Abstract
The glycoside hydrolases encoded by the human gut microbiome play an integral role in processing a variety of exogenous and endogenous glycoconjugates. Here we present three structurally and functionally distinct β-glucuronidase (GUS) glycoside hydrolases from a single human gut commensal microbe, Bacteroides uniformis. We show using nine crystal structures, biochemical, and biophysical data that whereas these three proteins share similar overall folds, they exhibit different structural features that create three structurally and functionally unique enzyme active sites. Notably, quaternary structure plays an important role in creating distinct active site features that are hard to predict via structural modeling methods. The enzymes display differential processing capabilities toward glucuronic acid–containing polysaccharides and SN-38-glucuronide, a metabolite of the cancer drug irinotecan. We also demonstrate that GUS-specific and nonselective inhibitors exhibit varying potencies toward each enzyme. Together, these data highlight the diversity of GUS enzymes within a single Bacteroides gut commensal and advance our understanding of how structural details impact the specific roles microbial enzymes play in processing drug-glucuronide and glycan substrates.
Keywords: microbiome, glycoside hydrolase, crystallography, enzyme structure, structure-function, bacteroides, carbohydrate metabolism, drug reactivation, GI tract, glucuronidase
Introduction
The carbohydrates and glycoconjugates that reach the human gastrointestinal (GI)3 tract are remarkably complex and sample a wide range of structural diversity. Despite the numerous and diverse carbohydrates humans consume, most of the enzymes required to process these molecules are not encoded by the human genome (1). Fortunately, a mutually beneficial relationship exists between the microbial inhabitants of the GI tract and the human host, in which the human gut microbiota (HGM) expand the host's metabolic capabilities via carbohydrate-active enzymes (CAZymes) (2). These CAZymes include glycoside hydrolases (GHs) and polysaccharide lyases (PLs) that mediate the fermentation of nondigestible carbohydrates and glycosides (3, 4). The major products of these processes, short chain fatty acids, account for up to 10% of the dietary energy in humans (5) and have been associated with a myriad of health benefits (6–8). In return, the HGM gain a stable energy source, which is crucial for microbial survival and maintaining balance within the HGM.
The Gram-negative phylum Bacteroidetes, one of two dominant bacterial phyla in the human gut microbiome, is a key metabolizer of diverse glycans in the GI tract. Members of the Bacteroidetes, the majority belonging to the genus Bacteroides, degrade both dietary and host-derived carbohydrates, and many species ferment multiple different polysaccharides (1, 9). Bacteroides thetaiotaomicron, for example, forages both host mucus glycans and plant polysaccharides, depending on their availability (10). Accordingly, Bacteroides encode genes for large numbers of CAZymes, particularly GHs (3), that are organized in polysaccharide utilization loci (PULs), a distinctive feature of their genomes (11).
Given the thousands of CAZymes that occur in Bacteroides, the functional and structural diversity of GHs within individual Bacteroides species still remain largely unexplored. Although the CAZyme classification system, which groups GHs into families based on their amino acid sequences, is reliable for the prediction of catalytic mechanisms and overall structural folds, substrate specificity, and unique structural features are difficult to predict. For example, the GH2 family comprises β-glucuronidases, β-glucosidases, β-galactosidases, and β-mannosidases, all of which possess an (α/β)8 TIM barrel-fold (3). Thus, it is important to experimentally characterize GHs to understand their structure and to assign function.
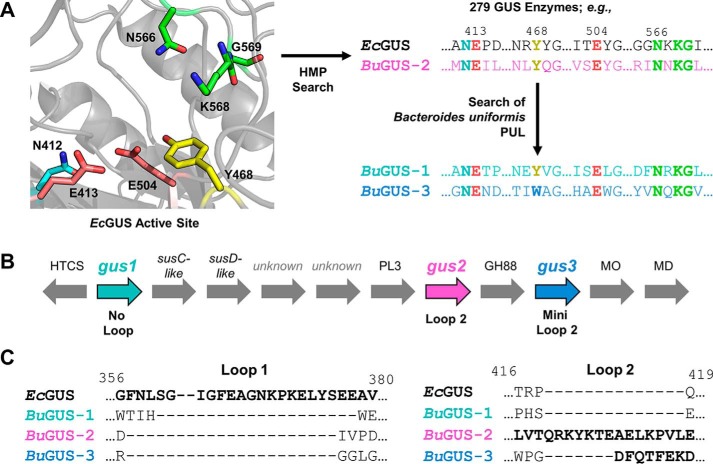
Recently, we presented a structure-guided approach to differentiate β-glucuronidase (GUS) proteins from their GH2 family members (12). As reported, 279 unique GUS enzymes were identified from the 4.8 million unique genes present in the stool sample database of the Human Microbiome Project (HMP) (12). This provided the first atlas of GUS enzymes in the human gut microbiome. Within that effort, we identified and characterized a GUS from the human gut bacterium Bacteroides uniformis, which has been reported to be highly abundant in the human GI tract (13). We demonstrated that it acts as a β-glucuronidase and is able to process both a small-molecule glucuronide and a polysaccharide with a terminal glucuronic acid moiety (12). As outlined below, in an attempt to gain further insight into its role in polysaccharide degradation, we searched the genomic region surrounding this GUS (Fig. 1A). We found two additional GH2 enzymes in the same PUL that retain sequence features previously identified as unique to GUS enzymes (Fig. 1A). To our knowledge, no previously characterized PUL contains three potential GUS enzymes. For this reason, we were interested in their differential structural properties and their abilities to cleave diverse glucuronic acid (GlcA)-containing substrates.
Figure 1.
Discovery and analysis of GUS genes in B. uniformis strain 3978 T3 i. A, schematic for the discovery of GUS enzymes from B. uniformis. B, genetic organization of a PUL from B. uniformis reveals 3 glycoside hydrolases with sequence features indicating GUS function, as well as other glucuronic acid-metabolizing enzymes. C, sequence alignment of EcGUS, BuGUS-1, BuGUS-2, and BuGUS-3 reveals distinct loop classes between these putative GUS enzymes. HTCS, hybrid two-component system; MO, mannonate oxidase; MD, mannonate dehydratase; susC/D, starch utilization system C/D.
Here we demonstrate that three putative GUS proteins from a single B. uniformis microbe share the TIM barrel structural fold but exhibit distinct tertiary and quaternary structures, not obvious from sequence analysis, and harbor unique structural features within their actives sites that likely afford them specific substrate processing capabilities. Indeed, these GUS enzymes displayed differential activities toward a variety of glucuronide substrates, including GlcA-containing polysaccharides and SN-38-G, a metabolite of the cancer drug irinotecan. Additionally, we tested the ability of both selective bacterial GUS inhibitors and a pan GUS inhibitor to inhibit the three GUSs from this microbe, which reveal distinct propensities for inhibition. We further examined the potential for these glucuronidases to act on other sugar acid-containing substrates, such as those that contain galacturonic acid, iduronic acid, or mannuronic acid. These results highlight the broad structural and functional diversity among GUS enzymes within a single human gut microbe. Furthermore, the data presented here provide a foundation for understanding the specialized roles of GUS enzymes in the deconstruction of a sugar acid-containing carbohydrate and the ability of the HGM to reactivate drug-glucuronide conjugates.
Results
Discovery and sequence analysis of GUS enzymes from a B. uniformis PUL
A GUS from the human gut bacterium B. uniformis strain 3978 T3i (BuGUS) was previously discovered in the HMP database (12). Further inspection of the genomic region flanking this GUS gene revealed a hallmark of PULs, a nearby susC/susD-like gene pair. These two genes are involved in the binding of polysaccharides on the outer membrane (SusD) and transport into the periplasm (SusC) (Fig. 1B) (11). The presence of the susC/susD homologs indicates that BuGUS is located in a PUL, which means BuGUS likely contributes to the orchestrated degradation of a GlcA-containing polysaccharide. Two additional enzymes predicted to belong to the GH2 family were also identified adjacent to BuGUS and the susC/susD-like pair (Fig. 1B). Each of these proteins possess key sequence features that are characteristic of GUS enzymes, including the asparagine-X-lysine (NXK) motif and catalytic glutamates that recognize and cleave glucuronides, respectively (12, 14) (Fig. 1A). Only BuGUS-1 and BuGUS-2, however, possess the GUS-specific tyrosine residue (Tyr-480 and Tyr-495, respectively) that hydrogen bonds with the nucleophilic glutamate. In BuGUS-3, a tryptophan (Trp-483) replaces the tyrosine (Fig. 1A and Fig. S1).
Sequence alignments with the previously characterized BuGUS (now termed BuGUS-2) and these two new GUS enzymes, termed BuGUS-1 and BuGUS-3, revealed sequence identity of 27 and 29%, respectively, whereas BuGUS-1 and BuGUS-3 share 18% sequence identity (Fig. S1). Sequence analysis also revealed that BuGUS-1, BuGUS-2, and BuGUS-3 fall into the previously defined No Loop (NL), Loop 2 (L2), and Mini Loop 2 (mL2) classes, respectively; these classifications are related to the size and location of loops at the active site of gut microbial GUS enzymes, and have been shown to play key roles in substrate specificity (12) (Fig. 1C). Utilizing the signal peptide prediction tool, SignalP 4.1 Server (15), we found that BuGUS-1, BuGUS-2, and BuGUS-3 have a signal peptide and are thus expected to be periplasmic. Together, this sequence analysis indicates that a PUL from B. uniformis contains three putative GUS enzymes with distinct sequence features.
