Abstract
Polyamines are organic polycations that bind to a variety of cellular molecules, including nucleic acids. Within cells, polyamines contribute to both the efficiency and fidelity of protein synthesis. In addition to directly acting on the translation apparatus to stimulate protein synthesis, the polyamine spermidine serves as a precursor for the essential post-translational modification of the eukaryotic translation factor 5A (eIF5A), which is required for synthesis of proteins containing problematic amino acid sequence motifs, including polyproline tracts, and for termination of translation. The impact of polyamines on translation is highlighted by autoregulation of the translation of mRNAs encoding key metabolic and regulatory proteins in the polyamine biosynthesis pathway, including S-adenosylmethionine decarboxylase (AdoMetDC), antizyme (OAZ), and antizyme inhibitor 1 (AZIN1). Here, we highlight the roles of polyamines in general translation and also in the translational regulation of polyamine biosynthesis.
Keywords: spermidine, translation, translation regulation, polyamine, eukaryotic initiation factor 5A (eIF5A), AdoMetDC, antizyme, antizyme inhibitor, autoregulation, ornithine decarboxylase
Introduction
Because of their cationic properties, polyamines bind to negatively charged molecules in cells. Nucleic acids, including both DNA and RNA, are prominent binding sites for polyamines with nearly 90% of the spermidine in Escherichia coli cells bound to RNA (1). A substantial fraction (∼15%) of the polyamines in E. coli are stably associated with ribosomes (2). Polyamines stimulate general protein synthesis and also impact the fidelity of translation. The importance of polyamines to translation is highlighted by the mRNA-specific translational control mechanisms that link polyamine levels to the synthesis of key polyamine biosynthetic enzymes and their regulators. In this Minireview, we will highlight the roles of polyamines in general translation and in the translational regulation of polyamine biosynthesis. In honor of Herb Tabor, we will also highlight his contributions to this area of polyamine research.
Overview of the translation pathway
To review the roles of polyamines in translation, it is important to first discuss the mechanism of cellular protein synthesis. As this Minireview will discuss both eukaryotic and bacterial translation, we will focus the description on eukaryotic translation but highlight differences in the bacterial translation mechanism that are relevant to polyamines. Ribosomes are ribonucleotide–protein particles consisting of 3–4 rRNA molecules and ∼53–79 ribosomal proteins depending on the organism. These components are organized into two subunits: the small subunit (40S in eukaryotes and 30S in bacteria) and the large subunit (60S in eukaryotes and 50S in bacteria). The small subunit contains the decoding center with binding sites for aminoacyl-tRNAs and the mRNA, and the large subunit contains the peptidyltransferase active site of the ribosome, which catalyzes peptide bond formation. There are three tRNA-binding sites on the ribosome: the aminoacyl (A)3 site, the peptidyl (P) site, and the exit (E) site. The protein synthesis pathway can be broken into four steps: initiation, elongation, termination, and ribosome recycling. Translation initiation culminates with the formation of a complex in which the initiator methionyl-tRNA (Met-tRNAiMet; note that in bacteria formylated fMet-tRNAfMet is used for initiation) is bound in the P site of the ribosome with its anticodon base-paired with the start codon of the mRNA. The translation initiation pathways are distinct in bacteria and eukaryotes and require distinct sets of trans-acting initiation factors (3). In bacteria, the 30S subunit with bound formylated fMet-tRNAfMet is recruited to the start codon of an mRNA via base-pairing interactions between the 3′ end of the 15S rRNA within the subunit and the Shine-Dalgarno sequence located a few bases upstream of the start codon in the mRNA. Following formation of this preinitiation complex (PIC), the large (50S) subunit joins to form the functional 70S ribosome (4). In contrast to this direct recruitment of the ribosome to the start codon in bacterial translation initiation, eukaryotic translation initiation relies primarily on the scanning mechanism to select the start codon (reviewed in Refs. 3, 5, 6). The eukaryotic translation initiation factor eIF2 binds GTP and Met-tRNAiMet to form a ternary complex that then binds to the 40S subunit along with several other translation initiation factors. This PIC then binds near the 5′ end of an mRNA with the assistance of the mRNA 5′ m7G cap-binding factor eIF4E and associated factors, including eIF4G. The PIC then scans the mRNA in a 3′ direction inspecting the sequence for a start codon. Base-pairing interactions between the anticodon of the tRNAiMet and an AUG codon in the mRNA triggers a sequence of events including: 1) release of the factor eIF1; 2) completion of GTP hydrolysis by eIF2 and release of inorganic phosphate (Pi); 3) structural rearrangements in the ribosome to strengthen Met-tRNAiMet binding in the P site and to prevent further scanning; and 4) release of eIF2–GDP. Like many G proteins, eIF2 has a higher affinity for GDP than for GTP. The guanine nucleotide exchange factor eIF2B recycles inactive eIF2–GDP to functional eIF2–GTP. Stress-responsive protein kinases, including the antiviral kinase PKR, the endoplasmic reticulum stress-responsive kinase PERK, and the amino acid starvation-activated kinase GCN2, phosphorylate the α subunit of eIF2 to convert eIF2 into an inhibitor of eIF2B and thereby block recycling and inhibit translation (7). Following release of eIF2–GDP from the small ribosomal subunit poised on the start codon, the 60S subunit joins to form an 80S ribosome that is ready for translation elongation.
The translation elongation pathway is conserved between bacteria and eukaryotes (8). The eukaryotic translation elongation factor (eEF) 1A like its bacterial homolog EF-Tu forms a ternary complex with GTP and an aminoacyl-tRNA. Binding of this ternary complex to the A site of the ribosome and base-pairing of the anticodon of the aminoacyl-tRNA to the mRNA codon present in the A site trigger GTP hydrolysis by eEF1A. Following release of the eEF1A–GDP complex, the aminoacyl-tRNA is accommodated into the A site. By positioning the aminoacyl-tRNA in the A site in close proximity to the peptidyl-tRNA in the P site, the ribosome promotes peptide bond formation. Of particular relevance when discussing the roles of polyamines in translation, the translation factor eIF5A (formerly called eIF-4D or IF-M2Bα), which is post-translationally modified by spermidine, binds in the E site and promotes peptide bond formation (9–12). The function of eIF5A is especially important for poor substrates like proline (13–15). Following peptide bond formation, the factor eEF2 (EF-G in bacteria) promotes translocation of the tRNAs from the P and A sites to the E and P sites, respectively, and the mRNA is translocated with the tRNAs. Following release of eEF2, the A site is vacant and available to receive the next aminoacyl-tRNA. Translation elongation will continue with cycles of peptide bond formation, translocation, and aminoacyl-tRNA binding until a termination codon enters the A site.
Translation termination is promoted by release factors (16). In eukaryotes, the release factor eRF1 in complex with the factor eRF3 interacts with a stop codon in the A site and promotes hydrolysis of the aminoacyl bond linking the completed peptide chain to the P-site tRNA. Recent studies have revealed that eIF5A, in addition to its role in elongation, is generally required for termination (14). Following hydrolysis of the peptidyl-tRNA bond, the nascent peptide is released from the ribosome. Additional factors, including ABC protein ABCE1, ligatin (eIF2D), density-regulated protein, and multiple copies in T-cell lymphoma-1 (MCT1), in eukaryotes or ribosome recycling factor in bacteria release the ribosomal subunits and deacylated tRNA from the mRNA for use in additional rounds of translation (16).
Polyamine stimulation of translation in vitro
Polyamines were initially linked to protein synthesis following the finding that polyamines were stably associated with ribosomes in the cell. In E. coli cells, ∼12–15% of polyamines are stably associated with ribosomes such that the polyamines do not readily exchange with free polyamines in cell extracts (2). The notion of specific polyamine-binding sites on ribosomes is supported by cross-linking studies using photoreactive analogs of spermine (17); however, the large number of bound polyamines (>500/ribosome) (18) raises questions about the specificity of some sites. High-resolution structural studies on the bacterial ribosome have identified only two well-positioned polyamines (Protein Data Bank code 4YBB (19)). One possibility is that only a few polyamines bind to critical defined sites required for ribosome function, and other sites are differentially occupied in a stochastic manner. In support of the notion that polyamines are important for ribosome function, the Tabor lab identified mutations in the small ribosomal subunit protein S12 of E. coli that conferred an essential requirement for polyamines for bacterial growth (20), although it is not clear whether the polyamine requirement was for general ribosome function or translation fidelity.
