyhypusine/hypusine modification in eukaryotic cell growth and animal development. Deletion of both eIF5A genes (14) or a single DHPS gene (64, 65) caused loss of viability in yeast. But a S. cerevisiae strain lacking the DOHH gene is viable (11), suggesting that the deoxyhypusine-containing eIF5A, eIF5A(Dhp), can support growth in yeast. In contrast, the disruption of any of the eIF5A, DHPS, or DOHH genes leads to embryonic lethality in mice (66, 67). Loss of DOHH is recessively lethal in Caenorhabditis elegans (68) and Drosophila (69).
The essential requirement for polyamines in S. cerevisiae cell growth (70) was attributed to the role of spermidine in hypusine synthesis in eIF5A (71). The question of whether hypusine formation is the most critical function of polyamines in yeast was addressed by Dr. Herbert Tabor's group in collaboration with our group (72). The S. cerevisiae spe2Δ mutant lacking SAM decarboxylase, and thereby spermidine biosynthesis, could grow at a nearly normal rate when cellular spermidine was ∼0.2% of that in the parental strain (72). In the WT strain, only ∼1.4% of cellular spermidine was converted to hypusine in eIF5A. However, increasing portions of total cellular spermidine (as much as ∼50%) were mobilized for hypusine synthesis in the spe2Δ mutant, as spermidine became severely limiting (72). This finding indicates that hypusination of eIF5A is the most critical function of polyamines in yeast cell growth.
In contrast to S. cerevisiae, mammalian cells depend on a high level (millimolar) of intracellular polyamine for cell proliferation. A decrease in either the polyamine or hypusinated eIF5A level independently led to a significant inhibition of growth in FM3A cells treated with inhibitors of polyamine biosynthesis or GC7 (73). Furthermore, a drastic inhibition of protein synthesis and cell growth was observed in 293T cells depleted of spermidine and spermine upon overexpression of a polyamine catabolic enzyme, spermidine spermine acetyltransferase 1, before any significant decrease in hypusinated eIF5A was detected (74). Polysome profiles of these polyamine-depleted 293T cells and also of DFMO-treated NIH3T3 cells (75) showed loss of polysomes, suggesting a block in translation initiation in these polyamine-depleted cells.
Evidence for the indispensability of eIF5A hypusination in mammalian cell growth was first reported in L1210 cells depleted of spermidine by treatment with 5′-([(Z)-4-aminobut-2-enyl]methylamino)-5′-deoxyadenosine (AbeAdo), an inhibitor of SAM decarboxylase (76, 77). Cytostasis in the AbeAdo-treated cells could be reversed by spermidine, spermine, or the polyamine analogs that function directly or indirectly as a precursor for hypusine or a hypusine analog (76, 77). Consistent with these results, in DU145 cells depleted of spermidine by treatment with α-DFMO (α-difluoromethylornithine, an irreversible inhibitor of ornithine decarboxylase), long-term growth could be supported only by the polyamine analogs that could be converted to hypusine in eIF5A (78). However, the early acute phase of growth inhibition could be rescued by several nonnatural polyamine analogs.
The role of spermidine in deoxyhypusine/hypusine modification is vital for cell growth in both yeast and mammalian cells. A question arises then as to why there is a huge difference (several hundred-fold) in the minimum polyamine requirement between the yeast and mammalian cells. In the cellular environment, polyamines and Mg2+ bind to ATP, DNA, RNA, and ribosomes and thereby contribute to the regulation of macromolecular synthesis. The ability of the yeast mutant strains to grow with drastically reduced cellular polyamines4 appears to be due to the substitution of polyamines by increased cellular Mg2+. In contrast to yeast, mammalian cell growth is sensitive to a reduction in cellular polyamine levels, probably because such a Mg2+-dependent compensatory mechanism does not operate in mammals. Thus, mammalian cells rely on the intricate mechanisms of polyamine homeostasis to sustain normal cell growth.
Mode of action of eIF5A in translation
Eukaryotic translation initiation factor 5A (old nomenclature, IF-M2Bα; renamed to eIF-4D and then to eIF5A) was initially isolated from rabbit reticulocyte ribosomes and was classified as an initiation factor, based on its stimulating activity on methionyl-puromycin synthesis (79, 80), a model assay for the first peptide bond formation. The hypusine residue was shown to be important for eIF5A activity in this assay; the unmodified eIF5A precursor was inactive (12, 15), whereas the deoxyhypusine-containing eIF5A was partially active in this assay (13). Furthermore, the eIF5A mutant with Ala or Arg substitution at the hypusine modification site (K51A or K51R in the yeast eIF5A) fails to support growth of yeast (14, 24), indicating the vital importance of hypusinated eIF5A in eukaryotic cell growth. The precise role of eIF5A in translation initiation remained questionable, as it did not enhance various steps leading to the formation of the 80S initiation complex (17). However, definitive evidence for its role in elongation was provided by an increase in the polysome fraction and in the ribosome transit time upon depletion of eIF5A in the S. cerevisiae mutant strains (81, 82).