BuGUS-1 and BuGUS-2 exhibit β-glucuronidase activity with 4-MUG
To begin to elucidate the substrate specificities of these three putative GUS enzymes, we performed in vitro activity assays with their purified protein products. BuGUS-2 has been previously shown to exhibit GUS activity (12). To confirm that BuGUS-1 and BuGUS-3 are also GUS enzymes, we synthesized, cloned, expressed, and purified their protein products. We then utilized the standard substrates 4-methylumbelliferyl-β-d-glucuronide (4-MUG) and p-nitrophenyl-β-d-glucuronide to assess the pH profile and kinetic parameters of GUS activity, respectively. BuGUS-1 (kcat/Km = 3.4 × 105 s−1 m−1) and BuGUS-2 (kcat/Km = 3.8 × 105 s−1 m−1) both efficiently processed these standard substrates, indicating that they are GUS enzymes (Table 1 and Fig. S2). However, BuGUS-3 was unable to catalyze the hydrolysis of these substrates (Table 1). Thus, whereas BuGUS-1 and BuGUS-2 can hydrolyze glucuronides, BuGUS-3 may have a distinct but related activity despite its GUS-like sequence features.
Table 1.
Kinetic parameters of 4-MUG hydrolysis for GUS enzymes at optimal pH
Errors represent S.D. of n = 3 biological replicates.
| Protein | kcat | Km | kcat/Km |
|---|---|---|---|
| s−1 | μm | s−1 m−1 | |
| BuGUS-1 | 8.6 ± 0.5 | 25 ± 8 | 3.4 × 105 |
| BuGUS-2 | 31 ± 5 | 80 ± 20 | 3.8 × 105 |
| BuGUS-3 | NAa | NA | NA |
| BMSP GUS | 1.2 ± 0.2 | 120 ± 30 | 1.0 × 104 |
| BuGUS-1 Δloop | 1.2 ± 0.2 | 74 ± 8 | 1.7 × 104 |
a NA, no activity.
BuGUS enzymes exhibit distinct structural features
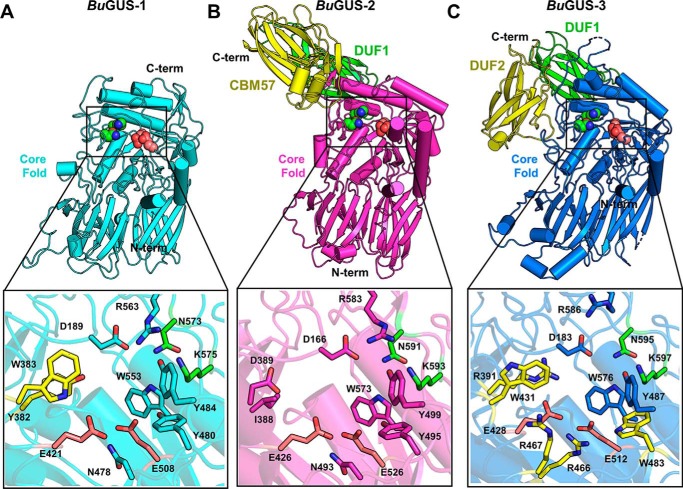
To evaluate the structural diversity of the three GUS enzymes present in B. uniformis, we determined the crystal structures of BuGUS-1 (space group: C2, molecules in asymmetric unit: 2) and BuGUS-3 (space group: C2, molecules in asymmetric unit: 2) and compared them to the previously reported BuGUS-2 structure (Table S1) (12). BuGUS-1 and BuGUS-3 share a similar TIM barrel core fold with BuGUS-2, with 3.2-Å root mean square deviation across 512 Cα positions and 3.4-Å root mean square deviation over 648 Cα positions, respectively (Fig. 2 and Fig. S3). BuGUS-2 and BuGUS-3 encode C-terminal extensions not found in the shorter BuGUS-1 enzyme (Fig. 2, A–C).
Figure 2.
Structural analysis of BuGUS-1, BuGUS-2, and BuGUS-3 reveals distinct tertiary and active site structure. A, tertiary structure of BuGUS-1 with the sugar acid-recognizing NXK motif highlighted as green spheres and catalytic glutamates as deep salmon spheres and zoom-in of active site with unique active site residues highlighted in yellow. B, tertiary structure of BuGUS-2 with core fold highlighted in magenta and additional C-terminal domains in green (DUF1) and yellow (CBM57) and zoom-in of the active site. C, tertiary structure of BuGUS-3 with core fold in blue, the sugar acid-recognizing NXK motif highlighted as green spheres, and catalytic glutamates as deep salmon spheres and additional C-terminal domains in green (DUF1) and yellow (DUF2) and zoom-in of the active site with unique active side residues highlighted in yellow.
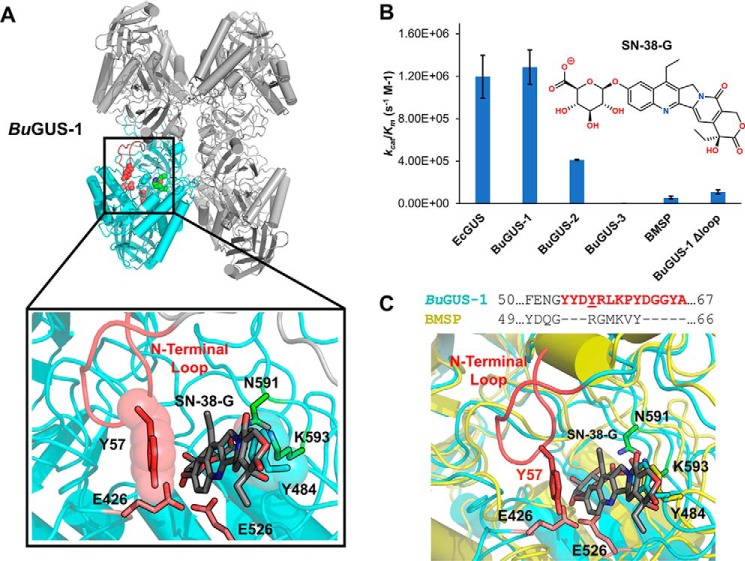
Although BuGUS-1 maintains a similar tertiary structure to other GUS enzymes, it possesses unique active site residues, particularly Tyr-382 and Trp-383 (Fig. 2A, highlighted in yellow). These positions are generally occupied by smaller residues in previously characterized GUS enzymes, such as BuGUS-2 (Fig. 2B). In addition to these unique active site residues, BuGUS-1 is only the second tetrameric bacterial GUS characterized that does not contain a Loop 1 by sequence analysis. Instead, remarkably, and unpredictably by sequence analysis alone, an N-terminal loop (NTL) (Fig. S1) is donated from an adjacent protomer and resembles the loop-based active sites of previously characterized Loop 1 GUS structures, as outlined below (Fig. 8A).
Figure 8.
Kinetic and structural analysis of SN-38-G hydrolysis reveals the importance of the N-terminal loop in BuGUS-1. A, BuGUS-1 tetramer with adjacent N-terminal loop highlighted in red, catalytic glutamates in deep salmon, and the NXK motif in green. Zoom-in of active site with SN-38-G manually docked in the active site of BuGUS-1 based on the PTG-bound structure. B, catalytic efficiencies kcat/Km for EcGUS, BuGUS-1, BuGUS-2, BuGUS-3, BMSP, and BuGUS-1 Δloop with the substrate SN-38-G. C, sequence alignment of BuGUS-1 and BMSP GUS N-terminal loop regions and overlay of BuGUS-1 and BMSP active sites with SN-38-G manually docked. Error bars represent S.D. of n = 3 biological replicates.
Compared with previously characterized GUS enzymes, BuGUS-3 deviates the most in its active site composition, containing five unique residues (yellow) (Fig. 2C). Most notably, three arginines in BuGUS-3 replace the small, polar residues, such as asparagine, which are conserved in previously characterized GUS enzymes, and Arg-391 and Arg-466 are positioned to form ionic interactions with the catalytic glutamates (Fig. 2C). Furthermore, the BuGUS-3 active site contains Trp-483, which replaces the conserved tyrosine in all other GUS enzymes characterized. Finally, Trp-431 and Arg-391, localized across the active site from the NXK motif (green), have not been observed in any other GUS enzymes (Fig. 2C). These distinct active site features may explain why BuGUS-3 does not process the standard glucuronide substrates despite the presence of the NXK motif and catalytic glutamates necessary for the recognition and cleavage of glucuronides.
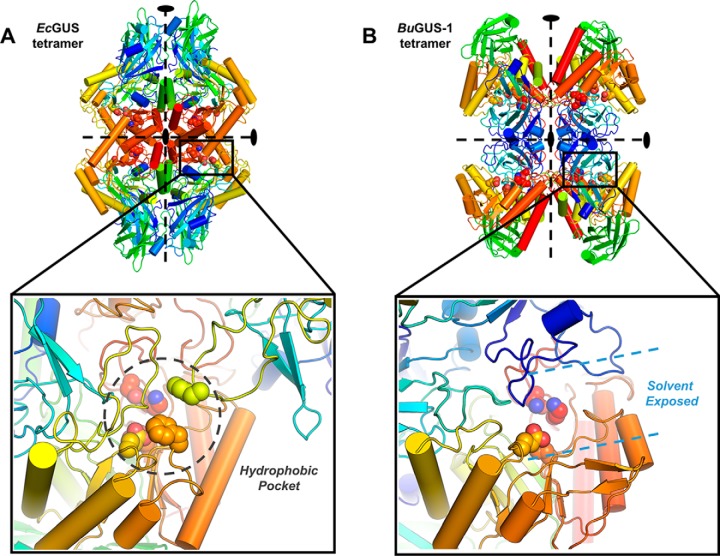
BuGUS-1 displays a unique quaternary structure despite having similar tertiary structure to EcGUS (Fig. 3A and Fig. S3). BuGUS-1 forms a unique inverted tetramer compared with the previously determined structures of E. coli GUS (EcGUS), Streptococcus agalactiae GUS, Clostridium perfringens GUS, and the human GUS ortholog (Fig. 3, A and B) (14, 16, 17). In BuGUS-1, individual protomers interact via their N termini compared with the previously examined GUS enzymes outlined above, in which the interface of protomers is formed by their C termini (Fig. 3, A and B). The consequence of this oligomeric organization is a solvent-exposed active site.