Many studies have reported stimulation of translation in vitro by polyamines. Early studies using cell-free extracts from bacteria or mammalian cells employed high concentrations of divalent cations, typically Mg2+, to stimulate protein synthesis. Subsequent studies revealed that inclusion of polyamines, especially spermidine or spermine, lowered the Mg2+ optimum for peptide synthesis (21–24). In addition to lessening the Mg2+ requirement for peptide synthesis, other studies showed that addition of polyamines increased the activity of in vitro translation systems and the total yield of protein (25–27). The use of gel-filtered in vitro translation systems revealed the distinct stimulatory properties of Mg2+ and polyamines. Addition of Mg2+ alone to filtered reticulocyte lysates failed to restore translation activity to the rate obtained in unfiltered lysates; however, supplementation with polyamines plus Mg2+ restored nearly full activity (28). Again, the inclusion of polyamines lowered the optimum Mg2+ concentration for protein synthesis. The ability of polyamines to stimulate translation in vitro was independent of the mRNA used and thus the protein synthesized (29), indicating that the polyamines were likely acting on the translational machinery.
Although polyamines could stimulate the rate and extent of in vitro protein synthesis by enhancing the initiation or elongation steps of translation, most studies have linked polyamines to translation elongation. Addition of polyamines to wheat germ extracts increased the rate of amino acid incorporation into lengthening polypeptides, as opposed to newly initiated proteins, indicating that polyamines were stimulating translation elongation (28). Moreover, polyamines stimulated methionyl-puromycin (Met-puro) synthesis in rabbit reticulocyte lysates (30). Although the Met-puro assay, which monitors the transfer of methionine from Met-tRNAiMet to the aminoacyl-tRNA analog puromycin, was originally thought to mimic first peptide bond formation and thereby monitor translation initiation, it is now clear that this assay reports on ribosomal peptidyltransferase activity and thus is a readout of translation elongation. Consistent with this interpretation, the translation factor eIF5A, which functions in translation elongation (14, 31), was originally identified based on its stimulation of Met-puro synthesis (32–34). In further support for polyamine stimulation of translation elongation, synthesis of the simple peptide Met–Phe–Phe–Phe (MFFF) in a fully reconstituted yeast in vitro assay system was highly dependent on added polyamines. Following pre-assembly of initiation complexes on the mRNAs, addition of spermine, spermidine, or putrescine was required for peptide synthesis (15). In these reactions, spermine (K1/2 = 0.08 mm) and spermidine (K1/2 = 0.24 mm) were substantially more effective than putrescine (K1/2 = 17 mm) (15). In kinetic assays using a reconstituted bacterial translation assay system, addition of spermine or spermidine stimulated the rate of codon recognition in the A site of the ribosome (35) without impacting the fidelity of translation. Perhaps the binding of polyamines to tRNA (36, 37) enhances the association of the EF-Tu–GTP–aminoacyl-tRNA complexes with the ribosome. The Tabor lab reported that addition of polyamines to an auxotrophic strain of E. coli enhanced the production of a full-length protein dependent on a nonsense suppressor tRNA (38). Although the simple interpretation of this result is that polyamines are enhancing the utilization of the suppressor tRNA during elongation, additional experiments are needed to determine whether the polyamines are affecting the levels or aminoacylation of the suppressor tRNA. Taken together, these in vitro and in vivo studies demonstrate that polyamines stimulate translation elongation; however, they do not rule out a role for polyamines in translation initiation. In fact, studies of mitochondrial translation initiation, which uses a bacteria-like translation system, reported stimulation of PIC formation by spermine, perhaps due to stabilization of fMet-tRNAfMet binding to the small ribosomal subunit (39).
In a fully reconstituted mammalian in vitro translation assay system, addition of the factor eIF5A, which as described above is modified by spermidine, or of polyamines decreased the Mg2+ optimum required for globin synthesis (34). Consistent with this overlap in eIF5A and polyamine function, eIF5A could functionally substitute for polyamines to stimulate MFFF synthesis in the reconstituted yeast in vitro translation assay system (15). In assays lacking polyamines, addition of either spermidine or eIF5A (interestingly, with or without its polyamine-derived modification) was sufficient to restore translational activity. As eIF5A binds in the ribosomal E site (10, 11, 13) and is thought to interact with both the body and acceptor stem of the P-site tRNA, this functional overlap of eIF5A and polyamines suggests that polyamines may interact with the peptidyl-tRNA, at least in eukaryotes, to promote translation elongation.
Translation factor eIF5A, hypusine, and polyamine stimulation of translation
As mentioned earlier, the translation factor eIF5A is at the interface of polyamines and cellular protein synthesis. eIF5A is the sole cellular protein that contains the amino acid hypusine. The hypusine residue in eIF5A is formed post-translationally in two steps (9, 12). First, the n-butylamine group from spermidine is transferred to the ϵ-amino group of a conserved Lys residue in eIF5A to form deoxyhypusine. A subsequent hydroxylation reaction on the second carbon of the transferred moiety completes the modification to form hypusine. Both eIF5A and its hypusine modification are essential for cell viability in yeast (40–42). Several studies, including work from the Tabor lab (43), demonstrated that spermidine, and not spermine, was the source of the n-butylamine moiety for hypusine formation (44–47). Although originally thought to function in translation initiation based on its ability to stimulate Met-puro synthesis (32, 33), subsequent studies revealed that eIF5A was generally required for translation elongation and termination (14, 31). The elongation function of eIF5A was especially noted for difficult substrates like polyproline (13, 15).
Several studies have sought to examine the role of eIF5A, hypusine, and polyamines in translation in vivo. By growing polyamine auxotrophic strains of yeast in varying concentrations of spermidine, the Tabor lab correlated hypusine formation in eIF5A with cellular growth rate (48). Although not definitive, this study suggested that hypusine formation on eIF5A is one of the most critical functions of spermidine in yeast cells. Depleting mammalian cells of polyamines using the drug difluoromethylornithine, an inhibitor of ornithine decarboxylase (ODC), or by overexpressing the polyamine catabolic enzyme N1-spermidine/spermine acetyltransferase 1 (SSAT1) inhibited cell growth (49, 50). Polysome profile analyses in these cells revealed a loss of large polysomes suggesting a defect in translation initiation that is at odds with the in vitro studies indicating that polyamines play a more prominent role in translation elongation. Interestingly, depletion of hypusine using the drug N1-guanyl-diaminoheptane, an inhibitor of deoxyhypusine synthase, likewise caused a loss of large polysomes (49). The discovery that polyamine depletion triggers activation of the stress-responsive kinase PERK and phosphorylation of translation initiation factor eIF2α provides a plausible explanation for the observed translation initiation defect in cells depleted for polyamines or hypusine (51). Accordingly, a reduction in polyamine levels activates a cellular stress response leading to phosphorylation of eIF2α and inhibition of translation initiation that is more rate-limiting than the impairment in translation elongation associated with loss of polyamines and hypusine. Of note, depletion of polyamines inhibited growth of cells in which eIF2α phosphorylation was prevented by mutation of the phosphorylation site to alanine (S51A) (51), consistent with the notion that polyamines play additional roles in cell proliferation. It will be of interest to determine whether translation initiation or elongation is impaired in the S51A cells following polyamine depletion. Finally, depletion of polyamines in mammalian cells using difluoromethylornithine plus N1-(3-aminopropyl)-cyclohexylamine, an inhibitor of spermine synthase, was reported to inhibit cell growth prior to impacts on hypusine levels (52). Although it is not clear whether this proliferation defect is due to impaired protein synthesis, this result suggests that in addition to their critical role in hypusine formation, polyamines can also contribute to cell growth independently of eIF5A and hypusine.