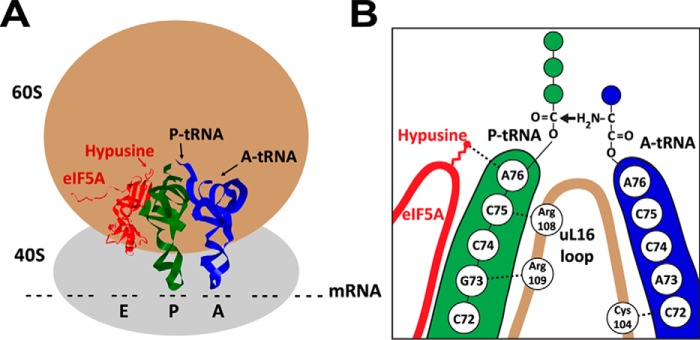
New insights into the mechanism of eIF5A action have been gleaned from studies of EF-P (for reviews, see Refs. 17–19). During translation elongation, peptide bond formation between the acceptor, aminoacyl-tRNA (A-tRNA), and the donor, peptidyl-tRNA (P-tRNA), does not occur easily at certain amino acids, such as proline and glycine, that are poor substrates for peptide bond formation, resulting in a drop-off of the P-tRNA from ribosome or ribosome stalling. Groundbreaking studies from two laboratories (83, 84) demonstrated the role of EF-P in alleviating ribosome stalling at the proline repeats. Gutierrez et al. (85) also reported evidence that eIF5A similarly stimulates peptide bond formation at consecutive proline sequences in S. cerevisiae. Additional evidence for the importance of eIF5A in translation of polyproline proteins was reported in T. brucei (86). Recent ribosome profile data demonstrate that eIF5A works more generally at many ribosome-stalled sites and that it also enhances translation termination (87, 88). The cryo-EM of yeast eIF5A bound to 80S ribosome (89) revealed the binding of eIF5A between the P-tRNA site and exit tRNA site, adjacent to the P-tRNA (Fig. 4). The hypusine side chain contacts the A76 of the CCA-end of the P-tRNA. Based on these findings, a model was proposed in which eIF5A facilitates peptide bond formation by stabilizing and orienting the CCA-end of the P-tRNA (Fig. 4) and its nascent peptide chain. The crystal structure of the eIF5A-yeast 80S ribosome complex suggests a possible connection between eIF5A–ribosome association and the conformational changes of the ribosome during protein synthesis (90). The hypusination of eIF5A not only confers its activity in translation elongation and termination, but also dictates its subcellular localization in the cytoplasm (91), the proper compartment for protein synthesis, with the aid of the nuclear export factor, exportin 4, which recognizes the hypusinated form of eIF5A (92).
Figure 4.
Model of eIF5A bound to 80S ribosome (A) and proposed mode of eIF5A action in translation (B). A, eIF5A (red; PDB code 5GAK), is bound to the 80S ribosome at the exit tRNA site adjacent to the P-tRNA (green; PDB code 5GAK). The A-tRNA is shown in blue (PDB code 5GAK). The hypusine side chain is indicated by a bright red arrow. B, the hypusine side chain of eIF5A (red) contacts A76 of the CCA end of P-tRNA to stabilize it and its nascent peptide chain and also promotes interactions between the ribosomal protein uL16 with both A- and P-tRNA and thereby stimulates peptide bond formation (modified from Ref. 89).
eIF5A isoforms and human diseases
Different organisms express different sets of closely related eIF5A isoforms. In mammals, there are two isoforms of eIF5A, eIF5A-1 and a highly similar eIF5A-2 (93, 94). eIF5A-1 (referred to as eIF5A) is the major protein constitutively expressed in all cells and tissues, whereas the eIF5A-2 protein is not normally detectable. High expression of eIF5A-2 was reported in certain cancer cells or tissues (95), and the eIF5A isoforms have been associated with human cancers (for reviews, see Refs. 44 and 96). eIF5A has been implicated in several other pathological conditions, including diabetes (97) and HIV-1 and retroviral infection (54, 55). Recent human genetic data from exome sequencing reveal the association of defects in the hypusine pathway with human disease.5 In five patients with a neurodevelopmental disorder, four independent biallelic variants in the DHPS gene were identified; three variants result in lack of or inactive DHPS, and a fourth variant results in a drastically reduced DHPS activity. Combination of one inactive allele with a partially active allele led to clinical phenotypes in these patients, suggesting pathogenesis due to defects in eIF5A hypusination. Furthermore, similar clinical phenotypes were observed in individuals carrying chromosome deletions in the 17p.13.1 region (98), which encompasses the EIF5A gene, supporting the importance of eIF5A in human brain development.