Figure 3.
Analysis of quaternary structures and their influence on the active site architecture of EcGUS and BuGUS-1. A, tetramer of EcGUS with zoom-in of the tetramer interface that reveals the hydrophobic pocket around the active site situated at the interface of C-terminal regions. B, tetramer of BuGUS-1 with zoom-in of the tetramer interface revealing a solvent-exposed active site.
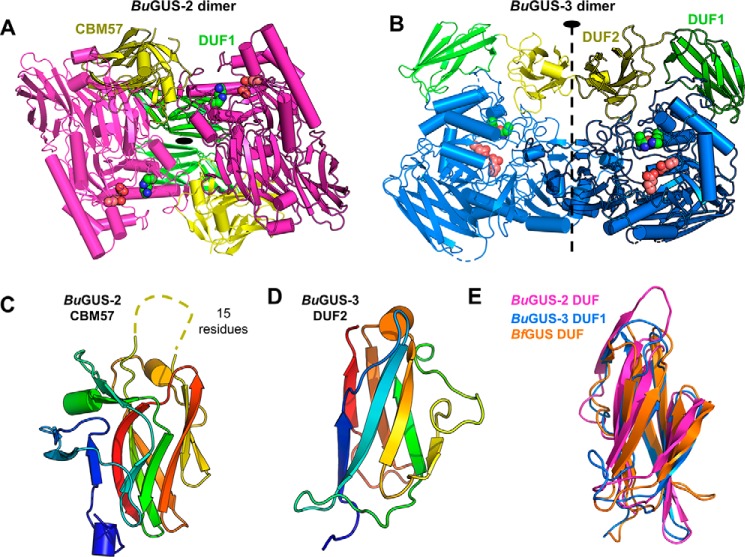
Unlike the tetrameric BuGUS-1, BuGUS-2 and BuGUS-3 both form dimers (Fig. 4, A and B, and Fig. S4) and contain extra domains at their C termini (Fig. 2, B and C, and Fig. S5). Excluding these additional domains, the core tertiary structures of BuGUS-2 and BuGUS-3 are TIM barrel folds with two β-sandwich-like domains, similar to previously characterized GUS enzymes (Fig. 2, B and C, and Fig. S3). Although BuGUS-2 and BuGUS-3 share similar core folds, they have distinct quaternary structures, likely a result of how their additional C-terminal domains are positioned that preclude similar homodimeric organizations (Figs. 2, B and C, and 4, A and B, and Fig. S5). Sequence and structural analysis of the C-terminal domains of BuGUS-2 revealed that the most C-terminal (yellow) is a member of the CBM 57 family, based on malectin that binds to developing glycans in the endoplasmic reticulum (Fig. 4C) (18). The remaining domains in BuGUS-2 and BuGUS-3 are “domains of unknown function” (DUF) and are not formally defined as, and simply may not be, CBMs (Fig. 2, B and C, and Fig. S5). Both sequence (NCBI BLAST) and structure-based (PDBeFold) searches of these additional C-terminal DUFs revealed hits for the C-domains of antibodies. These domains may only serve a role in the oligomeric organization of these proteins. The DUFs from the BuGUS enzymes are similar in structure and have been observed once previously in the structure of Bacteroides fragilis GUS (BfGUS), which was also previously designated as a DUF (Fig. S5). Collectively, the unique C-terminal domains of BuGUS-2 and BuGUS-3 may play roles in carbohydrate-binding and quaternary structure.
Figure 4.
Quaternary structure of BuGUS-2 and BuGUS-3 and structural analysis of C-terminal domains. A, BuGUS-2 dimer with core fold shown in magenta, DUF1 in green, and CBM57 domains in yellow with active site glutamates and the NXK motif shown as deep salmon and green spheres, respectively. B, BuGUS-3 dimer with core fold in blue, DUF1 in green, and DUF2 in yellow with catalytic glutamates and NxK motif in deep salmon and green spheres, respectively. C, CBM57 of BuGUS-2 shown with disordered loop shown as dotted line. D, structure of BuGUS-3 DUF2. E, structural alignment of BuGUS-2 DUF, BuGUS-2 DUF1, and BfGUS DUF.
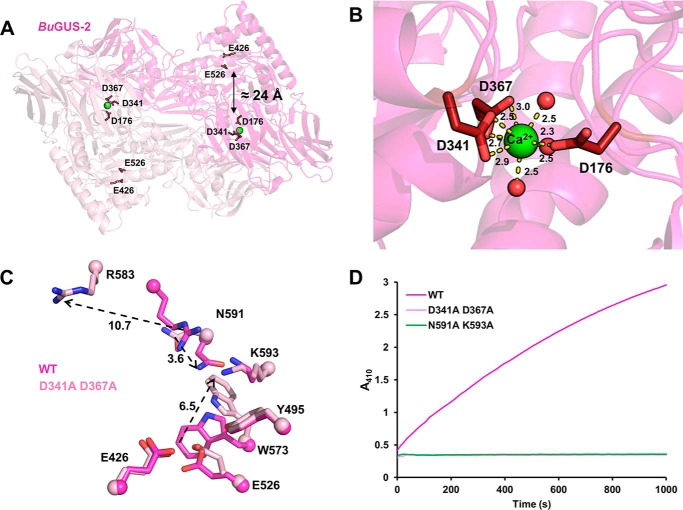
We also find that BuGUS-2 contains a well-organized predicted calcium-binding site (Fig. 5, A and B) that is unique to this GUS both in B. uniformis and in GUS enzymes of known structure to date. Approximately 24 Å from the active site of BuGUS-2 are three aspartic acids and three ordered water molecules that coordinate a predicted calcium ion (Fig. 5, A and B). Site-directed mutagenesis of Asp-341 and Asp-367 to alanines led to a complete loss of GUS activity and the crystal structure of this mutant (space group: P212121, molecules in asymmetric unit: 2, Table S1) revealed significant structural changes at the enzyme active site (Fig. 5, C and D). CD analysis also revealed a small loss in structural order for the predicted calcium-binding mutant compared with the wildtype (WT), but an equivalent melting temperature indicated no significant change in overall protein stability (Fig. S6). Sequence analysis of the 279 previously discovered GUS enzymes (12) revealed 17 additional GUS proteins with a predicted calcium-binding site (Table S2 and Fig. S7). Thus, it appears that the predicted calcium-binding site plays a key role in the structure and function of BuGUS-2 and is conserved among other GUS proteins in the human gut microbiome.
Figure 5.
Predicted calcium-binding site key for structural and functional integrity of BuGUS-2. A, BuGUS-2 dimer with predicted calcium-binding site (green sphere) 24 Å away from active site glutamates. B, the predicted calcium ion is contacted by Asp-176, Asp-341, Asp-367, and three water molecules. Distances are shown in angstroms. C, active site overlay of WT and calcium-binding mutant of BuGUS-2 reveals conformational changes that preclude functional activity. Distances are shown in angstroms. D, progress curves of BuGUS-2 activity reveal that mutation of the predicted calcium-binding site results in the same loss of function as mutation of essential active site residues.
BuGUS enzymes differentially process GlcA-containing polysaccharides
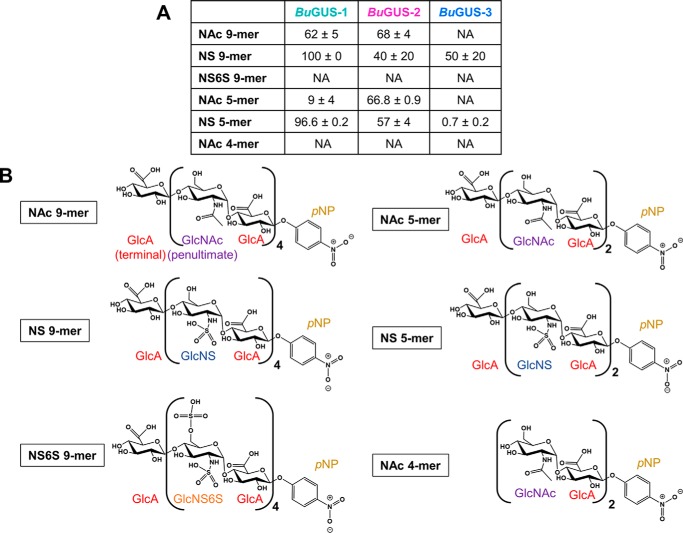
Given the distinct active site architectures of the three GUS enzymes examined here, as well as their differential processing of standard glucuronide substrates, we examined a set of pure synthetic polysaccharide substrates (Fig. 6, A and B). We chose heparin-like nonamers (9-mers) that contain GlcA and are either acetylated or sulfated. We also examined shorter polysaccharides (5-mers) and a substrate with GlcA at the penultimate rather than the terminal (nonreducing end) position (NAc 4-mer) (Fig. 6B). Both BuGUS-1 and BuGUS-2 were able to process the acetylated heparin-like nonamer substrate (NAc 9-mer), but BuGUS-3 showed no activity (Fig. 6A). However, all three GUS enzymes, including BuGUS-3, were able to process the terminal ends of a sulfated heparin-like substrate (NS 9-mer; Fig. 6A). We next examined a 9-mer with a doubly sulfated glucosamine moiety at the penultimate position (NS6S 9-mer). We found that this change eliminated activity with all three enzymes (Fig. 6A).
Figure 6.
Polysaccharide cleavage by BuGUS-1, BuGUS-2, and BuGUS-3. A, percent cleavage for BuGUS-1, BuGUS-2, and BuGUS-3 with an acetylated and sulfated heparin-like substrates for 3 h, pH 6.5. n = 3, ± S.D. NA, no activity. B, schematic structures of the pure synthetic polysaccharides utilized to measure polysaccharide processing by BuGUS enzymes. GlcA, glucuronic acid; GlcNS, N-sulfoglucosamine; GlcNS, N-sulfoglucosamine-6-sulfate; p-PNP, p-nitrophenol.