Impacts of polyamines on translational fidelity
Although translation elongation typically proceeds at high fidelity with the ribosome incorporating the proscribed amino acid and advancing by three nucleotides at each translocation step, errors due to misincorporation of the wrong amino acid or due to shifting of reading frames can occur. The frequency of both of these errors is influenced by polyamines. Multiple studies have linked lower levels of polyamines with increased misreading during translation elongation. In both bacterial and wheat germ extracts, increasing the concentration of spermidine from 1 to 3 mm decreased the rate of misincorporation of leucine in place of phenylalanine by ribosomes programmed with poly(U) mRNA-encoding polyphenylalanine (53, 54). Consistent with these findings, the relatively high level of misreading observed in E. coli strains that were defective or auxotrophic for polyamine synthesis was partially suppressed by addition of putrescine to the media (55). In contrast to these studies linking increased polyamines to enhanced fidelity during translation elongation, high polyamines were linked to stop codon readthrough on the β-globin mRNA in rabbit reticulocyte lysate (56). It is intriguing to recall that the translation factor eIF5A is generally required for termination (14), and as will be described below, high polyamines have been found to interfere with eIF5A function during translation of the antizyme inhibitor 1 (AZIN1) mRNA (57). Perhaps high polyamine stimulation of stop codon readthrough does not reflect decreased fidelity of aminoacyl-tRNA binding to the ribosomal A site but rather impaired termination at the stop codon due to interference with eIF5A function. Additional experiments are needed to distinguish between these possibilities. In a series of studies, the Tabor lab highlighted the impact of polyamines on frameshifting. Whereas translation of the yeast retrovirus Ty1 relies on a programmed +1 frameshift, translation of the endogenous yeast L-A virus RNA depends on a −1 programmed frameshift (58). Interestingly, increased +1 frameshifting was observed when a strain lacking S-adenosylmethionine decarboxylase (AdoMetDC), and thus unable to synthesize spermidine or spermine, was grown in low concentrations of spermidine (59). Surprisingly, this enhanced frameshifting was not simply due to decreased spermidine levels, but instead it depended on the combination of low spermidine levels and high putrescine levels due to the absence of AdoMetDC (60). Thus, not only total polyamine levels but the relative amounts of the different polyamines can impact the fidelity of translation.
Translational control of polyamine homeostasis
In addition to impacts on general protein synthesis, polyamines control the translation of specific mRNAs. The translational autoregulation of polyamine biosynthetic and regulatory proteins highlights the intriguing interplay between polyamines and protein synthesis. The regulatory schemes employed to maintain polyamine homeostasis identify steps in the translation process that are impacted by polyamines and provide additional insights into the roles of polyamines in translation. In this section, we will summarize the current understanding of how polyamines control the translation of mRNAs encoding ODC, ODC antizyme (OAZ), antizyme inhibitor 1 (AZIN1) and AdoMetDC. Intriguingly, the mRNAs encoding spermine synthase and SSAT1 contain conserved upstream open reading frames (uORFs) (61), and polyamines have been implicated in translational regulation of SSAT1 (62–64). However, the mechanism by which polyamines regulate translation of these mRNAs is not well understood, and SSAT1 is also regulated by polyamines through the alternative splicing of its mRNA (65).
Ornithine decarboxylase and antizyme
The first step in the biosynthesis of polyamines in eukaryotes is the decarboxylation of ornithine to putrescine, a reaction catalyzed by the enzyme ODC (see 66, 67). ODC functions as a homodimer with both monomers contributing residues to the catalytic center. Considering its central role in polyamine biosynthesis, ODC is, not surprisingly, subject to extensive regulation. Some of this regulation is part of negative feedback mechanisms that have evolved to maintain polyamine homeostasis in the cell. Much of the feedback regulation of ODC is post-translational and indirect. Important examples are discussed further below. There is, however, much evidence that mammalian ODC mRNA is subject to translational autoregulation by polyamines, and this regulation has been shown for mammalian ODC both in vitro and in vivo (68–70). The nature of the cis-acting elements and the mechanism of regulation are obscure at present; however, mutational analyses indicate that the translational control elements are contained within the 300-nucleotide 5′ leader of the ODC mRNA (reviewed in Ref. 71). A different and better understood translational autoregulation of the ODC mRNA is found in many nonmammalian animals and separately in filamentous fungi. This latter autoregulation is discussed in more detail below.
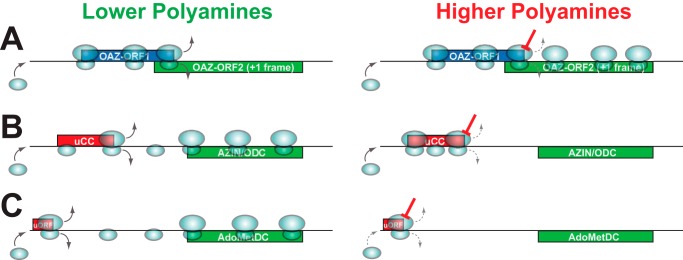
OAZ is a protein inhibitor of ODC, whose synthesis is induced by the end product of ODC, i.e. polyamines (72). As described in detail in another Minireview in this series, OAZ binds to ODC monomers preventing formation of functional ODC homodimers (67, 73). Once bound to ODC, OAZ presents ODC for ubiquitin-independent degradation by the 26S proteasome (74, 75). The first cloned OAZ was rat OAZ1. OAZ1 has orthologs in most or all vertebrates (76). Of note, work from the Tabor lab studying ODC turnover in Saccharomyces cerevisiae first predicted the presence of yeast OAZ (77). Depending on the species, vertebrates have up to four different OAZ paralogs. All mammals apparently have three OAZ paralogs (78, 79). The vertebrate OAZ genes identified to date are encoded by two partially overlapping reading frames, and +1 ribosomal frameshifting is required for synthesis of the functional full-length protein (Fig. 1A) (76, 80, 81). The frameshift occurs while ribosomes are decoding the stop codon of the first ORF (ORF1). Following its initial discovery, OAZ homologs were identified in many kingdoms of the eukaryotic domain of life, including most animals, fungi, and many protists (76, 80–82). The requirement for +1 frameshifting is nearly universally conserved; however, the exact frameshift site is not conserved. At least nine naturally occurring variants of OAZ frameshift sites have been identified. Interestingly, a bewildering array of cis-acting sequences that stimulate ribosomal frameshifting has evolved in different evolutionary branches. The originally cloned rat OAZ1, for example, contains a stimulatory RNA pseudoknot structure that is located 3′ of the shift site and is conserved throughout vertebrates (81). A different stimulatory RNA pseudoknot has evolved independently in invertebrates and is similarly well conserved (83). In other evolutionary branches, conserved RNA stem–loop structures located 3′ of the frameshift site have been confirmed, or are suspected, to act as frameshifting stimulators (80, 84). The nascent peptide near the C terminus of ORF2 in S. cerevisiae OAZ has been proposed to regulate the frameshifting (85); however, the conservation of this sequence is unclear even within the Saccharomyces clade. A frameshifting stimulatory sequence was also mapped downstream of the frameshift site in Schizosaccharomyces pombe OAZ; however, the nature of the stimulatory sequence is unclear (82). Although not well understood, cis-acting stimulators of frameshifting have also been mapped to the 5′ side of the shift site. Most of these 5′-stimulatory elements appear to act through their primary nucleotide sequence, although at least one element acts through the sequence of the encoded nascent peptide (80, 84).
Figure 1.
Schematic representation of polyamine control of OAZ, AZIN, and AdoMetDC mRNA translation. A, OAZ is encoded by two partially overlapping ORFs linked through ribosomal +1 frameshifting. Under lower polyamine conditions (left), ribosomes terminate at the stop codon of ORF1; under higher polyamine conditions (right), ribosomes frameshift at the ORF1 stop codon and synthesize full-length OAZ. B, AZIN and some ODC mRNAs contain a uCC element (in red, an inhibitory uORF with conserved peptide sequence and initiated at a near cognate start codon) upstream of the main ORF (green). In lower polyamine conditions, many scanning ribosomes bypass the uCC and translate the main ORF, and ribosomes that translate the uCC disengage from the mRNA after terminating. Under higher polyamine conditions, translating ribosomes pause during translation of the uCC and cause queuing of subsequent scanning ribosomes and enhanced initiation on the uCC. The increased translation of the uCC results in decreased translation of the main ORF. C, AdoMetDC mRNAs of vertebrates contain an inhibitory uORF (red) encoding a peptide with the sequence MAGDIS; the main ORF is in green. Under conditions of lower polyamines, many ribosomes bypass the uORF and translate the main ORF. The occasional ribosome that translates the uORF disengages from the mRNA after termination. Under conditions of higher polyamines, pausing of translating ribosomes at the uORF stop codon blocks additional ribosomes from loading on the mRNA and impairs main ORF translation.