Conclusions
The primary function of polyamines in mammalian cell growth is to promote translation initiation mediated by polycations and to promote translation elongation and termination mediated by hypusinated eIF5A. Hypusine synthesis clearly defines a most critical function of polyamines in eukaryotic cell proliferation and provides a solid connection between polyamines and cell growth/animal development, through the eIF5A-mediated regulation of translation. It is intriguing that hypusine modification occurs exclusively in a single protein, eIF5A. Why did nature devise such an elaborate mechanism involving two novel enzymes, DHPS and DOHH, to modify and activate just one protein? Structures of ribosome-bound eIF5A show the long, polyamine-derived, basic side chain of hypusine, reaching the peptidyl transferase center, suggesting its role in stabilizing P-tRNA and facilitating peptide bond formation at stalled ribosomes.
Whereas EF-P, aIF5A, and eIF5A represent universally conserved translation factors with significant structural and functional similarities (17–19), their specificity, function, and essentiality appear to have diverged during evolution. Whereas EF-P exhibits strong specificity toward proline repeat motifs, including PPP or PPG, eIF5A acts more generally at a broad range of ribosome-pausing sites to enhance elongation, and it also stimulates translation termination. EF-P or its β-lysylation enzymes are not essential for basal bacterial growth but are necessary for bacterial fitness, survival under stress conditions, or virulence (18). Only eIF5A and DHPS are essential in archaea and yeast, whereas eIF5A, DHPS, and DOHH are all required in higher eukaryotes. Thus, the stringent requirement for the hypusinated eIF5A seems to have evolved in higher eukaryotes with greater complexities of their genome and proteome. It is interesting that the frequencies of typical ribosome stalling motifs, such as PPP or PPG (potential targets of EF-P and eIF5A), have increased dramatically in proteomes of higher organisms (99). In this regard, it is tempting to speculate that the function of eIF5A has evolved to meet the demand to optimize translation rate by relieving a broad range of ribosome stalling and also by enhancing termination in higher eukaryotes (87).
Considering that eIF5A is generally abundant and stable in mammalian cells, it is curious that haploinsufficiency of eIF5A (98) or certain mutations in DHPS are associated with a rare human neurodevelopmental disorder.5 It is possible that some factors important for brain development become limiting when the level of active eIF5A or DHPS is reduced. However, the association of a neurological disorder may not be unique to eIF5A, as multiple neurological disorders have been linked to mutations in other genes encoding the translation machinery, suggesting an exceptional sensitivity of neurons to translational defects or errors (63). Future efforts will be directed toward elucidation of the cellular and molecular pathways linking deficiency in eIF5A and DHPS to neurodevelopmental disorders.
Acknowledgments
We thank Drs. Hans Johansson (LGC Biosearch Technologies) and Achim Werner (NIDCR, National Institutes of Health) for critical reading of the manuscript and helpful suggestions and Dr. Ashleigh Hanner (NIDCR, National Institutes of Health) for assistance with the preparation of the figures.
This work was supported by the intramural program of NIDCR, National Institutes of Health. This article is part of a series on “Polyamines,” written in honor of Dr. Herbert Tabor's 100th birthday. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
There exist two or more isoforms of eIF5A, in diverse eukaryotic organisms. We use eIF5A to represent all of the eIF5A isoforms or the major form (eIF5A-1).
A. Hanner and M. H. Park, unpublished results.
M. Ganapathi, L. R. Padgett, K. Yamada, O. Devinsky, R. Willaert, R. Person, P. Y. B. Au, J. Tagoe, M. McDonald, D. Karlowicz, B. Wolf, J. Lee, Y. Shen, V. Okur, L. Deng, et al. Recessive rare variants in deoxyhypusine synthase (DHPS), an enzyme involved in the synthesis of hypusine, are associated with a neurodevelopmental disorder. Submitted for publication.
- eIF5A
- eukaryotic translation initiation factor
- eIF5A(Lys)
- eIF5A precursor containing lysine
- eIF5A(Dhp)
- eIF5A intermediate containing deoxyhypusine
- DHPS
- deoxyhypusine synthase
- DOHH
- deoxyhypusine hydroxylase
- EF-P
- bacterial elongation factor P
- GC7
- N1-guanyl-1,7-diaminoheptane
- A-tRNA
- aminoacyl tRNA
- P-tRNA
- aminoacyl tRNA
- PDB
- Protein Data Bank
- aa
- amino acids
- SAM
- S-adenosylmethionine
- AbeAdo
- 5′-([(Z)-4-aminobut-2-enyl]methylamino)-5′-deoxyadenosine.