We tested the effect of polysaccharide length on activity by examining shorter 5-mer substrates. Our results were similar to the 9-mer data outlined above, BuGUS-1 and BuGUS-2 processed the NAc 5-mer, whereas BuGUS-3 did not, and all three GUS enzymes processed the NS 5-mer (Fig. 6A). Interestingly, although, BuGUS-3 displayed much weaker activity with the NS 5-mer than it did with the NS 9-mer (Fig. 6A). Finally, to confirm that these proteins act as exolytic enzymes toward substrates with terminal GlcA moieties, we examined a 4-mer polysaccharide with GlcA at the penultimate position (Fig. 6B). As expected, the three enzymes examined failed to process this compound, indicating that they do not act as endolytic enzymes toward this particular substrate (Fig. 6A). Taken together, these data using six distinct polysaccharide substrates related to compounds found in humans reveal that all three BuGUS enzymes are able to process sulfated 9-mers and sulfated 5-mers, whereas only BuGUS-1 and BuGUS-2 cleaved the acetylated heparin-like 9- and 5-mers. Moreover, the enzyme activity appears limited to removing terminal GlcA groups. Such data provide an initial molecular framework to understand the potential for microbial GUS enzymes to utilize polysaccharide substrates within the human GI tract.
BuGUS enzymes may process additional uronic acid-containing substrates
Given the diversity of uronate-containing polysaccharides, we considered the possibility that these GUS enzymes would process uronic acid conjugates beyond glucuronides. Thus, we docked into the three BuGUS enzymes the following four uronic acids: glucuronic acid (GlcA), galacturonic acid (GalA), mannuronic acid (ManA), and iduronic acid (IdoA). These sugar monosaccharides were identified from the PDB and docked manually in PyMOL based on the glucuronate-bound structure of BuGUS-1 (PDB 6D6W). Despite the differences in stereochemistry between these sugar acids, docking suggests that each may be accommodated within all three GUS active sites (Fig. S8). Galacturonate appeared to be the most sterically strained sugar, which has an axial hydroxyl at the 4-position that could clash with the aspartic acid side chain conserved in all three BuGUS enzymes (Fig. S8). To test the hypothesis that substrates with terminal sugar acids beyond GlcA could be utilized as substrates, we examined the ability of p-nitrophenyl-β-d-galacturonide (pNP-GalA) to act as a substrate for BuGUS-1, BuGUS-2, and BuGUS-3 (Fig. S9a). We found that only BuGUS-1 was able to process this galacturonide (Fig. S9b). Kinetic analysis of BuGUS-1 with both pNP-GlcA and pNP-GalA revealed catalytic efficiencies (kcat/Km) of 2.2 × 105 and 3.1 × 104, respectively, suggesting that whereas BuGUS-1 can hydrolyze galacturonides, it does so less efficiently than the analogous glucuronide (Fig. S9c). A model of galacturonic acid docked in the active site of BuGUS-1 shows that the aspartic acid (green) that could clash with the hydroxyl at the 4 position may cause this weaker efficiency (Fig. S9d). Taken together, docking studies and kinetics suggest that the GUS enzymes considered here may act on polysaccharide substrates containing terminal sugar acids beyond glucuronate, including mannuronate, iduronate, and galacturonate.
BuGUS structures in complex with substrate analogs
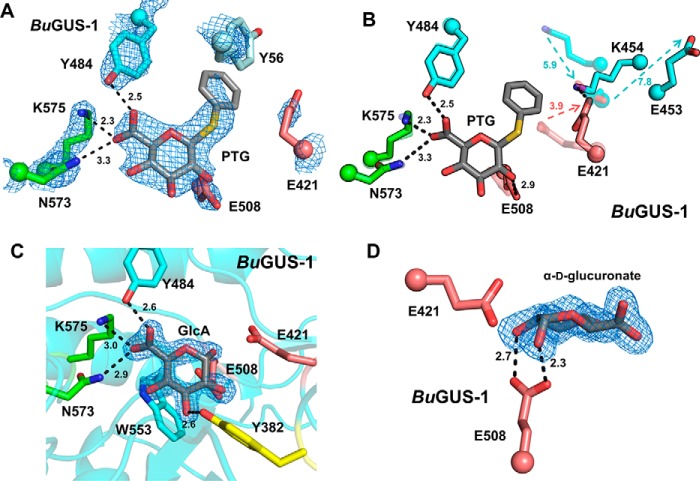
To gain a better understanding of substrate recognition by these novel GUS enzymes, we incubated them with the nonhydrolyzable substrate analog phenyl-thio-β-d-glucuronide (PTG) and attempted co-crystallization. Co-crystallization of a PTG-BuGUS-1 complex was successful (space group: P1211, molecules in asymmetric unit: 6, Table S1), and the crystal structure revealed a conformational shift in the active site in which the catalytic acid/base Glu-421 shifts away from the active site (Fig. 7, A and B). This conformational change is accommodated by additional shifts adjacent to the active site, in which Glu-453 and Lys-454 undergo 7.8- and 5.9-Å changes in position, respectively, relative to the unliganded structure (Fig. 7B). In line with previous studies, the carboxylate of PTG is recognized by Asn-573 and Lys-575 (NXK motif), as well as Tyr-484 (Fig. 7A). In addition to a PTG complex, we also determined the structure of BuGUS-1 in complex with GlcA (space group: C2, molecules in asymmetric unit: 4, Table S1). GlcA was bound to BuGUS-1 as the α-anomer (Fig. 7, C and D), and much like PTG, the carboxylate of GlcA is recognized by the NXK motif and other residues that contact its hydroxyl groups (Fig. 7C). Additionally, Trp-533 participates in C-H π interactions with the nonpolar face of GlcA (Fig. 7C). The anomeric hydroxyl group forms a hydrogen bond with Glu-508, the catalytic nucleophile (Fig. 7D). Together, these structural data highlight how GUS specifically recognizes its glucuronide substrate.
Figure 7.
Structural analysis of liganded BuGUS-1 and BuGUS-2 reveal chemical complementarity to GlcA. A, PTG bound to BuGUS-1 with mFo − DFc simple omit density shown at 2.5 σ with NXK motif shown in green, catalytic glutamates in deep salmon, and Tyr-56 from an adjacent monomer in pale cyan. B, overlay of PTG bound (opaque) and apo (transparent) BuGUS-1 active site reveals significant conformational shifts to catalytic acid/base Glu-421 as well as two nearby residues Lys-454 and Glu-453 to accommodate the large sulfur atom present in PTG. C, BuGUS-1 bound to GlcA with mFo − DFc simple omit density shown at 2.5 σ. D, BuGUS-1 active site with GlcA shown in the plane of the ring reveals an α configuration that forms a hydrogen bond with the catalytic nucleophile Glu-508. Distances shown are in units of angstroms.
Differential SN-38-G processing by BuGUS enzymes
GUS enzymes are promiscuous and can hydrolyze a variety of glucuronides related to mammalian gut toxicity (12, 14, 16, 19–25). Thus, we sought to determine whether these GUS enzymes are capable of reactivating the inactive metabolite SN-38-G of the cancer drug irinotecan. Despite their localization in a PUL, BuGUS-1 and BuGUS-2 hydrolyzed the small-molecule glucuronide SN-38-G (Fig. 8B). Strikingly, BuGUS-1 hydrolyzed SN-38-G with an efficiency that rivals previously characterized Loop 1 GUS enzymes that are not located in PULs and have been shown to prefer only small molecule glucuronides over polysaccharides (Fig. 8B) (12). We hypothesized that the NTL identified in the structure of BuGUS-1 may play a key role in recognizing the aglycone moiety of SN-38-G (Fig. 8A). The NTL is defined as residues Tyr-54 through Ala-67, and forms a loop that sits by the active site of an adjacent protomer (Fig. 8C). Indeed, the NTL loop deletion (Δloop BuGUS-1) displayed much slower processing with both 4-MUG and SN-38-G compared with the WT BuGUS-1 (Table 1 and Fig. 8B). We solved the structure of Δloop BuGUS-1 (space group: P1211, molecules in asymmetric unit: 4, Table S1), which shows the absence of this key loop structure (Fig. S10).
As an additional control to test the importance of the NTL for SN-38-G processing by BuGUS-1, we cloned, expressed, and purified a Bacteroides multispecies (BMSP) GUS that is similar to BuGUS-1 but, importantly, lacks the NTL sequence necessary for efficient processing of small molecule glucuronides (Fig. 8C). The 2.65-Å structure of BMSP GUS (space group: I41, molecules in asymmetric unit: 4, Table S1) reveals the same tetrameric organization as BuGUS-1 but lacks the N-terminal loop that forms the aglycone-binding site of BuGUS-1 (Fig. 8C and Fig. S11a). Importantly, BMSP displayed similar 4-MUG and SN-38-G processing efficiencies compared with the Δloop variant of BuGUS-1 (Fig. 8B and Table 1). These data suggest that an N-terminal sequence feature in the previously identified No Loop BuGUS-1 allows it to process SN-38-G with activity similar to Loop 1 GUS enzymes.
d-Glucaro-1,4-lactone inhibits BuGUS-1 and BuGUS-2
Due to their ability to process SN-38-G, we tested whether we could inhibit BuGUS-1 and BuGUS-2 with our selective bacterial GUS inhibitors as well as the nonspecific GUS inhibitor d-glucaro-1,4-lactone (Fig. 9C and Fig. S10). The GUS-specific inhibitors Inh1 and UNC10201652 did not inhibit either BuGUS-1 or BuGUS-2 up to 100 μm (Fig. S12) However, d-glucaro-1,4-lactone displayed mid-micromolar potency against both BuGUS-1 and BuGUS-2 (Table 2). Together, these data show that previously characterized selective inhibitors of GUS are not effective against BuGUS-1 and BuGUS-2, whereas the nonspecific GUS inhibitor d-glucaro-1,4-lactone is a mid-micromolar inhibitor of BuGUS-1 and BuGUS-2.