Because the sequences of both the shift sites and cis-acting elements are very diverse, it is likely that the molecular details of the frameshifting mechanism(s) will also vary widely. Experimental evidence also supports this conclusion. There are at least two central features to OAZ frameshifting. The first is slow recognition of the ORF1 stop codon. In characterized examples of +1 frameshifting on other mRNAs, inhibiting recognition of the A-site codon, for example by having a rare codon in that position, stimulates the change in reading frame (86, 87). As ribosomal frameshifting is in competition with standard decoding, any feature, like slow decoding of the A-site codon, that inhibits standard decoding will enhance the probability of the alternative event, frameshifting. Two features of the ORF1 stop codon suggest that termination will be inefficient. In greater than 95% of OAZ genes identified to date, the stop codon of ORF1 is UGA, the least efficient stop codon. Mutating this stop codon to UAA or UAG to enhance termination reduced frameshifting (81, 88). In both bacteria and eukaryotes, the 3′ nucleotide context of a stop codon affects the efficiency of termination (89, 90), and intriguingly, the termination codon of ORF1 of OAZ genes is almost invariably in poor termination context (80). The second critical feature of OAZ frameshifting is the identity of the P-site codon at the shift site. The apparent ancestral and most common shift site is UCC-U. This site is conserved from fungi to animals (82). Of the at least six other known shift sites (80), some, like the recurrent site UUU-U, where the P-site tRNA can form identical base-pairing in the 0 and +1 frames, are obviously conducive to +1 frameshifting; the function of other shift sites is less obvious. An important clue for the selection of other shift site sequences came from analysis of the GCG-U frameshift site in S. cerevisiae (91). This sequence is highly reminiscent of the +1 frameshift site, GCG-A, required for expression of the GagPol polyprotein of the retrotransposon Ty3 in S. cerevisiae (92). In the case of Ty3, the cognate tRNA (one that can form full Watson-Crick, or wobble, pairing) for GCG is missing from the yeast genome, and recognition by the obligatory substitute tRNAAla with anticodon 3′-CGI-5′, requires an unusual G:I (purine:purine) apposition during decoding (93). This apposition makes codon–anticodon pairing inherently unstable when present in the P site. Budding yeast that lack cognate tRNA for the GCG codon all use GCG-U as a shift site for their OAZ genes, indicating that the alternative OAZ shift sites might take advantage of the translational environment within the organism (94). Curiously, the shift site UCC-U, accompanied by the cis-acting elements of mammalian OAZ1, which have evolved separately from the yeast translational machinery, nevertheless supports ribosomal frameshifting into the +1 frame in yeast, although predominantly via a −2 shift event with only a minority of ribosomes shifting via a +1 event (95). Based on these results and a similar finding in S. pombe (96), it has been argued that mobilization of the tRNA in the P site of the OAZ shift site is essential (i.e. the P-site codon-anticodon pairing is inherently compromised in OAZ shift sites), whereas strong pairing of the-P site tRNA in the new frame is not crucial (80, 97).
How polyamines stimulate the regulatory frameshifting on the OAZ mRNA is an area of active investigation. Experiments with the mammalian OAZ1 sequence indicate that neither the 5′ nor 3′ cis-acting elements are required for polyamine regulation of frameshifting; these sequences merely enhance the levels of frameshifting across a gradient of polyamine concentrations. Replacing the stop codon of ORF1 with a sense codon; however, not only reduces frameshifting but also completely abolishes polyamine regulation (81, 88). These experiments implicate the process of translation termination in the regulation and suggest that the sensor for polyamine is a molecule that interacts with the stop codon. In vitro experiments with mammalian OAZ1 showed that enhancing termination at the ORF1 stop codon by adding excess eRF1 decreased frameshifting (98). Conversely, an inactivating mutation in eRF3 increased frameshifting (98).
The picture that has emerged from studies on the OAZ frameshifting in S. cerevisiae is considerably more complicated. One study has reported that a sequence comprising the shift site plus 17 nucleotides upstream and 10 nucleotides downstream is sufficient to confer robust polyamine regulation of yeast OAZ +1 frameshifting (99). This result is consistent with the idea, outlined above, that the stop codon is the essential component of the polyamine regulation. In support of this notion, conversion of yeast eRF3 to its inactive prion state [PSI+] enhanced frameshifting (100). However, another study identified two additional cis-acting sequences essential for polyamine regulation of yeast OAZ frameshifting (85). The first is a codon 5′ of the shift site that dampens frameshifting, and the second is part of the nascent peptide near the C terminus of ORF2. In the absence of polyamines, as the nascent peptide is synthesized it is proposed to jam the exit tunnel of the ribosome, causing the ribosomes to stall and fail to complete synthesis of OAZ. High polyamines are proposed to relieve this jamming and simultaneously enhance frameshifting at the upstream site by relieving the ribosomal queue. The apparent contradiction between the models focused on the shift site versus the nascent peptide has remained unresolved. The nascent peptide sequence responsible for the latter mechanism is not conserved beyond the Saccharomycotina lineage, indicating that this feature is unlikely to figure in the polyamine-induced +1 frameshifting of other OAZ mRNAs, even if it plays an important role in S. cerevisiae.
The features of polyamines required for stimulation of OAZ frameshifting are not well understood. In vitro experiments have shown that putrescine, spermidine, and spermine can all stimulate the frameshifting of mammalian OAZ1; although the optimum concentration of each polyamine to stimulate an equivalent level of frameshifting is different, putrescine is required at the largest concentration; spermidine is required at a lower concentration, and spermine is required at the lowest concentration (81, 101). This gradation correlates with the number of amine groups in each of polyamine. Superficially, this implies that the role of polyamines in antizyme frameshifting is to neutralize clusters of negatively charged residues in a target molecule(s). Similar results were observed in vivo in yeast using the minimal S. cerevisiae OAZ shift site sequence (99). This latter study also showed that, although putrescine and spermine act cooperatively to promote frameshifting, spermidine does not act cooperatively with the other polyamines. These results suggest the presence of multiple and distinct polyamine-binding sites that affect frameshifting, yet the different polyamines apparently in some cases compete for the same binding site. The most likely polyamine target molecule(s) is the ribosome machinery and, more specifically, its RNA components with their ample negatively charged phosphate groups. One conceivable mechanism is that polyamine binding in the A site of the ribosome inhibits the binding and function of the release factors. As mentioned previously, in vitro experiments in mammalian cells performed more that 30 years ago showed that the polyamines spermidine and spermine stimulate readthrough of a UGA, but not a UAA, stop codon, implying that polyamines can interfere with termination at select stop codons in cells (56). Experiments testing a modified OAZ2 frameshift site or the murine leukemia virus (MuLV) readthrough site in mammalian COS-7 cells showed that polyamines can stimulate stop codon readthrough (88). Further studies testing all three stop codons and all three physiologically important polyamines revealed that the combination of spermidine and the least efficient termination codon UGA resulted in the highest polyamine-stimulated readthrough, especially when the UGA was followed by cytidine (C), the worst context for termination in mammals (88). These results are consistent with the idea that polyamines, especially spermidine, interfere with termination at particularly inefficient stop codons, and this property is exploited by the unknown sensor of polyamines to promote OAZ frameshifting. Although the precise molecular mechanism for polyamine stimulation of frameshifting is unclear, polyamine interaction with rRNA may underlie the two proposed mechanisms. Polyamine binding in the A site may interfere with termination factor function to induce frameshifting on the minimal frameshift site, whereas polyamine binding in the polypeptide exit tunnel may mask regulatory interactions with the nascent peptide in the unusual mechanism described for the frameshifting of yeast OAZ.
Antizyme inhibitor
AZINs are a group of protein homologs of ODC that have lost the ability to decarboxylate ornithine but have retained the ability to bind to OAZ, often with higher affinity than ODC (67, 102–104). AZIN function is to bind and sequester OAZ away from ODC, thus enhancing the activity of ODC. AZINs have evolved independently at least three separate times, apparently through duplication of ODC with a subsequent loss of the decarboxylase activity. Such an event occurred during early vertebrate evolution resulting in AZIN1. Another duplication of ODC and loss of catalytic activity resulted in the emergence of AZIN2 in the mammalian lineage (104).
Sequence comparisons of the 5′ leaders of vertebrate AZIN1 mRNAs revealed the presence of upstream conserved coding regions (uCCs) (Fig. 1B) (105). These uCCs lack canonical translation initiation codons yet encode well-conserved peptide sequences. The uCC of AZIN1 was shown to direct polyamine translational regulation of reporters fused downstream of the 5′ leader of the AZIN1 mRNA (105). High levels of polyamines inhibit expression of the main ORF (mORF), whereas low levels of polyamines enhance expression. The polyamine regulation of AZIN1 depends on both the AUU near-cognate start codon and the most highly conserved C-terminal 10 amino acids of the uCC. Consistent with the notion that uCC translation is inversely correlated with mORF translation, polyamines were found to enhance initiation at the near-cognate start codon of a uCC-firefly luciferase reporter (105).