References
- 1. Tabor C. W., and Tabor H. (1984) Polyamines. Annu. Rev. Biochem. 53, 749–790 10.1146/annurev.bi.53.070184.003533 [DOI] [PubMed] [Google Scholar]
- 2. Pegg A. E., and Casero R. A. Jr. (2011) Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35 10.1007/978-1-61779-034-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Igarashi K., and Kashiwagi K. (2015) Modulation of protein synthesis by polyamines. IUBMB Life 67, 160–169 10.1002/iub.1363 [DOI] [PubMed] [Google Scholar]
- 4. Pegg A. E. (2016) Functions of polyamines in mammals. J. Biol. Chem. 291, 14904–14912 10.1074/jbc.R116.731661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerner E. W., and Meyskens F. L. Jr. (2004) Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer 4, 781–792 10.1038/nrc1454 [DOI] [PubMed] [Google Scholar]
- 6. Park M. H., Cooper H. L., and Folk J. E. (1981) Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. U.S.A. 78, 2869–2873 10.1073/pnas.78.5.2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park M. H. (2006) The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 139, 161–169 10.1093/jb/mvj034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolff E. C., Lee Y. B., Chung S. I., Folk J. E., and Park M. H. (1995) Deoxyhypusine synthase from rat testis: purification and characterization. J. Biol. Chem. 270, 8660–8666 10.1074/jbc.270.15.8660 [DOI] [PubMed] [Google Scholar]
- 9. Joe Y. A., Wolff E. C., and Park M. H. (1995) Cloning and expression of human deoxyhypusine synthase cDNA: structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem. 270, 22386–22392 10.1074/jbc.270.38.22386 [DOI] [PubMed] [Google Scholar]
- 10. Abbruzzese A., Park M. H., and Folk J. E. (1986) Deoxyhypusine hydroxylase from rat testis: partial purification and characterization. J. Biol. Chem. 261, 3085–3089 [PubMed] [Google Scholar]
- 11. Park J.-H., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., and Park M. H. (2006) Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. U.S.A. 103, 51–56 10.1073/pnas.0509348102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park M. H. (1989) The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D): purification of eIF-4D and its precursors and comparison of their activities. J. Biol. Chem. 264, 18531–18535 [PubMed] [Google Scholar]
- 13. Park M. H., Wolff E. C., Smit-McBride Z., Hershey J. W., and Folk J. E. (1991) Comparison of the activities of variant forms of eIF-4D. The requirement for hypusine or deoxyhypusine. J. Biol. Chem. 266, 7988–7994 [PubMed] [Google Scholar]
- 14. Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., and Hershey J. W. (1991) Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 11, 3105–3114 10.1128/MCB.11.6.3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smit-McBride Z., Schnier J., Kaufman R. J., and Hershey J. W. (1989) Protein synthesis initiation factor eIF-4D. Functional comparison of native and unhypusinated forms of the protein. J. Biol. Chem. 264, 18527–18530 [PubMed] [Google Scholar]
- 16. Shiba T., Mizote H., Kaneko T., Nakajima T., and Kakimoto Y. (1971) Hypusine, a new amino acid occurring in bovine brain: isolation and structural determination. Biochim. Biophys. Acta 244, 523–531 10.1016/0304-4165(71)90069-9 [DOI] [PubMed] [Google Scholar]
- 17. Dever T. E., Gutierrez E., and Shin B.-S. (2014) The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 49, 413–425 10.3109/10409238.2014.939608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lassak J., Wilson D. N., and Jung K. (2016) Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol. Microbiol. 99, 219–235 10.1111/mmi.13233 [DOI] [PubMed] [Google Scholar]
- 19. Rossi D., Kuroshu R., Zanelli C. F., and Valentini S. R. (2014) eIF5A and EF-P: two unique translation factors are now traveling the same road. Wiley Interdiscip. Rev. RNA 5, 209–222 10.1002/wrna.1211 [DOI] [PubMed] [Google Scholar]
- 20. Nakajima T., Matsubayashi T., Kakimoto Y., and Sano I. (1971) Distribution of hypusine, N6-(4-amino-2-hydroxybutyl)-2,6-diaminohexanoic acid, in mammalian organs. Biochim. Biophys. Acta 252, 92–97 10.1016/0304-4165(71)90095-X [DOI] [PubMed] [Google Scholar]
- 21. Imaoka N., and Nakajima T. (1973) Hypusine, N6-(4-amino-2-hydroxybutyl)-2,6-diaminohexanoic acid, in tissue proteins of mammals. Biochim. Biophys. Acta 320, 97–103 10.1016/0304-4165(73)90170-0 [DOI] [PubMed] [Google Scholar]