Figure 9.
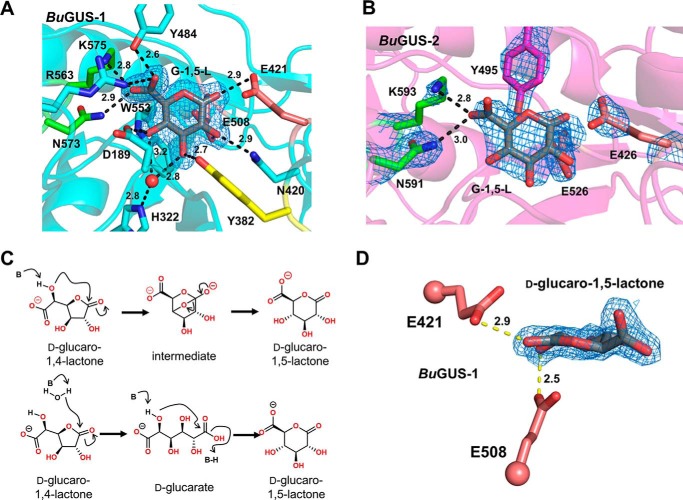
Structural analysis of BuGUS-1 and BuGUS-2 inhibition by d-glucaro-1,4-lactone reveals d-glucaro-1,5-lactone bound instead. A, BuGUS-1 bound to d-glucaro-1,5-lactone with mFo − DFc simple omit density shown at 2.5 σ. B, BuGUS-2 bound to d-glucaro-1,5-lactone with mFo − DFc simple omit density shown at 1.5 σ. C, proposed mechanisms for the conversion of d-glucaro-1,4-lactone to d-glucaro-1,5-lactone. D, close-up view in the plane of d-glucaro-1,5-lactone reveals planarity at α-carbon and key contact with catalytic acid/base Glu-421. All distances shown are in units of angstroms.
Table 2.
IC50 values of Inh1, UNC10201652, and d-glucaro-1,4-lactone for BuGUS-1, -2, -3, and BMSP at optimal pH
Values shown are in units of μm and errors are S.D. of n = 3 biological replicates.
| Protein | Inh1 | UNC10201652 | d-Glucaro-1,4-lactone |
|---|---|---|---|
| BuGUS-1 | >100 | >100 | 8.3 ± 0.6 |
| BuGUS-2 | >100 | >100 | 9 ± 2 |
| BuGUS-3 | NAa | NA | NA |
| BMSP | >100 | >100 | 27 ± 1 |
a NA, no activity.
Next, we co-crystalized BuGUS-1 and BuGUS-2 in the presence of d-glucaro-1,4-lactone. Successful crystals were grown for both BuGUS-1 and BuGUS-2 upon incubation with d-glucaro-1,4-lactone, and structures at 2.0 Å (space group: C2, molecules in asymmetric unit: 2) and 2.5 Å (space group: P212121, molecules in asymmetric unit: 2) were determined, respectively (Table S1). Analysis of the active site of these structures revealed d-glucaro-1,5-lactone bound instead of d-glucaro-1,4-lactone (Fig. 9, A, B, and D). Previous studies have shown the spontaneous conversion of d-glucaro-1,5-lactone to d-glucaro-1,4-lactone, as it is the thermodynamic product (26). The reverse process is also chemically possible (Fig. 9C). As expected, the carboxylate of d-glucaro-1,5-lactone interacts with the NXK motif of both BuGUS-1 and BuGUS-2 (Fig. 9, A and B). The remaining hydroxyl groups are recognized by other residues, including the catalytic acid/base and nucleophile glutamates (Fig. 9, A, B, and D).
Discussion
The GHs encoded by Bacteroides species play key roles in the processing of carbohydrates and glycosides that reach the GI tract. Here we present three unique GUS enzymes from the human gut microbe B. uniformis that advance our understanding of the structural and functional diversity within this particular GH family. By analyzing the genes adjacent to a previously characterized B. uniformis GUS (12), we discovered two additional GUS enzymes from a B. uniformis PUL (Fig. 1, A and B). One of these GHs we termed BuGUS-1, as it retained the GUS-specific features previously used to identify GUS enzymes in the HMP (12). We demonstrated that it is a GUS capable of processing a variety of GlcA-containing substrates (Table 1, Fig. 6, and Fig. S2). BuGUS-3 also possessed several GUS-specific features, including the core fold, catalytic residues, and NXK motif; however, a tryptophan replaced the tyrosine that hydrogen bonds to and structurally stabilizes the nucleophilic glutamate (Fig. 2C). Although BuGUS-3 was unable to process 4-MUG, it did exhibit GUS activity toward the heparin sulfate 9-mer (Fig. 6A). This discovery suggests that the initial GUS rubric defined previously could allow either a tyrosine or a tryptophan at this sequence position (12). Indeed, a tryptophan is present at this position in the GUS module of BT0996, one of the enzymes responsible for the degradation of rhamnogalacturonan-II in B. thetaiotaomicron (27). This information indicates that the 279 GUS proteins previously identified represent an initial GUS atlas and should be reexamined and updated as new structural and functional data are determined regarding this enzyme family. Indeed, a preliminary analysis of the HMP identified 10 additional GUS proteins with a tryptophan residue in this position (Table S3); these novel proteins will be the subject of future studies.
As previously discussed by Pollet et al. (12), GUS enzymes with longer loops adjacent to the active site (e.g. Loop 1 GUS enzymes) were shown to process small glucuronides, and those possessing open active sites were able to process larger GlcA-containing polysaccharides. In previously determined GUS structures, the tetrameric interface between GUS protomers have been formed by their C termini, and active site adjacent loop structures (Loop 1) from these adjacent protomers formed the aglycone-binding site (Fig. 3A), limiting the access of larger substrates (12, 14, 16). In contrast, BuGUS-2 was shown to form a dimer, leaving its active site open and solvent exposed to accommodate larger polysaccharides (12) (Figs. 2B and 4A). BuGUS-1, which exhibits an open active site via a unique N-terminal–mediated tetrameric arrangement (Fig. 3B), processed 4-MUG with a similar efficiency to BuGUS-2 (Table 1).
BuGUS-1 was also shown to process SN-38-G faster than both BuGUS-2 and BuGUS-3 (Fig. 8B). Surprisingly, the NL BuGUS-1 processed SN-38-G at an efficiency that rivaled that of L1 EcGUS, despite the lack of an active site loop at the canonical position in its amino acid sequence (Fig. 1C). Further examination of the crystal structure of BuGUS-1 revealed the presence of an N-terminal loop (NTL) donated from an adjacent protomer (Fig. 8A). This donated loop mimics the Loop 1 present in L1 GUS enzymes and appears to enable BuGUS-1 to process the small molecule glucuronide 4-MUG (Table 1) and SN-38-G (Fig. 8B) at efficiencies similar to those of characterized Loop 1 GUS enzymes (Fig. 8B) (14, 16). Indeed, kinetic analysis of BMSP GUS and the Δloop variant of BuGUS-1 suggest that SN-38-G and 4-MUG processing by BuGUS-1 is greatly facilitated by its N-terminal loop (Table 1 and Fig. 8B). Examination of the GUS proteins present in the GI tracts of healthy humans (12) revealed that six additional enzymes beyond BuGUS-1 maintain an NTL (Table S4). Collectively, these data indicate that the determination of novel crystal structures of GUS enzymes will continue to enhance our understanding of the structural and functional variations present in this family of proteins.
To further investigate how the BuGUS-1 active site may interact with SN-38-G, we manually docked SN-38-G based on the PTG-bound structure of BuGUS-1. Our analysis shows that the planar, nonpolar aglycone of SN-38-G could interact favorably with the BuGUS-1 active site (Fig. 8A). Notably, Tyr-57 located in the donated loop participates in π-π interactions with the aromatic scaffold of SN-38-G in the binding mode modeled (Fig. 8A). This may explain its ability to efficiently hydrolyze this substrate. Docking of SN-38-G into the active sites of BuGUS-2 and BuGUS-3 demonstrates that they do not harbor the same active site features of BuGUS-1 that would allow them to recognize SN-38-G (Fig. S9). Specifically, the tyrosine in BuGUS-1 is replaced by an arginine and a tryptophan in BuGUS-2 and BuGUS-3, respectively, which do not appear to favorably interact with the aromatic scaffold of SN-38-G (Fig. S9, c and d).
In addition to small glucuronides, we demonstrated that all three GUS enzymes differentially processed GlcA-containing polysaccharides. Although bioinformatic analysis of the genes in this PUL do not reveal a clear polysaccharide substrate for these enzymes to act on, we showed that BuGUS-2 was capable of processing a sulfated heparin-like 9-mer and an acetylated heparin-like 9-mer, and BuGUS-3 processed the sulfated heparin-like 9-mer (Fig. 6A). Given the unique nature of the BuGUS-3 active site (Fig. 2C) compared with previously characterized GUS enzymes, this likely is a key feature that leads to its lack of activity with most of the glucuronide-containing polysaccharides. The differences in polysaccharide processing may also be explained by differences in quaternary structures. Although both BuGUS-2 and BuGUS-3 are dimers and contain extra C-terminal domains, the positioning of these domains are distinct and influence protomer organization (Fig. 4, A and B). Taken together, a combination of unique active site residues and quaternary structures likely dictate the specific substrates of these GHs.