The uCC of AZIN1 evolved early in the animal lineage, and homologous sequences are widespread in metazoans. However, in invertebrates, and even some vertebrates, the uCC elements are most often located on mRNAs encoding ODC, indicating that the uCC first evolved on the ODC mRNA and was transferred to AZIN1 as a consequence of the duplication event at the root of AZIN1 evolution. Notably, the ODC mRNA in mammals has lost any vestiges of the metazoan ODC/AZIN uCC, and as discussed above, the mRNA sequences directing polyamine translational regulation are not known. Careful examination of uCC sequences from diverse animal species shows remarkable conservation of the peptide motif PPW within the C-terminal region of the AZIN1 mRNA that is required for polyamine regulation. Examination of ODC mRNA sequences from other eukaryotic lineages, specifically from fungi, showed that uCC sequences have evolved independently several times. The fungal sequences fall in two classes. The first class, like the uCCs in animals, is characterized by a conserved PPW motif. The second class has a conserved (R)PP-stop motif. Curiously, a PP-stop motif is also present in the uCC-like uORF of mammalian AZIN2, which has evolved from a classical uCC but has acquired an AUG start codon in poor initiation context. The recurrent selection for PPW and PP-stop motifs in uCC elements suggests that these motifs might play essential roles in the polyamine regulation. Consistent with this idea, polyamine repression of ODC (spe-1) synthesis in Neurospora crassa was shown to depend on nucleotide sequences within the 5′ leader of the spe-1 mRNA (106), which we now know includes a uCC element (105).
A recent study has defined the mechanism of regulation and the role played by polyamines and the PPW motif of the AZIN1 uCC more precisely (57). Ribosomal profiling analyses to monitor the positions of ribosomes on mRNAs in cells revealed enhanced ribosome occupancy on the uCC in the presence of high polyamines, and this correlated with low translation of the mORF. In contrast, decreased occupancy of the uCC in the presence of low polyamines correlated with derepressed expression and increased ribosome occupancy of the mORF. One of the codons showing high ribosome occupancy in the presence of high polyamines is the Trp codon of the PPW motif. This finding is consistent with a translation elongation pause in which the second Pro codon occupies the ribosomal P site and the Trp codon is in the A site. A second pronounced peak in the ribosomal profiling under high polyamine conditions corresponds to ribosomes pausing at termination codon of the uCC. Interestingly, mutating either the PP or the W of the PPW motif significantly impaired regulation by polyamines, although regulation was completely abolished if a second conserved motif “PS-stop” was also mutated. Mutating the PS-stop motif alone had a smaller effect than mutating “PPW.” Notably, in some organisms either the PPW or the PS-stop motif, but not both, is conserved.
Translation of PPW motifs is known to be inefficient and to require the assistance in bacteria of the translation factor EF-P, which is the homolog of the eukaryotic factor eIF5A. Experiments using fully reconstituted in vitro translation systems showed that eIF5A is required for the synthesis of the peptide MEPPWK and that this stimulatory activity of eIF5A was inhibited by increasing concentrations of spermidine. Addition of excess eIF5A overcame the inhibition by spermidine, suggesting that polyamines and eIF5A compete for a functional site on the ribosome.
A complete model for polyamine regulation of AZIN1 via its uCC has been developed (Fig. 1B). Under lower polyamine conditions, most scanning ribosomes skip over the uCC without translating. These ribosomes then translate AZIN1. The occasional ribosome that translates the uCC disengages from the mRNA at the uCC stop codon and thus cannot synthesize AZIN1. Under higher polyamine conditions, the occasional ribosome that initiates at the weak start site of the uCC pauses when translating the PPW motif. This pausing is due to polyamine inhibition of eIF5A function in translation elongation. Subsequent scanning ribosomes, most of which will skip over the weak start site of the uCC, form a queue upstream of the stalled ribosome on the PPW motif. As the queue lengthens, eventually a scanning ribosome is positioned in the vicinity of the uCC start codon, leading to more frequent initiation. The pausing-induced enhanced utilization of the uCC start codon and greater translation of the uCC will result in a reinforcing positive feedback to enhance uCC translation and repress AZIN1 synthesis.
SAM decarboxylase
Production of decarboxylated SAM, the aminopropyl donor for the biosynthesis of larger polyamines spermidine and spermine, is catalyzed by the enzyme AdoMetDC. Like ODC, OAZ, and AZIN, synthesis of AdoMetDC is regulated at the translational level. In vertebrates, the mRNA encoding AdoMetDC contains a short conserved uORF that starts just 13–14 nucleotides downstream of the 5′ cap (Fig. 1C) (107, 108). This uORF encodes the peptide MAGDIS, which is absolutely conserved in all sequenced vertebrate AdoMetDC genes. Although not conserved in the yeast AdoMetDC mRNA, the MAGDIS peptide mediates polyamine-dependent translational regulation in yeast (109). Extensive molecular and biochemical studies have revealed the basic outlines for polyamine regulation through MAGDIS. Saturation mutagenesis of the last three codons of the uORF showed that polyamine regulation depends on aspartic acid at the fourth position and isoleucine, or its close relative valine, at the fifth position (110). Increased polyamine levels lead to enhanced ribosomal stalling during termination on the uORF, and the amino acids at the fourth and fifth positions facilitate the most prolonged stall (111–113). However, elevated polyamine levels were also found to increase stalling at the termination codon on a mutant peptide sequence that is otherwise less inhibitory on downstream translation (111). These results suggest that translation of the MAGDIS peptide results in inherently inefficient termination that is exacerbated in the presence of high polyamines. The inefficient termination and ribosome stalling on the uORF stop codon may impair reinitiation. In addition, a ribosome stalled at the stop codon of the MAGDIS uORF will, by virtue of its bulk and proximity to the 5′ end, prevent loading of incoming 40S PICs. Together, these two effects of polyamines on uORF translation will block AdoMetDC synthesis. Like OAZ, the molecular sensor of polyamines in regulation via the MAGDIS uORF is not known.
AdoMetDC is also translationally regulated in plants (114). In land plants, the 5′ leader of AdoMetDC typically contains two AUG-initiated uORFs. The first uORF is very short, often just three codons, and its stop codon overlaps the start codon of a much longer uORF that is about 45–60 codons in length. The longer uORF shows some conservation at the amino acid level with the C-terminal motif PS-stop invariably conserved in land plants. The mechanism of polyamine translational regulation of AdoMetDC in plants is presently not known. Deletion of the tiny uORF by mutating its start codon leads to constitutive repression. Eliminating both uORFs leads to constitutive derepression. Placing the coding sequence of the longer uORF in a different reading frame also causes derepression and substantial loss of polyamine regulation. These results suggest that the tiny uORF may facilitate access to the longer inhibitory uORF in the presence of polyamines, and conversely, under low polyamines the short uORF may deflect ribosomes from translating the longer uORF. As the longer uORF is less inhibitory if its amino acid sequence is changed, the nascent peptide might be contributing to polyamine inhibition in cis. Consistent with this idea, the PS-stop motif at the C terminus of the long uORF of plant AdoMetDC was, as described above, also found in the AZIN1 uCC and shown to confer polyamine regulation (57). Perhaps, analogous to the PPW motif in elongation, termination at PS-stop is hyperdependent on eIF5A and thus sensitive to polyamine inhibition of eIF5A function. AdoMetDC mRNAs from other evolutionary branches also include conserved uORFs. The uORF in nematode AdoMetDC mRNAs includes the absolutely conserved sequence RPP-stop, which is identical to the absolutely conserved C-terminal sequence of the uCC in the mRNA of ODC from Pezizomycotina (filamentous fungi) (61). It is tempting to speculate that the RPP-stop motif imposes a heightened eIF5A requirement for termination, especially because eIF5A is known to be required for efficient translation of poly-Pro stretches.
Conclusion
The polyamine regulation of ODC, OAZ, AZIN, and AdoMetDC has provided an extraordinary wealth of information on the roles of polyamines in translation and revealed new paradigms for translational control. These translational control pathways also serve as a testament to the innate ability of polyamines to affect different aspects of translation–initiation, elongation, and termination–which have been harnessed time and again by the forces of evolution in the regulation of genes in the polyamine biosynthetic pathway.