- 22. Cooper H. L., Park M. H., Folk J. E., Safer B., and Braverman R. (1983) Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc. Natl. Acad. Sci. U.S.A. 80, 1854–1857 10.1073/pnas.80.7.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolff E. C., Kang K. R., Kim Y. S., and Park M. H. (2007) Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids 33, 341–350 10.1007/s00726-007-0525-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cano V. S., Jeon G. A., Johansson H. E., Henderson C. A., Park J. H., Valentini S. R., Hershey J. W., and Park M. H. (2008) Mutational analyses of human eIF5A-1–identification of amino acid residues critical for eIF5A activity and hypusine modification. FEBS J. 275, 44–58 10.1111/j.1742-4658.2007.06172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kyrpides N. C., and Woese C. R. (1998) Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U.S.A. 95, 224–228 10.1073/pnas.95.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bartig D., Schümann H., and Klink F. (1990) The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the Archebacterial kingdom. Syst. Appl. Microbiol. 13, 112–116 10.1016/S0723-2020(11)80156-6 [DOI] [Google Scholar]
- 27. Glick B. R., and Ganoza M. C. (1975) Identification of a soluble protein that stimulates peptide bond synthesis. Proc. Natl. Acad. Sci. U.S.A. 72, 4257–4260 10.1073/pnas.72.11.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tong Y., Park I., Hong B. S., Nedyalkova L., Tempel W., and Park H. W. (2009) Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins 75, 1040–1045 10.1002/prot.22378 [DOI] [PubMed] [Google Scholar]
- 29. Kim K. K., Hung L. W., Yokota H., Kim R., and Kim S. H. (1998) Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 95, 10419–10424 10.1073/pnas.95.18.10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanawa-Suetsugu K., Sekine S., Sakai H., Hori-Takemoto C., Terada T., Unzai S., Tame J. R., Kuramitsu S., Shirouzu M., and Yokoyama S. (2004) Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. U.S.A. 101, 9595–9600 10.1073/pnas.0308667101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yanagisawa T., Sumida T., Ishii R., Takemoto C., and Yokoyama S. (2010) A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 17, 1136–1143 10.1038/nsmb.1889 [DOI] [PubMed] [Google Scholar]
- 32. Navarre W. W., Zou S. B., Roy H., Xie J. L., Savchenko A., Singer A., Edvokimova E., Prost L. R., Kumar R., Ibba M., and Fang F. C. (2010) PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 39, 209–221 10.1016/j.molcel.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bullwinkle T. J., Zou S. B., Rajkovic A., Hersch S. J., Elgamal S., Robinson N., Smil D., Bolshan Y., Navarre W. W., and Ibba M. (2013) (R)-β-lysine-modified elongation factor P functions in translation elongation. J. Biol. Chem. 288, 4416–4423 10.1074/jbc.M112.438879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park J. H., Johansson H. E., Aoki H., Huang B. X., Kim H. Y., Ganoza M. C., and Park M. H. (2012) Post-translational modification by β-lysylation is required for activity of Escherichia coli elongation factor P (EF-P). J. Biol. Chem. 287, 2579–2590 10.1074/jbc.M111.309633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park M. H., Cooper H. L., and Folk J. E. (1982) The biosynthesis of protein-bound hypusine (Nϵ-(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (Nϵ-(4-aminobutyl)lysine). J. Biol. Chem. 257, 7217–7222 [PubMed] [Google Scholar]
- 36. Wolff E. C., Folk J. E., and Park M. H. (1997) Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J. Biol. Chem. 272, 15865–15871 10.1074/jbc.272.25.15865 [DOI] [PubMed] [Google Scholar]
- 37. Wolff E. C., Wolff J., and Park M. H. (2000) Deoxyhypusine synthase generates and uses bound NADH in a transient hydride transfer mechanism. J. Biol. Chem. 275, 9170–9177 10.1074/jbc.275.13.9170 [DOI] [PubMed] [Google Scholar]
- 38. Park J. H., Wolff E. C., Folk J. E., and Park M. H. (2003) Reversal of the deoxyhypusine synthesis reaction. Generation of spermidine or homospermidine from deoxyhypusine by deoxyhypusine synthase. J. Biol. Chem. 278, 32683–32691 10.1074/jbc.M304247200 [DOI] [PubMed] [Google Scholar]
- 39. Ober D., and Hartmann T. (1999) Deoxyhypusine synthase from tobacco. cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J. Biol. Chem. 274, 32040–32047 10.1074/jbc.274.45.32040 [DOI] [PubMed] [Google Scholar]
- 40. Liao D. I., Wolff E. C., Park M. H., and Davies D. R. (1998) Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure 6, 23–32 10.1016/S0969-2126(98)00004-5 [DOI] [PubMed] [Google Scholar]
- 41. Umland T. C., Wolff E. C., Park M. H., and Davies D. R. (2004) A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme·NAD·inhibitor ternary complex. J. Biol. Chem. 279, 28697–28705 10.1074/jbc.M404095200 [DOI] [PubMed] [Google Scholar]