Interestingly, BuGUS-1 was also shown to process both GlcA-containing polysaccharides tested (Fig. 6A). Compared with traditional L1 GUS enzymes, the active site of BuGUS-1 is more open due to its N-terminal-mediated tetrameric interface (Fig. 3B), which allows larger polysaccharides to access the active site. In addition to its unique tetrameric state, the flexible nature of its active site, as evidenced by the PTG-bound structure, may also explain the ability of BuGUS-1 to process bulkier polysaccharides. Upon PTG binding, several conformational shifts occur, including that exhibited by the catalytic base Glu-421, which appears to conflict with the large sulfur atom of PTG (Fig. 7, A and B). Although this conformation would preclude function, as Glu-421 is far from the position it would need to be to serve as an acid/base, the structure demonstrates that there is enough mobility in the active site to accommodate this shift and suggests that the active site is also capable of accommodating larger polysaccharide substrates.
We further found via docking that other sugar acids, like galacturonate, mannuronate, and iduronate, are likely to be accommodated in the active sites of these GUS enzymes (Fig. S8), and we confirm that BuGUS-1 can utilize a small-molecule galacturonide as a substrate (Fig. S9). This finding expands our understanding of the substrate-utilization capacities of the gut microbial GUS enzymes, and suggests that these enzymes may coordinate the degradation of polysaccharides that contain uronic acids beyond glucuronate.
Given the importance of quaternary structure relative to GUS function, we were interested in whether computational approaches would provide this critical information. We used Rosetta modeling to predict the tertiary and quaternary structures of the three GUS enzymes reported here. Although the core fold was predicted with a high degree of accuracy for all GUS enzymes analyzed, the critical loop structures as well as the orientation of C-terminal domains were more difficult to position and were heavily influenced by extant structures (Fig. S13). These results highlight the importance of using experimental structures to further refine modeling approaches to accurately predict protein quaternary structures.
Upon determining that BuGUS-1 and BuGUS-2 are targets to prevent GI side effects via SN-38-G processing, we tested whether they are susceptible to inhibition. Our GUS-specific inhibitors Inh1 and UNC10201652 did not inhibit BuGUS-1 and BuGUS-3 (Table 2). Previously, we have shown that the loop in L1 GUS enzymes stabilizes Inh1 (16). Although BuGUS-1 contains an N-terminal loop that replaces L1 in the active site, it is distinct from classic Loop 1 GUS enzymes that form deep hydrophobic pockets constructed from two loops from adjacent monomers (Fig. 3, A and B) (16). Thus, the active site in BuGUS-1 is more hydrophilic and solvent accessible, making it unfavorable for binding to the hydrophobic scaffold of Inh1 and UNC10201652.
Although Inh1 and UNC10201652 did not inhibit GUS activity, we showed that the nonspecific GUS inhibitor d-glucaro-1,4-lactone did inhibit BuGUS-1 and BuGUS-2 (Table 2). The crystal structure of BuGUS-1 incubated with the inhibitor revealed d-glucaro-1,5-lactone bound instead of d-glucaro-1,4-lactone (Fig. 7, A and B). We hypothesize that d-glucaro-1,5-lactone is spontaneously generated in solution over the time scale of crystal formation, upon which it is stabilized by binding to the GUS active site. Previous studies indicate that hydrolases in general, and GUS specifically, binds more tightly to d-glucaro-1,5-lactone than d-glucaro-1,4-lactone (28) and may explain its presence in the active site. The same result was observed for BuGUS-2, with d-glucaro-1,5-lactone apparent in the active site instead of the administered d-glucaro-1,4-lactone (PDB 6D50). Importantly, this pan-GUS inhibitor exhibited mid-micromolar potency against BuGUS-1 and BuGUS-2. These data suggest that other inhibitor chemotypes could be employed to prevent the actions of non-Loop 1 GUS enzyme-mediated reactivation of SN-38-G in the intestinal lumen.
The presence of three structurally and functionally unique GUS enzymes within a single B. uniformis PUL suggests that they have evolved to cleave distinct bonds in a uronic acid-rich polysaccharide. However, the action of GUS enzymes is not sufficient to carry out the complete catabolism of a complex polysaccharide. Thus, it is likely that these GUS enzymes act in concert with the glucuronyl hydrolase 88 enzyme, mannonate oxidase, mannonate dehydratase, and PL enzymes found in the same PUL to deconstruct a complex uronate-containing glycan found in the human gut. Additionally, the hallmark of SusC/SusD proteins likely mediate the transportation of the polysaccharide into the periplasmic space of B. uniformis for subsequent catabolism. Further studies are needed to determine the true polysaccharide associated with this PUL, but the data presented provides a basis for understanding the roles these GUS enzymes play in polysaccharide processing as well as their more established roles in drug-glucuronide reactivation.
Experimental procedures
Enzyme cloning
The full-length BuGUS-1, BuGUS-3, and BMSP genes were purchased from BioBasic in the pUC57 vector. Protein sequences were analyzed for signal peptide cleavage sites using the online SignalP 4.1 server (15). The mature gene lacking the signal peptide was amplified and inserted into the pLIC-His vector using the primers in Table S5.
Site-directed mutagenesis
The BuGUS-1 NTL deletion, BuGUS-2 D341A/D367A (BuGUS-2 ΔCa2+), and BuGUS-2 N591A/K593A mutants were created using site-directed mutagenesis. Primers were synthesized by Integrated DNA Technologies and are shown in Table S5. The mutant plasmids were sequenced to confirm the mutations. The mutants were produced and purified using Escherichia coli BL21 (DE3) Gold as described above. The NTL deletion for BuGUS-1 encompassed residues Tyr54–Ala67, which were replaced by a 6-residue linker (RGMKVY) based on the structure of BMSP GUS to maintain protein stability.
Protein expression and purification
Each β-glucuronidase expression plasmid was transformed into BL21 (DE3) Gold cells for enzyme expression. Cells were grown in the presence of ampicillin in LB medium with shaking at 225 rpm at 37 °C to an A600 of 0.5, at which point the temperature was reduced to 18 °C. At A600 of 0.8, protein expression was induced by the addition of 0.1 mm isopropyl 1-thio-d-galactopyranoside (IPTG) and incubation continued overnight. Cells were collected by centrifugation at 4500 × g for 20 min at 4 °C in a Sorvall (model RC-3B) swinging bucket centrifuge. Cell pellets were resuspended in Buffer A (20 mm potassium phosphate, pH 7.4, 50 mm imidazole, 500 mm NaCl), DNase, lysozyme, and a Roche Complete EDTA-free protease inhibitor tablet. Resuspended cells were sonicated and clarified via centrifugation at 17,000 × g for 60 min in a Sorvall (model RC-5B). The lysate was flowed over a nickel-nitrilotriacetic acid HP column (GE Healthcare) loaded onto the Aktaxpress FPLC system (Amersham Bioscience) and washed with Buffer A. Protein was eluted with Buffer B (20 mm potassium phosphate, pH 7.4, 500 mm imidazole, 500 mm NaCl). For BuGUS-1 used for crystallography, the His tag was removed by tobacco etch virus cleavage in the presence of 1 mm DTT and incubated overnight at 4 °C. This sample was then applied to the nickel-nitrilotriacetic acid column again and the flow through was collected. Fractions containing the protein of interest were combined and passed over a HiLoadTM 16/60 SuperdexTM 200 gel filtration column. Proteins were eluted in S200 buffer (20 mm HEPES, pH 7.4, 50 mm NaCl), except for BMSP and the BuGUS-1 ΔN-terminal loop mutant, which were eluted in S200 buffer that contained 300 mm NaCl. Fractions were analyzed by SDS-PAGE and those with >95% purity were combined and concentrated for long-term storage at −80 °C.
Crystallization and structure determination
Protein crystals were formed at 20 °C via the hanging drop vapor diffusion method with 15-well EasyXtal Qiagen trays with 300 μl of well solution in the reservoir and 3 μl of total drop volumes. Drop conditions were as follows: BuGUS-1-apo, 2 μl of 15 mg/ml of BuGUS-1, and 1 μl of 20% PEG 1000 and 0.1 m Tris, pH 8.5; BuGUS-1-G-1,5-L crystals, 2 μl of 10 mg/ml of BuGUS-1 in 1 μl of 16% PEG 1000, Tris, pH 8.5, and 2 mm glucaro-1,4-lactone; BuGUS-1-GlcA, 2 μl of 10 mg/ml of BuGUS-1 in 1 μl of 0.1 m sodium citrate, pH 5.5, 20% PEG 3000, and 20 mm glucuronic acid; BuGUS-2-G-1,5-L crystals, 2 μl of 12 mg/ml of BuGUS-2 in 1 μl of 0.2 m potassium chloride, 20% PEG 3350, and 10 mm glucaro-1,4-lactone; BuGUS-2 ΔCa2+, 2 μl of 12 mg/ml of the BuGUS-2 D341A/D367A mutant in 1 μl of 0.2 m potassium chloride and 20% PEG 3350; BuGUS-3, 2 μl of 15 mg/ml of BuGUS-3 in 1 μl of 8% PEG 3350, 0.2 m potassium thiocyanate, and 0.1 m HEPES, pH 8.0. The following three structures were derived from crystals made at 20 °C via the sitting drop method in Hampton Research 3-well Midi Crystallization Plates (Swissci) by an Art Robbins Instruments Crystal Phoenix robot with the following drop conditions: BMSP, 100 nl of 8 mg/ml of BMSP in 100 nl of 0.2 m magnesium chloride, 0.1 m MES:NaOH, pH 5.5, and 20% PEG 4000; BuGUS-1-PTG, 100 nl of 10 mg/ml of BuGUS-1 in 100 nl of 0.2 m potassium sulfate, 20% PEG 3350, and 10 mm phenyl-thio-β-d-glucuronide; BuGUS-1 ΔN-terminal loop, 100 nl of 12 mg/ml of the BuGUS-1 ΔN-terminal loop mutant in 100 nl of 0.1 m sodium citrate, pH 5.5, and 20% PEG 3000. Prior to crystallization, the N-terminal histidine tag was removed from BuGUS-1 as described in the purification methods above.