This work was supported by the Intramural Research Program of the NICHD, National Institutes of Health. This article is part of a series on “Polyamines,” written in honor of Dr. Herbert Tabor's 100th birthday. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- A
- aminoacyl
- P
- peptidyl
- E
- exit
- AdoMetDC
- S-adenosylmethionine decarboxylase
- ODC
- ornithine decarboxylase
- OAZ
- ODC antizyme
- AZIN1
- antizyme inhibitor 1
- Met-tRNAiMet
- initiator methionyl-tRNA
- fMet-tRNAfMet
- formylated methionyl-tRNA
- PIC
- preinitiation complex
- eIF
- eukaryotic translation initiation factor
- eEF
- eukaryotic translation elongation factor
- Met-puro
- methionyl-puromycin
- SAT1
- N1-spermidine/spermine acetyltransferase 1
- uCC
- upstream conserved coding region
- uORF
- upstream open reading frame
- SSAT1
- spermine/spermidine N1-acetyltransferase 1
- SAM
- S-adenosylmethionine.
References
- 1. Igarashi K., and Kashiwagi K. (2015) Modulation of protein synthesis by polyamines. IUBMB Life 67, 160–169 10.1002/iub.1363 [DOI] [PubMed] [Google Scholar]
- 2. Cohen S. S., and Lichtenstein J. (1960) Polyamines and ribosome structure. J. Biol. Chem. 235, 2112–2116 [PubMed] [Google Scholar]
- 3. Hinnebusch A. G., and Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodnina M. V. (2018) Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10, a032664 10.1101/cshperspect.a032664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 10.1128/MMBR.00008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- 7. Pavitt G. D. (2018) Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wiley Interdiscip. Rev. RNA 9, e1491 10.1002/wrna.1491 [DOI] [PubMed] [Google Scholar]
- 8. Dever T. E., Dinman J. D., and Green R. (2018) Translation elongation and recoding in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032649 10.1101/cshperspect.a032649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dever T. E., Gutierrez E., and Shin B. S. (2014) The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 49, 413–425 10.3109/10409238.2014.939608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Melnikov S., Mailliot J., Shin B. S., Rigger L., Yusupova G., Micura R., Dever T. E., and Yusupov M. (2016) Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J. Mol. Biol. 428, 3570–3576 10.1016/j.jmb.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt C., Becker T., Heuer A., Braunger K., Shanmuganathan V., Pech M., Berninghausen O., Wilson D. N., and Beckmann R. (2016) Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 44, 1944–1951 10.1093/nar/gkv1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park M. H., and Wolff E. C. (2018) Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 293, 18710–18718 10.1074/jbc.TM118.003341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., Buskirk A. R., and Dever T. E. (2013) eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuller A. P., Wu C. C., Dever T. E., Buskirk A. R., and Green R. (2017) eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shin B. S., Katoh T., Gutierrez E., Kim J. R., Suga H., and Dever T. E. (2017) Amino acid substrates impose polyamine, eIF5A, or hypusine requirement for peptide synthesis. Nucleic Acids Res. 45, 8392–8402 10.1093/nar/gkx532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hellen C. U. (2018) Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032656 10.1101/cshperspect.a032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xaplanteri M. A., Petropoulos A. D., Dinos G. P., and Kalpaxis D. L. (2005) Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acids Res. 33, 2792–2805 10.1093/nar/gki557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amarantos I., Xaplanteri M. A., Choli-Papadopoulou T., and Kalpaxis D. L. (2001) Effects of two photoreactive spermine analogues on peptide bond formation and their application for labeling proteins in Escherichia coli functional ribosomal complexes. Biochemistry 40, 7641–7650 10.1021/bi010010s [DOI] [PubMed] [Google Scholar]
- 19. Noeske J., Wasserman M. R., Terry D. S., Altman R. B., Blanchard S. C., and Cate J. H. (2015) High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 10.1038/nsmb.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tabor H., Tabor C. W., Cohn M. S., and Hafner E. W. (1981) Streptomycin resistance (rpsL) produces an absolute requirement for polyamines for growth of an Escherichia coli strain unable to synthesize putrescine and spermidine [Δ(speA-speB) ΔspecC]. J. Bacteriol. 147, 702–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hershko A., Amoz S., and Mager J. (1961) Effect of polyamines and divalent metals on in vitro incorporation of amino acids into ribonucleoprotein particles. Biochem. Biophys. Res. Commun. 5, 46–51 10.1016/0006-291X(61)90078-X [DOI] [PubMed] [Google Scholar]
- 22. Martin R. G., and Ames B. N. (1962) The effect of polyamines and of poly U size on phenylalanine incorporation. Proc. Natl. Acad. Sci. U.S.A. 48, 2171–2178 10.1073/pnas.48.12.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nathans D., and Lipmann F. (1961) Amino acid transfer from aminoacyl-ribonucleic acids to protein on ribosomes of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 47, 497–504 10.1073/pnas.47.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeda Y. (1969) Polyamines and protein synthesis. I. The effect of polyamines on cell free polyphenylalanine synthesis in Escherichia coli. J. Biochem. 66, 345–349 10.1093/oxfordjournals.jbchem.a129152 [DOI] [PubMed] [Google Scholar]
- 25. Igarashi K., Hikami K., Sugawara K., and Hirose S. (1973) Effect of polyamines on polypeptide synthesis in rat liver cell-free system. Biochim. Biophys. Acta 299, 325–330 10.1016/0005-2787(73)90356-0 [DOI] [PubMed] [Google Scholar]
- 26. Igarashi K., Sugawara K., Izumi I., Nagayama C., and Hirose S. (1974) Effect of polyamines of polyphenylalanine synthesis by Escherichia coli and rat-liver ribosomes. Eur. J. Biochem. 48, 495–502 10.1111/j.1432-1033.1974.tb03790.x [DOI] [PubMed] [Google Scholar]
- 27. Konecki D., Kramer G., Pinphanichakarn P., and Hardesty B. (1975) Polyamines are necessary for maximum in vitro synthesis of globin peptides and play a role in chain initiation. Arch. Biochem. Biophys. 169, 192–198 10.1016/0003-9861(75)90332-X [DOI] [PubMed] [Google Scholar]
- 28. Hunter A. R., Farrell P. J., Jackson R. J., and Hunt T. (1977) The role of polyamines in cell-free protein synthesis in the wheat-germ system. Eur. J. Biochem. 75, 149–157 10.1111/j.1432-1033.1977.tb11512.x [DOI] [PubMed] [Google Scholar]
- 29. Atkins J. F., Lewis J. B., Anderson C. W., and Gesteland R. F. (1975) Enhanced differential synthesis of proteins in a mammalian cell-free system by addition of polyamines. J. Biol. Chem. 250, 5688–5695 [PubMed] [Google Scholar]
- 30. Ogasawara T., Ito K., and Igarashi K. (1989) Effect of polyamines on globin synthesis in a rabbit reticulocyte polyamine-free protein synthetic system. J. Biochem. 105, 164–167 10.1093/oxfordjournals.jbchem.a122633 [DOI] [PubMed] [Google Scholar]
- 31. Saini P., Eyler D. E., Green R., and Dever T. E. (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 10.1038/nature08034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benne R., and Hershey J. W. (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253, 3078–3087 [PubMed] [Google Scholar]
- 33. Kemper W. M., Berry K. W., and Merrick W. C. (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 251, 5551–5557 [PubMed] [Google Scholar]
- 34. Schreier M. H., Erni B., and Staehelin T. (1977) Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J. Mol. Biol. 116, 727–753 10.1016/0022-2836(77)90268-6 [DOI] [PubMed] [Google Scholar]
- 35. Hetrick B., Khade P. K., Lee K., Stephen J., Thomas A., and Joseph S. (2010) Polyamines accelerate codon recognition by transfer RNAs on the ribosome. Biochemistry 49, 7179–7189 10.1021/bi1009776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nilsson L., Rigler R., and Wintermeyer W. (1983) The influence of spermine on the structural dynamics of yeast tRNAPhe. Biochim. Biophys. Acta 740, 460–465 10.1016/0167-4781(83)90095-7 [DOI] [PubMed] [Google Scholar]
- 37. Quigley G. J., Teeter M. M., and Rich A. (1978) Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 75, 64–68 10.1073/pnas.75.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tabor H., and Tabor C. W. (1982) Polyamine requirement for efficient translation of amber codons in vivo. Proc. Natl. Acad. Sci. U.S.A. 79, 7087–7091 10.1073/pnas.79.23.7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Christian B. E., Haque M. E., and Spremulli L. L. (2010) The effect of spermine on the initiation of mitochondrial protein synthesis. Biochem. Biophys. Res. Commun. 391, 942–946 10.1016/j.bbrc.2009.11.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park M. H., Joe Y. A., and Kang K. R. (1998) Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273, 1677–1683 10.1074/jbc.273.3.1677 [DOI] [PubMed] [Google Scholar]