- 42. Lee C. H., Um P. Y., and Park M. H. (2001) Structure-function studies of human deoxyhypusine synthase: identification of amino acid residues critical for the binding of spermidine and NAD. Biochem. J. 355, 841–849 10.1042/bj3550841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jakus J., Wolff E. C., Park M. H., and Folk J. E. (1993) Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies: effective inhibition by bis- and mono-guanylated diamines and polyamines. J. Biol. Chem. 268, 13151–13159 [PubMed] [Google Scholar]
- 44. Nakanishi S., and Cleveland J. L. (2016) Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48, 2353–2362 10.1007/s00726-016-2275-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park M. H., Wolff E. C., Lee Y. B., and Folk J. E. (1994) Antiproliferative effects of inhibitors of deoxyhypusine synthase: inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J. Biol. Chem. 269, 27827–27832 [PubMed] [Google Scholar]
- 46. Schultz C. R., Geerts D., Mooney M., El-Khawaja R., Koster J., and Bachmann A. S. (2018) Synergistic drug combination GC7/DFMO suppresses hypusine/spermidine-dependent eIF5A activation and induces apoptotic cell death in neuroblastoma. Biochem. J. 475, 531–545 10.1042/BCJ20170597 [DOI] [PubMed] [Google Scholar]
- 47. Jasiulionis M. G., Luchessi A. D., Moreira A. G., Souza P. P., Suenaga A. P., Correa M., Costa C. A., Curi R., and Costa-Neto C. M. (2007) Inhibition of eukaryotic translation initiation factor 5A (eIF5A) hypusination impairs melanoma growth. Cell Biochem. Funct. 25, 109–114 10.1002/cbf.1351 [DOI] [PubMed] [Google Scholar]
- 48. Park M. H., Mandal A., Mandal S., and Wolff E. C. (2017) A new non-radioactive deoxyhypusine synthase assay adaptable to high throughput screening. Amino Acids 49, 1793–1804 10.1007/s00726-017-2477-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim Y. S., Kang K. R., Wolff E. C., Bell J. K., McPhie P., and Park M. H. (2006) Deoxyhypusine hydroxylase is a Fe(II)-dependent, HEAT-repeat enzyme: identification of amino acid residues critical for Fe(II) binding and catalysis [corrected]. J. Biol. Chem. 281, 13217–13225 10.1074/jbc.M601081200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han Z., Sakai N., Böttger L. H., Klinke S., Hauber J., Trautwein A. X., and Hilgenfeld R. (2015) Crystal structure of the peroxo-diiron(III) intermediate of deoxyhypusine Hydroxylase, an oxygenase involved in hypusination. Structure 23, 882–892 10.1016/j.str.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 51. Vu V. V., Emerson J. P., Martinho M., Kim Y. S., Münck E., Park M. H., and Que L. Jr. (2009) Human deoxyhypusine hydroxylase, an enzyme involved in regulating cell growth, activates O2 with a nonheme diiron center. Proc. Natl. Acad. Sci. U.S.A. 106, 14814–14819 10.1073/pnas.0904553106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jasniewski A. J., Engstrom L. M., Vu V. V., Park M. H., Que L. Jr. (2016) X-ray absorption spectroscopic characterization of the diferric-peroxo intermediate of human deoxyhypusine hydroxylase in the presence of its substrate eIF5A. J. Biol. Inorg. Chem. 21, 605–618 10.1007/s00775-016-1373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kang K. R., Kim Y. S., Wolff E. C., and Park M. H. (2007) Specificity of the deoxyhypusine hydroxylase-eukaryotic translation initiation factor (eIF5A) interaction: identification of amino acid residues of the enzyme required for binding of its substrate, deoxyhypusine-containing eIF5A. J. Biol. Chem. 282, 8300–8308 10.1074/jbc.M607495200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olsen M. E., and Connor J. H. (2017) Hypusination of eIF5A as a target for antiviral therapy. DNA Cell Biol. 36, 198–201 10.1089/dna.2016.3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoque M., Hanauske-Abel H. M., Palumbo P., Saxena D., D'Alliessi Gandolfi D., Park M. H., Pe'ery T., and Mathews M. B. (2009) Inhibition of HIV-1 gene expression by ciclopirox and deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology 6, 90 10.1186/1742-4690-6-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mémin E., Hoque M., Jain M. R., Heller D. S., Li H., Cracchiolo B., Hanauske-Abel H. M., Pe'ery T., and Mathews M. B. (2014) Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res. 74, 552–562 10.1158/0008-5472.CAN-13-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thompson G. M., Cano V. S., and Valentini S. R. (2003) Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 555, 464–468 10.1016/S0014-5793(03)01305-X [DOI] [PubMed] [Google Scholar]