Crystal specimens were cryo-protected in the crystallization conditions as described above containing 20% glycerol, and diffraction data were collected for all crystals at 100 K at APS Beamline 23-ID-D, except for BuGUS-2 ΔCa2+, which was collected at APS Beamline 23-ID-B. The data were processed with XDS and all structures were solved via molecular replacement in Phenix (29) using the E. coli GUS structure (PDB 5CZK) as a search model for BuGUS-1-apo, the B. uniformis GUS structure (PDB 5UJ6) as a search model for BuGUS-2-ΔCa2+, the B. fragilis structure (PDB 3CMG) as a search model for BuGUS-3, and the BuGUS-1-apo structure (PDB 6D1N) for the remaining structures. The resulting starting model and maps from molecular replacement were then used in the AutoBuild function of Phenix. Structures were refined in Phenix and visually inspected and manually built using COOT (30). Final PDB coordinates for all structures have been deposited to the RCSB Protein Data Bank with corresponding PDB codes in parentheses: BuGUS-1 (6D1N), BuGUS-3 (PDB 6D1P), BMSP (PDB 6D8K), BuGUS-1 + G-1,5-L complex (PDB 6D41), BuGUS-2 + G-1,5-L complex (PDB 6D50), BuGUS-1 + GlcA complex (PDB 6D6W), BuGUS-1 Δloop (PDB 6D89), BuGUS-2 calcium-binding mutant (PDB 6D8G), and BuGUS-1 + TPG complex (PDB 6D7F).
GUS activity assay of 4-MUG hydrolysis
Initial pH screening was performed with PNPG, as described previously. Because PNPG is not amenable for continuous kinetic studies at pH below 6.5, we utilized an analogous but fluorescent GUS substrate 4-MUG for subsequent kinetic investigations. In vitro assays of GUS activity with 4-MUG were carried out in Costar black 96-well clear flat bottom plates. Total reaction volume was 50 μl with 5 μl of GUS and 5 μl of 10× buffer (250 mm HEPES, 250 mm NaCl, pH 7.0) mixed and preincubated at 37 °C before reaction initiation by addition of 40 μl of 4-MUG. Concentration of enzyme was specific to each GUS: 5 nm EcGUS, 5 nm BuGUS-1, 20 nm BuGUS-2, 40 nm BuGUS-1 Δloop, 80 nm BMSP GUS, and 320 nm BuGUS-3. Reactions were monitored continuously in a BMG lab tech PHERAstar plate reader with an excitation wavelength of 350 nm and an emission wavelength of 450 nm. Resultant progress curves were fit by a custom linear regression analysis program in MATLAB. Initial velocities were then analyzed in the enzyme kinetics module of SigmaPlot 13.0 by Michaelis-Menten fit to determine the catalytic turnover (kcat) and Michaelis constant (Km).
GUS activity assay of SN-38-G hydrolysis
In vitro assays of GUS activity with the substrate SN-38-G were carried out in Costar black 96-well clear flat bottom plates. Total reaction volume was 50 μl with 5 μl of SN-38-G at a range of low substrate concentrations (15, 10, 7.5, 5, and 2.5 μm final), 5 μl of 10× buffer (250 mm HEPES, 250 mm NaCl, pH 7.0), and 35 μl of water mixed and preincubated at 37 °C before reaction initiation by addition of 5 μl of GUS. Concentration of enzyme was specific to each GUS: 5 nm EcGUS, 5 nm BuGUS-1, 20 nm BuGUS-2, 40 nm BuGUS-1 Δloop, 80 nm BMSP GUS, and 320 nm BuGUS-3. Reactions were monitored continuously by fluorescence with an emission wavelength of 420 nm and an excitation wavelength of 230 nm. Resultant progress curves were fit by a custom linear regression analysis program in MATLAB. Initial velocities were then plotted against substrate concentration and fit with linear regression in Microsoft Excel to determine catalytic efficiency (kcat/Km).
GlcA-containing polysaccharide processing assay
The sulfated heparin-like nonasaccharide (GlcA-(GlcNS-GlcA)4-PNP (where GlcA is glucuronic acid, GlcNS is N-sulfated glucosamine)) and the acetylated heparin-like nonasaccharide (GlcA-(GlcNAc-GlcA)4-PNP (where GlcA is glucuronic acid, GlcNAc is GlcNAc)) substrates were from Glycantherapeutics. The additional polysaccharides employed were synthesized in-house (37). Putative polysaccharide substrates were digested with each GUS enzyme for 3 h. Digestion reactions were composed of 0.5 μm GUS enzyme and 10 μg of oligosaccharide. Reactions were terminated by heating for 5 min at 95 °C. Aliquots of the resultant solutions were analyzed by polyamine-based anion exchange (PAMN)-HPLC. Sugars were eluted from the PAMN column (0.46 × 25 cm from Waters) with a linear gradient of KH2PO4 from 0 to 1 m in 40 min at a flow rate of 0.5 ml/min. The eluent was monitored by a UV detector at 310 nm. Aliquots of the digestion reactions were analyzed by electrospray ionization-MS (ESI-MS) by first purifying the reaction mixture by C18 column eluted with a linear gradient of methanol with 1% TFA from 0 to 1 m in 60 min at a flow rate of 0.5 ml/min. The purified oligosaccharides were then dried. Electrospray ionization-MS analysis was performed on a Thermo LCQ-Deca in negative ionization mode. A syringe pump (Harvard Apparatus) was used to introduce the sample by direct infusion (50 ml/min). The purified oligosaccharides were diluted in 200 ml of H2O with the electrospray source set to 3 KV and 150 °C. The automatic gain control was set to 1 × 107 for full scan MS. The MS data were acquired and processed using Xcalibur 1.3.
GUS inhibition assay
In vitro assays of GUS activity with the substrate 4-MUG were carried out in Costar black 96-well clear flat bottom plates. Total reaction volume was 50 μl with 5 μl of GUS, 10 μl of 5× buffer (125 mm HEPES, 125 mm NaCl, pH 7.0), and 5 μl of inhibitor mixed and preincubated at 37 °C before reaction initiation by addition of 30 μl of 4-MUG. Reactions were monitored continuously in a PHERAstar plate reader at 410 nm. End point absorbance values after 1 h were converted to % inhibition values via the following equation,
| (Eq. 1) |
where Aexp is the end point absorbance at a particular inhibitor concentration, Amax is the absorbance of the uninhibited reaction, and Abg is the background absorbance. Percent inhibition values were subsequently plotted against the log of inhibitor concentration and fit with a four-parameter logistic function in SigmaPlot 13.0 to determine the concentration at which 50% inhibition (IC50) is observed.
SEC-MALS analysis of BuGUS enzymes
BuGUS-1, BuGUS-2, and BuGUS-3 were analyzed on a Superdex 200 size exclusion column connected to an Agilent FPLC system, Wyatt DAWN HELEOS II multi-angle light scattering instrument, and a Trex refractometer. The injection volume was 50 μl, and each protein was assessed at 10 mg/ml in 50 mm HEPES and 150 mm NaCl, pH 7.4, buffer. A flow rate of 0.5 ml/min was used. Light scattering and refractive index data were collected and analyzed using Wyatt ASTRA (version 6.1) software. A dn/dc value of 0.185 was used for calculations. Approximately 99% of BuGUS-1 eluted in a single peak with a molar mass of 275 kDa, indicating that it forms a tetramer in solution. In contrast, 99% of BuGUS-2 and 95% BuGUS-3 eluted in single peaks with molar masses of 189 and 175 kDa, respectively, indicating that they form dimers in solution.
CD analysis of BuGUS-2 calcium-binding mutant
The protein stabilities of WT BuGUS-2 and the calcium-binding mutant were determined using the circular dichroism method (31). Enzyme (1 μm) in CD buffer containing 10 mm potassium phosphate, pH 7.4, and 100 mm potassium fluoride was loaded into a 1-mm cuvette. Using a Chirascan-plus instrument (Applied Photophysis Limited), spectra from 185 to 280 nm were recorded at 20 ± 1.0 °C. Measurements were corrected for background signal using a CD buffer sample. The melting profile of the sample (5 μm) was monitored at 218 nm from 25 to 94 °C.
Manual docking of monosaccharide in PyMOL
Galacturonate, mannonate, and iduronate monosaccharides were accessed from the PDB in previously solved crystal structures (PDB 1KCC for galacturonate, PDB 3VLW for mannuronate, and PDB 4OBR for iduronate). These were then imported into PyMOL and manually aligned to the GlcA-bound BuGUS-1 structure (PDB 6DW6) with the 3-button editing tool. After manual alignment of the sugar monosaccharides, structures of BuGUS-1 and BuGUS-3 were aligned to the GlcA-bound BuGUS-2 structure. Visual inspection and final figures after alignment were generated in PyMOL.
Rosetta modeling
The full-length amino acid sequences BuGUS-1, BuGUS-3, and BMSP GUS were submitted to the Robetta modeling server (32–34) to produce 3D homology models of these proteins, including their oligomeric complexes, based on template protein structures available in the Protein Data Bank from December 2017 to January 2018. The BuGUS-1 and BMSP GUS Robetta homology models were based on the E. coli β-glucuronidase structure (PDB 3LPF). For the BuGUS-3 homology model, the Robetta selected template was a β- galactosidase from Bacillus circulans ATCC 31382 (PDB 4YPJ). Backbone C-α coordinates of the homology model protein structures were then superimposed onto X-ray crystal structures using TM-align algorithms (35).
Identification of predicted calcium-binding sites
To identify calcium-binding sites in GUS enzymes from the HMP dataset, the 279 GUS protein sequences previously identified (12) were aligned pairwise to BuGUS-2 using NCBI BLASTp (36). These alignments were then probed for the three aspartic acid residues in BuGUS-2 (Asp-176, Asp-341, and Asp-367) deemed necessary for calcium binding.