- 41. Sasaki K., Abid M. R., and Miyazaki M. (1996) Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 384, 151–154 10.1016/0014-5793(96)00310-9 [DOI] [PubMed] [Google Scholar]
- 42. Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., and Hershey J. W. (1991) Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3105–3114 10.1128/MCB.11.6.3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chattopadhyay M. K., Tabor C. W., and Tabor H. (2003) Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase. Proc. Natl. Acad. Sci. U.S.A. 100, 13869–13874 10.1073/pnas.1835918100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Byers T. L., Lakanen J. R., Coward J. K., and Pegg A. E. (1994) The role of hypusine depletion in cytostasis induced by S-adenosyl-l-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine. Biochem. J. 303, 363–368 10.1042/bj3030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gerner E. W., Mamont P. S., Bernhardt A., and Siat M. (1986) Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem. J. 239, 379–386 10.1042/bj2390379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee C. H., and Park M. H. (2000) Human deoxyhypusine synthase: interrelationship between binding of NAD and substrates. Biochem. J. 352, 851–857 10.1042/0264-6021:3520851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park M. H., Cooper H. L., and Folk J. E. (1981) Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. U.S.A. 78, 2869–2873 10.1073/pnas.78.5.2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chattopadhyay M. K., Park M. H., and Tabor H. (2008) Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. U.S.A. 105, 6554–6559 10.1073/pnas.0710970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Landau G., Bercovich Z., Park M. H., and Kahana C. (2010) The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 285, 12474–12481 10.1074/jbc.M110.106419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mandal S., Mandal A., Johansson H. E., Orjalo A. V., and Park M. H. (2013) Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 2169–2174 10.1073/pnas.1219002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Landau G., Ran A., Bercovich Z., Feldmesser E., Horn-Saban S., Korkotian E., Jacob-Hirsh J., Rechavi G., Ron D., and Kahana C. (2012) Expression profiling and biochemical analysis suggest stress response as a potential mechanism inhibiting proliferation of polyamine-depleted cells. J. Biol. Chem. 287, 35825–35837 10.1074/jbc.M112.381335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishimura K., Murozumi K., Shirahata A., Park M. H., Kashiwagi K., and Igarashi K. (2005) Independent roles of eIF5A and polyamines in cell proliferation. Biochem. J. 385, 779–785 10.1042/BJ20041477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Igarashi K., Hashimoto S., Miyake A., Kashiwagi K., and Hirose S. (1982) Increase of fidelity of polypeptide synthesis by spermidine in eukaryotic cell-free systems. Eur. J. Biochem. 128, 597–604 [DOI] [PubMed] [Google Scholar]
- 54. Igarashi K., Kashiwagi K., Aoki R., Kojima M., and Hirose S. (1979) Comparative studies on the increase by polyamines of fidelity of protein synthesis in Escherichia coli and wheat germ cell-free systems. Biochem. Biophys. Res. Commun. 91, 440–448 10.1016/0006-291X(79)91541-9 [DOI] [PubMed] [Google Scholar]
- 55. McMurry L. M., and Algranati I. D. (1986) Effect of polyamines on translation fidelity in vivo. Eur. J. Biochem. 155, 383–390 10.1111/j.1432-1033.1986.tb09502.x [DOI] [PubMed] [Google Scholar]
- 56. Hryniewicz M. M., and Vonder Haar R. A. (1983) Polyamines enhance readthrough of the UGA termination codon in a mammalian messenger RNA. Mol. Gen. Genet. 190, 336–343 10.1007/BF00330661 [DOI] [PubMed] [Google Scholar]
- 57. Ivanov I. P., Shin B. S., Loughran G., Tzani I., Young-Baird S. K., Cao C., Atkins J. F., and Dever T. E. (2018) Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell 70, 254–264.e6 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harger J. W., Meskauskas A., and Dinman J. D. (2002) An “integrated model” of programmed ribosomal frameshifting. Trends Biochem. Sci. 27, 448–454 10.1016/S0968-0004(02)02149-7 [DOI] [PubMed] [Google Scholar]
- 59. Balasundaram D., Dinman J. D., Wickner R. B., Tabor C. W., and Tabor H. (1994) Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 91, 172–176 10.1073/pnas.91.1.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balasundaram D., Dinman J. D., Tabor C. W., and Tabor H. (1994) SPE1 and SPE2: two essential genes in the biosynthesis of polyamines that modulate +1 ribosomal frameshifting in Saccharomyces cerevisiae. J. Bacteriol. 176, 7126–7128 10.1128/jb.176.22.7126-7128.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ivanov I. P., Atkins J. F., and Michael A. J. (2010) A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res. 38, 353–359 10.1093/nar/gkp1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parry L., Balaña Fouce R., and Pegg A. E. (1995) Post-transcriptional regulation of the content of spermidine/spermine N1-acetyltransferase by N1N12-bis(ethyl)spermine. Biochem. J. 305, 451–458 10.1042/bj3050451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Perez-Leal O., Barrero C. A., Clarkson A. B., Casero R. A. Jr., and Merali S. (2012) Polyamine-regulated translation of spermidine/spermine-N1-acetyltransferase. Mol. Cell. Biol. 32, 1453–1467 10.1128/MCB.06444-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Butcher N. J., Broadhurst G. M., and Minchin R. F. (2007) Polyamine-dependent regulation of spermidine-spermine N1-acetyltransferase mRNA translation. J. Biol. Chem. 282, 28530–28539 10.1074/jbc.M701265200 [DOI] [PubMed] [Google Scholar]
- 65. Hyvönen M. T., Uimari A., Keinänen T. A., Heikkinen S., Pellinen R., Wahlfors T., Korhonen A., Närvänen A., Wahlfors J., Alhonen L., and Jänne J. (2006) Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. RNA 12, 1569–1582 10.1261/rna.39806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pegg A. E. (2009) Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894 10.1002/iub.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kahana C. (2018) The antizyme family for regulating polyamines. J. Biol. Chem. 293, 18730–18735 10.1074/jbc.TM118.003339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kameji T., and Pegg A. E. (1987) Inhibition of translation of mRNAs for ornithine decarboxylase and S-adenosylmethionine decarboxylase by polyamines. J. Biol. Chem. 262, 2427–2430 [PubMed] [Google Scholar]
- 69. Persson L., Holm I., and Heby O. (1986) Translational regulation of ornithine decarboxylase by polyamines. FEBS Lett. 205, 175–178 10.1016/0014-5793(86)80892-4 [DOI] [PubMed] [Google Scholar]
- 70. Ito K., Kashiwagi K., Watanabe S., Kameji T., Hayashi S., and Igarashi K. (1990) Influence of the 5′-untranslated region of ornithine decarboxylase mRNA and spermidine on ornithine decarboxylase synthesis. J. Biol. Chem. 265, 13036–13041 [PubMed] [Google Scholar]
- 71. Shantz L. M., and Pegg A. E. (1999) Translational regulation of ornithine decarboxylase and other enzymes of the polyamine pathway. Int. J. Biochem. Cell Biol. 31, 107–122 10.1016/S1357-2725(98)00135-6 [DOI] [PubMed] [Google Scholar]
- 72. Heller J. S., Fong W. F., and Canellakis E. S. (1976) Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc. Natl. Acad. Sci. U.S.A. 73, 1858–1862 10.1073/pnas.73.6.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matsufuji S., Miyazaki Y., Kanamoto R., Kameji T., Murakami Y., Baby T. G., Fujita K., Ohno T., and Hayashi S. (1990) Analyses of ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J. Biochem. 108, 365–371 10.1093/oxfordjournals.jbchem.a123207 [DOI] [PubMed] [Google Scholar]
- 74. Li X., and Coffino P. (1992) Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol. Cell. Biol. 12, 3556–3562 10.1128/MCB.12.8.3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., and Ichihara A. (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 10.1038/360597a0 [DOI] [PubMed] [Google Scholar]