- 58. Lee Y. B., Joe Y. A., Wolff E. C., Dimitriadis E. K., and Park M. H. (1999) Complex formation between deoxyhypusine synthase and its protein substrate, the eukaryotic translation initiation factor 5A (eIF5A) precursor. Biochem. J. 340, 273–281 10.1042/bj3400273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Joe Y. A., and Park M. H. (1994) Structural features of the eIF-5A precursor required for posttranslational synthesis of deoxyhypusine. J. Biol. Chem. 269, 25916–25921 [PubMed] [Google Scholar]
- 60. Cooper H. L., Park M. H., and Folk J. E. (1982) Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell 29, 791–797 10.1016/0092-8674(82)90441-X [DOI] [PubMed] [Google Scholar]
- 61. Gerner E. W., Mamont P. S., Bernhardt A., and Siat M. (1986) Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem. J. 239, 379–386 10.1042/bj2390379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Z. P., and Chen K. Y. (1997) Marked elevation of hypusine formation activity on eukaryotic initiation factor 5A in v-HA-RAS transformed mouse NIH3T3 cells. Cancer Lett. 115, 235–241 10.1016/S0304-3835(97)04741-1 [DOI] [PubMed] [Google Scholar]
- 63. Kapur M., and Ackerman S. L. (2018) mRNA translation gone awry: translation fidelity and neurological disease. Trends Genet. 34, 218–231 10.1016/j.tig.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Park M. H., Joe Y. A., and Kang K. R. (1998) Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 1677–1683 10.1074/jbc.273.3.1677 [DOI] [PubMed] [Google Scholar]
- 65. Sasaki K., Abid M. R., and Miyazaki M. (1996) Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 384, 151–154 10.1016/0014-5793(96)00310-9 [DOI] [PubMed] [Google Scholar]
- 66. Nishimura K., Lee S. B., Park J. H., and Park M. H. (2012) Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino acids 42, 703–710 10.1007/s00726-011-0986-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sievert H., Pällmann N., Miller K. K., Hermans-Borgmeyer I., Venz S., Sendoel A., Preukschas M., Schweizer M., Boettcher S., Janiesch P. C., Streichert T., Walther R., Hengartner M. O., Manz M. G., Brümmendorf T. H., et al. (2014) A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis. Model. Mech. 7, 963–976 10.1242/dmm.014449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sugimoto A. (2004) High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation 72, 81–91 10.1111/j.1432-0436.2004.07202004.x [DOI] [PubMed] [Google Scholar]
- 69. Patel P. H., Costa-Mattioli M., Schulze K. L., and Bellen H. J. (2009) The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J. Cell Biol. 185, 1181–1194 10.1083/jcb.200904161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chattopadhyay M. K., Tabor C. W., and Tabor H. (2002) Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomyces pombe). Proc. Natl. Acad. Sci. U.S.A. 99, 10330–10334 10.1073/pnas.162362899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chattopadhyay M. K., Tabor C. W., and Tabor H. (2003) Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc. Natl. Acad. Sci. U.S.A. 100, 13869–13874 10.1073/pnas.1835918100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chattopadhyay M. K., Park M. H., and Tabor H. (2008) Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. U.S.A. 105, 6554–6559 10.1073/pnas.0710970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nishimura K., Murozumi K., Shirahata A., Park M. H., Kashiwagi K., and Igarashi K. (2005) Independent roles of eIF5A and polyamines in cell proliferation. Biochem. J. 385, 779–785 10.1042/BJ20041477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mandal S., Mandal A., Johansson H. E., Orjalo A. V., and Park M. H. (2013) Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 2169–2174 10.1073/pnas.1219002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Landau G., Bercovich Z., Park M. H., and Kahana C. (2010) The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 285, 12474–12481 10.1074/jbc.M110.106419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Byers T. L., Ganem B., and Pegg A. E. (1992) Cytostasis induced in L1210 murine leukaemia cells by the S-adenosyl-l- methionine decarboxylase inhibitor 5′-([(Z)-4-amino-2- butenyl]methylamino)-5′-deoxyadenosine may be due to hypusine depletion. Biochem. J. 287, 717–724 10.1042/bj2870717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Byers T. L., Lakanen J. R., Coward J. K., and Pegg A. E. (1994) The role of hypusine depletion in cytostasis induced by S-adenosyl-l-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine. Biochem. J. 303, 363–368 10.1042/bj3030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hyvönen M. T., Keinänen T. A., Cerrada-Gimenez M., Sinervirta R., Grigorenko N., Khomutov A. R., Vepsäläinen J., Alhonen L., and Jänne J. (2007) Role of hypusinated eukaryotic translation initiation factor 5A in polyamine depletion-induced cytostasis. J. Biol. Chem. 282, 34700–34706 10.1074/jbc.M704282200 [DOI] [PubMed] [Google Scholar]