Identification of tryptophan substitutions
To identify additional GUS enzymes in the HMP Clustered genes (HMGC) dataset that possess a tryptophan rather than a tyrosine at position Trp-483 in BuGUS-3, the ∼267,000 sequences previously determined to share 25% identify with EcGUS, SaGUS, CpGUS, and BfGUS (12) were aligned pairwise to the these GUS enzymes and BuGUS-3 using NCBI BLASTp (36). The sequences were then probed for the presence of the NXKG motif, catalytic E residues, and N and W motifs.
Identification of N-terminal loops
To identify N-terminal loops in GUS enzymes from the HMP dataset, the 279 GUS protein sequences previously identified (12) were aligned pairwise to BuGUS-1 using NCBI BLASTp (36). These alignments were then probed for the N-terminal loop in BuGUS-1.
Author contributions
S. J. P., W. G. W., and M. R. R. conceptualization; S. J. P., W. G. W., D. T.-R., and Y. X. data curation; S. J. P. and W. G. W. formal analysis; S. J. P., W. G. W., D. T.-R., B. C. C., Y. X., A. T., and L. J. S. investigation; S. J. P. and K. A. B. writing-original draft; S. J. P., W. G. W., K. A. B., and M. R. R. writing-review and editing; W. G. W. and M. R. R. supervision; W. G. W. methodology; J. L. and M. R. R. funding acquisition; M. R. R. project administration.
Supplementary Material
Acknowledgments
We thank Samantha Ervin and Jerry Wei for experimental assistance.
This work was supported by the National Institutes of Health Grants CA098468 and CA207416 (to M. R. R.) and GM102137 and HL094463 (to J. L.). M. R. R. is the scientific founder of Symberix, Inc., which is developing microbiome-targeted therapeutics. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S13 and Tables S1–S5.
- GI
- gastrointestinal
- HGM
- human gut microbiota
- GH
- glycoside hydrolase
- CAZymes
- carbohydrate-active enzymes
- GlcA
- glucuronic acid
- NL
- No Loop
- L2
- Loop 2
- mL2
- mini Loop 2
- 4-MUG
- 4-methylumbelliferyl-β-d-glucuronide
- PTG
- phenyl-thio-β-d-glucuronide
- NTL
- N-terminal loop
- BMSP
- Bacteroides multispecies
- PL
- polysaccharide lyase
- HMP
- human microbiome project
- PUL
- polysaccharide utilization loci
- GUS
- β-glucuronidase
- PDB
- Protein Data Bank
- CBM
- carbohydrate binding module
- DUF
- domains of unknown function
- PNPG
- 4-nitrophenyl β-d-glucuronide.
References
- 1. El Kaoutari A., Armougom F., Gordon J. I., Raoult D., and Henrissat B. (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 2. Flint H. J., Scott K. P., Duncan S. H., Louis P., and Forano E. (2012) Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 3, 289–306 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lombard V., Bernard T., Rancurel C., Brumer H., Coutinho P. M., and Henrissat B. (2010) A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 10.1042/BJ20101185 [DOI] [PubMed] [Google Scholar]
- 5. McNeil N. I. (1984) The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 39, 338–342 10.1093/ajcn/39.2.338 [DOI] [PubMed] [Google Scholar]
- 6. Maslowski K. M., Vieira A. T., Ng A., Kranich J., Sierro F., Yu D., Schilter H. C., Rolph M. S., Mackay F., Artis D., Xavier R. J., Teixeira M. M., and Mackay C. R. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pluznick J. L., Protzko R. J., Gevorgyan H., Peterlin Z., Sipos A., Han J.,Brunet I., Wan L. X., Rey F., Wang T., Firestein S. J., Yanagisawa M., Gordon J. I., Eichmann A., et al. (2013) Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 4410–4415 10.1073/pnas.1215927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samuel B. S., Shaito A., Motoike T., Rey F. E., Backhed F., Manchester J. K., Hammer R. E., Williams S. C., Crowley J., Yanagisawa M., and Gordon J. I. (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 105, 16767–16772 10.1073/pnas.0808567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salyers A. A., Vercellotti J. R., West S. E., and Wilkins T. D. (1977) Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sonnenburg J. L., Xu J., Leip D. D., Chen C., Benjamin P., Weatherford J., Buhler J. D., and Gordon J. I. (2005) Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science (80-.). 307, 1955–1959 10.1126/science.1109051 [DOI] [PubMed] [Google Scholar]
- 11. Martens E. C., Koropatkin N. M., Smith T. J., and Gordon J. I. (2009) Complex glycan catabolism by the human gut microbiota: the bacteroidetes sus-like paradigm. J. Biol. Chem. 284, 24673–24677 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pollet R. M., Agostino E. H., Walton W. G., Xu Y., Little M. S., Biernat K. A., Pellock S. J., Patterson L. M., Creekmore B. C., Isenberg H. N., Bahethi R. R., Bhatt A. P., Liu J., Gharaibeh R. Z., and Redinbo M. R. (2017) An atlas of β-glucuronidases in the human intestinal microbiome. Structure 25, 967–977 10.1016/j.str.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zitomersky N. L., Coyne M. J., and Comstock L. E. (2011) Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect. Immun. 79, 2012–2020 10.1128/IAI.01348-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wallace B. D., Roberts A. B., Pollet R. M., Ingle J. D., Biernat K. A., Pellock S. J., Venkatesh M. K., Guthrie L., O'Neal S. K., Robinson S. J., Dollinger M., Figueroa E., McShane S. R., Cohen R. D., Jin J., et al. (2015) Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249 10.1016/j.chembiol.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 16. Wallace B. D., Wang H., Lane K. T., Scott J. E., Orans J., Koo J. S., Venkatesh M., Jobin C., Yeh L.-A., Mani S., and Redinbo M. R. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science (80-.). 330, 831–835 10.1126/science.1191175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain S., Drendel W. B., Chen Z. W., Mathews F. S., Sly W. S., and Grubb J. H. (1996) Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 3, 375–381 10.1038/nsb0496-375 [DOI] [PubMed] [Google Scholar]
- 18. Schallus T., Jaeckh C., Fehér K., Palma A. S., Liu Y., Simpson J. C., Mackeen M., Stier G., Gibson T. J., Feizi T., Pieler T., and Muhle-Goll C. (2008) Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 19, 3404–3414 10.1091/mbc.e08-04-0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. LoGuidice A., Wallace B. D., Bendel L., Redinbo M. R., and Boelsterli U. (2012) A pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 341, 447–454 10.1124/jpet.111.191122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boelsterli U. A., Redinbo M. R., and Saitta K. S. (2013) Multiple NSAID-induced hits injure the small intestine: underlying mechanisms and novel strategies. Toxicol. Sci. 131, 654–667 10.1093/toxsci/kfs310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saitta K. S., Zhang C., Lee K. K., Fujimoto K., Redinbo M. R., and Boelsterli U. A. (2014) Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica 44, 28–35 10.3109/00498254.2013.811314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mani S., Boelsterli U. A., and Redinbo M. R. (2014) Understanding and modulating mammalian-microbial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 54, 559–580 10.1146/annurev-pharmtox-011613-140007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roberts A. B., Wallace B. D., Venkatesh M. K., Mani S., and Redinbo M. R. (2013) Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 84, 208–217 10.1124/mol.113.085852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wallace B. D., and Redinbo M. R. (2013) The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol. 17, 379–384 10.1016/j.cbpa.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pellock S. J., and Redinbo M. R. (2017) Glucuronides in the gut: sugar-driven symbioses between microbe and host. J. Biol. Chem. 292, 8569–8576 10.1074/jbc.R116.767434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andberg M., Maaheimo H., Boer H., Penttilä M., Koivula A., and Richard P. (2012) Characterization of a novel Agrobacterium tumefaciens galactarolactone cycloisomerase enzyme for direct conversion of d-galactarolactone to 3-deoxy-2-keto-l-threo-hexarate. J. Biol. Chem. 287, 17662–17671 10.1074/jbc.M111.335240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ndeh D., Rogowski A., Cartmell A., Luis A. S., Baslé A., Gray J., Venditto I., Briggs J., Zhang X., Labourel A., Terrapon N., Buffetto F., Nepogodiev S., Xiao Y., Field R. A., et al. (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70 10.1038/nature21725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Conchie J., Hay A. J., Strachan I., and Levvy G. A. (1967) Inhibition of glycosidases by aldonolactones of corresponding configuration: preparation of (1→5)-lactones by catalytic oxidation of pyranoses and study of their inhibitory properties. Biochem. J. 102, 929–941 10.1042/bj1020929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 1948–1954 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 31. Sreerama N., and Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 10.1006/abio.2000.4880 [DOI] [PubMed] [Google Scholar]
- 32. Park H., Ovchinnikov S., Kim D. E., DiMaio F., and Baker D. (2018) Protein homology model refinement by large-scale energy optimization. Proc. Natl. Acad. Sci. U.S.A. 115, 3054–3059 10.1073/pnas.1719115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park H., Kim D. E., Ovchinnikov S., Baker D., and DiMaio F. (2018) Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins 86, 283–291 10.1002/prot.25387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ovchinnikov S., Park H., Kim D. E., DiMaio F., and Baker D. (2018) Protein structure prediction using Rosetta in CASP12. Proteins 86, 113–121 10.1002/prot.25390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong R., Peng Z., Zhang Y., and Yang J. (2018) mTM-align: an algorithm for fast and accurate multiple protein structure alignment. Bioinformatics 34, 1719–1725 10.1093/bioinformatics/btx828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 37. Xu Y., Cai C., Chandarajoti K., Hsieh P. H., Li L., Pham T. Q., Sparkenbaugh E. M., Sheng J., Key N. S., Pawlinski R., et al. (2014) Homogeneous low-molecular-weight heparins with reversible anticoagulant activity. Nat. Chem. Biol. 10, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.