- 76. Ivanov I. P., Gesteland R. F., and Atkins J. F. (2000) Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 28, 3185–3196 10.1093/nar/28.17.3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gupta R., Hamasaki-Katagiri N., White Tabor C., and Tabor H. (2001) Effect of spermidine on the in vivo degradation of ornithine decarboxylase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 10620–10623 10.1073/pnas.181341298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ivanov I. P., Gesteland R. F., and Atkins J. F. (1998) A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics 52, 119–129 10.1006/geno.1998.5434 [DOI] [PubMed] [Google Scholar]
- 79. Ivanov I. P., Rohrwasser A., Terreros D. A., Gesteland R. F., and Atkins J. F. (2000) Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc. Natl. Acad. Sci. U.S.A. 97, 4808–4813 10.1073/pnas.070055897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ivanov I. P., and Atkins J. F. (2007) Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 35, 1842–1858 10.1093/nar/gkm035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J. F., Gesteland R. F., and Hayashi S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80, 51–60 10.1016/0092-8674(95)90450-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ivanov I. P., Matsufuji S., Murakami Y., Gesteland R. F., and Atkins J. F. (2000) Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 19, 1907–1917 10.1093/emboj/19.8.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ivanov I. P., Anderson C. B., Gesteland R. F., and Atkins J. F. (2004) Identification of a new antizyme mRNA +1 frameshifting stimulatory pseudoknot in a subset of diverse invertebrates and its apparent absence in intermediate species. J. Mol. Biol. 339, 495–504 10.1016/j.jmb.2004.03.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yordanova M. M., Wu C., Andreev D. E., Sachs M. S., and Atkins J. F. (2015) A nascent peptide signal responsive to endogenous levels of polyamines acts to stimulate regulatory frameshifting on antizyme mRNA. J. Biol. Chem. 290, 17863–17878 10.1074/jbc.M115.647065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kurian L., Palanimurugan R., Gödderz D., and Dohmen R. J. (2011) Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 477, 490–494 10.1038/nature10393 [DOI] [PubMed] [Google Scholar]
- 86. Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., and Baranov P. V. (2016) Ribosomal frameshifting and transcriptional slippage: from genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Belcourt M. F., and Farabaugh P. J. (1990) Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7-nucleotide minimal site. Cell 62, 339–352 10.1016/0092-8674(90)90371-K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Petros L. M., Howard M. T., Gesteland R. F., and Atkins J. F. (2005) Polyamine sensing during antizyme mRNA programmed frameshifting. Biochem. Biophys. Res. Commun. 338, 1478–1489 10.1016/j.bbrc.2005.10.115 [DOI] [PubMed] [Google Scholar]
- 89. Poole E. S., Brown C. M., and Tate W. P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 14, 151–158 10.1002/j.1460-2075.1995.tb06985.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McCaughan K. K., Brown C. M., Dalphin M. E., Berry M. J., and Tate W. P. (1995) Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 92, 5431–5435 10.1073/pnas.92.12.5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Palanimurugan R., Scheel H., Hofmann K., and Dohmen R. J. (2004) Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 23, 4857–4867 10.1038/sj.emboj.7600473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Farabaugh P. J., Zhao H., and Vimaladithan A. (1993) A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell 74, 93–103 10.1016/0092-8674(93)90297-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sundararajan A., Michaud W. A., Qian Q., Stahl G., and Farabaugh P. J. (1999) Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell 4, 1005–1015 10.1016/S1097-2765(00)80229-4 [DOI] [PubMed] [Google Scholar]
- 94. Ivanov I. P., Gesteland R. F., and Atkins J. F. (2006) Evolutionary specialization of recoding: frameshifting in the expression of S. cerevisiae antizyme mRNA is via an atypical antizyme shift site but is still +1. RNA 12, 332–337 10.1261/rna.2245906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Matsufuji S., Matsufuji T., Wills N. M., Gesteland R. F., and Atkins J. F. (1996) Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J. 15, 1360–1370 10.1002/j.1460-2075.1996.tb00478.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ivanov I. P., Gesteland R. F., Matsufuji S., and Atkins J. F. (1998) Programmed frameshifting in the synthesis of mammalian antizyme is +1 in mammals, predominantly +1 in fission yeast, but −2 in budding yeast. RNA 4, 1230–1238 10.1017/S1355838298980864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ivanov I. P., and Matsufuji S. (2010) in Recoding: Expansion of Decoding Rules Enriches Gene Expression (Atkins J. F., and Gesteland R. F., eds) pp. 281–300, Springer, New York [Google Scholar]
- 98. Karamysheva Z. N., Karamyshev A. L., Ito K., Yokogawa T., Nishikawa K., Nakamura Y., and Matsufuji S. (2003) Antizyme frameshifting as a functional probe of eukaryotic translational termination. Nucleic Acids Res. 31, 5949–5956 10.1093/nar/gkg789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rato C., Amirova S. R., Bates D. G., Stansfield I., and Wallace H. M. (2011) Translational recoding as a feedback controller: systems approaches reveal polyamine-specific effects on the antizyme ribosomal frameshift. Nucleic Acids Res. 39, 4587–4597 10.1093/nar/gkq1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Namy O., Galopier A., Martini C., Matsufuji S., Fabret C., and Rousset J. P. (2008) Epigenetic control of polyamines by the prion [PSI+]. Nat. Cell Biol. 10, 1069–1075 10.1038/ncb1766 [DOI] [PubMed] [Google Scholar]
- 101. Howard M. T., Shirts B. H., Zhou J., Carlson C. L., Matsufuji S., Gesteland R. F., Weeks R. S., and Atkins J. F. (2001) Cell culture analysis of the regulatory frameshift event required for the expression of mammalian antizymes. Genes Cells 6, 931–941 10.1046/j.1365-2443.2001.00477.x [DOI] [PubMed] [Google Scholar]
- 102. Murakami Y., Ichiba T., Matsufuji S., and Hayashi S. (1996) Cloning of antizyme inhibitor, a highly homologous protein to ornithine decarboxylase. J. Biol. Chem. 271, 3340–3342 10.1074/jbc.271.7.3340 [DOI] [PubMed] [Google Scholar]
- 103. Kahana C. (2009) Antizyme and antizyme inhibitor, a regulatory tango. Cell. Mol. Life Sci. 66, 2479–2488 10.1007/s00018-009-0033-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ivanov I. P., Firth A. E., and Atkins J. F. (2010) Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function. J. Mol. Evol. 70, 289–302 10.1007/s00239-010-9331-5 [DOI] [PubMed] [Google Scholar]
- 105. Ivanov I. P., Loughran G., and Atkins J. F. (2008) uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 105, 10079–10084 10.1073/pnas.0801590105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hoyt M. A., Broun M., and Davis R. H. (2000) Polyamine regulation of ornithine decarboxylase synthesis in Neurospora crassa. Mol. Cell. Biol. 20, 2760–2773 10.1128/MCB.20.8.2760-2773.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ruan H., Hill J. R., Fatemie-Nainie S., and Morris D. R. (1994) Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J. Biol. Chem. 269, 17905–17910 [PubMed] [Google Scholar]
- 108. Ruan H., Shantz L. M., Pegg A. E., and Morris D. R. (1996) The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J. Biol. Chem. 271, 29576–29582 10.1074/jbc.271.47.29576 [DOI] [PubMed] [Google Scholar]
- 109. Mize G. J., and Morris D. R. (2001) A mammalian sequence-dependent upstream open reading frame mediates polyamine-regulated translation in yeast. RNA 7, 374–381 10.1017/S1355838201001972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mize G. J., Ruan H., Low J. J., and Morris D. R. (1998) The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Biol. Chem. 273, 32500–32505 10.1074/jbc.273.49.32500 [DOI] [PubMed] [Google Scholar]
- 111. Law G. L., Raney A., Heusner C., and Morris D. R. (2001) Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 276, 38036–38043 [DOI] [PubMed] [Google Scholar]
- 112. Raney A., Baron A. C., Mize G. J., Law G. L., and Morris D. R. (2000) In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J. Biol. Chem. 275, 24444–24450 10.1074/jbc.M003364200 [DOI] [PubMed] [Google Scholar]
- 113. Raney A., Law G. L., Mize G. J., and Morris D. R. (2002) Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 277, 5988–5994 10.1074/jbc.M108375200 [DOI] [PubMed] [Google Scholar]
- 114. Hanfrey C., Elliott K. A., Franceschetti M., Mayer M. J., Illingworth C., and Michael A. J. (2005) A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J. Biol. Chem. 280, 39229–39237 10.1074/jbc.M509340200 [DOI] [PubMed] [Google Scholar]