- 79. Kemper W. M., Berry K. W., and Merrick W. C. (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 251, 5551–5557 [PubMed] [Google Scholar]
- 80. Benne R., Brown-Luedi M. L., and Hershey J. W. (1978) Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J. Biol. Chem. 253, 3070–3077 [PubMed] [Google Scholar]
- 81. Gregio A. P., Cano V. P., Avaca J. S., Valentini S. R., and Zanelli C. F. (2009) eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380, 785–790 10.1016/j.bbrc.2009.01.148 [DOI] [PubMed] [Google Scholar]
- 82. Saini P., Eyler D. E., Green R., and Dever T. E. (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 10.1038/nature08034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., and Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- 84. Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., and Jung K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85 10.1126/science.1228985 [DOI] [PubMed] [Google Scholar]
- 85. Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., Buskirk A. R., and Dever T. E. (2013) eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nguyen S., Leija C., Kinch L., Regmi S., Li Q., Grishin N. V., and Phillips M. A. (2015) Deoxyhypusine modification of eukaryotic translation initiation factor 5A (eIF5A) is essential for Trypanosoma brucei growth and for expression of polyprolyl-containing proteins. J. Biol. Chem. 290, 19987–19998 10.1074/jbc.M115.656785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schuller A. P., Wu C. C., Dever T. E., Buskirk A. R., and Green R. (2017) eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pelechano V., and Alepuz P. (2017) eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338 10.1093/nar/gkx479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schmidt C. B., Becker T., Heuer A., Brauger K., Shanmuganathan V., Pech M., Berninghausen O., Wilson D. N., and Beckmann R. (2016) Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 44, 1944–1951 10.1093/nar/gkv1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Melnikov S., Mailliot J., Shin B.-S., Rigger L., Yusupova G., Micura R., Dever T. E., and Yusupov M. (2016) Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J. Mol. Biol. 428, 3570–3577 10.1016/j.jmb.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lee S. B., Park J. H., Kaevel J., Sramkova M., Weigert R., and Park M. H. (2009) The effect of hypusine modification on the intracellular localization of eIF5A. Biochem. Biophys. Res. Commun. 383, 497–502 10.1016/j.bbrc.2009.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Aksu M., Trakhanov S., and Görlich D. (2016) Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat. Commun. 7, 11952 10.1038/ncomms11952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jenkins Z. A., Hååg P. G., and Johansson H. E. (2001) Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 71, 101–109 10.1006/geno.2000.6418 [DOI] [PubMed] [Google Scholar]
- 94. Clement P. M., Henderson C. A., Jenkins Z. A., Smit-McBride Z., Wolff E. C., Hershey J. W., Park M. H., and Johansson H. E. (2003) Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 270, 4254–4263 10.1046/j.1432-1033.2003.03806.x [DOI] [PubMed] [Google Scholar]
- 95. Guan X. Y., Sham J. S., Tang T. C., Fang Y., Huo K. K., and Yang J. M. (2001) Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 61, 3806–3809 [PubMed] [Google Scholar]
- 96. Mathews M. B., and Hershey J. W. (2015) The translation factor eIF5A and human cancer. Biochim. Biophys. Acta 1849, 836–844 10.1016/j.bbagrm.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Maier B., Tersey S. A., and Mirmira R. G. (2010) Hypusine: a new target for therapeutic intervention in diabetic inflammation. Discov. Med. 10, 18–23 [PubMed] [Google Scholar]
- 98. Zeesman S., Kjaergaard S., Hove H. D., Kirchhoff M., Stevens J. M., and Nowaczyk M. J. (2012) Microdeletion in distal 17p13.1: a recognizable phenotype with microcephaly, distinctive facial features, and intellectual disability. Am. J. Med. Genet. A 158A, 1832–1836 10.1002/ajmg.a.35508 [DOI] [PubMed] [Google Scholar]
- 99. Mandal A., Mandal S., and Park M. H. (2014) Genome-wide analyses and functional classification of proline repeat-rich proteins: potential role of eIF5A in eukaryotic evolution. PLoS One 9, e111800 10.1371/journal.pone.0111800 [DOI] [PMC free article] [PubMed] [Google Scholar]